Abstract
A role for caspase-10, previously implicated in the autoimmune lymphoproliferative syndrome, in death receptor signaling has not been directly shown. Here we show that caspase-10 can function independently of caspase-8 in initiating Fas- and tumor necrosis factor-related apoptosis-inducing ligand-receptor-mediated apoptosis. Moreover, Fas crosslinking in primary human T cells leads to the recruitment and activation of caspase-10. Fluorescent resonance energy transfer analysis indicates that the death-effector domains of caspase-8 and -10 both interact with the death-effector domain of FADD. Nonetheless, we find that caspase-8 and -10 may have different apoptosis substrates and therefore potentially distinct roles in death receptor signaling or other cellular processes.
Death receptors (DRs) of the tumor necrosis factor receptor superfamily contain a conserved “death domain” in their intracellular region (1). Fas associates with an adaptor molecule, FADD, through homotypic death domain interactions (2, 3). FADD then recruits caspase-8 through homotypic interactions of death-effector domains (DEDs), leading to caspase-8 activation and apoptosis (4, 5). Other DRs may also signal by recruiting caspase-8 through FADD. Among at least 14 mammalian caspases identified so far, only caspase-10 shares homologous DEDs with caspase-8 (6, 7), suggesting that caspase-10 may also function by interacting with DRs. However, recent studies have failed to detect caspase-10 in DR signaling complexes (8, 9).
We have studied patients with autoimmune lymphoproliferative syndrome, a genetic disorder of lymphocyte homeostasis and immune tolerance. Some of these patients harbor a dominant interfering mutation in caspase-10 (10). Caspase-10 abnormalities are associated with defective DR-mediated apoptosis in T cells and dendritic cells. Another homozygous-altered allele was found to be associated with autoimmune lymphoproliferative syndrome (10). Recently, this same mutant allele (V367I in caspase-10S or V410I in caspase-10L) has been found to be present at high frequency in heterozygous form in certain populations (11). Given that this allele encodes a protein with reduced caspase activity, the wide occurrence in this allele may suggest a broader role as a susceptibility factor in immune diseases. Here we show that caspase-10 enters DR signaling complexes in primary T cells and that caspase-10 can functionally substitute for caspase-8, providing direct evidence for a physiological role of caspase-10 in DR signal transduction.
Experimental Procedures
Cell Line, Abs, and Plasmids.
Wild-type Jurkat cells and caspase-8-deficient Jurkat mutant, I9.2, were kindly provided by John Blenis (12). Monoclonal anti-caspase-10 Ab specific for caspase-10 DEDs was from the Medical and Biological Laboratories (Nagoya, Japan). MAbs to caspase-3, caspase-8, RIP, and FADD were from PharMingen. Anti-APO1.3 (anti-Fas) was obtained from Kamiya Biomedical (Thousand Oaks, CA). Polyclonal rabbit anti-Fas Ab was from Marcus Peter (University of Chicago). Full-length caspase-8 (caspase-8a/FLICE/MACHα) was used in transfection studies. Long (caspase-10L) and short (caspase-10S) isoforms of caspase-10/Mch4 were described (10). The cysteine at the protease-active site of caspase-8/-10 was mutated by site-specific mutagenesis (Stratagene) to serine for the protease-inactive mutants caspase-8 C360S, caspase-10S C358S, and caspase-10L C401S. Tagged caspases, TRAF1, and FADD were generated by fusing cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) in-frame to the C terminus of full-length caspase-8a, caspase-10L, the caspase-8a prodomain (amino acids 1–207), the caspase-10L prodomain (amino acids 1–272), TRAF1, or the FADD DED (amino acids 1–121).
Preparation of Human T, B, and Dendritic Cells (DCs), Immunoprecipitation, and Western Blot Analysis.
T cells were enriched from human peripheral blood by using FITC-conjugated Abs to CD1a, CD16, and CD19 (PharMingen), and anti-FITC-conjugated magnetic beads (≈90% positive for CD3 by flow cytometry). The cells were stimulated with 2 μg/ml of phytohemagglutinin (PHA) for 2 days followed by culturing with 100 units/ml of IL-2 or 100 ng/ml of IL-4 for 7 days. B cells were enriched with neuraminidase-treated sheep red blood cells (Cedarlane, Ontario, Canada) to rosette T lymphocytes and were >85% positive by CD19 staining. B cells were activated by 1:10,000 Staphylococcus aureus Cowen I (Calbiochem) plus 100 units/ml of IL-2. DCs were prepared as described (10). Cells were lysed (13) and analyzed by Western blots with anti-caspase-8 or -10. To detect caspase-10 in Fas signaling complexes, primary T cells were activated by 2 μg/ml of PHA for 2 days followed by 100 units/ml of IL-2 for 7 days and incubated with 1 μg/ml of anti-Fas at 37°C for 0, 5, 10, and 15 min. Cell lysates were immunoprecipitated with anti-Fas followed by Western blotting with anti-caspase-10. The blots were then stripped (13) and sequentially probed with anti-caspase-8, anti-FADD, and rabbit polyclonal anti-Fas.
