Activation of the adaptive response to cellular stress orchestrated by heat shock factor 1 (HSF1), which is an evolutionarily conserved transcriptional regulator of chaperone response and cellular bioenergetics in diverse model systems, is a central feature of organismal defense from environmental and cellular stress. HSF1 activity, induced by proteostatic, metabolic, and growth factor signals, is regulated by posttranscriptional modifications, yet the mechanisms that regulate HSF1 and particularly the functional significance of these modifications in modulating its biological activity in vivo remain unknown.
KEYWORDS: posttranslational HSF1 regulation, phosphorylation of HSF1 (Ser303A/Ser307A), knock-in mouse model, obesity, insulin resistance
ABSTRACT
Activation of the adaptive response to cellular stress orchestrated by heat shock factor 1 (HSF1), which is an evolutionarily conserved transcriptional regulator of chaperone response and cellular bioenergetics in diverse model systems, is a central feature of organismal defense from environmental and cellular stress. HSF1 activity, induced by proteostatic, metabolic, and growth factor signals, is regulated by posttranscriptional modifications, yet the mechanisms that regulate HSF1 and particularly the functional significance of these modifications in modulating its biological activity in vivo remain unknown. HSF1 phosphorylation at both Ser303 (S303) and Ser307 (S307) has been shown to repress HSF1 transcriptional activity under normal physiological growth conditions. To determine the biological relevance of these HSF1 phosphorylation events, we generated a knock-in mouse model in which S303 and S307 were replaced with alanine (HSF1303A/307A). Our results confirmed that loss of phosphorylation in HSF1303A/307A cells and tissues increases protein stability but also markedly sensitizes HSF1 activation under normal and heat- or nutrient-induced stress conditions. Interestingly, the enhanced HSF1 activation in HSF1303A/307A mice activates a supportive metabolic program that aggravates the development of age-dependent obesity, fatty liver diseases, and insulin resistance. Thus, these findings highlight the importance of a posttranslational mechanism (through phosphorylation at S303 and S307 sites) of regulation of the HSF1-mediated transcriptional program that moderates the severity of nutrient-induced metabolic diseases.
INTRODUCTION
The protective adaptive response of organisms to cellular and exogenous stress involves upregulation of transcriptional programs, which are required to maintain cellular protein homeostasis and optimal regulation of metabolic bioenergetics. In particular, a primary feature of this protective response is the activation of heat shock factor 1 (HSF1), which is triggered by a variety of environmental and cellular stressors. Besides acting as a key transactivator of the classical heat shock response genes, HSF1 potentiates the expression of a wide network of stress-protective genes that are involved in a variety of cellular processes, including metabolism, cell growth, cell survival, and tumorigenesis (1–8).
The stress-dependent transcription program by HSF1 is a highly regulated multistep process that is incompletely understood. HSF1 consists of several functional domains, including the N-terminal helix-turn-helix DNA-binding domain, the heptad repeat enriched oligomerization domain, the regulatory domain, and the C-terminal transactivation domain (9, 10). Under normal physiological growth conditions, HSF1 exists in a monomeric form and kept inactive in a protein complex with heat shock protein 90 (HSP90), HSP70, and their cofactors. Upon heat shock or nutrient stimulation, the activation of HSF1 is displayed through its trimerization, nuclear translocation, and acquisition competence for binding to heat shock elements (HSEs) in the promoter regions of target genes encoding HSPs and other proteins (9–11). A complex network of posttranslational modifications, such as phosphorylation, acetylation, and sumoylation, are additionally involved to regulate different steps of HSF1 activation and stability (8). Thus, acetylation occurring on different subunits may impair chromatin binding ability but on the other hand may serve to fine-tune HSF1 activation by stabilizing protein against proteasomal degradation (12–14). Small ubiquitin-like modifier (SUMO) modifications on many different lysine residues of HSF1, directly or through an S303 phosphorylation-dependent step, are also thought to be involved in its repression (15–17). Although it is widely considered that hyperphosphorylation of HSF1 upon stress represents an activation state, several residues are also phosphorylated under basal growth conditions and modulate its activity. Indeed, basal and stress-induced phosphorylation of HSF1 by multiple kinases on multiple serine and threonine residues that can have both stimulatory and inhibitory effects on its activation is considered to be a critical posttranslational mechanism modulating HSF1 transcriptional programs (8, 10, 18, 19). However, emerging evidence suggests an alternative model of HSF1 regulation, in which phosphorylation may not control the interaction of HSF1 with targets; rather, altering HSF1 expression level acts as a fine-tuning mechanism for activation (20, 21). For example, S303/S307 phosphorylation by glycogen synthase kinase 3β (GSK3β), casein kinase 2 (CK2), and mitogen-activated protein (MAP) kinases MEK1 and extracellular signal-regulated kinase (ERK) is thought to promote degradation and downregulate HSF1 activity under normal physiological growth conditions (22–27). Along this line, it was reported that phosphorylation of S303/S307 residues is required for FBXW7 ubiquitin ligase binding, ubiquitination, and degradation of HSF1 (28). In cancer, reduced FBXW7 expression via mutations or transcriptional downregulation seems to increase malignant transformation and metastatic potential by stabilizing HSF1 protein levels. In contrast, pathogenic polyglutamine (polyQ)-conjugated huntingtin protein expression in neurons and brain tissues of patients with Huntington disease seems to increase FBXW7- and CK2α-dependent HSF1 phosphorylation on S303 and S307, and this may aggravate disease progression by driving HSF1 degradation (29). Thus, an increasing body of experimental evidence from cell-based studies and animal models highlights the important role of posttranslational modifications, in particular, phosphorylation events, in fine-tuning the regulation of HSF1 activity during cellular adaptation and disease (cancer or neuronal disorders) progression. Despite these advances, however, the functions of these phosphorylation events in vivo, although important to complete HSF1 activation, are not well understood.
In this study, we presented conclusive evidence, using a knock-in mouse model in which S303 and S307 were replaced with alanine residues, that a tight posttranslational modification program via phosphorylation events on these sites is involved to regulate at the organismal level HSF1 activity and adjust age-dependent metabolic homeostasis under normal physiological conditions.
RESULTS
Generation of hsf1303A/307A knock-in mice.
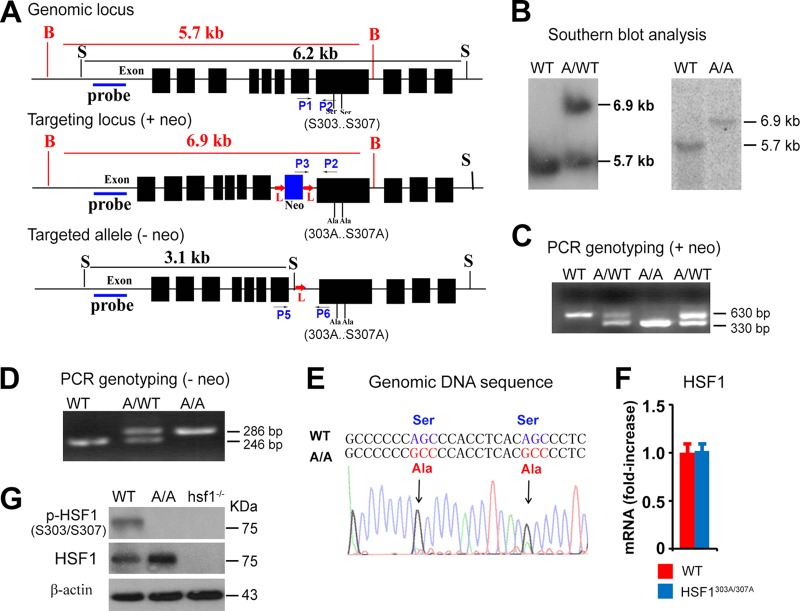
Phosphorylation of HSF1 at S303 and S307, located in the regulatory domain of murine and human HSF1 proteins, by GSK3β and ERK, respectively, has been suggested to negatively impact its transcriptional activity and decrease protein stability by priming ubiquitin ligation and degradation (8). To examine the potential impact of these phosphorylation sites on the physiological function of HSF1, we generated a knock-in mouse line in which S303 and S307 were deleted by introducing alanine substitutions (HSF1303A/307A) (see the targeting strategy in Fig. 1A). Normal splicing of the hsf1303A/307A knock-in allele was confirmed by Southern blotting (Fig. 1B) and by PCR performed on genomic DNA before (Fig. 1C) and after (Fig. 1D) removal of the neomycin gene. Genomic sequencing confirmed the desired coding changes at S303/S307 (changes of AGC to GCC) and revealed no unexpected differences in the noncoding or protein-coding sequences in this region (Fig. 1E). The real-time (RT) PCR analyses performed on mouse embryo fibroblasts (MEFs) derived from embryonic day 13 (E13) and E14 littermates from HSF1303A/307A and wild-type (WT) C57BL/6 mice did not reveal a significant difference in mRNA expression levels between the genotypes. However, a marked increase in HSF1 protein expression was evident in Hsf1303A/307A compared to WT MEFs (Fig. 1F and G). In addition, using HSF1S303/S307 phosphorylation-specific antibody, we confirmed the deletion of S303 and S307 by immunoblot analysis. Of note, hsf1−/− MEFs were used in this analysis as a control for the specificity of the antibody (Fig. 1G). The hsf1303A/307A/WT heterozygous and hsf1303A/307A homozygous mice were viable, born at the expected Mendelian distribution, and were fertile, with no apparent morphological or microscopic abnormalities detected in different tissues analyzed at 2 months of age in both genders (data not shown).
FIG 1.
Generation of HSF1 (S303A/S307A) knock-in mouse model. (A) Scheme illustrating the strategy used to generate HSF1 (S303A/S307A) (HSF1303A/307A) mutant mice. The genomic hsf1 locus, the targeted locus with the neomycin (neo) gene, and the targeted allele after neomycin removal are shown. The neomycin gene is presented in blue, and exons are in black. Locations of S303 and S307 sites in the genomic hsf1 locus, probes for Southern blot analysis, and genotyping primers (P1 to P6) for PCR are indicated. Notably, neo is flanked by loxP sites and can be removed using Cre recombinase transgenic mice. B, BglII; S, SacI restriction enzyme sites; L, loxP site. (B) Southern blot analysis using genomic tail DNA prepared from WT, heterozygous hsf1303A-307A/WT (A/WT), and homozygous hsf1303A/307A (A/A) mutant mice. DNA digested with BglII was hybridized with the probe indicated in panel A to generate a fragment of 5.7 kb for the WT locus and one of 6.9 kb for the mutant locus. (C) PCR-based genotyping and verification of WT, hsf1303A-307A/WT (A/WT), and hsf1303A/307A (A/A) mice before removal of the neo gene, performed with tail DNA, of the indicated genotypes. The combination of primers P1, P2, and P3 amplifies PCR fragments of 630 and 330 bp for WT and mutant (with neo) hsf1 alleles, respectively. (D) PCR-based genotyping of WT, heterozygous hsf1303A-307A/WT (A/WT), and homozygous hsf1303A/307A (A/A) mice after removal of the neo gene, performed on tail DNA of the indicated genotypes. The combination of primers P5 and P6 amplifies PCR fragments of 246 and 286 bp for WT and mutant (without neo) hsf1 alleles, respectively. (E) Sequence of PCR-amplified 450-bp DNA fragment including the S303 and S307 coding sequence from WT and hsf1303A/307A (A/A) mice. The coding substitutions replacing serine with alanine are indicated in the mutant genomic locus. (F) Relative mRNA levels of hsf1 in WT and hsf1303A/307A MEF cells determined by RT-PCR. Bars are means ± SD (n = 5 per group). (G) Immunoblot analysis of total and phosphorylated S303/S307 HSF1 in MEF lysates prepare from WT, hsf1303A/307A (A/A), and hsf1−/− mice. β-Actin was used as a loading control.
Loss of S303/S307 phosphorylation leads to increased cytosolic and nuclear accumulation of HSF1 and expression of downstream targets under physiological growth conditions and stress exposure.
