The rapid detection of Mycobacterium tuberculosis complex (MTUBC) in clinical samples is essential for successful treatment. New techniques such as real-time PCR have been developed in order to facilitate rapid diagnosis, but their sensitivity is low in extrapulmonary specimens, due to the low bacillary load in such samples.
KEYWORDS: extrapulmonary tuberculosis, molecular diagnosis, Mycobacterium tuberculosis, Xpert MTB/RIF Ultra
ABSTRACT
The rapid detection of Mycobacterium tuberculosis complex (MTUBC) in clinical samples is essential for successful treatment. New techniques such as real-time PCR have been developed in order to facilitate rapid diagnosis, but their sensitivity is low in extrapulmonary specimens, due to the low bacillary load in such samples. A next-generation assay has recently been developed to try to overcome this limitation. The aim of this study was to analyze the effectiveness of the Xpert MTB/RIF Ultra (GX-Ultra) for the detection of MTUBC DNA in 108 smear-negative extrapulmonary specimens that were MTUBC culture positive. In addition, 40 extrapulmonary culture-negative samples and 20 samples with nontuberculous mycobacteria were tested to evaluate the specificity of the assay. All samples were collected between May 1999 and May 2017. The GX-Ultra detected DNA of MTUBC in 82 extrapulmonary specimens that were MTUBC culture positive (75.9% sensitivity; 95% confidence interval [CI], 66.6 to 83.4%). The assay was negative for all clinical specimens that were MTUBC culture negative and the samples with nontuberculous mycobacteria (100% specificity). Furthermore, two (1.8%) samples presented mutations related to rifampin resistance. The highest sensitivity was obtained in samples of lymph nodes (94.1%) and nonsterile fluids (93.7%), followed by tissue specimens (86.6%), stool material (80%), abscess aspirates (64.7%), and sterile fluids (60.5%). Pleural fluids, one of the least optimal samples for detecting DNA of MTUBC, were GX-Ultra positive in 10/21 (47.6%) of cases. In summary, GX-Ultra showed excellent specificity and high sensitivity in paubacillary specimens, making it a useful tool for rapid diagnosis of extrapulmonary tuberculosis.
INTRODUCTION
Tuberculosis (TB) is a major public health problem in terms of morbidity and mortality worldwide, with 6.3 million new cases reported and an estimated incidence of 10.4 million in 2016. The extrapulmonary form of the disease represented 15% of the notified cases (1), and its frequency of presentation markedly increased in cases of immunodeficiency.
The microbial diagnosis of TB has traditionally been carried out using two different procedures: (i) direct smear microscopy of the sample (Ziehl-Neelsen and/or auramine-rhodamine stain), which is quick, inexpensive, and simple but has poor sensitivity, especially in nonrespiratory samples (2), and (ii) mycobacterial culture, which despite being considered the gold standard technique for TB diagnosis can take several weeks to provide a confirmation (3, 4). New molecular methods have thus been developed to improve TB control, with the most sensitive ones being those based on nucleic acid amplification (5).
One of these methods is the Xpert MTB/RIF assay (Cepheid, Sunnyvale, CA), which is based on a real-time PCR that utilizes molecular beacon technology to simultaneously detect DNA of Mycobacterium tuberculosis complex (MTUBC) and rifampin (RIF) resistance conferred by the rpoB gene mutation. This molecular system, endorsed by the WHO in 2010 (6), has been demonstrated to be a useful tool for the detection of pulmonary TB (7, 8); however, its sensitivity for extrapulmonary samples remains a challenge due to their low bacillary load (9).