Transfection of Jurkat Cells, Fluorescent Resonance Energy Transfer (FRET) Analysis, Retroviral-Mediated Gene Transfer, and Chemically Induced Dimerization.
I9.2 cells were transfected by electroporation with different constructs and a green fluorescent protein (GFP) vector at the ratio of 3:1. Twelve hours later, cells were treated with FLAG-tagged Fas ligand (FasL) or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) plus 1 μg/ml of anti-FLAG (Alexis, San Diego), and the percentage of loss of GFP+ cells that were negative for annexin V-staining was calculated (13). I9.2 cells transfected with 10 μg of various CFP, YFP, or CFP plus YFP fusion plasmids by electroporation and cultured for 16 h and used for FRET analysis as described (14). 293T cells semiconfluent in 12-well plates were transfected with 2.5 μg of various CFP, YFP, or CFF plus YFP fusion constructs and 10 μl of Fugene (Roche Diagnostics). The cells were cultured for 16 h and collected for FRET analysis. Retrovirus was produced by transfecting 293T cells with HIV-long terminal repeat (LTR)-caspase-10 mutant-internal ribosome entry site (IRES)-GFP or HIV-LTR-IRES-GFP plus a packaging vector, Δ8.2, followed by infection of T cells (15). The cells were cultured overnight and used for apoptosis assays. FK509 binding protein (FKBP; ref. 16) was fused to the N terminus of caspase-8 and -10. I9.2 cells were transfected with 10 μg of FKBP-caspase-8, FKBP-caspase-8 mutant, FKBP-caspase-10, or FKBP-caspase-10 mutant together with 3 μg of GFP plasmid by electroporation. Cells were treated with AP20187 (16), a dimeric FK506 analog as the chemical inducer of dimerization (CID) for 24 h. Loss of annexin V-negative and GFP+ live cells were quantitated by flow cytometry 24 h later (13).
Results
Caspase-10 in Fas- and TRAIL Receptor-Signaling and Expression Pattern of Caspase-10.
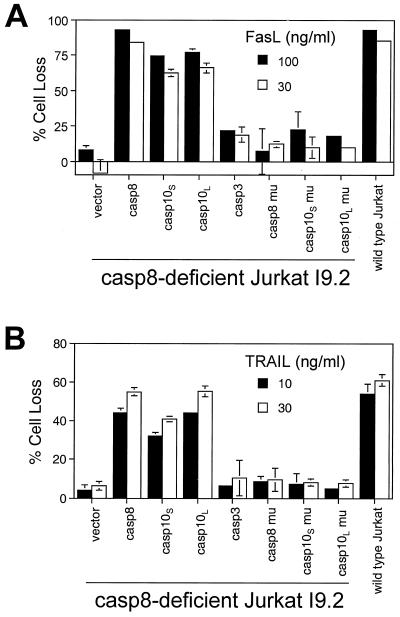
Caspase-8-deficient I9.2 Jurkat cells resist Fas-induced apoptosis (ref. 12; Fig. 1A). We observed that I9.2 cells, like parental Jurkat cells, have very low endogenous caspase-10 (Fig. 2A). We therefore reconstituted I9.2 cells with caspase-8 or -10. Consistent with previous studies, replacement of caspase-8 in I9.2 cells restores Fas-dependent death (Fig. 1A). Interestingly, expression of either short or long isoforms of caspase-10 (caspase-10S and -10L), but not caspase-3, also recovered Fas-mediated apoptosis. No effect was seen by using caspase-8 or -10 with mutated inactive protease domains. Therefore, either DED-containing caspase rescues Fas-dependent apoptosis (Fig. 1A). Similarly, I9.2 cells were defective in TRAIL-induced apoptosis but could be reconstituted with either caspase-8 or -10 (Fig. 1B).
Figure 1.
Reconstitution of DR-mediated apoptosis in I9.2 Jurkat cells. I9.2 cells were transfected with 12 μg of vector or various constructs and 4 μg of a GFP construct. Caspase-8 (casp8), short (casp10S) and long (casp10L) isoforms of caspase-10, caspase-3 (casp3), and mutants (mu) of caspase-8 and -10 containing mutated protease-active sites were used. Cells were treated 8 h later with FasL (A) or TRAIL (B) for 24 h, and the percentage cell loss of GFP+ cells that are negative for annexin V was quantitated (13). Transfected cells positive for annexin V staining without FasL or TRAIL treatment are below 5% (data not shown).
