In principle, whole-genome sequencing (WGS) can predict phenotypic resistance directly from a genotype, replacing laboratory-based tests. However, the contribution of different bioinformatics methods to genotype-phenotype discrepancies has not been systematically explored to date.
KEYWORDS: Staphylococcus aureus, antibiotic resistance, bioinformatics, whole-genome sequencing
ABSTRACT
In principle, whole-genome sequencing (WGS) can predict phenotypic resistance directly from a genotype, replacing laboratory-based tests. However, the contribution of different bioinformatics methods to genotype-phenotype discrepancies has not been systematically explored to date. We compared three WGS-based bioinformatics methods (Genefinder [read based], Mykrobe [de Bruijn graph based], and Typewriter [BLAST based]) for predicting the presence/absence of 83 different resistance determinants and virulence genes and overall antimicrobial susceptibility in 1,379 Staphylococcus aureus isolates previously characterized by standard laboratory methods (disc diffusion, broth and/or agar dilution, and PCR). In total, 99.5% (113,830/114,457) of individual resistance-determinant/virulence gene predictions were identical between all three methods, with only 627 (0.5%) discordant predictions, demonstrating high overall agreement (Fleiss' kappa = 0.98, P < 0.0001). Discrepancies when identified were in only one of the three methods for all genes except the cassette recombinase, ccrC(b). The genotypic antimicrobial susceptibility prediction matched the laboratory phenotype in 98.3% (14,224/14,464) of cases (2,720 [18.8%] resistant, 11,504 [79.5%] susceptible). There was greater disagreement between the laboratory phenotypes and the combined genotypic predictions (97 [0.7%] phenotypically susceptible, but all bioinformatic methods reported resistance; 89 [0.6%] phenotypically resistant, but all bioinformatics methods reported susceptible) than within the three bioinformatics methods (54 [0.4%] cases, 16 phenotypically resistant, 38 phenotypically susceptible). However, in 36/54 (67%) cases, the consensus genotype matched the laboratory phenotype. In this study, the choice between these three specific bioinformatic methods to identify resistance determinants or other genes in S. aureus did not prove critical, with all demonstrating high concordance with each other and phenotypic/molecular methods. However, each has some limitations; therefore, consensus methods provide some assurance.
INTRODUCTION
Staphylococcus aureus causes both superficial infections (such as boils) and life-threatening disease, including septicemia (1). There were 11,405 S. aureus bacteremias in England in 2015 and 2016 (2); 7.2% were methicillin-resistant S. aureus (MRSA), which has increased costs and poorer patient outcomes (3). Fast accurate resistance prediction is key to managing S. aureus infections. Molecular-based methods directed at detecting specific genes, e.g., through rapid multiplex PCR and microarrays, can reduce the time to identify resistance determinants and the time on broad-spectrum antibiotics (4–6). However, they require specific primers that impact sensitivity and specificity.
In principle, whole-genome sequencing (WGS) has the potential to predict phenotypic resistance directly from the genotype, replacing laboratory-based phenotypic tests (7). Several studies report high concordance between genotypic predictions based on known or novel resistant determinants and phenotypic methods (8–13). However, these studies used various sequence-processing pipelines and bioinformatics methods to identify in silico resistance determinants. Without formal comparisons between the various methods, it is unclear whether the underlying differences affect results or whether differences in methodology could cause some of the observed discrepancies between genotypic predictions and phenotype.
Therefore, we compared three WGS-based bioinformatics methods (Genefinder [read based], Mykrobe [de Bruijn graph based], and Typewriter [BLAST based]) in terms of predictions of the presence/absence of different resistance determinants and the overall prediction of antimicrobial susceptibility and the presence/absence of virulence genes from short-read Illumina WGS.
MATERIALS AND METHODS
Three sets of S. aureus isolates with known high-quality phenotypes were analyzed: derivation, n = 501, and validation, n = 491, sets (denoted Oxford derivation/validation) from blood cultures and nasal swab isolates at the Oxford Radcliffe Hospitals NHS Trust and Brighton and Sussex University Hospitals NHS Trust, spanning a period of 13 years, sequenced for an initial assessment of genotypic prediction of susceptibility phenotype in S. aureus (9, 10) and 397 isolates that had been referred to the Public Health England (PHE) reference laboratory for investigation (denoted Colindale 397; NCBI BioProject number PRJNA445516). The Oxford derivation set had previously been used in the development of Typewriter and Mykrobe but not Genefinder; the former methods were then applied to the Oxford validation set.
The phenotypes for Oxford derivation/validation isolates were determined by disc diffusion and/or automated broth diffusion (BD Phoenix), with discrepancies between phenotype and genotype resolved as described previously (11). All PHE isolates (n = 397) were subjected to MIC testing by the PHE Staphylococcal Reference Laboratory using the agar dilution method (14). In addition, the mecA-mecC status and virulence gene profile of the PHE isolates was determined by PCR or microarray testing as described previously (15). The European Committee on Antimicrobial Susceptibility Testing (EUCAST) thresholds were used to determine the sensitivity or resistance for each phenotype (http://www.eucast.org/clinical_breakpoints).
All Oxford derivation/validation isolates were sequenced using the Illumina HiSeq 2000 platform as previously described (16). PHE samples were sequenced in an Illumina HiSeq 2500 platform as described previously (17) (both 150-bp reads). Samples determined as mixed based on WGS were excluded from further analysis. For quality control of sequences at PHE, trimmomatic software was used (Illumina adapter removed, leading and trailing quality threshold set to 30, and minimum length of read set to 50 bases) (18). Isolates from Oxford analyzed by Typewriter were mapped and de novo assembled with exclusion parameters of <70% coverage of the reference genome for mapping and <50% of the genome in contigs >1 kb (10). Mykrobe processes raw sequence data with no prior cleaning of the data. The isolates came from 111 sequence types, including 29 new sequence types (STs)/alleles, covering the range of S. aureus genomic diversity as previously described in Oxfordshire.