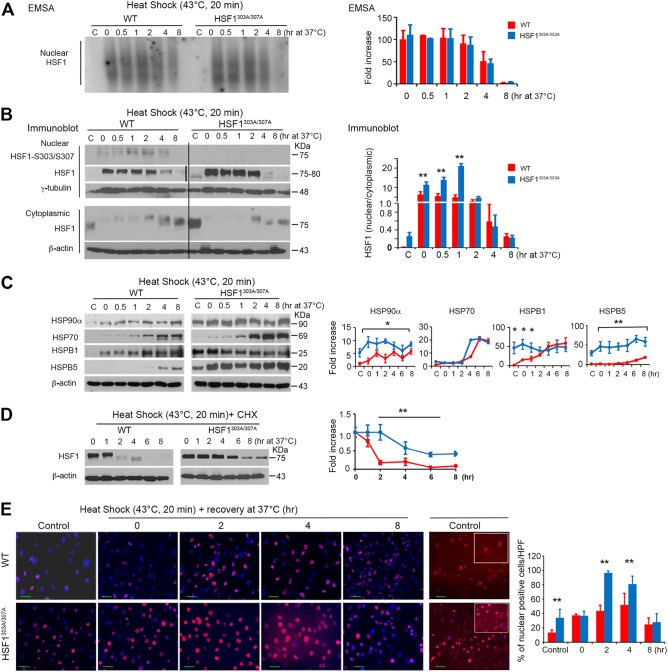
As noted earlier, HSF1 localizes in both the cytoplasm and nucleus, but upon stress it accumulates in the nucleus, where upon trimerization and posttranslational modifications it binds to DNA to induce its downstream targets. In particular, HSF1 phosphorylation on S303 and S307 has been suggested to play a role in repressing hsf1 transcriptional activity during recovery from heat challenge (23, 25–27). To examine the binding abilities of WT and HSF1303A/307A proteins under basal control conditions or following thermal stress, MEFs prepared from the two genotypes were left untreated or exposed to a moderate heat shock (43°C) for 20 min and recovered for the indicated times. Notably, exposure of cells to heat did not evidently impair cell viability, at least for the 8-h period of this analysis. Electrophoretic mobility shift assays (EMSA) performed on nuclear extracts confirmed enhanced HSF1 binding to HSE elements following heat shock, but this interaction with respect to its duration and intensity did not significantly differ between HSF1303A/307A and WT cells (Fig. 2A). Strikingly, subcellular fractionation analysis revealed increased basal (37°C) cytoplasmic and even nuclear levels of HSF1 in mutant compared with WT cells (Fig. 2B). While exposure of both of the cell lines to heat shock markedly increased nuclear HSF1 levels, we observed significantly higher levels of nuclear HSF1 accumulation in HSF1303A/307A cells. In contrast, we noted that the reduction of nuclear HSF1 levels during the recovery period was identical for the two cell types. Nuclear fractions analyzed by immunoblotting also confirmed a marked increase of phosphorylation on S303/S307 HSF1 levels in WT cells after exposure to heat shock and during the recovery period (Fig. 2B).
FIG 2.
Mutation of S303 and S307 results in HSF1 stability, nuclear accumulation, and upregulated expression of its downstream targets under basal conditions and upon exposure to exogenous stress. (A) HSF1 DNA binding activity in nuclear fractions visualized by EMSA using a radiolabeled HSE oligonucleotide. WT and HSF1303A/307A MEFs were analyzed in the absence of heat (C, control) or after heat shock (43°C for 20 min) and following recovery at 37°C for the indicated times. Blots were quantified by densitometry (right). Levels of HSF1 binding activity were expressed as relative fold increases compared to WT control (C, at 37°C). (B) Representative immunoblot analysis performed on the samples indicated in panel A to show HSF1 and phosphorylated HSF1 (S303/S307) expression levels in the cytosolic and nuclear fractions (n = 2 experiments). β-Actin and γ-tubulin were used as loading controls for cytoplasmic and nuclear fractions, respectively. The graph shows the quantification of HSF1 protein ratio in the cytosolic and nuclear fractions normalized to the loading control level (right). The second thin vertical line in the nuclear HSF1 image indicates that the blots were run in parallel and exposed to the same film. (C) Heat stress-induced expression of HSF1 target proteins. Immunoblot analysis and quantification of the indicated proteins in total extracts from WT and HSF1303A/307A MEFs treated as described for panel A. For quantification (right), the level of HSP protein normalized to the loading control (β-actin) was expressed as relative fold increase compared to control (WT, at 37°C), which was arbitrarily set at 1 (100%) (n = 3 analyses). (D) WT and HSF1303A/307A MEFs cultured in the presence of 20 μg/ml of cycloheximide (CHX) were heat shocked at 43°C for 20 min and analyzed by immunoblotting for the indicated length of recovery time. Samples were blotted for the indicated proteins. Quantification of the data is presented in the right panel (n = 2 analyses). (E) Representative immunofluorescence staining of HSF1 in WT and HSF1303A/307A MEFs. Cells were left untreated (37°C) or exposed to heat (43°C for 20 min). After recovery from heat shock at 37°C (0 to 8 h), cells were fixed and processed for staining. HSF1 was detected using red-fluorescence-conjugated secondary antibody. Cells were stained with DAPI (blue) to detect the nuclei. Additional immunofluorescence images from untreated cells acquired with higher resolution and contrast adjustment are presented (right-hand panels) to visualize the cytoplasmic distribution of HSF1. The quantification of HSF1-positive cells per high-power field (HPF) is presented (right panel). An average of 10 HPF (at a magnification of ×200 or ×400) randomly selected were counted for each section. For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01. Bars represent WT (red) or HSF1303A/307A (blue) cells.
Next, we tested whether enhanced accumulation of cytoplasmic and nuclear HSF1 levels in mutant cells might affect HSF1 target gene expression under basal conditions or after heat shock exposure. We monitored the inducible expression of HSPs in WT or HSF1303A/307A MEFs following exposure to heat shock (43°C for 20 min) and during 8-h recovery at 37°C (Fig. 2C). In the mutant cells, the expression of HSP90α, HSPB1 (HSP25), and HSPB5 (αB-crystalline) was markedly higher under basal conditions than in WT cells, as well as slightly elevated after heat shock and recovery. However, we could not detect significant differences in HSP70 expression levels during heat shock and recovery between the cell lines (Fig. 2C).
Subsequently, we tested whether lack of S303/S307 phosphorylation affects HSF1 stability, turnover, and nuclear accumulation following heat shock exposure. To address these issues, cells were cultured with eukaryotic translation inhibitor cycloheximide (CHX), and HSF1 protein levels were analyzed after heat shock (43°C, 20 min) or during a 6-h period of recovery from stress. We observed a marked reduction in the degradation of HSF1 in mutant cells compared to WT cells (Fig. 2D), indicating that the lack of phosphorylation affects protein stability. Quantification of the data revealed the increase in HSF1 half-time of decay from ∼1.5 h in WT cells to ∼4 h in mutant cells. This is consistent with a previous report showing that S303/S307 phosphorylation of HSF1 has a priming role on FBXW7-mediated direct control of HSF1 ubiquitylation and degradation (28).
Unlike human cell lines, which show HSF1 nuclear stress bodies following exposure to heat shock (26, 30), murine cells normally show a diffuse staining pattern. HSF1 activation, evident by its nuclear accumulation, was investigated by immunofluorescence staining of MEFs cultured at 37°C or following heat exposure (43°C, 20 min). Under basal conditions (37°C) or heat stress, there was a marked increase (in the amount and intensity) of nuclear HSF1 staining in mutant cells compared to WT cells (Fig. 2E), consistent with the HSF1 subcellular protein analyses (Fig. 2B).
Together, our data suggest that loss of HSF1 S303/S307 sites resulted in its protein stability and enhanced nuclear accumulation under basal and heat stress conditions but did not result in defective attenuation of the heat shock response. This altered HSF1 stability and nuclear expression pattern positively impact the expression of specific downstream HSP targets under normal physiological growth conditions.
Loss of S303/S307 phosphorylation reduces the HSF1 transcriptional activation threshold in response to thermal stress.
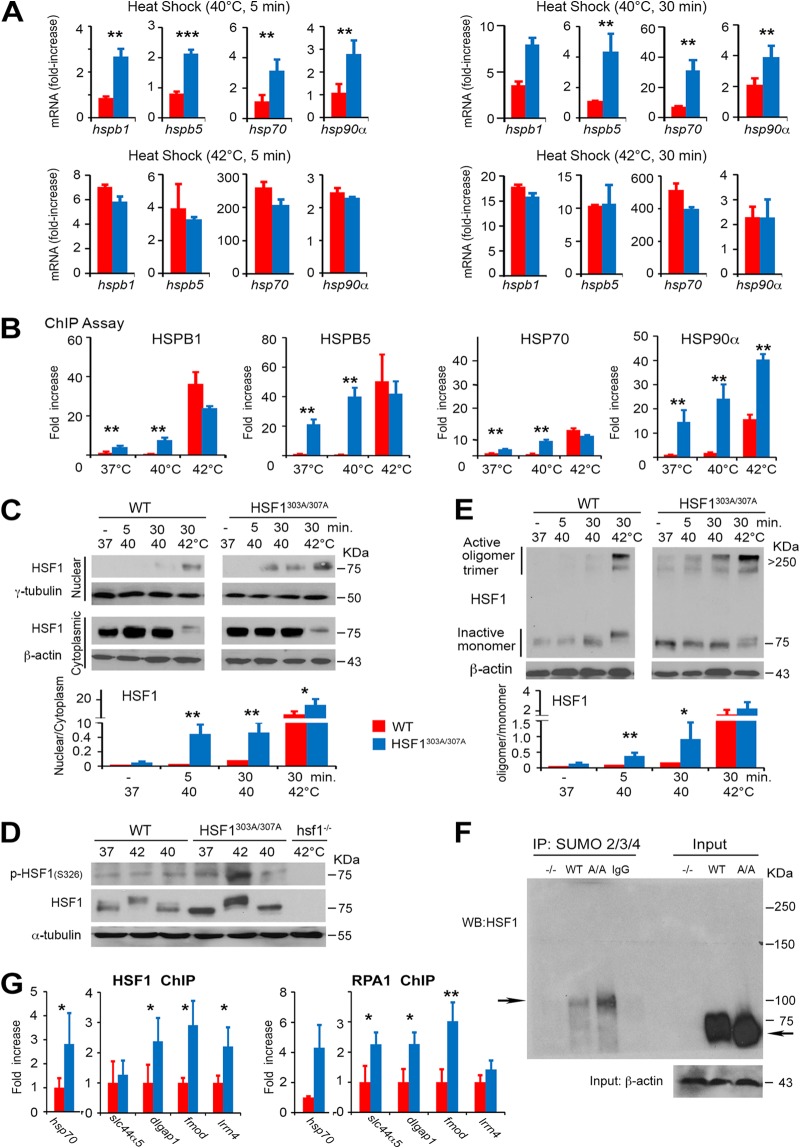
At a relatively moderate heat shock (43°C for 20 min), we could not detect any significant difference in the HSF1 binding between WT and HSF1303A/307A cells by EMSA (Fig. 2A). However, this result does not preclude the possibility that loss of S303/S307 phosphorylation affects the threshold of HSF1 activation, which apparently depends on the severity of exogenous stress. To test this prediction, we exposed both cell lines to a different intensity of heat stress (40°C or 42°C for 5 or 30 min) and monitored the changes in the mRNA expression of HSF1 targets (hspb1, hspb5, hsp70, or hsp90α) after 1 h of recovery at 37°C, which reflects its activation (Fig. 3A). Heat stress at 40°C for 5 min induced in WT cells low mRNA levels of HSF1 target genes; prolonged exposure of cells to 40°C for 30 min resulted only in a moderate increase in the expression levels. However, heat stress at 42°C induced maximal mRNA expression levels of HSF1 target genes. Strikingly, a marked increase of mRNA levels of positively regulated HSF1 targets that was even readily detectable after heat stress at 40°C for 5 min was observed in HSF1303A/307A cells. Notably, the levels in mutant cells significantly exceeded those of HSF1 targets in WT cells (Fig. 3A). The mRNA response was further enhanced after exposure of cells to 42°C, but the peak level of HSF1 targets that apparently reached maximal induction did not significantly differ between the mutant and WT cells. These data are consistent with our model, by which S303/S307 phosphorylation critically adjusts HSF1 activity in response to varying degrees of exogenous stress.
FIG 3.