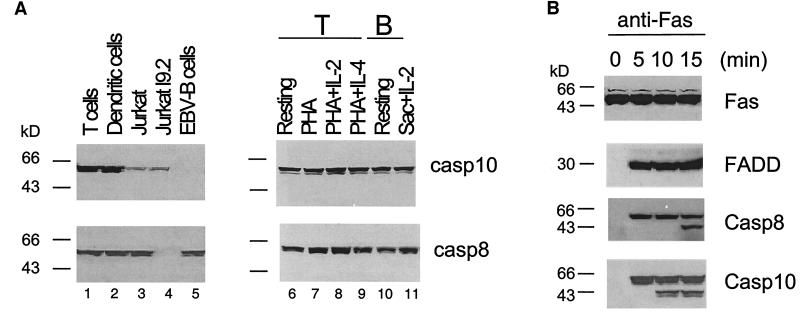
Figure 2.
Caspase-10 expression and recruitment to the Fas signaling complex. (A Left) T cells were stimulated with PHA for 2 days followed by culture in IL-2 for 7 days, and DCs were generated as described (10). Lysates (100 μg) of primary human T cells and DCs or cell lines were used for Western blot analysis of caspase-10 (casp10) and -8 (casp8). (Right) Primary human T cells (resting T cells) were stimulated with PHA for 2 days with or without further culture in IL-2 or IL-4 for 2 days. Resting human T and B cells stimulated with S. aureus Cowen I plus IL-2 for 2 days were also prepared. Cell lysates (100 μg) were used for Western blot analysis with anti-caspase-10 and -8. (B) T cells were cultured in PHA for 2 days and in IL-2 for 7 days. The cells were incubated with anti-Fas (anti-APO1.3) at 37°C for indicated time, and lysates were immunoprecipitated with anti-Fas followed by Western blot analysis, using anti-caspase-10, -8, anti-FADD, and anti-Fas Abs.
We examined the expression of caspase-10 in primary T cells, DCs, and several cell lines. Interestingly, caspase-10 is highly expressed in primary T cells, B cells, and DCs (Fig. 2A). By contrast, caspase-10 is significantly lower in the T lymphoma cell line Jurkat and undetectable in B cells transformed with Epstein–Barr virus (EBV; Fig. 2A). The absence of caspase-10 in EBV-transformed cells is common to other EBV-transformed cell lines, including Raji cells (data not shown). By contrast, caspase-8 is abundantly expressed in primary T cells, DCs, Jurkat cells, and EBV cells, but absent in I9.2, a laboratory-selected Jurkat subline lacking caspase-8 (ref. 12; Fig. 2A). Caspase-10 expression is thus quite variable with the highest expression in primary cells, whereas caspase-8 is ubiquitously expressed. Because primary T cells and DCs were stimulated and cultured in cytokines (Fig. 2A Left), we tested whether high levels of caspase-10 in primary cells are due to cell activation. However, we found that freshly isolated T and B cells from human peripheral blood abundantly express caspase-10, which was unchanged by stimulation of T cells by PHA with or without IL-2 or IL-4, or activation of B cells with S. aureus Cowen I plus IL-2 (Fig. 2A Right). Therefore, caspase-10 is constitutively expressed in primary lymphocytes.
Association of Caspase-10 with the Fas Signaling Complex in Primary T Lymphocytes.
We therefore examined caspase-10 recruitment after Fas engagement in primary T cells. Both FADD and caspase-8 are detected in the Fas signaling complex after 5 min of crosslinking Fas (Fig. 2B). Processed caspase-8 was also observed after 15 min of Fas-crosslinking, indicating productive signaling. We also found caspase-10 in the same Fas immunoprecipitates by using a monoclonal anti-caspase-10 specific for caspase-10 DEDs (Fig. 2B). Processed forms of caspase-10 can be seen after 10 min of Fas-crosslinking (Fig. 2B). Therefore, caspase-10 is recruited into the Fas signaling complex and becomes activated like caspase-8 with slightly faster kinetics under these conditions (Fig. 2B). Similar results were obtained when FasL instead of anti-Fas was used to stimulate Fas (data not shown).
Caspase-10 Activation by Chemically Induced Dimerization.