Three programs, Genefinder (M. Doumith, PHE, not published), Mykrobe (P. Bradly, version v0.3.13-2-gd5880fa, open-source at https://github.com/iqbal-lab/Mykrobe-predictor), and Typewriter (T. Golubchik, MMM group, Oxford University; version 2.0, https://github.com/tgolubch/typewriter) (Table 1), were compared to determine presence/absence of resistance determinants (genes or variants) and toxin genes (Tables 2, 3, and 4). Mykrobe is part of the automated processing with the Complete Pathogen Software Solution (COMPASS) developed at the University of Oxford. This returns quality and depth of sequence metrics, maps against a reference (MRSA 252, GenBank accession no. BX571856.1) using Stampy (19) and performs de novo assembly using Velvet v1.0.18 (20). These de novo assemblies formed the basis for the Typewriter program, whereas Genefinder used the raw sequencing reads.
TABLE 1.
Overview of Genefinder, Mykrobe, and Typewriter methods and requirements
| Characteristics | Genefinder | Mykrobe (9) | Typewriter (10) |
|---|---|---|---|
| Method | Maps raw reads to list of target alleles using Bowtie | Looks for list of target alleles in de Bruijn assembly graph | Blasts list of target alleles against de novo assembliesa |
| Input | Fastq file | Fastq file | Genome assembly output (Velvet) |
| Required homology to declare gene presence/absence | >90% to target allele | Based on kmer recovery: K is minimum percentage expected to be recovered for a gene; K = 0.3 for blaZ, K = 0.6 for fusB and fusC, K = 0.8 otherwise | >90% relative coverage (homology by length) (80% for blaZ) |
| Required homology to declare SNP | >90% to target: can be modified | 100% of 63-kmers required to call a variant present | >90% to target: can be modified |
| Prediction of stop codons in genes present | Yes | No: there is no assembly | Yes |
| Reads can be mapped to | Multiple targets | Single target | Single target |
| Speed/processor | 1 to 3 min on laptop with 2.3-GHz processor and 16-GB memoryb | 2 min on laptop with 2.3-GHz processor and 16-GB memory | 3 h for assemblies on cloud computational system, then a few minutes for BLAST |
| Sequence quality control | Threshold adjusted if gene has multiple reference sequences or variable level of diversity; can detect potential contamination by comparing avg depth of coverage | Can identify mixtures of different species and same species | Thresholds for N50 and parallel reference-based mapping: nothing reported if below these thresholds |
Using blastn for sequence identity and tblast for mutations.
Genefinder speed is relative to the number of genes present in the database.
TABLE 2.
Predicted antibiotic susceptibility phenotypes from WGS by Genefinder, Mykrobe, and Typewriter
| Antibiotic | Susceptibility prediction for Genefinder, Mykrobe, and Typewriter (n)a |
Discordant across methods (n [%]) | |||||
|---|---|---|---|---|---|---|---|
| RRR | SSS | RRS | RSR | RSS | SRS | ||
| Ciprofloxacin | 304 | 1,072 | 0 | 2 | 0 | 1 | 3 (0.2) |
| Clindamycin | 338 | 1,024 | 7 | 0 | 0 | 10 | 17 (1.2) |
| Erythromycin | 354 | 1,011 | 6 | 0 | 0 | 8 | 14 (1.2) |
| Fusidic acid | 151 | 1,221 | 4 | 0 | 0 | 3 | 7 (0.5) |
| Gentamicin | 76 | 1,300 | 1 | 0 | 0 | 2 | 3 (0.2) |
| Methicillin | 393 | 984 | 2 | 0 | 0 | 0 | 2 (0.1) |
| Mupirocin | 15 | 1,362 | 0 | 0 | 2 | 0 | 2 (0.1) |
| Penicillin | 1,161 | 211 | 3 | 0 | 0 | 4 | 7 (0.5) |
| Rifampin | 23 | 1,354 | 0 | 1 | 0 | 1 | 2 (0.1) |
| Tetracycline | 121 | 1,249 | 4 | 0 | 0 | 5 | 9 (0.7) |
| Trimethoprim | 175 | 1,199 | 3 | 1 | 0 | 1 | 5 (0.4) |
| Vancomycin | 0 | 1,379 | 0 | 0 | 0 | 0 | 0 (0.0) |
| Total (% of 16,548) | 3,111 (18.8) | 13,366 (80.8) | 30 (0.2) | 4 (0.02) | 2 (0.01) | 35 (0.2) | 71 (0.4) |
n = 1,379. R, resistant; S, susceptible.
TABLE 3.
Predicted phenotype for antimicrobial susceptibility
| Laboratory phenotypea | Antimicrobial susceptibility prediction from Genefinder, Mykrobe, Typewriter (n)b |
Total (n) | |||||
|---|---|---|---|---|---|---|---|
| RRR | SSS | RRS | RSR | RSS | SRS | ||
| R | 2,720 | 89 | 9 | 3 | 0 | 4 | 2,825 |
| S | 97 | 11,504 | 13 | 1 | 2 | 22 | 11,639 |
| Total | 2,817 | 11,593 | 22 | 4 | 2 | 26 | 14,464 |
R, resistant; S, susceptible.