The recently developed next-generation Xpert MTB/RIF Ultra (GX-Ultra; Cepheid) assay aims to overcome the limitations of its predecessor by increasing the sensitivity for detection of MTUBC DNA when few bacilli are present in a clinical sample (10). This new and fully automated nested real-time PCR assay differs from its predecessor in several ways: the larger PCR chamber with a total capacity of 50 μl, in contrast to 25 μl in the previous cartridge (10), the incorporation of two different multicopy targets (IS1081 and IS6110 insertion sequences), and the optimization of PCR and thermal-cycling parameters. The results are provided automatically in 77 min if the genetic material is amplified or in 66 min if it is not. The system classifies MTUBC detection in the following semiquantitative results: high, medium, low, very low, and a new category named trace, and it classifies RIF resistance as detected, not detected, or indeterminate.
The aim of the present work was thus to evaluate the effectiveness of GX-Ultra for direct detection of MTUBC DNA in smear-negative extrapulmonary samples.
MATERIALS AND METHODS
Samples and mycobacterial identification.
A total of 168 smear-negative extrapulmonary samples (Table 1), obtained from 148 adult patients and collected between May 1999 and May 2017, were analyzed in the Department of Microbiology at the Hospital Universitari de Bellvitge (Barcelona, Spain). Nonsterile samples were pretreated using an N-acetyl-l-cysteine-NaOH digestion-decontamination procedure (11), with a final volume of 2 ml, whereas sterile fluids were directly processed. Biopsy samples were previously disaggregated and resuspended in 2 ml of saline solution. In all cases 1 ml of the samples was frozen at −80°C until later use. The remaining volume was processed as follows: (i) smear microscopy for acid-fast organisms (Ziehl-Neelsen and/or auramine-rhodamine stain) and (ii) mycobacterial culture in solid (Löwenstein-Jensen) and liquid media (Bactec MGIT 960; Becton Dickinson, Towson, MD). The smear microscopy and the culture tests were done at the time of collection of the specimens. Mycobacterial identification was carried out using the DNA AccuProbe (Gen-Probe Inc., San Diego, CA), BIO-LINE SD Ag MPT64 TB test (Standard Diagnostics, Yongin, South Korea), and Genotype Mycobacterium CM/MTBC (Hain Lifescience, Nehren, Germany).
TABLE 1.
Results obtained by the Xpert MTB/RIF Ultra assay according to the source and MTUBC culture of the specimens
| Clinical sample | Total no. of samples | Samples MTUBC culture positive |
Sensitivity (%) | Samples MTUBC culture negative |
Specificity (%) | ||
|---|---|---|---|---|---|---|---|
| GXU+a | GXU−b | GXU+ | GXU− | ||||
| Sterile fluids | 44 | 60.5 | 100 | ||||
| Pleural fluid | 24 | 10 | 11 | 47.6 | 0 | 3 | |
| Cerebrospinal fluid | 4 | 3 | 0 | 100 | 0 | 1 | |
| Joint fluid | 9 | 7 | 1 | 87.5 | 0 | 1 | |
| Ascitic fluid | 3 | 1 | 2 | 33.3 | 0 | 0 | |
| Pericardial fluid | 4 | 2 | 1 | 66.6 | 0 | 1 | |
| Nonsterile fluids | 29 | 93.7 | 100 | ||||
| Gastric aspirate | 5 | 3 | 1 | 75 | 0 | 1 | |
| Urine | 24 | 12 | 0 | 100 | 0 | 12 | |
| Lymph nodes | 25 | 16 | 1 | 94.1 | 0 | 8 | 100 |
| Abscess aspirates | 20 | 64.7 | 100 | ||||
| Cervical abscess | 6 | 4 | 1 | 80 | 0 | 1 | |
| Skin abscess | 6 | 2 | 2 | 50 | 0 | 2 | |
| Paravertebral abscess | 3 | 2 | 1 | 66.6 | 0 | 0 | |
| Osteitis pus | 5 | 3 | 2 | 60 | 0 | 0 | |
| Tissues | 24 | 86.6 | 100 | ||||
| Skin biopsy | 8 | 2 | 0 | 100 | 0 | 6 | |
| Intervertebral disc biopsy | 2 | 2 | 0 | 100 | 0 | 0 | |
| Bone biopsy | 4 | 2 | 0 | 100 | 0 | 2 | |
| Pleural biopsy | 2 | 2 | 0 | 100 | 0 | 0 | |
| Rectal biopsy | 1 | 0 | 1 | 0 | 0 | 0 | |
| Costal cartilage biopsy | 1 | 1 | 0 | 100 | 0 | 0 | |
| Liver biopsy | 1 | 1 | 0 | 100 | 0 | 0 | |
| Cervical tissue | 1 | 1 | 0 | 100 | 0 | 0 | |
| Mediastinal tissue | 1 | 0 | 1 | 0 | 0 | 0 | |
| Synovial tissue | 3 | 2 | 0 | 100 | 0 | 1 | |
| Joint biopsy | 18 | 0 | 0 | 0 | 18 | ||
| Stool | 8 | 4 | 1 | 80 | 0 | 3 | 100 |
| Total | 168 | 82 | 26 | 75.9 | 0 | 60 | 100 |
GXU+, positive results from GeneXpert MTB/RIF Ultra.