Phosphorylation on S303/S307 defines the activation threshold of HSF1 to heat stress. (A) Heat stress-induced HSF1 target gene expression is enhanced upon mutation of S303/S307 phosphorylation. mRNA transcript levels of selected HSF1 target genes (hsp70, hspb1, hspb5, and hsp90α) in WT and HSF1303A/307A MEFs exposed to a different intensity of heat stress (40°C or 42°C for 5 or 30 min) and following recovery at 37°C for 1 h. Values are presented as relative mRNA expression compared to controls (WT and HSF1 mutant cells at 37°C) (n = 5 assays). (B) Increase in the DNA-binding capacity of HSF1 upon loss of S303/S307 phosphorylation. ChIP quantitative PCR (qPCR) assessing HSF1 occupancy at the HSE containing promoter regions of hspb1, hspb5, hsp70, or hsp90α in WT or HSF1303A/307A MEFs. Cells were either left untreated or exposed to heat shock (37°C, 40°C, or 42°C for 5 min). For quantification of HSF1 binding capacity, the values of qPCR normalized to the input values were expressed relative to WT control at 37°C, which was arbitrarily set at 1 (n = 4 assays). (C) Heat-induced HSF1 nuclear translocation increased following loss of phosphorylation on S303/S307. Immunoblot analysis of HSF1 in cytoplasmic and nuclear fractions prepared from WT and HSF1303A/307A MEFs and exposed to heat stress (37°C, 40°C, or 42°C for 5 or 30 min.). β-Actin and γ-tubulin were used as loading controls for cytoplasmic and nuclear fractions, respectively. The graph shows quantification of the HSF1 protein ratio in cytosolic and nuclear fractions normalized to the loading control level (right) (n = 3 analyses). (D) WT and HSF1303A/307A MEFs were exposed to heat shock (37°C, 40°C, or 42°C for 30 min), followed by recovery for 30 min at 37°C. Total cell lysates were analyzed by immunoblotting as indicated. α-Tubulin was used as a loading control. Lysates from hsf1−/− MEFs exposed to heat shock (42°C for 30 min) were used as a control for antibody specificity (n = 3 analyses). (E) High-molecular-weight oligomer of HSF1 formed during heat shock is stabilized by loss of S303/S307 phosphorylation. WT and HSF1303A/307A MEFs were exposed to heat shock (37°C, 40°C, or 42°C for 5 or 30 min) as indicated. Cell lysates prepared immediately after heat shock were subjected to cross-linking and blotted for HSF1. Ratios of active HSF1 (oligomers at 250 to 300 kDa) to inactive HSF1 monomer are shown (n = 3 analyses). (F) Representative endogenous analysis of SUMO-2/3/4-modified HSF1 in MEFs. Sumo-conjugated HSF1 was immunoprecipitated (IP) from MEF extracts from WT and mutant HSF1 (A/A) mice with an anti-SUMO-2/3/4 antibody and analyzed by Western blotting with antibody to HSF1. The arrows indicate high molecular mass of endogenous SUMO-modified HSF1 and unmodified native WT or mutant HSF1. β-Actin was used as a loading control. MEFs from whole-body HSF1 deletion mice (−/−) were included as a control (n = 2 analyses). (G) ChIP quantitative PCR assessing HSF1 (WT and HSF1303A/307A) and RPA1 binding at the putative HSE containing promoter region of hsp70, slc44a5, dlgap, fmod, and lrrn4 in MEFs of WT and HSF1 mutant mice. Analyses were performed using HSF1 or RPA1 antibody (n = 3 analyses). For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT cells; blue, HSF1303A/307A cells.
To investigate the physiological relevance of the above-described findings and directly determine whether HSF1 stability can control target gene occupancy by this transcription factor, we performed chromatin immunoprecipitation (ChIP) assays. For this analysis, MEFs of both genotypes were exposed to 37°C, 40°C, or 42°C for 5 min. Our analysis revealed that under basal growth conditions (37°C), HSF1303A/307A binds to the promoters of hspb1, hspb5, hsp70, and hsp90α, and this interaction was significantly enhanced compared to what was seen in WT cells (Fig. 3B). We also observed a further differentially increased binding of HSF1303A/307A (above the basal level at 37°C) to the promoters of hspb1, hspb5, hsp70, and hsp90α in cells exposed to a mild heat shock (40°C for 5 min). We could detect that the HSF1 binding to the promoters in mutant cells was significantly higher than in WT cells. In cells exposed to 42°C for 5 min, we found that the binding of HSF1 to the promoters of hsp targets was further increased, but we could detect a significant difference only for hsp90α promoter occupancy, when comparing HSF1303A/307A cells with WT cells (Fig. 3B).
Finally, we tested the stress-dependent subcellular localization and oligomerization of HSF1. Moderate heat stress at 40°C for 5 min induced nuclear localization and accumulation of HSF1 in mutant cells. In contrast, WT cells required a more severe heat stress to display nuclear HSF1 accumulation, which occurred only at 42°C for a 5-min exposure (Fig. 3C). HSF1 phosphorylation on S326 within the regulatory domain has been widely used as a surrogate marker for its transcriptional activity, especially in cancer cells (11, 31). In a heat stress dose experiment (Fig. 3D), we confirmed a superior potent transcriptional activator ability of HSF1303A/307A protein, as indicated by the strong expression of p-HSF1 (S326) in the mutant cells at 42°C for a 30-min exposure. However, we could not detect significant differences in the accumulation of slower-migrating species of HSF1 representing hyperphosphorylated HSF1 between the genotypes (Fig. 3D). In protein-protein cross-linking analysis, we also monitored the formation of a high-molecular-weight oligomer of HSF1 reflecting trimeric HSF1 species and transcriptional activity. HSF1 complexes migrating at 250 to 300 kDa were readily detectable in mutant cells under basal conditions (37°C), and during heat shock their levels were further stabilized (Fig. 3E). In contrast, HSF1 oligomers were reliably detected in WT cells only upon exposure to severe heat stress (42°C for 30 min). These data suggest that fine-tuning of the HSF1 activity may be facilitated by the enhanced oligomer formation in mutant cells. Taken together, these results suggest that lack of phosphorylation of S303/S307 stabilizes HSF1, possibly by blocking the lysine residues for ubiquitination. By altering conformational properties, silencing of this posttranslational modification probably promotes the formation and stability of trimeric HSF1 species, which is considered a prerequisite for DNA binding and HSF1 activation.
Ablation of S303/S307 preserves the interaction of HSF1 with replication protein A (RPA) and recruitment to the target promoters.
Previous studies have shown that activating and inhibitory posttranslational modifications, including phosphorylation, acetylation, and sumoylation, occurring at multiple sites, can have a complex role in regulating HSF1 activity and stability. For example, acetylation and phosphorylation of HSF1 on several sites can coordinately function as a timing mechanism, regulating its stability and activation/deactivation during heat stress response. In addition, it has been proposed that sumo modifications at multiple sites modulate HSF1-driven gene expression in an orchestrated manner. While the role of putative posttranslational sites of HSF1 has been evaluated using plasmid transfection-based assays, it has been a challenge, however, to clarify how ablation of phosphorylation at 303/307 may alter individual posttranslational modifications and assess their impact on the overall transcriptional function of endogenous HSF1. Based on the observation that the phosphorylation of S303 has a priming function for K298 sumoylation of HSF1, we have evaluated the potential impact of S303A/S307A mutation on the overall sumoylation level of mutant and WT HSF1. However, using SUMO-2/3/4 antibody for immunoprecipitation (IP) assays, we did not observe the predicted decrease in the overall sumoylation level of mutant HSF1 in MEFs. In contrast, the normalized sumo-modified level of mutant HSF1 under normal cell culture conditions increased slightly (about 20%) above the wild-type HSF1 control level (Fig. 3F). Notably, recent evidence has elucidated several additional sumo-modified sites on the HSF1 protein that may explain our result (17). IP assays performed with liver extracts confirmed this result and further provided corroborating evidence that the ablation of S303/S307 phosphorylation has an insignificant effect on the overall level of endogenous HSF1 acetylation (data not shown).
We next asked to what extent the ablation of S303/S307 phosphorylation affects the interaction with other proteins stimulating the recruitment of HSF1 and other components of the transcription machinery to the promoter of target genes. HSF1 access to nucleosomal DNA and target gene expression has been demonstrated to require the interaction of HSF1 through its wing domain, especially glycine at amino acid 87, with RPA1, which plays a pivotal role in DNA metabolism (32). Under normal growth conditions, the ability of mutant HSF1 to interact with RPA1 and form a complex was preserved in HSF1 mutant MEFs. ChIP assay showed that both HSF1 and RPA1 were occupied in the promoter of HSP70 and other selected non-HSP target genes (32, 33). However, the occupancy was significantly increased in mutant cells compared to WT HSF1-expressing cells (Fig. 3G). Taken together, these results emphasize the superior ability of the HSF1 mutant-RPA1 complex to occupy DNA and recruit other chromatin-remodeling and histone chaperone proteins, under control conditions, to regulate the expression of target genes.
Regulation of HSF1 and downstream target genes in tissues of HSF1303A/307A mice during chronological aging under basal conditions or upon exposure to heat-induced stress.
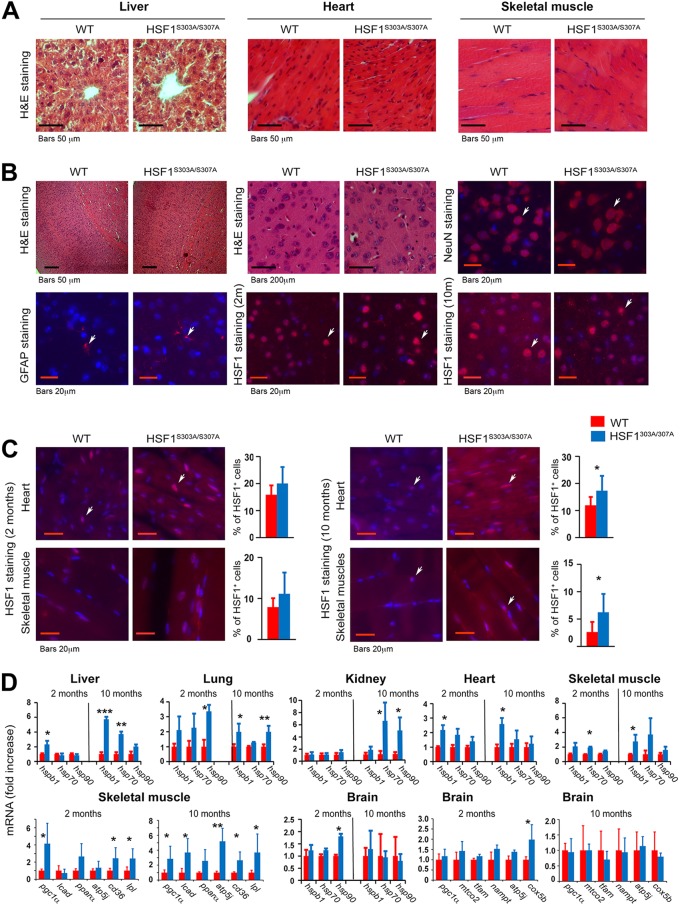
An age-related decline in stress response and HSF1 activity has been documented (8, 34), but the underlying mechanism remains elusive. Histological sections of organs (cardiac and skeletal muscles, liver, kidney, and brain) from young and aged mice, stained for hematoxylin and eosin (H&E), did not reveal apparent morphological changes (Fig. 4A). In addition, immunofluorescence imaging of brain tissues revealed no difference in fluorescence intensity of NeuN (a marker for neurons) and GFAP (a marker for astrocytes) cells or in intensity of nuclear HSF1 between the genotypes (Fig. 4B). Under basal conditions, however, there was a marked increase (numerical) of nuclear HSF1 staining in cardiac and skeletal muscles of young and aged mutant mice compared to WT mice (Fig. 4C). Furthermore, despite variation, the overall mRNA profiles of major hsp target genes revealed significant increases in several tissues of mutant mice, the only exception being the brain, where not one marked effect was observed (Fig. 4D). Interestingly, expression of mutant HSF1 in skeletal muscle resulted in strong induction of genes related to metabolism and lipid oxidation. These changes may be compensatory to an increase in the organismal anabolic capacity of aged mutant HSF1 mice.
FIG 4.