Not all isolates were phenotyped for all antimicrobials; therefore, total with phenotypes (14,464) is less than the total with genotypic predictions (16,548) in Table 2. Boldface font shows complete concordance, and italic font indicates a majority concordance between predictions.
TABLE 4.
Predicted genotype for virulence genes, ccr genes, and mecA-mecC
| PCRa | Prediction from Genefinder, Mykrobe, and Typewriter (n)b |
Total (n) | |||
|---|---|---|---|---|---|
| AAA | PPP | APA | PPA | ||
| A | 3,362 | 82 | 10 | 17 | 3,475 |
| P | 14 | 618 | 2 | 10 | 643 |
| Total | 3,376 | 700 | 12 | 27 | 4,115 |
A, absent; P, present.
Only PHE isolates had PCR results for some virulence genes. Boldface font shows complete concordance, and italic font indicates a majority concordance between predictions.
Although all three methods search for matches to a predefined list of alleles, they have different approaches to their identification (further details below). Genefinder and Mykrobe required fastq files, whereas Typewriter used BLAST use de novo assemblies. All used preset thresholds to detect genes. Thresholds are adapted for certain genes (e.g., blaZ, which can be chromosomally integrated or carried on plasmids) to improve the prediction and for quality control. Both Typewriter and Mykrobe identified the presence or absence of each target singly, whereas Genefinder identified which of closely related homologs is most plausibly present. Genefinder and Mykrobe were very fast, between 1 and 3 min, and can be used on a standard desktop computer (specification of a 2.3-GHz processor and 16-GB memory). Typewriter, as it requires de novo assembly, took up to 3 h and used cloud computing or high-capacity servers.
Genefinder was written by M. Doumith. It used a mapping approach (similar to SRST2, https://github.com/katholt/srst2) to detect the presence or absence of predefined genes or variations in predefined genes using Bowtie. The thresholds were defined at 90% overall, but amended where required in order to distinguish between both variants where genes were represented with multiple reference sequences and the level of diversity expected for each gene sought. Genefinder also checked for premature stop codons and compared the average depth of read coverage to identify any potential sequence contamination.
Mykrobe was written by P. Bradley and Z. Iqbal (9). A threshold frequency was generated for each gene (K minimum percentage) based on the empirical level of diversity observed in the training set described by Bradley (K = 0.3 for blaZ, K = 0.6 for fusB and fusC, K = 0.8 otherwise). The maximum likelihood from 3 models (gene absent, gene present in minor proportion, gene present) was chosen. The models took into account the expected proportion of kmers based on the depth of coverage and empirical level of diversity (described in reference 9). Mutations were genotyped by choosing the maximum likelihood model from 3 Poisson models comparing the depths of coverage across 63-bp reference and alternate alleles while demanding 100% coverage across the allele, also described in reference 9.
Typewriter was developed by T. Golubchik (described in reference 10). It considered BLAST results over a query reference (blastn for sequence identity, tblastn for mutations). It used a “relative coverage” to determine presence/absence of a gene, a metric that gives equal weights to coverage and sequence identity. Typewriter reported this value for each query gene of interest, and cutoffs were adjusted to optimize the specificity/sensitivity for different genes. In this study, a relative cutoff of 90% for resistance and toxin genes was used except blaZ, for which a cutoff of 80% was used. For variant reporting, mutations were reported above a given threshold of relative coverage (e.g., 90%); however, this could be changed or set to 0% to report all identified differences from the query sequence. Stop codons were predicted, as were novel mutations.
Eighty-four genes were included in the analysis; 46 acquired resistance genes, 5 sets of chromosomal variants within resistance-associated genes, 5 cassette chromosome recombinase genes (ccr), and 28 virulence genes (Tables 2, 3, and 4). Acquired resistance genes were classified as present (p, P) or absent (a, A), setting 3 missing Genefinder predictions (“ND” or “X”) to absent. Chromosomal resistance variants were those listed in Table S4 in the supplemental material; 23 other mutations were reported in the relevant genes but were not compared, as they are not considered resistance-determinants (Table S4 in the supplemental material). For all methods, genotype predictions of susceptibility phenotypes were based on the presence of any relevant resistance determinant as shown in Tables 2, 3, and 4 (as described in reference 10 with minor modifications and updates from reference 9). Intermediate phenotype results were excluded from analysis (80 cases [0.5%]).
RESULTS
Short-read Illumina WGS data were available from 1,389 samples; 992 from a collection held in Oxford (previously described by Gordon et al. [9, 10]) and 397 from Public Health England (PHE) Staphylococcus Reference Service, Colindale. Ten samples were excluded due to mixed/contaminated WGS results, leaving 1,379 for analysis. Samples were analyzed by Genefinder and Typewriter (Table 1) after sequence mapping and variant calling and by Mykrobe from raw fastq reads.