GXU−, negative results from GeneXpert MTB/RIF Ultra.
Among the samples studied, 108 were MTUBC culture positive (including three Mycobacterium bovis bacillus Calmette-Guérin). Sixty clinical samples (one sample per patient) were also included to test the specificity of the assay, divided into 40 clinical samples that were culture negative and 20 that were culture positive for nontuberculous mycobacteria (NTM) covering a total of 18 species.
Xpert MTB/RIF Ultra assay.
The GX-Ultra assay was performed according to the manufacturer's instructions (12). Briefly, frozen samples were thawed and 2 ml of the sample reagent was added to 1 ml of the clinical specimens (2:1 ratio) in order to inactivate them. The tube containing the sample was then incubated at room temperature for 15 min and vortexed once for 7 to 8 min. Then 2 ml of the volume was transferred to the test cartridge and inserted into the GeneXpert instrument.
Time to culture positivity and effect of freezing duration.
This study also examined the relationship between the results obtained by GX-Ultra and (i) the prolonged preservation of frozen samples, which were divided into specimens frozen for less than 5 years (freezing starting at 2013 to 2017), between 5 and 10 years (2008 to 2012), between 10 and 15 years (2003 to 2007), and more than 15 years (1999–2002), and (ii) bacillary load, analyzing the time to positivity of cultures in days for extrapulmonary samples in liquid media.
Statistical analysis.
A chi-squared test and a Student t test were performed to determine whether the duration of freezing of the samples and the time to growth of the mycobacteria had any impact on the results obtained, respectively. A P value of less than 0.05 was considered statistically significant. Sensitivity and specificity values were calculated at the 95% confidence interval (CI) for GX-Ultra, using the mycobacterial culture as the reference standard.
RESULTS
GX-Ultra detected DNA of MTUBC in 82 smear-negative extrapulmonary specimens that were MTUBC culture positive (75.9% sensitivity; 95% CI, 66.6 to 83.4%). The positive results obtained using this technique were classified as follows: medium (n = 15), low (n = 19), very low (n = 29), and trace (n = 19). There were no specimens in the high category. Two samples (1.8%; included in the low and very low groups) presented mutations related to RIF resistance and showed multidrug resistance (MDR).
The sensitivities of GX-Ultra for different sample groups were as follows (Table 1): 94.1% in lymph nodes, 93.7% in nonsterile fluids, 86.6% in tissue specimens, 80% in stool material, 64.7% in abscess aspirates, and 60.5% in sterile fluids. Ten of 21 (47.6%) pleural fluid samples were GX-Ultra positive for the detection of MTUBC DNA.
GX-Ultra presented a specificity of 100% for the 40 clinical specimens with a negative MTUBC culture and the 20 that were NTM culture-positive (Table 1).
Evaluation of the effect of the duration of freezing of the samples revealed that 16 (80%) of those stored since 2013 to 2017 presented a GX-Ultra-positive result. A positive result was also obtained from 6 (85.7%) samples stored since 2008 to 2012, 36 (75%) since 2003 to 2007, and 24 (72.7%) since 1999 to 2002. There was no significant difference in time to culture positivity between groups (P > 0.05).