Tissue morphology and expression of HSF1 and its downstream target genes in tissues of HSF1303A/307A mice maintained on a normal chow during chronological aging. (A) Representative images of H&E staining of liver, heart, and skeletal muscle tissue sections from WT and HSF1303A/307A mice maintained on an ND at different ages (2 and 10 months) (n = 3 mice per group). (B) Representative H&E and immunofluorescence staining of cortical brain sections for neuronal (NeuN) and astrocytes (GFAP) markers or nuclear HSF1 from WT and HSF1303A/307A mice fed on an ND, at different ages (2 and 10 months). Notably, no apparent histological alterations or significant changes in HSF1 expression or number of brain cell markers were observed between the genotypes (n = 3 mice per group). (C) Representative immunofluorescence staining of nuclear HSF1 in heart and skeletal muscle of HSF1303A/307A mice indicating a significant age-dependent increased nuclear HSF1 expression in tissues of mutant mice. Quantification of HSF1-positive cells is presented on the right of the micrographs (n = 3 or 4 mice per group). (D) Relative mRNA levels of genes associated with mitochondrial function, metabolism, and heat shock response in different tissues (lung, heart, brain, kidney, liver, muscle, WAT) of HSF1 mutant and WT control mice fed with ND at different ages (n = 4 mice per group). For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
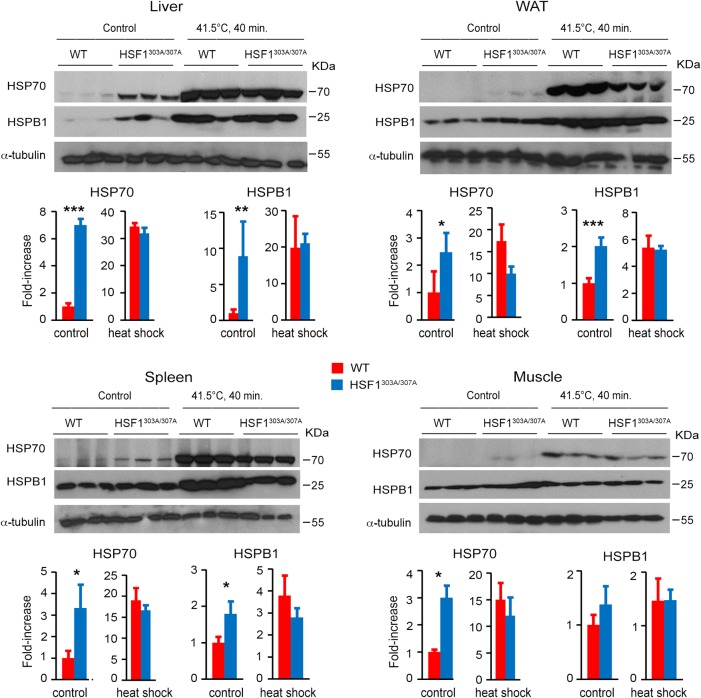
We next examined the effects of heat shock on expression of HSPs in tissues of adult mice (Fig. 5). HSF1303A/307A and WT mice were exposed to whole-body thermal stress (41.5°C), and levels of HSP70 and HSPB1 were assessed following recovery for 6 h at room temperature. Interestingly, we found that S303/S307 abrogation significantly elevated the basal expression of HSP70 and HSPB1 in the HSF1303A/307A mouse tissues. Heat stress of mice induced a greater effect on HSP70 and HSPB1 expression, but the protein levels did not significantly differ between the genotypes. These data demonstrate that loss of S303/S307 phosphorylation leads to persistent HSF1 activation and downstream target expression in different tissues.
FIG 5.
Elevated expression of HSF1 targets in tissues of HSF1303A/307A mice maintained on a normal diet. WT and HSF1303A/307A mice were untreated or exposed to whole-body hyperthermia at 41.5°C for 40 min and allowed to recover for 6 h. Protein extracts from different tissues were assessed by immunoblotting with antibodies specific for HSP70 and HSPB1. Quantification of the HSP protein levels (normalized by WT mice) is shown (n = 3 mice for each genotype). For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
Effects of S303A/S307A mutation on nutrient-induced obesity and insulin resistance.
To determine whether altered HSF1 stability and activity can have a significant impact on protein homeostasis and bioenergetics, we performed metabolic and signaling analyses in WT and HSF1303A/307A mice under basal conditions or upon exposure to nutrient-induced stress.
A causal link between metabolic bioenergetics and protein homeostasis driven by HSF1 with respect to the adaptive response of organisms to energy and nutrient stress has been well established (4, 7, 35–37). Particularly, the activation of HSF1 has a positive effect on the nutrient- and energy-regulated anabolic pathways, thus providing forces driving enhanced lipid deposition, obesity, and systemic insulin resistance. This information prompted us to investigate the metabolic consequences of S303/S307 phosphorylation loss, focusing on the effects of age- and nutrient-induced obesity and the associated development of insulin resistance.
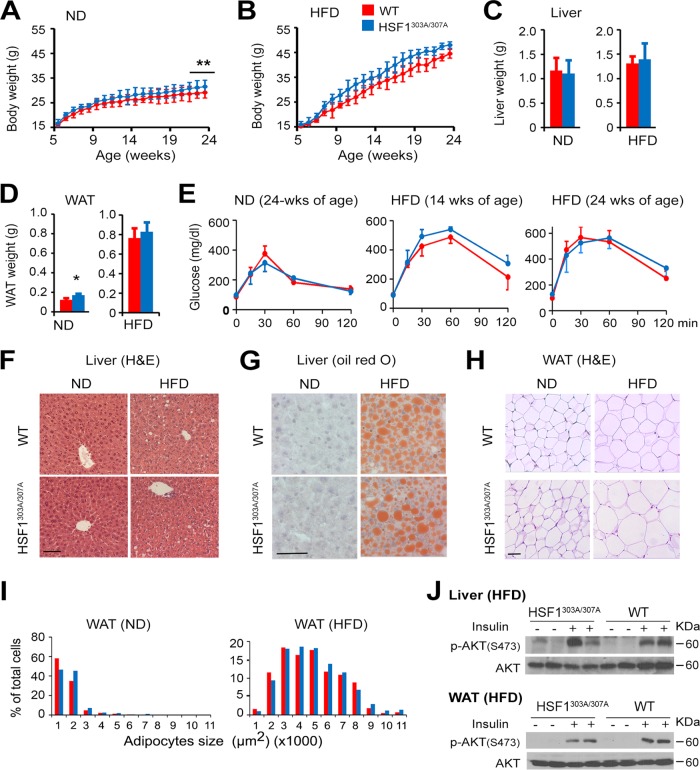
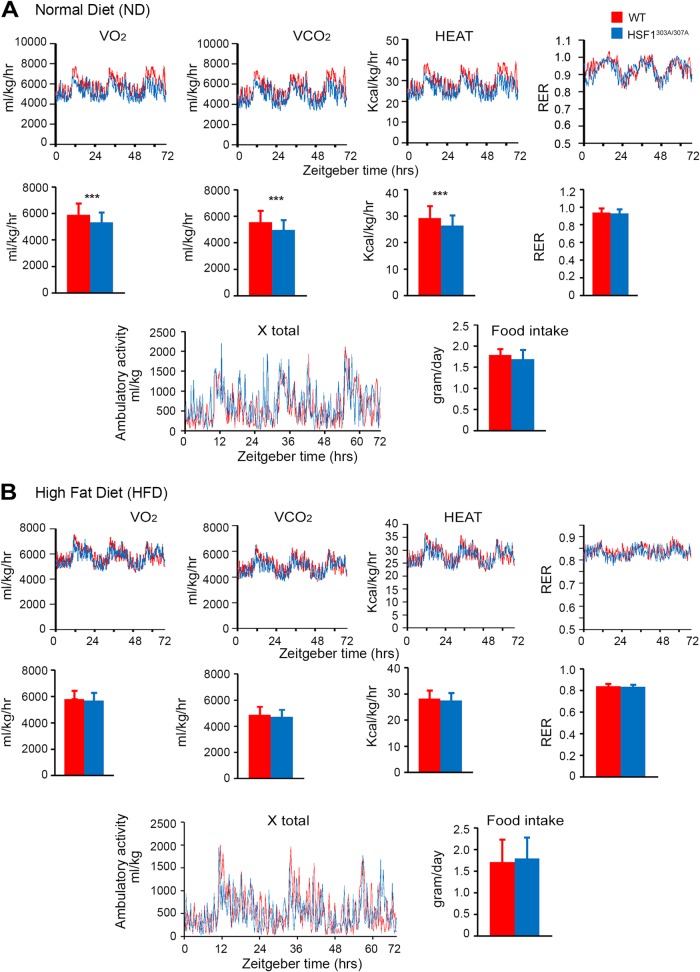
To more sensitively test the potential effects of the S303/S307 mutation on organ metabolism, we examined the effects of nutrient availability on lipid metabolism in mice fed with a normal diet (ND) or a high-fat diet (HFD) for a period of 20 weeks. The rate at which HSF1303A/307A mice gained body weight on ND was significantly higher than that seen in WT mice (Fig. 6A). Moreover, HSF1303A/307A mice displayed a slightly better response to body weight gain upon HFD than did WT mice, although this did not reach statistical significance (Fig. 6B). Examination of organ mass at necropsy indicated that the majority of the increase in weight gain in the mutant mice on ND feeding was associated with increased epididymal white adipose tissue (WAT) mass, without changes in liver weight (Fig. 6C and D). A glucose tolerance test (GTT) performed at 24 (ND) or at 14 and 24 (HFD) weeks of age did not differ between HSF1303A/307A and WT mice (Fig. 6E). Liver tissue sections demonstrated increased hepatic steatosis upon HFD feeding; however, we did not observe markedly different patterns between the genotypes (Fig. 6F and G). Similarly, analysis of sections prepared from WAT did not reveal significant differences in adipocyte size between HSF1303A/307A and WT mice fed an ND or HFD for 20 weeks (Fig. 6H and I). Further, no significant difference in insulin stimulation of AKT phosphorylation, which reflects its activation, was observed between genotypes on HFD feeding (Fig. 6J). In order to assess the potential impact of energy balance on the increase body gain observed in ND-fed HSF1303A/307A mice, we measured energy expenditure, food intake, and activity using a calorimetric caging system (Fig. 7). At 25°C, there were no differences in food intake and activity level between HSF1303A/307A mice and WT mice and feeding conditions (ND or 2 weeks on HFD). However, HSF1303A/307A mice had significantly reduced O2 consumption and CO2 production as well as heat production compared to WT mice. The respiratory exchange rate (RER) indicated that the preferential nutrient (fat or carbohydrate) utilization was similar between the genotypes. On the high-fat diet, metabolic recording did not reveal any significant changes in energy expenditure parameters between the mutant and WT mice. Taken together, the results revealed that the positive impact of S303/S307 mutation on body weight and lipid accumulation under ND feeding conditions possibly reflects a downshift in oxygen consumption and energy expenditure.
FIG 6.
Mutation of S303/S307 does not have measurable effects on HFD-induced obesity and insulin resistance. (A and B) Body weight gain in WT and HSF1303A/307A male mice maintained on standard normal diet (ND) or on a high-fat-diet (HFD) beginning at 4 weeks of age and for a period of 20 weeks. Scale bars are means ± SD (n = 10 mice per group for ND; n = 13 mice per group for HFD). **, P < 0.01. (C and D) Weights of liver and adipose tissue (WAT) of 24-week-old WT and HSF1303A/307A mice maintained on ND or HFD as described for panels A and B. Bars are means ± SD. *, P < 0.05. (E) Glucose tolerance test (GTT) on WT and HSF11303A/307A mice maintained on ND (24 weeks of age) or HFD for 10 or 20 weeks beginning at 4 weeks of age. Bars are means ± SD (n = 5 mice per genotype). (F and G) Representative liver sections from 24-week-old WT and HSF1303A/307A mice on ND or HFD as indicated in panels A and B. Sections were stained with H&E (F) or oil red O (G). Bars, 50 μm. (H) Representative WAT tissue sections from 24-week-old WT and HSF1303A/307A mice as indicated for panels A and B, stained with H&E. (I) Quantification of the data from sections in panel H. Whisker plot of adipocyte sizes from evaluation of 300 adipocytes in 10 randomly selected microscopic fields expressed as percentage of the total analyzed cells (bar, 50 μm). Bars are means ± SD (n = 4 mice per genotype). (J) Immunoblot analysis of insulin-induced AKT signaling in liver and WAT of 24-week-old WT and HSF1303A/307A mice fed with HFD for 20 weeks. Mice were fasted for 16 h and injected with saline or insulin (2 mU/g) i.p. Insulin-induced signaling in tissue extracts was examined 10 min after insulin or saline injection. The level of p-AKT (Ser473) was examined. Bars are means ± SD (n = 4 mice per genotype). For all panels, red represents WT (red) and blue represents HSF1303A/307A mice.
FIG 7.
The lack of S303/S307 phosphorylation of HSF1 impacts metabolic mouse phenotype under normal diet but not HFD feeding conditions The metabolic activity of 5-month-old HSF1303A/307A and WT mice maintained on an ND (A) or exposed to a short period of 2 weeks on HFD (B) was determined using a calorimetric caging system. The animals were acclimatized for 3 days at 25°C before data were recorded for 4 light/dark cycles. Food intake, O2 consumption, CO2 production, and ambulatory activity of the mice were recorded every 2 min. The respiratory exchange rate (RER) (CO2/O2 ratio) and rate of energy expenditure (heat) were calculated. Graphs show quantification of metabolic parameters. Error bars are means ± SD (n = 6 mice for each genotype). ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice. The corresponding Zeitgeber hours are indicated at the bottom (0 is the time at which light is on).
Effects of S303A/S307A mutation on age-dependent metabolic adaptation and reprogramming.
Our previous study revealed that whole-body deletion of hsf1 has a negative impact on diet-induced obesity and insulin resistance (4). Based on these findings, we predict that the metabolic effects of HSF1 loss could be reversed by loss of S303/S307 phosphorylation, which alters HSF1 stability and activity. This is confirmed by our observation on the greater fat accumulation and development of insulin resistance in HSF1303A/307A mice of 24 weeks of age kept on ND feeding. Because metabolic dysfunction associated with the declining of bioenergetics resources is a hallmark of aging (38–40), we further investigated the potential metabolic consequences in ND-fed HSF1303A/307A and WT mice of different ages. For this analysis, we focused on highly metabolic active tissues such as liver, skeletal muscle, and WAT.