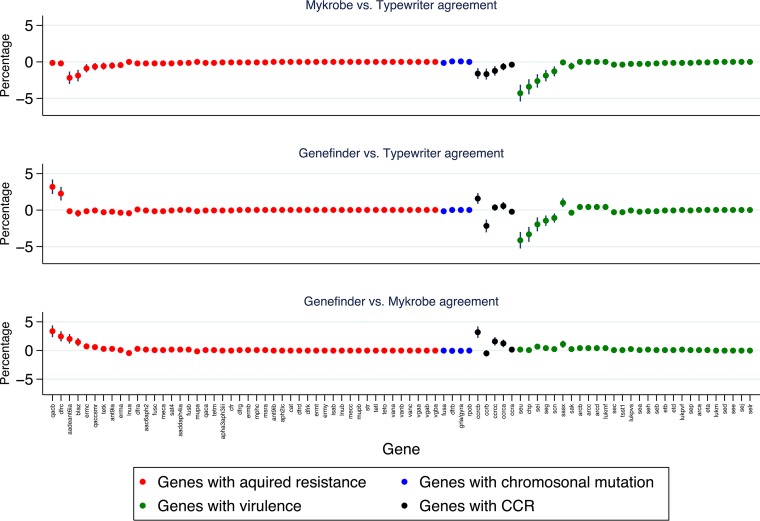
Eighty-four genes were included: 46 acquired resistance genes, 5 sets of chromosomal variants within genes associated with resistance, 3 cassette chromosome recombinase, ccrA, ccrB, and ccrC, including three variants of ccrC [ccrC(a), ccrC(b), and ccrC(c)], and 28 virulence genes (see Table S1 in the supplemental material). Overall, 99.5% (113,830/114,457) of the individual resistance determinant/virulence gene predictions were identical between all three methods (Fig. 1; Table S1), with only 627 (0.5%) discordant predictions, demonstrating high overall agreement (Fleiss' kappa = 0.98, P < 0.0001). Overall, one method disagreed with both other methods in 0.23% for Typewriter (263/114,457 predictions), 0.16% for Mykrobe (183/114,457), and 0.16% for Genefinder (181/114,457). The three most common discrepancies for Typewriter were the nondetection of virulence genes identified by other methods (seu, 57 samples; chp, 46 samples; sei, 33 samples). Similarly, for Genefinder, the three most common discrepancies were nondetection of resistance genes (qacB, 44 samples; dfrC, 34 samples) or other genes (ccBb, 22 samples) identified by other methods. Genefinder reported the presence of dfrA, qacA, or ccrC(b) genes in these samples. In contrast, Typewriter and Mykrobe reported the presence of two dfr, two qac, and three ccrC genes, where the detected variants for each of these three genes shared more than 90% nucleotide identity. The most common discrepancies for Mykrobe were identifying resistance/other genes as present when the other two methods called them absent (aadE–ant(6)-Ia, 28 samples; blaZ, 19 samples; ccrCB, 22 samples). No gene was ever identified as present by Typewriter alone. Fourteen of the 84 genes had >1% discrepancies (maximum, 4.3% for seu), but the majority of discrepancies were in only one method for all genes except ccrC(b).
FIG 1.
Determinant-by-determinant disagreements between methods. Each panel shows percentage differences in proportions of the detected presence of each determinant between the first method and the second.
Discrepancies were similar in acquired resistance genes (0.3% [221/63,434]) and chromosomal resistance genes (0.1% [8/5,516]), but slightly larger for ccr genes (1.8% [123/6,895]) and virulence genes (0.7% [275/38,612]) (see Table S2). The percentage discrepancies varied modestly across the different sample sets, being higher for the PHE set (1.1% [349/32,928]; particularly for ccr genes with 4.2% [83/1,960] discrepancies), intermediate for the Oxford derivation set (0.6% [233/42,084]) and lowest for the Oxford validation set (0.1% [45/40,824]) (Table S2).
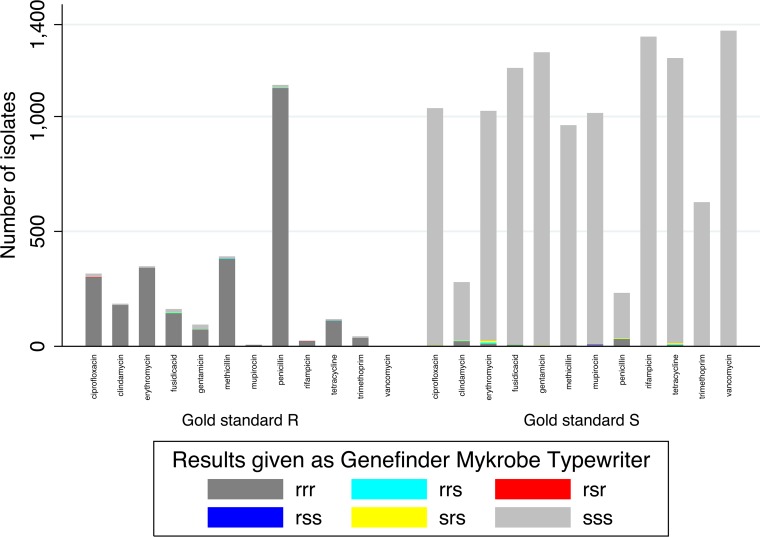
Genotypic predictions of antimicrobial susceptibility were also identical in 99.6% of cases (16,477/16,548 predictions) (Table 2). Of the 71 discrepancies in susceptibility prediction between the methods, 42% (30/71) occurred with Typewriter reporting susceptible when Genefinder and Mykrobe reported resistant, and 49% (35/71) occurred with Mykrobe reporting resistant where Genefinder and Typewriter reported susceptible.