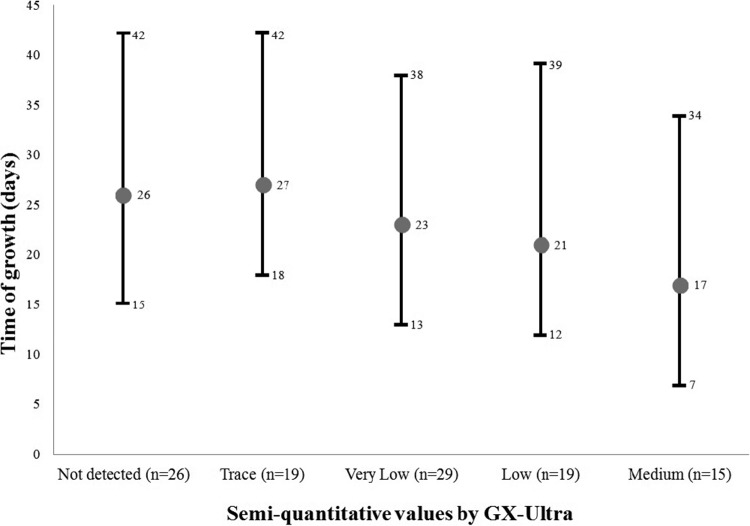
Regarding the time required for culture-positive growth (Fig. 1), significant differences were found when specimens from the trace category of the GX-Ultra were compared with those from the low (P = 0.04) and medium (P = 0.003) categories and when comparing the samples in the not-detected category with those in the medium category (P = 0.005). The comparison between the not-detected, trace, and very low categories presented few differences, none of which reached statistical significance (P = 0.7 for not detected and trace, P = 0.1 for not detected and very low, and P = 0.08 for trace and very low). Interestingly, the average times required for culture-positive growth between the not-detected and trace categories were very similar: 26 days (15 to 42) and 27 days (18 to 42), respectively.
FIG 1.
Time required for growth of smear-negative samples, including the average number of days, classified according to the semiquantitative value provided by the GX-Ultra.
DISCUSSION
GX-Ultra has previously been demonstrated to show high sensitivity for the rapid diagnosis of pulmonary TB in adults and children, in addition to HIV-infected patients (13–16). However, to date only one study has analyzed the sensitivity of GX-Ultra in extrapulmonary disease, specifically in tuberculous meningitis (17). In the present study, high sensitivity values were obtained with GX-Ultra in a large variety of extrapulmonary samples, and the results were superior to those of previous studies that applied Xpert MTB/RIF to similar specimens. The improvement was especially notable for lymph nodes, whose sensitivity increased between 17.1 and 23.5% more with GX-Ultra than with its predecessor (9, 18–20). Similarly, the nonsterile fluids obtained sensitivity values 15.7 to 27% higher with the novel assay than with Xpert MTB/RIF (9, 19, 20); in the case of tissue samples, the increase was 20 to 44.9% larger with the GX-Ultra (9, 19). It should be noted that pleural fluids, which are probably the least optimal sample used to detect DNA in MTUBC, presented interesting results. In fact, several studies that used the previous version of the assay, Xpert MTB/RIF, only achieved sensitivity values of 25 to 37% for pleural fluid samples (9, 20–23), while in the current study GX-Ultra detected about half of the culture-positive specimens of MTUBC (Table 1), representing a remarkable improvement.