First, we noted that at the age of 2 months, hepatic insulin-stimulated AKT phosphorylation and mRNA levels of key lipogenic factors, including peroxisome proliferator-activated receptor gamma (PPARγ1/γ2), sterol regulatory element-binding protein (SREBP1c), CCAAT/enhancer-binding protein beta (C/EBPβ), and liver X receptor (LXR), were not different between HSF1303A/307A and WT mice on ND feeding (Fig. 8A and B). A transition to insulin resistance associated with a reduced AKT (S308) phosphorylation level was evident in WAT of mutant mice at this age (Fig. 8E). However, robust evidence of insulin resistance associated with diminished p-AKT was clearly observed in mutant mice at 10 months of age, with significant changes in hepatic mRNA levels of lipogenic genes srebp1c, fas, and acc1 (Fig. 8C, D, and F). Notably, the mRNA level of NAD-synthesizing enzyme nampt increased significantly in the liver of old mutant mice. Similar to our observations in liver and WAT, insulin-signaling attenuation was also observed for skeletal muscle (Fig. 8G and H).
FIG 8.
S303/S307 mutation of HSF1 progressively intensifies age-associated physiological decline leading to insulin resistance. (A and C) Immunoblot analysis of insulin-induced AKT signaling in livers of 2-month-old (A) or 10-month-old (C) WT and HSF1303A/307A mice fed with ND. Mice were fasted for 16 h and injected with saline or insulin (2 mU/g) i.p. Insulin-induced signaling in tissue extracts was examined 10 min after insulin or saline injection. The level of p-AKT (Ser473) was examined, and blots were quantified (right). Level of p-AKT normalized to total protein level was expressed as relative fold increase compared to control (WT without insulin). Bars are means ± SD (n = 4 mice per genotype). (B and D) Quantitative RT-PCR of mRNA transcript levels of selected genes that regulate lipid and NAD+ biosynthesis in livers of 2-month-old (B) or 10-month-old (D) WT and HSF1303A/307A mice fed with ND. Values are presented as relative mRNA expression (n = 5 mice of each genotype). (E and F) Insulin-stimulated signaling in WAT of 2-month-old (B) or 10-month-old (D) WT and HSF1303A/307A mice fed with ND. Analyses and quantification of blots (right) were performed as described for panels A and C (n = 4 mice of each genotype). (G and H) Insulin-stimulated signaling in skeletal muscle of different ages WT and HSF1303A/307A mice. Analyses were performed as described for panels A and C (n = 4 mice per group). For all panels, bars are means ± SD. *, P < 0.05; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
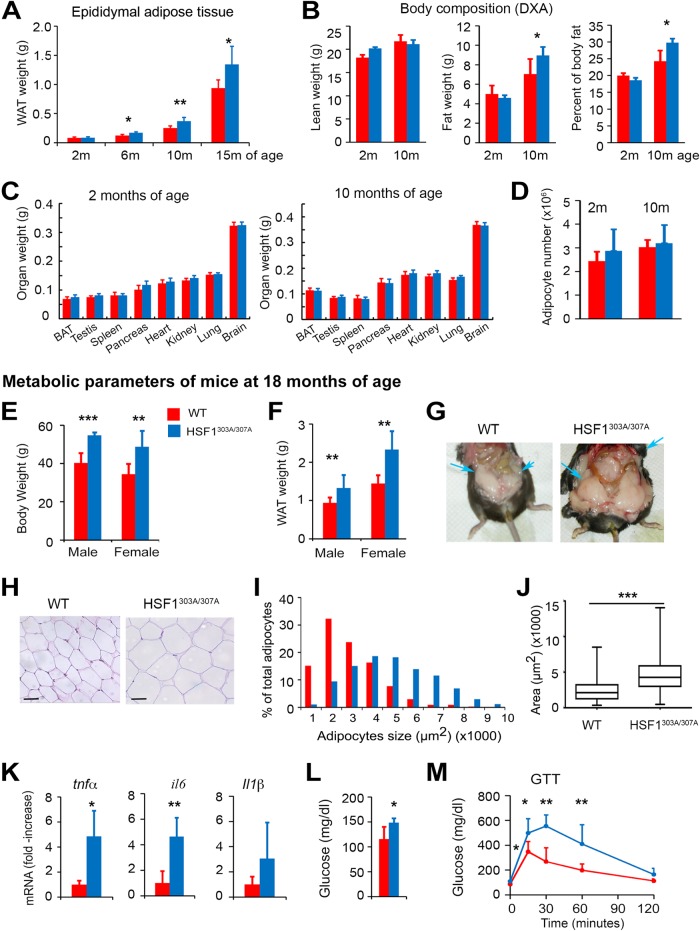
Energy expenditure decline is a hallmark of aging and has been proposed as a potential cause of age-associated metabolic impairment and development of obesity. We first confirmed that under normal feeding conditions the fat mass gradually increased in epididymal WAT with aging in both genotypes. However, HSF1303A/307A mice displayed a reduced metabolic performance, which apparently resulted in a greater increase in WAT depot (Fig. 9A). HSF1 mutant mice showed also an increased total body fat mass (Fig. 9B), while lean mass or organ weights did not differ between the genotypes at different ages (Fig. 9C). In addition, H&E staining revealed that epididymal adipocytes of mutant HSF1 mice were larger (hypertrophy), while a calculation of total adipocyte numbers in epididymal depots (hyperplasia) revealed no significant difference between WT and mutant HSF1 mice (Fig. 9D).
FIG 9.
HSF1303A/307A mice are profoundly susceptible to advanced age-dependent metabolic decline associated with development of obesity and insulin resistance. (A to C) Epididymal WAT weight (A), total body fat distribution measured by DXA (B), and weight of indicating organs (C) of mutant HSF1 and WT control mice of different ages maintained on ND (n = 4 to 10 mice per group). BAT, brown adipose tissue. (D) Evaluation of hyperplasia in epididymal WAT by extrapolating the total number of adipocytes from the size of adipocytes (via histology) and the weight of the fat depots in wild-type and mutant mice. Analyses were performed on mice maintained on ND, at an early (2 months) and advanced (10 months) age (n = 4 mice per group). (E and F) Body weight and epididymal WAT mass of 18-month-old WT and HSF1303A/307A mice fed an ND. Bars are means ± SD (n = 11 male and n = 10 female WT mice per group; n = 8 male and n = 13 female mutant mice per group). (G) Representative abdominal depots (arrows) of 18-month-old WT and HSF1303A/307A mice exhibiting a substantial increase in the amount of fat tissue in a mutant mouse. (H) Histological analysis (H&E staining) of epididymal WAT sections of 18-month-old WT and HSF1303A/307A mice on ND. The analysis revealed that HSF1303A/307A adipocytes were consistently larger than adipocytes from age-matched WT mice. Bars, 50 μm. (I and J) Data from whisker plots of adipocyte sizes from evaluation of 300 adipocytes in 10 randomly selected microscopic fields expressed as percentages of the total analyzed cells (I); averages of adipocyte areas (J); (n = 5 mice per genotype). (K) Increased expression of inflammatory cytokines in WAT of HSF1303A/307A aged mice. mRNA transcript levels of selected cytokine genes (tnfα, il6, and il1β) in WAT of 18-month-old WT and HSF1303A/307A mice measured by quantitative RT-PCR. Values are presented as relative mRNA expression (n = 5 mice per genotype). (L and M) Blood glucose levels and GTT in 16-h-fasted 18-month-old WT and HSF1303A/307A mice (n = 5 mice per genotype). For all panels, error bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
The age-related metabolic dysfunction was further enhanced in intensity in the liver and WAT at 18 months of age. Both male and female HSF1303A/307A mice displayed a significant increase in body weight, WAT mass, and adipocyte size compared to WT mice (Fig. 9E to J). Age-associated adipose tissue inflammation, a hallmark of obesity and insulin resistance (41), was also significantly enhanced in WAT of mutant mice. There was differential mRNA upregulation of selected cytokines, including tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), and IL-1β between the genotypes (Fig. 9K). Consistent with increased insulin resistance, we also found a significant increase of fasting blood glucose and glucose excursion in a GTT in WAT of 18-month-old HSF1303A/307A mice compared to WT mice (Fig. 9L and M).
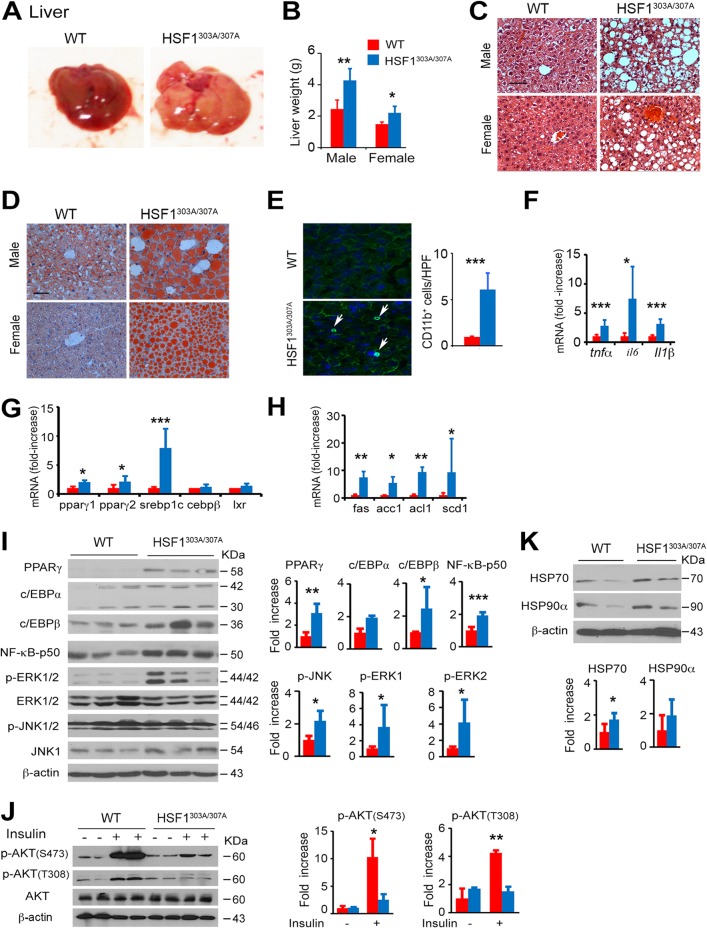
In subsequent comparisons, we tested the metabolic effects in liver of aged mice (18 months of age). Consistent with the adipose-tissue results, the hepatic metabolic dysfunctions and the low-grade inflammation occurring during aging were significantly enhanced in both male and female HSF1303A/307A mice compared to aged WT mice. The livers of the mutant mice exhibited severe hepatic steatosis, characterized by increased organ mass, large lipid deposition in hepatocytes, and severe hepatic inflammation associated with infiltration and accumulation of monocytes (CD11b+ cells) and upregulated mRNA levels of proinflammatory cytokine genes (tnfα, il6 and il-1β) (Fig. 10A to F). Immunoblot analysis confirmed the elevated expression of the p50 isoform of NF-κB as well as phosphorylated ERK1/2 and JNK1 in the liver of HSF1303A/307A mice, further supporting activation of inflammation-related signaling pathways (Fig. 10I) (42). Furthermore, the protein and mRNA expression levels of key transcription factors and their target genes involved in lipid synthesis, including PPARγ, SREBP1c, C/EBPβ, and LXR, as well as fatty acid synthase (FAS), acetyl coenzyme A (acetyl-CoA) carboxylase (ACC1), ATP synthase lyase (ACL), and stearoyl-CoA desaturase (SCD1) (43), were significantly upregulated in the liver of HSF1303A/307A mice compared to WT mice (Fig. 10G to I). These results suggest dramatically potentiated anabolic lipid metabolism in HSF1303A/307A mice. Finally, as expected, insulin-stimulated phosphorylation of AKT at Ser473 and Thr308 was significantly diminished in the liver of mutant mice, indicating the development of systemic insulin resistance (Fig. 10J) (44). Immunoblotting assays showed that the protein expression levels of HSF1 targets HSP70 and HSP90α were higher in the livers of aged HSF1303A/307A mice. This is consistent with the notion of a stabilizing role of S303A/S307A mutation and upmodulation of the HSF1 activity (Fig. 10K). These results suggest that S303/S307 phosphorylation is critical for HSF1 activity and has a potential regulatory role in the manifestation of age-related metabolic diseases.
FIG 10.