Comparing genetic predictions to laboratory phenotypes (restricted to samples either phenotypically resistant or susceptible), in 98.3% (14,224/14,464) of cases, all three bioinformatics methods and the gold standard laboratory results agreed completely (2720 [18.8%] resistant, 11,504 [79.5%] susceptible) (Table 3 and Fig. 2). There was greater disagreement between the laboratory phenotypic results and the combined genotypic predictions than within the three bioinformatics methods. In 97 (0.7%) instances, the laboratory phenotype was susceptible, but all bioinformatic methods reported resistance. Of these, 33% (32/97) were for penicillin, 23% (22/97) for clindamycin, and 11% (11/97) for erythromycin, with smaller numbers for fusidic acid (7), tetracycline (6), mupirocin (6), methicillin (5), ciprofloxacin (4), gentamicin (3), and rifampin (1), and none for trimethoprim. In 89 (0.6%) instances, the laboratory phenotype was resistant, but all three bioinformatics methods reported susceptible, most commonly to gentamicin (21% [15/89]), ciprofloxacin (17% [15/89]), and fusidic acid (15% [13/89]). The remaining 54 (0.4%) cases (16 phenotypically resistant, 38 phenotypically susceptible) had different genotypic predictions made from the different methods. However, in 36/54 (67%), the consensus genotype (predicted by two of the three methods) matched the laboratory phenotype.
FIG 2.
Antimicrobial susceptibility genotypic predictions compared to phenotype.
PCR/array results were available for some virulence genes (15) and mecA-mecC for all 397 PHE isolates. Compared with genetic predictions, in 96.8% (3,983/4,115) of cases, all three bioinformatics methods and the PCR/array results agreed completely (3,364 [81.7%] absent, 619 [15.0%] present) (Table 4; see also Fig. S1). As for antimicrobial resistance, there was greater disagreement between the laboratory PCR/array results and the combined genotypic predictions than within the three bioinformatics methods, with 81 (2.0%) cases where all three methods called a gene present that had not been detected by PCR/array and 12 (0.3%) where no method called a gene present that had been detected by PCR/array, in comparison with 39 (0.9%) discrepant predictions between the methods. In 20/39 (51%) cases, the consensus genotype matched the PCR/array result.
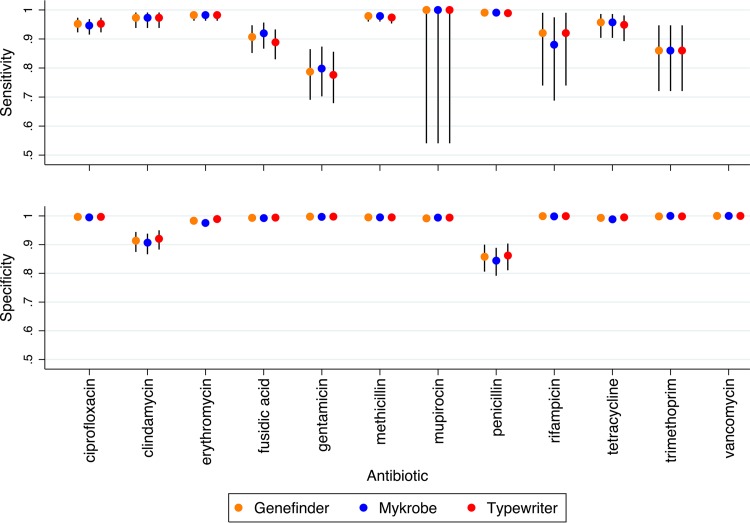
The sensitivity and specificity of all three bioinformatics methods compared to laboratory phenotypic methods in predicting antimicrobial susceptibility were very similar. Across the 14,464 genotypic predictions, Typewriter had the lowest overall sensitivity (0.964 [95% CI, 0.956 to 0.970]), but the highest specificity (0.992 [0.990 to 0.993]), while Mykrobe had higher sensitivity (0.967 [0.960 to 0.974]) and the lowest specificity (0.989 [0.987 to 0.990]). Genefinder's performance fell between that of Mykrobe and Typewriter for specificity (0.990 [0.988 to 0.992]), with a sensitivity equal to that of Mykrobe (0.967 [0.960 to 0.973]). Specificity and sensitivity varied across the different antibiotics (Fig. 3), but were broadly similar between the three methods, overall and within each data set (see Table S3). There were no vancomycin-resistant isolates identified by either phenotyping or bioinformatics methods. Similarly, specificity and sensitivity to identify PCR-detected virulence and other genes varied across the different genes, but were broadly similar between the three methods (see Fig. S2).
FIG 3.
Sensitivity and specificity of genotypic predictions of antimicrobial susceptibility.
DISCUSSION
While WGS is increasingly used to detect antibiotic resistance and virulence determinants, to our knowledge, this is the first study that compares three methods for predicting genotypes of large numbers of isolates. As discussed in the recent European Committee on Antimicrobial Susceptibility Testing (EUCAST) report (21), discordance can occur between phenotypic and genotypic resistance due to inadequate limits of detection for WGS methods, incomplete understanding of the genotypic basis of phenotypic resistance, flaws with the phenotypic or molecular (e.g., PCR) methods currently used to detect resistance, and/or WGS failures, including the lack of assembly caused by multiple operons or similar sequences, incomplete gene coverage, nonfunctional genes (e.g., due to the presence of stop codons/indels), or cropped contigs.