The increase in sensitivity observed with GX-Ultra might be due mainly to the incorporation of two new targets (IS1081 and IS6110) that can be found in MTUBC (24, 25). These allow detection of the new semiquantitative category named trace, enabling the assay to detect 16 CFU/ml, in contrast to the 114-CFU/ml limit of detection of Xpert MTB/RIF (26). To achieve this category result, at least one of the probes for IS1081 or IS6110 has to be positive with cycle thresholds less than 37 cycles and no more than one rpoB probe with cycle thresholds less than 40 cycles (10). Other modifications to the assay, such as a larger PCR chamber in the cartridge, appear to have improved its sensitivity, while the high-resolution melt technology seems to be able to differentiate silent mutations that confer RIF resistance more efficiently (10, 26). However, we stress that in the current study only two samples presented mutations related to RIF resistance (and showed multidrug resistance) and that as a result, it was impossible to check the new high-resolution melt technology incorporated into the Xpert MTB/RIF Ultra. Additionally, a trace result cannot provide information about RIF resistance, and the results are thus reported as “MTB detected, trace, RIF indeterminate.” About a quarter (23.8%) of the positive results in this study corresponded to the trace category. However, this situation probably is not as relevant in low-MDR-TB-incidence settings, such as our study setting, in which not one of the samples categorized as trace showed RIF resistance according to drug susceptibility testing.
The specificity values for specimens that were MTUBC culture negative and NTM samples were excellent (100%) in this study. It is important to note that these results were obtained from a limited number and variety of samples, as well as a few NTM species. In other studies in which GX-Ultra was used, the authors indicated that the increase in sensitivity with this new version came at the expense of a decrease in specificity, specifically in patients with a history of tuberculosis (13, 27) and patients with meningitis (17). They hypothesized that the presence of DNA from MTUBC or intact bacilli could explain the reduction in specificity. However, it is not known with certainty what causes this reduction in specificity. Thus, further studies are needed to evaluate the real specificity of this next-generation assay, especially in paubacillary samples.
With regard to the cryopreservation time of the specimens, GX-Ultra showed a nonsignificant trend for lower sensitivity values in samples frozen for a longer time. Many factors might influence the cryopreservation of clinical TB samples and therefore could explain any variations in sensitivity, e.g., the cooling rate, the sample container, volume of samples, and heat transfer conditions inside the freezer (28).
Although clearly showing an improvement in sensitivity, GX-Ultra is influenced by the bacillary load of the clinical samples. In those that were more paubacillary, the percent sensitivity provided by the technique was lower. This was supported by the finding that the differences between the categories provided by the system (high, medium, low, very low, and trace) and the times required for growth were statistically significant (Fig. 1). On the other hand, the similarity of results observed in time to culture-positive growth between the not-detected and trace categories could be explained by the new and lower analytic limit of detection (threshold) of the trace category. Therefore, as all samples studied were originally culture-positive, it appears that the not-detected category was also at the limit of detection or just below it.
Another important aspect of GX-Ultra is the rapidity of the results. The latest version has a shorter test time (33 min less for a negative result and 45 min less for a positive result if the genetic material is amplified) than the Xpert MTB/RIF. This could be due to the following: a trace and a negative result are provided in a shorter time because the assay analyzes only IS1081 to IS6110; the other semiquantitative categories and RIF resistance detection require more time to finish the test because the assay uses four probes to detect the rpoB gene.
In summary, GX-Ultra has proven to be a highly sensitive and specific test for the direct detection of MTUBC DNA in smear-negative extrapulmonary samples, which are a challenge due to their low bacillary burden. Therefore, this next-generation assay could be a useful tool for the rapid diagnosis of extrapulmonary TB.
ACKNOWLEDGMENTS
We are grateful to Cepheid, for providing us with the Xpert MTB/RIF Ultra reagents.
We declare that we have no conflicts of interest.