HSF1303A/307A mice exhibit increased hepatic steatosis caused by age-dependent metabolic decline. (A) Representative macroscopic images of livers from 18-month-old WT and HSF1303A/307A mice maintained on an ND. (B) Liver weights of 18-month-old WT and HSF1303A/307A mice. Bars are means ± SD (n = 11 male and n = 10 female WT mice per group; n = 8 male and n = 13 female mutant mice per group). (C and D) Representative histological analysis by H&E staining (C) and oil red O staining (D) of liver sections from 18-month-old WT and HSF1303A/307A mice maintained on an ND. Bar, 50 μm. (E) Representative immunofluorescence staining of monocytes using CD11b marker in liver tissue sections from 18-month-old WT and HSF1303A/307A mice. Nuclei were stained with DAPI. Quantification of CD11b+ cells (average number of CD11b+ cells in 10 randomly selected ×200 microscopic fields) is tabulated (right) (n = 5 mice for each genotype). (F to H) mRNA transcript levels of selected genes for cytokines (tnfα, il6, il1β) (F), transcription factors (G), and enzymes (H) involved in lipid synthesis in livers of 18-month-old WT and HSF1303A/307A mice measured by quantitative RT-PCR. Values are presented as relative mRNA expression (n = 6 mice for each genotype). (I) Immunoblot analysis of representative proteins for lipid synthesis, inflammatory signaling and MAPK/ERK and JNK pathways in liver extracts of 18-month-old WT and HSF1303A/307A mice. Quantification of indicated proteins (normalized to β-actin loading control) is shown (n = 4 mice for each genotype). (J) Immunoblot analysis of insulin-induced AKT signaling in liver of 18-month-old WT and HSF1303A/307A mice. Analyses and quantification of the blots is given (right) (n = 3 mice of each genotype). (K) Immunoblot analysis of HSP70 and HSP90α proteins in liver of 18-month-old WT and HSF1303A/307A mice. Quantification of the blots is given (right) (n = 4 mice for each genotype). For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
Effects of the S303/S307 mutation on the expression of nutrient-induced HSF1 targets.
In accordance with previous studies, the HSF1 activity is fluctuated in a nutrient-dependent manner (37, 45, 46). This prompted us to define the role of S303/S307 phosphorylation in regulating HSF1 activity in response to nutrient (fed-fasting) stress. Interestingly, we found that the basal nuclear HSF1 level in the liver was substantially elevated over that of WT controls in fasted HSF1303A/307A mice. Refeeding caused a stronger increase in nuclear steady-state levels of HSF1 in the mutant mice (Fig. 11A). This robust nutrient-induced response, which was significantly greater in mutant mice than WT mice, translated into significantly stronger increase of mRNA and protein levels of HSF1 targets (HSPB1, HSP70, and HSP90α) in liver and WAT tissues (Fig. 11B, C, D, and E). As expected, we did not observe markedly different mRNA levels of HSF1 target genes in HFD-fed HSF1303A/307A and WT mice (Fig. 11F and G).
FIG 11.
Loss of S303/S307 phosphorylation results in increased nutrient-induced nuclear accumulation of HSF1 and upregulated expression of its target genes. (A) Immunoblot analysis of HSF1 in cytoplasmic and nuclear fractions prepared from WT and HSF1303A/307A mouse livers. Mice maintained on an ND were fasted for 24 h or fasted (24 h) and refed for 1 h. β-Actin and γ-tubulin were used as loading controls for cytoplasmic and nuclear fractions, respectively. The graphs show quantification of the HSF1 protein in cytosolic and nuclear fractions normalized to loading control level and expressed as relative fold increases to control (WT fasted, arbitrarily set at 1) (n = 4 mice for each genotype). (B and C) mRNA transcript levels of HSF1 target genes (hspb1, hsp70, and hsp90α) in liver (B) and WAT (C) from 24-h fasted or from fasted and 3-h refed WT and HSF1303A/307A mice. Quantification of mRNA levels comparing refed with fasted mice (refed/fasted ratio) (n = 5 mice for each genotype). (D and E) Immunoblot analysis of HSF1 targets (HSPB1, HSP70, and HSP90α) in liver (D) and WAT (E) tissue extracts prepared from WT and HSF1303A/307A mice under fasting (24 h) or refeeding (3 h) conditions (n = 4 mice for each genotype). (F and G) mRNA transcript levels of HSF1 target genes (hspb1, hsp70, and hsp90α) in liver (F) and WAT (G) tissues of WT and HSF1303A/307A mice fed an HFD for 20 weeks beginning at 4 weeks of age (n = 5 mice for each genotype). For all panels, bars are means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Red, WT mice; blue, HSF1303A/307A mice.
Finally, we tested whether stabilization of HSF1 by loss of phosphorylation on S303/S307 may have a feed-forward regulatory role on nutrient-induced expression of GSK3β, CK2, and ERK kinases, which have been proposed to mediate the phosphorylation of these sites. Although there was an increase in the phosphorylation of GSK3β upon refeeding in the liver, no significant differences in their expression pattern were observed between WT and mutant mice. Similarly, the levels of CK2 and phosphorylated ERK kinases did not differ between the mouse lines (data not shown).
In summary, our results in animal assays indicate that the effects of physiological nutrient-induced response on HSF1 activation may be mediated, at least in part, through posttranslational events modulating the S303/S307 phosphorylation. However, under conditions of nutrient overload, for example, induced by HFD, the effects on HSF1 stability and activity were attenuated by mechanisms that need to be investigated.
DISCUSSION
The primary role of the stress response networks is to alleviate the immediate or chronic consequences of cellular and environmental stress, allowing them to ensure a balanced proteome and optimal cellular metabolic bioenergetic capacity. These adaptive and protective networks coordinate these cellular processes with the participation of a large set of molecular chaperones and other quality control elements as well as several transcriptional and translational regulators acting as sensors and information hubs (47). Given the age-related progressive decline of physiological and cellular functions (38, 48, 49), the critical question is how the cells sense and adjust the proteostatic and metabolic networks to balance the bioenergetic needs under physiological and permanently challenging environmental stress conditions. Undoubtedly, the functionally diverse HSF1 transcriptional programs that operate under basal, metabolic, and disease/stress conditions are a central node of this regulatory system that can impact age-related pathophysiology and life span. Indeed, there is strong evidence for a cross talk between the diverse cellular-stress events and elements of the adaptive response, with HSF1 playing a central role in rewiring gene expression and integrating signals from multiple protein homeostasis, metabolic, and energetic cellular processes (7, 8, 10, 37). Although the functions of HSF1 as regulator both of the classical response to changes in the temperature and of quality control to proteotoxic stressors are reasonably understood, the different molecular cues and structural modification that drive HSF1-specific transcriptional programs supporting a diverse array of cellular functions, including metabolic bioenergetics, neuronal functions, as well as initiation of malignant transformation and metastatic cancer development, remain poorly defined at the organismal level. Consistent with the notion that posttranslational modifications regulate HSF1 activity, the present study confirmed the significant role of HSF1 phosphorylation on both S303 and S307 residues in downmodulation of its transcriptional activity. Moreover, it presents direct evidence, using a physiologically relevant knock-in mouse model, that inactivation of these phosphorylation residues markedly increases HSF1 protein stability but, importantly, lowers the threshold of hsf1 transcriptional activation. Uncoupling HSF1 activity from the phosphorylation events occurring on S303/S307 residues drives an anabolic metabolic bioenergetic process featured by markedly enhanced lipid biosynthesis, hepatic steatosis, inflammation, and development of insulin resistance. As HSF1 has a critical role in cellular NAD+ and ATP production capacity, we propose that a tight posttranslational modification program via phosphorylation, described here, represents a bona fide regulatory mechanism for HSF1 function as a sensor of metabolic and energetic demands and as an information hub for upregulation of anabolic bioenergetic networks.
Through an extensive array of posttranslational modifications (8, 13, 17, 18), HSF1 activity is regulated and controls transcriptional programs that facilitate an adaptive response. However, the precise mechanistic links between many of these modifications and HSF1 activity and its functional properties remain to be defined. Phosphorylation of HSF1 on several serine and threonine residues, occurring mostly in the regulatory domain, is one common mechanism proposed to regulate HSF1 activity and can have both activating and inhibiting effects. In contrast to the prevailing paradigm that hyperphosphorylation reflects HSF1 activity, however, mutagenesis studies in which a subset or all the phosphorylation sites of human or yeast HSF1 were disrupted suggest that HSF1 phosphorylation intrinsically is not required for heat shock-induced activity; rather, phosphorylation, especially in the regulatory domain, may increase the ability of HSF1 to interact with elements of the transcriptional machinery, thus enabling the transcription of target genes with reduced activation threshold (20, 21). Contrary to this, phosphorylation events can affect HSF1 activity and stability under basal and disease states (8), indicating a more complex course of action. Our study provides direct in vivo evidence illustrating that alanine substitution in the S303/S307 motif profoundly increases the overall HSF1 protein stability upon heat shock or basal conditions but other functional aspects of the HSF1 activation cycle, such as shuttling between cytoplasm and nucleus, formation of active oligomers, and chromatin binding ability, remain largely intact. Remarkably, in the absence of elevated stress, the inactivating substitution of S303/S307 residues renders only a small fraction of HSF1 transactivation competent as demonstrated by the increased level of oligomer formation. This implies that the loss of this modification, additional signals such as thermal stress or fluctuations in nutrient and metabolic activity are required for full activation of the DNA-binding activity of HSF1 (23, 25). While changes in S303/S307 motifs can stabilize HSF1, preventing its interaction with FBXW7α (28, 29), it may also structurally modulate protein stability. Based on our findings that the S303A/S307A mutant potentiates HSF1 chromatin binding capacity above control levels, we also speculate that this modification might render it competent to interact more efficiently with other core elements of the transcription machinery. Notably, nuclear exclusion, thought to mediate inactivation of HSF1 during recovery from stress, is not affected by disrupting this posttranslational modification. This again indicates that the interaction of CUL1:FBXW7α with S303/S307 residues of HSF1 is unlikely to be solely responsible for its inactivation during recovery from stress. The nuclear proteasome activity and the interaction with highly expressed HSPs (8, 13, 50) might synergistically participate in HSF1 stability and activity during recovery from heat stress. Thus, according to our model, the S303/S307 phosphorylation regulates mainly HSF1 activity via protein stabilization, but the effects of other phosphorylation events, for example, on S326 (activating) (11), cannot be excluded. Another feature of HSF1 repression is the priming role of S303 phosphorylation for subsequent sumoylation at Lys298 (16, 17). The predicted loss of this modification and its impact on HSF1 activity could not be experimentally assessed in our model, because attempts to demonstrate Lys298 sumo-modification of endogenous HSF1 were unsuccessful (data not shown). Finally, the possibility that other compensatory changes in the acetylation profile further enhance HSF1 mutant stability deserves further investigation. Notably, acetylation of HSF1 by EP300 occurring on different sites can function as a timing mechanism, regulating its stability and activation/deactivation during the heat stress response (13).
One interesting finding of this study is the markedly increased baseline protein levels of HSP90α, HSPB1, and HSPB5 but not of HSP70 in HSF1 mutant MEFs under unstressed conditions (Fig. 2). The mRNA levels of these HSF1 targets remain low under basal conditions (Fig. 3). This finding is consistent with previous observations that hsp70 mRNA is unstable under control conditions and heat stress increases its stability (51, 52). Because HSP70 is also regulated at the translational level, the effects of other factors may account for this phenomenon. The differential effects of mutant HSF1 on the basal HSP levels also imply that this modification may affect HSF1 promoter accessibility on specific target genes, although several other phosphorylation sites remain intact and the chromatin structure can influence HSF1 binding and activity (53, 54). This raises an interesting area for further investigation aimed at defining how different signaling pathways and phosphorylation events can modulate HSF1 accessibility to its target genes in different tissues under physiological and disease conditions.