Here, we found that three different approaches for identifying genetic determinants of resistance and virulence (Genefinder, Mykrobe, and Typewriter) agreed in 99.5% of predictions. Genefinder and Mykrobe were fast, taking under 5 min, whereas Typewriter, while also taking a few minutes per sample, required an initial genome assembly that increased the turnaround time by up to 3 h. Mykrobe and Typewriter are freely available (https://github.com/iqbal-lab/Mykrobe-predictor and https://github.com/tgolubch/typewriter, respectively); Genefinder is not, but the underpinning methods are relatively straightforward, and the freely available SRST2 (https://github.com/katholt/srst2) follows an analogous mapping approach (22), which would likely provide very similar results with the same catalogue. Previous comparisons of bioinformatics methods relevant to the microbiology community are limited. Bradley et al. (9) found good concordance between Mykrobe and SeqSphere (23), an allele-based method that detects the presence/absence of a limited number of resistance and virulence markers. SeqSphere took longer than Mykrobe as, like Typewriter, it uses Velvet assemblies. Other previous studies have shown 100% concordance between the resistome and toxome in 14 MRSA isolates (24), 98.6% concordance across 5,288 susceptibility predictions in 308 S. aureus isolates (both MRSA and MSSA) (25), 100% concordance for selected resistance and toxin gene presence/absence in 18 MRSA strains (23), and 97% and 97% sensitivity and specificity, respectively, for Typewriter and 99.1% and 99.6% sensitivity and specificity, respectively, for Mykrobe for predicting phenotypic resistance in the Oxford validation samples used here (9, 10). A comparison between microarray and WGS in 154 isolates reported 1.7% discordancy in detecting resistance and virulence genes (26), mainly due to the failure of WGS to detect enterotoxins and super antigens (similar to that for Typewriter in this study).
Individually, the three programs demonstrated high concordance, but interestingly, in almost all genes, only one of the three bioinformatics methods did not identify a determinant that the other two methods did identify, or vice versa. The most common discrepancy with Typewriter was the failure to identify virulence genes identified by Mykrobe and Genefinder (namely, seu, chp, and sei). Two of these genes, sei and seu, are located on the enterotoxin gene cluster (egc) (27, 28), referred to as an enterotoxin gene nursery (29), and the other, chp, is located on a prophage (30). Such regions may be particularly susceptible to recombination (31, 32) and paralogs. As Typewriter uses BLAST, it may have a higher chance of detecting one of multiple closely related genes than the other two methods.
Similarly to Typewriter, the most common discrepancy with Genefinder was a failure to identify genes reported by Typewriter or Mykrobe, particularly, ccrB, qacB (quaternary ammonium compound B, conferring resistance to chlorhexidine [33] via an efflux drug pump, but differing from another gene, qacA, by only seven nucleotides [34]), and dfrC (a dihydrofolate conferring resistance to trimethoprim believed to be the origin of the more common transposon-associated drfA gene). The fact that Genefinder identified only one variant of acquired dfr and qac may indicate that the other two methods were misidentifying paralogs (35). Alternatively, as Genefinder detects predetermined alleles, the recombination of partial genes or differences in flanking sites or genomic variation alone may reduce its ability to detect some genes. One advantage of Genefinder is its ability to detect variations in multicopy genes such as the rRNA encoding genes associated with linezolid resistance in staphylococci.
In contrast, Mykrobe most commonly identified a determinant that other methods did not, particularly, aadE(ant6′)-Ia, an adenyltransferase encoding resistance to aminoglycosides. This gene is associated with small plasmids flanked by direct repeats of staphylococcal insertion sequence IS257 (36). Although Mykrobe is kmer based, it requires a high match across the whole gene, not just flanking sequences, and so the reason for this is unclear. Mykrobe also had a higher false-positive rate in blaZ, as reported previously (9). Although this was previously attributed to phenotypic errors, the fact that neither Genefinder nor Typewriter identified blaZ in these isolates suggests the algorithm/threshold may need adjusting for this gene. Mykrobe also had a high false-positive rate for the ccrCB gene, which is part of the cassette chromosome recombinase (ccr) associated with SSCmec (37). As all ccrC genes share >87% similarity and were not included in the original Mykrobe implementation, further investigation and modification of sequence identity thresholds may be required to accurately classify this gene, whose different alleles can have 60% to 82% sequence identity.
Overall, the comparison highlights key challenges inherent in all methods. First are the trade-off between specificity and sensitivity to detect specific genes/variants and the need for adjustment based on specific features, such as the proximity to repetitive elements or similarity with other alleles. Specific genes may also require different approaches, e.g., the ccr genes were the most discordant overall in the study. These genes were more often present in the staphylococcal reference laboratory isolates, increasing the overall error rates for this sample set. Reference libraries of genes/variants also require frequent updating with new alleles, and appropriate thresholds must be set to enable separate copies of closely related genes (e.g., qacA and qacB) to be detected if genuinely present. Taking the consensus prediction across the three different bioinformatics methods is one strategy for balancing these different trade-offs. As error rates were low overall, this only improved the genetic predictions slightly, but in samples where the susceptibility is unknown, it could be valuable, particularly if the two fast implementations (Genefinder and Mykrobe) are used, followed by the slower assembly-based method only if they disagree.
Our main findings were that the largest discordance occurred between phenotype and genotype regardless of the method used to predict genotype and that the “consensus” genotypic prediction agreed with the phenotype in two-thirds of the small number of cases where bioinformatics methods made different predictions. Where bioinformatics methods are concordant but disagree with phenotype, the unresolved question is which is “correct,” in terms of a drug achieving clinical cure in a patient infected with this strain. Penicillin and clindamycin/erythromycin were most likely to be called resistant by all methods but susceptible by phenotyping. Previous studies of erythromycin and clindamycin resistance have reported positive ermC PCR results from nondetectable resistance phenotypes (38) and have suggested that plasmids conferring resistance to these antibiotics may be lost in subculture (9, 39). Sensitivity to penicillin by phenotypic methods where genotype methods predict resistance has been reported previously (40, 41), and the evidence suggests that phenotyping underreports resistance. The EUCAST guidelines illustrate the challenges in distinguishing between penicillin-resistant and -susceptible isolates based on fuzzy versus sharp zones (42). Overall, therefore, it is plausible that the genetic detection of resistance may reflect more closely the impact of the strain on a patient.