REFERENCES
- 1.World Health Organization. 2017. Global tuberculosis report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Armand S, Vanhuls P, Delcroix G, Courcol R, Lemaître N. 2011. Comparision of the Xpert MTB/RIF test with an IS6110-TaqMan real-time PCR assay for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J Clin Microbiol 49:1772–1776. doi: 10.1128/JCM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewinsohn DM, Leonard MK, LoBue PA, Cohn DL, Daley CL, Desmond E, Keane J, Lewinsohn DA, Loeffler AM, Mazurek GH, O'Brien RJ, Pai M, Richeldi L, Salfinger M, Shinnick TM, Sterling TR, Warshauer DM, Woods GL. 2017. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention clinical practice guidelines: diagnosis of tuberculosis in adults and children. Clin Infect Dis 64:111–115. doi: 10.1093/cid/ciw778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pai M, Flores LL, Pai N, Hubbard A, Riley LW, Colford JM Jr. 2003. Diagnostic accuracy of nucleic acid amplification tests for tuberculous meningitis: a systematic review and meta-analysis. Lancet Infect Dis 3:633–643. doi: 10.1016/S1473-3099(03)00772-2. [DOI] [PubMed] [Google Scholar]
- 5.Alcaide F, Coll P. 2011. Advances in rapid diagnosis of tuberculosis disease and anti-tuberculous drug resistance. Enferm Infecc Microbiol Clin 29:34–40. doi: 10.1016/S0213-005X(11)70016-7. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. 2011. Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF system. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 7.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moure R, Martín R, Alcaide F. 2012. Effectiveness of an integrated real-time PCR method for detection of the Mycobacterium tuberculosis complex in smear-negative extrapulmonary samples in an area of low tuberculosis prevalence. J Clin Microbiol 50:513–515. doi: 10.1128/JCM.06467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization. 2017. WHO meeting report of a technical expert consultation: non-inferiority analysis of Xpert MTB/RIF Ultra compared to Xpert MTB/RIF. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 11.Kubica GP, Dye WE, Cohn ML, Middlebrook G. 1963. Sputum digestion and decontamination with N-acetyl-L-cysteine-sodium hydroxide for culture of mycobacteria. Am Rev Respir Dis 87:775–779. doi: 10.1164/arrd.1963.87.5.775. [DOI] [PubMed] [Google Scholar]
- 12.Cepheid. 2017. Xpert MTB/RIF Ultra: instruction manual. Cepheid, Sunnyvale, CA. [Google Scholar]
- 13.Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, Hall SL, Chakravorty S, Cirillo DM, Tukvadze N, Bablishvili N, Stevens W, Scott L, Rodrigues C, Kazi MI, Joloba M, Nakiyingi L, Nicol MP, Ghebrekristos Y, Anyango I, Murithi W, Dietze R, Lyrio Peres R, Skrahina A, Auchynka V, Chopra KK, Hanif M, Liu X, Yuan X, Boehme CC, Ellner JJ, Denkinger CM, Study Team. 2018. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis 18:76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zar HJ, Workman L, Nicol MP. 2017. Diagnosis of pulmonary tuberculosis in HIV-infected and uninfected children using Xpert MTB/RIF Ultra, abstr A7610. ATS 2017 International Conference. American Thoracic Society, Washington, DC. [Google Scholar]
- 15.Nicol MP, Workman L, Prins M, Bateman L, Ghebrekristos Y, Mbhele S, Denkinger CM, Zar HJ. 22 February 2018. Accuracy of Xpert MTB/RIF Ultra for the diagnosis of pulmonary tuberculosis in children. Pediatr Infect Dis J. doi: 10.1097/INF.0000000000001960. [DOI] [PubMed] [Google Scholar]