Changes in HSF1 activity have been linked to numerous diseases. The established role of HSF1 in driving malignant transformation makes its reduction/inhibition in level or activity a potential therapeutic target for cancer. On the other hand, activation of HSF1 coupled with enhanced chaperone expression can ameliorate the course of protein aggregate-related neurological diseases (55). From the organismal prospective, improved proteome stability and metabolic bioenergetics efficiency by increasing levels and activity of HSF1, for example, by modulating the posttranslational signals at S303/S307, are expected to be beneficial and may delay aging. Overexpression of HSF1 during development can prevent a general deterioration of the quality of the proteome and has been linked to life extension in worm models (49). While our study did not explore the possible impact on life span extension, it revealed that raising HSF1 activity during aging can function as a positive regulator of energy production and excessive lipid synthesis and organ fat storage. Conceivably, this anabolic effect might be explained by the concomitant increased supply in ATP, NAD+/NADH, and NADP/NADPH cofactors (37) combined with a powerful increase in transcriptional upregulation of several genes encoding proteins in the lipid biosynthetic pathway (e.g., PPARγ, c/EBPβ, FAS, and SCD1). The deterioration of metabolic function and the accompanying increased lipid accumulation, reduced energy expenditure, and development of insulin resistance and organ inflammation occur presumably as a consequence of nutrient overload. An age-related decline in metabolic fitness and dysregulated nutrient sensing (48) was expected to augment the organismal metabolic dysfunction and likely contributed to the metabolic syndrome progression in the HSF1 mutant mice. It seems intuitive to think that high levels of HSF1 activity support the high biosynthetic demand of cancer and may impact spontaneous malignant transformation, which is a hallmark of aging (56). Consistent with this prediction, exposure of HSF1 mutant mice to a chemical carcinogen, diethylnitroseamine (DEN), slightly accelerated liver cancer development (data not shown). Whether HSF1 mutant mice are prone to spontaneous tumor development deserves further investigation. Notably, stabilization of HSF1 by reduced expression or mutagenesis of the E3 ligase FBXW7, which interacts with S303/S307, has been shown to increase metastatic potential and disease progression in melanoma (28). Thus, as counterintuitive as it may seem, improving quality control networks through HSF1 stabilization may also have detrimental effects on metabolic homeostasis and malignant transformation at an advanced age. The specific role of elevating HSF1 activity in proteome stability, however, is expected to be therapeutically beneficial for neurodegenerative disorders. Certainly, the dichotomy in the roles of HSF1 in malignancy and neurodegenerative diseases requires an in-depth analysis of the multiple distinct regulatory programs, before pursuing clinical application of HSF1-based interventions. Perhaps most important would be to gain a precise insight into the mechanism of age-related physiological dysfunctions and how they can effectively be prevented by approaches that avoid possible detrimental effects on pathophysiological conditions. The direct in vivo effects of increased or reduced HSF1 level and activity on metabolic and protein quality control and disease progression at young and advanced ages are yet to be determined. This will provide information as to whether age-related physiological dysfunctions can be effectively prevented by interventions to manipulate HSF1 activity while avoiding possible detrimental effects on pathophysiological conditions. Finally, in terms of the relevance of our findings to human diseases, the conservation of S303/S307 phosphorylation motifs between mouse and human HSF1 suggests that manipulation of this posttranslational modification through alterations of the activities of the protein kinases phosphorylating these sites to maintain an appropriate level of HSF1 may be a realistic therapeutic approach in patients.
In summary, our findings suggest that phosphorylation on S303/S307 residues in coordination with other posttranslational modifications can effectively modulate the HSF1 transcription response signatures under physiological and environmental stress conditions. The results prompt expectations that identification and regulation of the HSF1 transcriptional programs under physiological and disease states and during aging will provide new, amenable approaches to counteract the actions of HSF1 under human pathological conditions, such as metabolic syndrome, cancer, and neurodegenerative and aging-associated pathologies.
MATERIALS AND METHODS
Animals.
To generate the Hsf1303A/307A mutant mice, a targeting vector containing the mutant S303Ala and S307Ala knock-in allele of hsf1 was constructed. Briefly, a 3.0-kb proximal fragment (including exons 1 to 8) and a 3.9-kb distal fragment (including exons 9 to 12 with the S303Ala and S307Ala mutations encoded in exon 9) of hsf1 allele were amplified by PCR from the bacterial artificial chromosome (BAC) clone PR23-266H9. The DNA fragments amplified by PCR were sequenced to verify the absence of any undesired mutations. The proximal and distal DNA fragments and the tk-neomycin (1.3-kb) selectable marker flanked by Cre recombinase recognition (loxP) sequences were introduced into the XhoI site of the phage DNA vector λDASHII-254-2TK, which was flanked by two thymidine kinase genes for drug-negative selection (57). The final vector was linearized at the unique NotI site for embryonic stem (ES) cell transfection. ES cells (D3; Incyte Genomics, St. Louis, MO) were derived from 129S2/SvPas mice. Positive ES clones identified by Southern blotting were microinjected into C57BL/6 blastocysts, and germ line-transmitting chimeric mice were crossed with C57BL/6 mice to generate heterozygous Hsf1303A/307A/WT mice. To avoid potential interference of the tk-neo marker gene on the expression of mutant hsf1 allele, mice were bred with C57BL/6J mice expressing the cre gene (Jackson Lab.). Homozygous Hsf1303A/307A mice on a C57BL/6J genetic background for at least 10 generations were used for experiments. Animal care and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and National Institutes of Health guidelines.
For Southern blot analysis, genomic DNA digested with BglII was hybridized with a PCR-amplified genomic probe (578 bp) indicated in Fig. 1A. The amplification of the probe was performed using genomic tail DNA and a combination of the following primers: forward, 5′-GGGAAGAATGGGGACTAAAC-3′; reverse, 5′-TACATGTGGCACTCACATAT-3′. Hybridization of BglII-digested genomic DNA generates 5.7 kb for the WT locus and 6.9 kb for the mutant allele.
PCR-based genotyping of mice before removal of the neo gene was performed on tail DNA, and the combination of primers P1 and P2 (forward, 5′-CCATGGGACTGCCAGTAAGT-3′; reverse, 5′-CTTGTCTAGGCAGGCTACGC-3′) to generate a fragment of 630 bp and the combination of P3 and P2 (forward, 5′-CTCGACATCGGAAGATCCAT-3′; reverse, 5′-CTTGTCTAGGCAGGCTACGC-3′) to generate a fragment of 330 bp for the mutant locus were used. For PCR-based genotyping of mice after neo removal, the primer combination P5 (forward, 5′-GCAGGACCTTTATCCCTCCT-3′) and P6 (reverse, 5′-GTTGGGGACAAAGGGGTATC-3′) generates fragments of 246 and 286 bp for the WT and mutant loci, respectively.
Unless otherwise indicated, Hsf1303A/307A homozygous mutant mice and wild-type (WT) littermates of both genders were used.
Cell culture.
Mouse embryo fibroblasts (MEFs) were prepared from embryonic day 13.5 following timed pregnancies of Hsf1303A/307A/WT heterozygous intercrosses. Primary MEFs at first or second passages were stably transformed using retroviral vectors containing E1A and were selected following culturing in 2 mg/ml of puromycin. Both primary and transformed MEFs were cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS).
Chemical cross-linking analyses, immunoblot analyses, and subcellular fractionation.
For HSF1 protein cross-linking studies, 2 mM ethylene glycol bis(succinimidyl succinate) (EGS) was added to MEF cell extracts (80 μg of protein), and after incubation for 2 h at 4°C the reaction was quenched by addition of 50 mM glycine. Cross-linked HSF1 was detected by 6% SDS-PAGE electrophoresis (Bio-Rad, Hercules, CA) and Western blot analysis as described previously (58).
For immunoblot analysis, tissue or cultured cells were homogenized in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris, pH 7.5) containing protease and phosphatase inhibitor cocktail (Pierce, Thermo Scientific, IL). Equal amounts (30 μg) of protein were loaded onto SDS-PAGE gels, transferred onto a polyvinylidene fluoride membrane, and probed with the indicated proteins as described previously (4). The following antibodies were used: HSPB5/αB-crystalline (SPA-822), HSPB1/HSP25 (SPA-801), HSP90α (SPA-840) (Enzo Life Sciences, Farmingdale, NY), HSP70 (heat inducible; SMC-100A/B; StressMarq Biosciences Inc., Victoria, BC, Canada), p-JNK (catalog number 9251), HSF1 (catalog number 4356), AKT (catalog number 9272), PPARγ (catalog number 2430), C/EBPα (catalog number 2295), C/EBPβ (catalog number 3082) (Cell Signaling, Danvers, MA), and phosphor-AKT (S473) (sc-7985-R), SUMO-2/3/4 (C3; sc-393144), γ-tubulin (sc-7396), β-actin (sc-47778) (Santa Cruz Biotechnology, Santa Cruz, CA), p-HSF1(S326) (ab115702; Abcam), CK2 (GTX 107897; Genetex). A polyclonal antibody that specifically recognizes phosphorylated S303/S307 sites of murine HSF1 was kindly provided by L. Sistonen (Abo Academi University, Finland).
Nuclear and cytoplasmic fractions were separated using NE-PER reagent according to the manufacturer's protocol (Thermo Fisher Scientific, Waltham, MA).
Histology and immunohistochemistry.
For histological analysis, liver or epididymal white adipose tissues (WAT) were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for at least 7 days and embedded in paraffin. Hematoxylin and eosin staining was performed on 7-μm paraffin sections. For oil red O staining, frozen tissue sections fixed in 10% formalin for 10 min at 25°C were stained with 0.5% oil red O solution (Thermo Fisher Scientific, Waltham, MA) for 8 min at 60°C. After rinsing with distilled water, sections were counterstained with Mayer's hematoxylin for 30 s. For immunohistochemistry (IHC) staining, frozen liver tissue sections were fixed in 4% PFA, processed, and stained with antibody to CD11b (Biolegend, San Diego, CA).
Adipose tissue morphology.
The evaluation of adipocyte hyperplasia was carried out by extrapolating the total number of adipocytes from the size of adipocytes (via histology) and the weight of fat depot of epididymal WAT as described previously (59, 60). Briefly, fat cell volume was obtained for each capture field using the formula π/6 × (3σ2 × d + d3) (where d is the mean diameter of 100 measured cells in the field and σ is the standard deviation [SD] of the diameter). Then, fat cell density was applied (0.92 g/ml) to determine fat cell weight. The total fat cell number in epididymal fat of each animal was calculated by dividing the total fat depot weight by the mean cell weight of all captured fields.
Immunofluorescence staining.
For immunofluorescence, MEFs untreated or exposed to heat shock (43°C for 20 min) were fixed in 4% PFA for 10 min at 25°C, permeabilized with 0.1% Triton X-100 in PBS (PBST) for 5 min at 4°C, blocked in 5% bovine serum albumin (BSA) in PBST for 1 h, and incubated with a primary antibody against HSF1 overnight at 4°C followed by Alexa Fluor 594 (A11007; Invitrogen) as a secondary anti-rabbit IgG antibody. The slides were stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) in PBS as a nuclear counterstaining and mounted with Vectashield H-1200 (Vectar). Image acquisition was performed on a AxioVision Imager fluorescent microscope (Zeiss) supported with an AxioCam MRC or HRC camera (Zeiss) using a 40× objective lens (numerical aperture [NA], 0.75). AxioVision REL 4.8 software was used for data analysis.
Electrophoretic mobility shift assays.
Detection of Hsf1 DNA binding activity determined by EMSA has been described in detail previously (26). A 15-μg amount of nuclear protein in extraction buffer (10 mM HEPES, pH 7.9, 0.4 mM NaCl, 0.1 mM EDTA, 0.5 mM dithiothreitol [DTT], 5% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) was added to the reaction mixture, which contained 4 μl of binding buffer (37.5 mM NaCl, 15 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 0.5 mM DTT, 5% glycerol), 10 μg of yeast tRNA, 1 μg of sheared Escherichia coli DNA, 10 μg of poly(dI-dC), and 1 ng of 32P-labeled HSE. The mixture was incubated for 15 min at 25°C and resolved on a 4.5% nondenaturing polyacrylamide gel. After electrophoresis, gels were fixed in 7% (vol/vol) acetic acid, rinsed once in distilled water, and exposed to X-ray film. The nucleotide sequence used for HSE was 5′-GTCGACGGATCCGAGCGCCTCGAATGTTCTAGAAAAGG-3′. The double-stranded oligonucleotide containing HSE was labeled with α-[32P]dCTP, Klenow fragment of DNA polymerase I, and deoxynucleotide triphosphates.
Quantitative RT-PCR for mRNA quantification.
Total RNA was isolated from WAT and liver tissue or cultured MEFs using TRIzol (Invitrogen, Carlsbad, CA) and reverse transcribed using the Iscript cDNA synthesis kit (170-8891; Bio-Rad Laboratories). Quantitative RT-PCR was performed using iQ SYBR green Supermix (Bio-Rad Laboratories, Hercules, CA) on an Eppendorf RealPlex 4s real-time PCR system (Eppendorf, Hauppauge, NY). Relative quantitative plots were constructed for quantity of RNA input and for each gene of interest. The gene-specific primer sequences are shown in Table S1 in the supplemental material.
Chromatin immunoprecipitation assay.