An interpretation where phenotyping reports resistance but WGS methods predict susceptibility is more difficult. One possibility is small colony variants (SCV) being present phenotypically but overgrown in WGS culture and thus not represented in the sequence. Resistance associated with gentamicin, fusidic acid, and ciprofloxacin, the main antibiotics where this phenomenon was observed, is observed with SCV phenotypes (43, 44). An alternative explanation is novel resistance mechanisms, for example, for ciprofloxacin (45), leading to false-negative WGS predictions. The need for a continuously updated curated database is a key challenge for WGS methods. As more sequencing occurs, novel mutations will be identified in resistance genes that may or may not confer phenotypic resistance, but these can at least be identified and tested; identifying entirely new resistance-conferring genes is more complex, and prediction software that can recognize new clinically important genes a priori would be a valuable addition to an analysis pipeline. However, we observed similar differences between concordant genotypic predictions and both phenotypic antimicrobial susceptibilities and single gene PCR results, suggesting that the underlying causes may not necessarily be related to resistance. As previously noted, the agreement between WGS and phenotyping is higher (98.6%) than between phenotyping undertaken by two separate laboratories (97.6%) (25); thus, at least some discrepancies are probably due to incorrect phenotyping results. In contrast, the concordance between genotypic predictions made using a single method but based on WGS generated from 5 different laboratories was recently shown to be >99.8% (46).
Limitations.
This comparison was based on a prespecified set of resistance- or virulence-associated genes: some genetic traits previously associated with resistance were omitted (e.g., IleS mutations linked to low-level mupirocin resistance). Despite this, we found good agreement between genotypic predictions and phenotype. Typewriter used Velvet de novo assemblies: other newer assemblers (e.g., SPADES [47]) might have improved predictions further. We included data which had been used in the development of two of the methods compared, which could potentially have led to overfitting, although the performances of all three methods were in fact similar on this data set (see Table S2 in the supplemental material). All analyses were undertaken on short-read Illumina data. The increasing use of long-read sequences will require further software testing, although Mykrobe has been successfully used for initial resistance calling in Mycobacterium tuberculosis from Nanopore sequencing in a small number of samples (48). However, it has not been comprehensively tested, nor have Typewriter and Genefinder, with long-read sequences generated using Nanopore or PacBio technology. The greatest differences detected in this study were between phenotype and genotype, which could be partly due to the method of phenotypic testing and recognized issues with reproducibility. We did not have resources to rephenotype all or a subset of the isolates; well-characterized sets of repeatedly phenotyped isolates would be useful for further studies. We found no suggestion that missing calls in one program were associated with scores just below a threshold but did not undertake a more detailed assessment of specific sequence coverage and quality around discrepant genetic predictions.
Conclusion.
In summary, in this study, the choice between three specific bioinformatics methods to identify resistance determinants or other genes in S. aureus did not prove critical. All demonstrated a high concordance with each other and with phenotypic methods and can be recommended for genotype prediction. However, each has some limitations; therefore, consensus methods provide at least some assurance. Due to computational speed, Mykrobe (de Bruijn graph based) and Genefinder (or equivalent mapping-based program such as SRST2 [22]) are a sensible combination to use as an initial consensus method, followed by Typewriter (BLAST based) if these two methods disagree. As a set of 34 diverse bacteria have been made available for whole-genome sequencing validation (49), the study strains and genotypic predictions are available as a resource for other studies investigating different bioinformatics analysis methods, which will become increasingly important as this technique is more widely used to inform clinical management, through bacterial identification, antimicrobial susceptibility prediction, and virulence profiling. External quality control of clinical laboratory performance in predicting antibiotic resistance is provided by UK proficiency testing schemes such as the United Kingdom National External Quality Assessment Service for Microbiology (UK NEQAS) (50); a similar set of standards will need to be created to accredit whole-genome sequencing methods.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at Oxford University in partnership with Public Health England ([PHE] grant HPRU-2012-10041) and the NIHR Oxford Biomedical Research Centre; D.C. and T.P. are NIHR senior investigators.
This report presents independent research funded by the National Institute for Health Research and the Department of Health. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, the Department of Health, or Public Health England.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.00813-18.