- 16.Kendall EA, Schumacher SG, Denkinger CM, Dowdy DW. 2017. Estimated clinical impact of the Xpert MTB/RIF Ultra cartridge for diagnosis of pulmonary tuberculosis: a modeling study. PLoS Med 14:e1002472. doi: 10.1371/journal.pmed.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahr NC, Nuwagira E, Evans EE, Cresswel FV, Bystrom PV, Byamukama A, Bridge SC, Bangdiwala AS, Meya DB, Denkinger CM, Muzoora C, Boulware DR, Trial Team ASTRO-CM. 2018. Diagnostic accuracy of Xpert MTB/RIF Ultra for tuberculous meningitis in HIV-infected adults: a prospective cohort study. Lancet Infect Dis 18:68–75. doi: 10.1016/S1473-3099(17)30474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullah I, Javaid A, Masud H, Ali M, Basit A, Ahmad W, Younis F, Yasmin R, Khan A, Jabbar A, Husain M, Butt ZA. 2017. Rapid detection of Mycobacterium tuberculosis and rifampicin resistance in extrapulmonary tuberculosis and sputum smear-negative pulmonary suspects using Xpert MTB/RIF. J Med Microbiol 66:412–418. doi: 10.1099/jmm.0.000449. [DOI] [PubMed] [Google Scholar]
- 19.Zeka AN, Tasbakan S, Cavusoglu C. 2011. Evaluation of the GeneXpert MTB/RIF assay for rapid diagnosis of tuberculosis and detection of rifampin resistance in pulmonary and extrapulmonary specimens. J Clin Microbiol 49:4138–4141. doi: 10.1128/JCM.05434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penz E, Boffa J, Roberts DJ, Fisher D, Cooper R, Ronksley PE, James MT. 2015. Diagnostic accuracy of the Xpert® MTB/RIF assay for extra-pulmonary tuberculosis: a meta-analysis. Int J Tuberc Lung Dis 19:278–284. doi: 10.5588/ijtld.14.0262. [DOI] [PubMed] [Google Scholar]
- 21.Vadwai V, Boehme C, Nabeta P, Shetty A, Alland D, Rodrigues C. 2011. Xpert MTB/RIF: a new pillar in diagnosis of extrapulmonary tuberculosis. J Clin Microbiol 49:2540–2545. doi: 10.1128/JCM.02319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich SO, von Groote-Bidlingmaier F, Diacon AH. 2011. Xpert MTB/RIF assay for diagnosis of pleural tuberculosis. J Clin Microbiol 49:4341–4342. doi: 10.1128/JCM.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusiba JK, Nakiyingi L, Kirenga BJ, Kiragga A, Lukande R, Nsereko M, Ssengooba W, Katamba A, Worodria W, Joloba ML, Mayanja-Kizza H. 2014. Evaluation of Cepheid's Xpert MTB/RIF test on pleural fluid in the diagnosis of pleural tuberculosis in a high prevalence HIV/TB setting. PLoS One 9:e102702. doi: 10.1371/journal.pone.0102702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankar S, Kuppanan S, Balakrishnan B, Nandagopal Balaji. 2011. Analysis of sequence diversity among IS6110 sequence of Mycobacterium tuberculosis: possible implications for PCR based detection. Bioinformation 6:283–285. doi: 10.6026/97320630006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Soolingen D, Hermans PW, de Haas PE, van Embden JD. 1992. Insertion element IS1081-associated restriction fragment length polymorphisms in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol 30:1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravorty S, Simmons AM, Rowneki M, Parmar H, Cao Y, Ryan J, Banada PP, Deshpande S, Shenai S, Gall A, Glass J, Krieswirth B, Schumacher SG, Nabeta P, Tukvadze N, Rodrigues C, Skrahina A, Tagliani E, Cirillo DM, Davidow A, Denkinger CM, Persing D, Kwiatkowski R, Jones M, Alland D. 2017. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampicin in an assay suitable for point-of-care testing. mBio 8:e00812-17. doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theron G, Venter R, Smith L, Esmail A, Randall P, Sood V, Oelfese S, Calligaro G, Warren R, Dheda K. 2018. False-positive Xpert MTB/RIF results retested patients with previous tuberculosis: frequency, profile, and prospective clinical outcomes. J Clin Microbiol 56:e01696-17. doi: 10.1128/JCM.01696-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu Z, Weigel KM, Soelberg SD, Lakey A, Cangelosi GA, Lee KH, Chung JH, Gao D. 2012. Cryopreservation of Mycobacterium tuberculosis complex cells. J Clin Microbiol 50:3575–3580. doi: 10.1128/JCM.00896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]