ChIP-PCR studies were performed using a ChIP assay kit (EMD Millipore, Billerica MA) according to the manufacturer's instructions and as described in a previous report (5). One million cells were cross-linked with 1% formaldehyde for 10 min at 37°C. Cells were washed twice with PBS containing protease inhibitor cocktail (1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml pepstatin), pelleted, resuspended in 200 μl lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1), and sonicated in an ultrasonic processor (Sonicator 3000; Misonix Inc., Newtown, CT) to a DNA size of 200 to 1,000 bp that was confirmed by agarose gel electrophoresis. Samples were precleared with protein A-agarose beads and immunoprecipitated with HSF1 antibody (ABE1044; Millipore), RPA1 (sc-28304; Santa Cruz Biotechnology), or a rabbit IgG antibody as a control, at 4°C for 16 h. After rinsing, chromatin protein/DNA complexes were eluted twice from the agarose beads by adding elution buffer (1% SDS, 0.1 M NaHCO3, pH 8.0) at room temperature for 15 min. Cross-linking was reversed by heating the samples at 65°C for 4 h. DNA fragments were purified by phenol-chloroform and ethanol precipitation, and the HSE sites present in the mouse hsp promoters were amplified by RT-PCR with the following primers: hsp90α forward (F), 5′-CGCTTCGTAATTACCGCATT-3′; hsp90α reverse (R), 5′-TCTGGAACACAGAGGACGACT-3′; hsp70 F, 5′-TCCAGAGACAAGCGAAGACA-3′; hsp70 R, 5′-GAGTAGGTGGTGCCCAGGT-3′; hspb1 F, 5′-CAAGCTTAGGGGGAGGAATG-3′; hspb1 R, 5′-GTCATGTTCTTGGCTGGTCA-3′; hspb5 F, 5′-CTGCCTGTGTTTCTC TTTTCT-3′; hspb5 R, 5′-CTCCGAAGAACTGGTCGAAGA-3′; mLrrm4 F, 5′-GAACTGAGGACAATAGCCCAGC-3′; mLrrm4 R, 5′-GCTTGAGAAAAGAAGCCAGCAG-3′; mFmod F, 5′-GCAGCAAGAGACTTCAGGCTG-3′; mFmod R, 5′-GGAGCCTAAGCTGGCTTTTTC-3′; mSlc44a5 F, 5′-GCACCAACTCTAACCCAGATAGG-3′; mSlc44a5 R, 5′-GAATTCCCACAAATGGCCTGTCA-3′; mdlgap1 F, 5′-CATTTTAAAACTAGGCCCAAACTCAGC-3′; dlgap1 R, 5′-AGAGAATATTTCTTCCCCAGGTAGGAT-3′.
High-fat diet and glucose tolerance test.
Male mice were fed with HFD (55 to 60% of calories from fat; Envigo) beginning at 4 weeks of age (4). Mice were weighed two times per week. At the termination of the study, the glucose tolerance test (GTT) was performed and tissues were excised, weighed, and frozen in OCT or fixed in 4% PFA. GTT was performed after overnight fasting (16 h) by intraperitoneal (i.p.) injection of glucose (2 g/kg body weight). Blood glucose levels were measured using an automated glucose monitor (Bayer HealthCare LLC, IN).
Measurement of body composition and energy expenditure.
The body composition (lean and fat percentage) was measured by dual-energy X-ray absorptiometry (DXA; Piximus system; GE Lunar, Madison, WI). Scan analysis was performed using GE Encore 11.10 software.
Metabolic rate was determined by placing 5-month-old male mice in an open-circuit calorimeter (CLAMs, Oxymax System; Columbus Instruments, Columbus, OH) for a period of 7 days. After a period of 3 days of adaptation, O2 consumption, CO2 production, and energy expenditure (heat) were measured. The respiratory exchange rate (RER) was calculated as the ratio of volume of CO2 to volume of O2 (VCO2/VO2). The cages were kept at 25°C, and the measurements were normalized to the body weight. Heat outflow was measured during the 7-day period in light and dark cycles and under fed conditions and is indicated as kilocalories/gram/day.
Statistical analysis.
Both male and female mice were used as indicated in the figure legends. Cohorts of 5 to 15 mice were used for the in vivo experiments. Data from three independent experiments are presented as means ± standard deviation (SD). Differences between groups were analyzed by Student's t test (two-tailed distribution with two sample equal variance), and P values of <0.05 were considered significant. For data sets with significant variant difference confirmed by Bartlett's test for equality of variance, statistical significance was evaluated by Welch's test.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by National Cancer Institute grants CA062130, CA06130S, and CA132640 (N. F. Mivechi and D. Moskophidis).
We thank Carlos Isales (Augusta University) for the DXA analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00095-18.
REFERENCES
- 1.Morimoto RI, Kroeger PE, Cotto JJ. 1996. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. EXS 77:139–163. [DOI] [PubMed] [Google Scholar]
- 2.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. 2007. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26:5086–5097. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 3.Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin X, Moskophidis D, Mivechi NF. 2011. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab 14:91–103. doi: 10.1016/j.cmet.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, Bronson RT, Whitesell L, Lindquist S. 2012. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest 122:3742–3754. doi: 10.1172/JCI62727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Labbadia J, Morimoto RI. 2017. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol 27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Pastor R, Burchfiel ET, Thiele DJ. 2018. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C. 1995. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 10.Anckar J, Sistonen L. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 11.Naidu SD, Dinkova-Kostova AT. 2017. Regulation of the mammalian heat shock factor 1. FEBS J 284:1606–1627. doi: 10.1111/febs.13999. [DOI] [PubMed] [Google Scholar]
- 12.Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raychaudhuri S, Loew C, Korner R, Pinkert S, Theis M, Hayer-Hartl M, Buchholz F, Hartl FU. 2014. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell 156:975–985. doi: 10.1016/j.cell.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 14.Zelin E, Freeman BC. 2015. Lysine deacetylases regulate the heat shock response including the age-associated impairment of HSF1. J Mol Biol 427:1644–1654. doi: 10.1016/j.jmb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276:40263–40267. doi: 10.1074/jbc.M104714200. [DOI] [PubMed] [Google Scholar]
- 16.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol 23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. 2017. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 24:325–336. doi: 10.1038/nsmb.3366. [DOI] [PubMed] [Google Scholar]
- 18.Guettouche T, Boellmann F, Lane WS, Voellmy R. 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem 6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batista-Nascimento L, Neef DW, Liu PC, Rodrigues-Pousada C, Thiele DJ. 2011. Deciphering human heat shock transcription factor 1 regulation via post-translational modification in yeast. PLoS One 6:e15976. doi: 10.1371/journal.pone.0015976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budzynski MA, Puustinen MC, Joutsen J, Sistonen L. 2015. Uncoupling stress-inducible phosphorylation of heat shock factor 1 from its activation. Mol Cell Biol 35:2530–2540. doi: 10.1128/MCB.00816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS, Pincus D. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife 5:e18638. doi: 10.7554/eLife.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mivechi NF, Giaccia AJ. 1995. Mitogen-activated protein kinase acts as a negative regulator of the heat shock response in NIH3T3 cells. Cancer Res 55:5512–5519. [PubMed] [Google Scholar]
- 23.Knauf U, Newton EM, Kyriakis J, Kingston RE. 1996. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev 10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- 24.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. 1996. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem 271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 25.Kline MP, Morimoto RI. 1997. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol 17:2107–2115. doi: 10.1128/MCB.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He B, Meng YH, Mivechi NF. 1998. Glycogen synthase kinase 3beta and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Mol Cell Biol 18:6624–6633. doi: 10.1128/MCB.18.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu B, Zhong R, Soncin F, Stevenson MA, Calderwood SK. 1998. Transcriptional activity of heat shock factor 1 at 37 degrees C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3 and protein kinases Calpha and Czeta. J Biol Chem 273:18640–18646. doi: 10.1074/jbc.273.29.18640. [DOI] [PubMed] [Google Scholar]
- 28.Kourtis N, Moubarak RS, Aranda-Orgilles B, Lui K, Aydin IT, Trimarchi T, Darvishian F, Salvaggio C, Zhong J, Bhatt K, Chen EI, Celebi JT, Lazaris C, Tsirigos A, Osman I, Hernando E, Aifantis I. 2015. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat Cell Biol 17:322–332. doi: 10.1038/ncb3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Pastor R, Burchfiel ET, Neef DW, Jaeger AM, Cabiscol E, McKinstry SU, Doss A, Aballay A, Lo DC, Akimov SS, Ross CA, Eroglu C, Thiele DJ. 2017. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington's disease. Nat Commun 8:14405. doi: 10.1038/ncomms14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biamonti G, Vourc'h C. 2010. Nuclear stress bodies. Cold Spring Harb Perspect Biol 2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, Voellmy R. 2004. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci U S A 101:4100–4105. doi: 10.1073/pnas.0304768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura S, Natsume T, Nakai A. 2012. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell 48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Zeng Q, Bhutkar A, Galvan JA, Karamitopoulou E, Noordermeer D, Peng MW, Piersigilli A, Perren A, Zlobec I, Robinson H, Iruela-Arispe ML, Hanahan D. 2018. GKAP acts as a genetic modulator of NMDAR signaling to govern invasive tumor growth. Cancer Cell 33:736–751 e735. doi: 10.1016/j.ccell.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnemolla A, Labbadia JP, Lazell H, Neueder A, Moussaoui S, Bates GP. 2014. Contesting the dogma of an age-related heat shock response impairment: implications for cardiac-specific age-related disorders. Hum Mol Genet 23:3641–3656. doi: 10.1093/hmg/ddu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minsky N, Roeder RG. 2015. Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1alpha. Proc Natl Acad Sci U S A 112:E5669–E5678. doi: 10.1073/pnas.1516219112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su KH, Cao J, Tang Z, Dai S, He Y, Sampson SB, Benjamin IJ, Dai C. 2016. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol 18:527–539. doi: 10.1038/ncb3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiao A, Jin X, Pang J, Moskophidis D, Mivechi NF. 2017. The transcriptional regulator of the chaperone response HSF1 controls hepatic bioenergetics and protein homeostasis. J Cell Biol 216:723–741. doi: 10.1083/jcb.201607091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdin E. 2015. NAD(+) in aging, metabolism, and neurodegeneration. Science 350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 39.Fontana L, Partridge L. 2015. Promoting health and longevity through diet: from model organisms to humans. Cell 161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshino J, Baur JA, Imai SI. 2018. NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab 27:513–528. doi: 10.1016/j.cmet.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregor MF, Hotamisligil GS. 2011. Inflammatory mechanisms in obesity. Annu Rev Immunol 29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 42.Grivennikov SI, Karin M. 2010. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev 21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr.. 2013. Cellular fatty acid metabolism and cancer. Cell Metab 18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manning BD, Cantley LC. 2007. AKT/PKB signaling: navigating downstream. Cell 129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn JS, Thiele DJ. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem 279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- 46.Dai S, Tang Z, Cao J, Zhou W, Li H, Sampson S, Dai C. 2015. Suppression of the HSF1-mediated proteotoxic stress response by the metabolic stress sensor AMPK. EMBO J 34:275–293. doi: 10.15252/embj.201489062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klaips CL, Jayaraj GG, Hartl FU. 2018. Pathways of cellular proteostasis in aging and disease. J Cell Biol 217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higuchi-Sanabria R, Frankino PA, Paul JW III, Tronnes SU, Dillin A. 2018. A futile battle? Protein quality control and the stress of aging. Dev Cell 44:139–163. doi: 10.1016/j.devcel.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silverman JS, Skaar JR, Pagano M. 2012. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci 37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banerji SS, Theodorakis NG, Morimoto RI. 1984. Heat shock-induced translational control of HSP70 and globin synthesis in chicken reticulocytes. Mol Cell Biol 4:2437–2448. doi: 10.1128/MCB.4.11.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theodorakis NG, Morimoto RI. 1987. Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol Cell Biol 7:4357–4368. doi: 10.1128/MCB.7.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guertin MJ, Lis JT. 2010. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet 6:e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. 2017. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun 8:255. doi: 10.1038/s41467-017-00151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neef DW, Jaeger AM, Thiele DJ. 2011. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov 10:930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campisi J. 2013. Aging, cellular senescence, and cancer. Annu Rev Physiol 75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. 2002. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem 86:376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
- 58.Goodson ML, Sarge KD. 1995. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem 270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- 59.Eriksson-Hogling D, Andersson DP, Backdahl J, Hoffstedt J, Rossner S, Thorell A, Arner E, Arner P, Ryden M. 2015. Adipose tissue morphology predicts improved insulin sensitivity following moderate or pronounced weight loss. Int J Obes (Lond) 39:893–898. doi: 10.1038/ijo.2015.18. [DOI] [PubMed] [Google Scholar]
- 60.Pascual-Serrano A, Arola-Arnal A, Suarez-Garcia S, Bravo FI, Suarez M, Arola L, Blade C. 2017. Grape seed proanthocyanidin supplementation reduces adipocyte size and increases adipocyte number in obese rats. Int J Obes (Lond) 41:1246–1255. doi: 10.1038/ijo.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.