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01815-17.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Public Health England. 2016. Annual epidemiological commentary: mandatory MRSA, MSSA and E. coli bacteraemia and C. difficile infection data 2015/16. Public Health England, London, United Kingdom: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/535635/AEC_final.pdf. [Google Scholar]
- 3.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol 26:166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, Shah ND, Mandrekar JN, Patel R. 2015. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 61:1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strauss C, Endimiani A, Perreten V. 2015. A novel universal DNA labeling and amplification system for rapid microarray-based detection of 117 antibiotic resistance genes in Gram-positive bacteria. J Microbiol Methods 108:25–30. doi: 10.1016/j.mimet.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Berthet N, Dickinson P, Filliol I, Reinhardt AK, Batejat C, Vallaeys T, Kong KA, Davies C, Lee W, Zhang S, Turpaz Y, Heym B, Coralie G, Dacheux L, Burguiere AM, Bourhy H, Old IG, Manuguerra JC, Cole ST, Kennedy GC. 2008. Massively parallel pathogen identification using high-density microarrays. Microb Biotechnol 1:79–86. doi: 10.1111/j.1751-7915.2007.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price JR, Didelot X, Crook DW, Llewelyn MJ, Paul J. 2013. Whole genome sequencing in the prevention and control of Staphylococcus aureus infection. J Hosp Infect 83:14–21. doi: 10.1016/j.jhin.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TE, Crook DW, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 13.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):S5–S16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 15.Dhup V, Kearns AM, Pichon B, Foster HA. 2015. First report of identification of livestock-associated MRSA ST9 in retail meat in England. Epidemiol Infect 143:2989–2992. doi: 10.1017/S0950268815000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahuerta-Marin A, Guelbenzu-Gonzalo M, Pichon B, Allen A, Doumith M, Lavery JF, Watson C, Teale CJ, Kearns AM. 2016. First report of lukM-positive livestock-associated methicillin-resistant Staphylococcus aureus CC30 from fattening pigs in Northern Ireland. Vet Microbiol 182:131–134. doi: 10.1016/j.vetmic.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden M, Hopkins KL, Iredell J, Kahlmeter G, Koser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen O, Woodford N. 2017. The role of whole genome sequencing (WGS) in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aanensen DM, Feil EJ, Holden MT, Dordel J, Yeats CA, Fedosejev A, Goater R, Castillo-Ramirez S, Corander J, Colijn C, Chlebowicz MA, Schouls L, Heck M, Pluister G, Ruimy R, Kahlmeter G, Ahman J, Matuschek E, Friedrich AW, Parkhill J, Bentley SD, Spratt BG, Grundmann H, European SRL Working Group. 2016. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. mBio 7:e00444-16. doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss L, Ruffing U, Abdulla S, Alabi A, Akulenko R, Garrine M, Germann A, Grobusch MP, Helms V, Herrmann M, Kazimoto T, Kern W, Mandomando I, Peters G, Schaumburg F, von Muller L, Mellmann A. 2016. Detecting Staphylococcus aureus virulence and resistance genes: a comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol 54:1008–1016. doi: 10.1128/JCM.03022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munson SH, Tremaine MT, Betley MJ, Welch RA. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun 66:3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letertre C, Perelle S, Dilasser F, Fach P. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J Appl Microbiol 95:38–43. doi: 10.1046/j.1365-2672.2003.01957.x. [DOI] [PubMed] [Google Scholar]
- 29.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol 166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 30.Bae T, Baba T, Hiramatsu K, Schneewind O. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol 62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A 98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2005. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol Lett 246:191–198. doi: 10.1016/j.femsle.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Poovelikunnel T, Gethin G, Humphreys H. 2015. Mupirocin resistance: clinical implications and potential alternatives for the eradication of MRSA. J Antimicrob Chemother 70:2681–2692. doi: 10.1093/jac/dkv169. [DOI] [PubMed] [Google Scholar]
- 34.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 93:3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dale GE, Broger C, Hartman PG, Langen H, Page MG, Then RL, Stuber D. 1995. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermidis ATCC 14990: the origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol 177:2965–2970. doi: 10.1128/jb.177.11.2965-2970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byrne ME, Gillespie MT, Skurray RA. 1991. 4′,4″ adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid 25:70–75. doi: 10.1016/0147-619X(91)90008-K. [DOI] [PubMed] [Google Scholar]
- 37.International Working Group on the Classification of Staphylococcal Cassette Chromosome E. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martineau F, Picard FJ, Lansac N, Menard C, Roy PH, Ouellette M, Bergeron MG. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 44:231–238. doi: 10.1128/AAC.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlickova H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramirez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nubel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaase M, Lenga S, Friedrich S, Szabados F, Sakinc T, Kleine B, Gatermann SG. 2008. Comparison of phenotypic methods for penicillinase detection in Staphylococcus aureus. Clin Microbiol Infect 14:614–616. doi: 10.1111/j.1469-0691.2008.01997.x. [DOI] [PubMed] [Google Scholar]
- 41.El Feghaly RE, Stamm JE, Fritz SA, Burnham CA. 2012. Presence of the bla(Z) beta-lactamase gene in isolates of Staphylococcus aureus that appear penicillin susceptible by conventional phenotypic methods. Diagn Microbiol Infect Dis 74:388–393. doi: 10.1016/j.diagmicrobio.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 42.EUCAST. 2017. Testing TECoAS. Breakpoints for interpretation of MICs and zone diameters; version 7.0.
- 43.Norström T, Lannergard J, Hughes D. 2007. Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob Agents Chemother 51:4438–4446. doi: 10.1128/AAC.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz FJ, von Eiff C, Gondolf M, Fluit AC, Verhoef J, Peters G, Hadding U, Heinz HP, Jones ME. 1999. Staphylococcus aureus small colony variants: rate of selection and MIC values compared to wild-type strains, using ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin. Clin Microbiol Infect 5:376–378. doi: 10.1111/j.1469-0691.1999.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 45.Piddock LJ, Jin YF, Webber MA, Everett MJ. 2002. Novel ciprofloxacin-resistant, nalidixic acid-susceptible mutant of Staphylococcus aureus. Antimicrob Agents Chemother 46:2276–2278. doi: 10.1128/AAC.46.7.2276-2278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mellmann A, Andersen PS, Bletz S, Friedrich AW, Kohl TA, Lilje B, Niemann S, Prior K, Rossen JW, Harmsen D. 2017. High interlaboratory reproducibility and accuracy of next-generation-sequencing-based bacterial genotyping in a ring trial. J Clin Microbiol 55:908–913. doi: 10.1128/JCM.02242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozyreva VK, Truong CL, Greninger AL, Crandall J, Mukhopadhyay R, Chaturvedi V. 2017. Validation and implementation of clinical laboratory improvements act-compliant whole-genome sequencing in the public health microbiology laboratory. J Clin Microbiol 55:2502–2520. doi: 10.1128/JCM.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White LO. 2000. UK NEQAS in antibiotic assays. J Clin Pathol 53:829–834. doi: 10.1136/jcp.53.11.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.