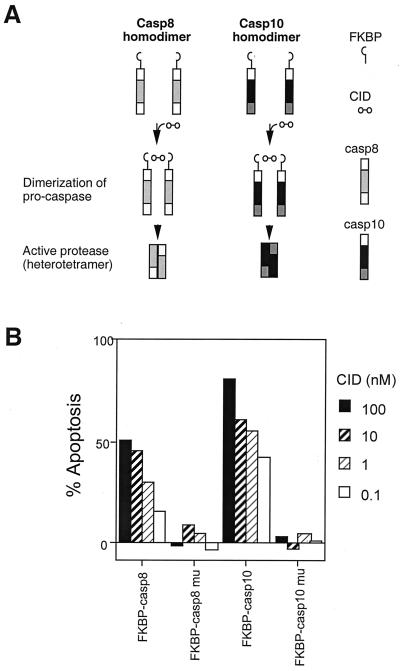
Initiator caspases, including caspase-8 and -9, typically undergo autoactivation when two or more caspases are brought together according to the proximity model (17). We hypothesized that caspase-10 also underwent autoactivation after dimerization. FKBP can form inducible dimers with a bivalent FK506 analog as a “CID” (Fig. 3A; ref. 16). Cells transfected with FKBP-caspase-8 were treated with CID; apoptosis was induced in I9.2 cells (refs. 16 and 18; Fig. 3B). CID was not able to induce apoptosis with an FKBP-caspase-8 fusion protein with a mutated protease-active site. We also observed that CID induced apoptosis in cells expressing the FKBP-casapse-10 fusion protein but not if the protease domain was inactive (Fig. 3B). Thus, dimerization of caspase-10 leads to caspase activation and apoptosis.
Figure 3.
Caspase-10 activation by chemically induced dimerization. (A) Caspase-8 or -10 fused to FKBP were crosslinked by AP20187, an FK506 analog as the CID and underwent activation according to the “proximity model” of caspase activation. (B) I9.2 cells were transfected with FKBP fused with wild-type caspase-8 and -10 or caspase-8 C360S and caspase-10 C401S mutants. The cells were treated with CID for 24 h, and the percentage of cells undergoing apoptosis was quantitated as described (13). Transfected cells positive for annexin V staining without CID treatment are below 5% (data not shown).
Measurement of DED Interactions Between Caspase-10 and FADD.
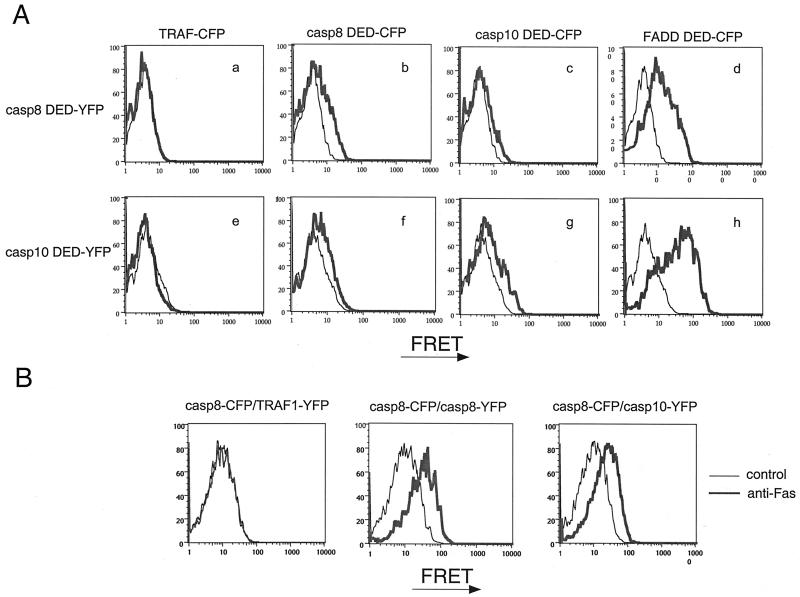
To measure the interactions between caspase-10 DEDs and FADD DED, we used a FRET flow cytometric assay between spectral variants of GFP (14). By using fusions of the DEDs of caspase-8, caspase-10, and FADD to either CFP or YFP, we found clear associations among the DEDs of caspase-8 or -10 and that of FADD as indicated by the peak FRET signal (Fig. 4A). We reproducibly detected a greater FRET shift between caspase-10 DED and FADD DED than with caspase-8 DED and FADD DED, indicating a potentially closer/stronger interaction between the caspase-10 and FADD DEDs. We also observed weak but clear FRET signals by using CFP/YFP pairs of caspase-8 and -10 DEDs with themselves and with each other, indicating a natural propensity of these domains to cause homotypic and heterotypic complexes (Fig. 4A). Single-color conventional flow cytometry showed that all of the fusion proteins were comparably expressed (data not shown). This finding provides biophysical support for the interactions between caspase-10 DEDs and FADD DED.
Figure 4.
Measurement of the interactions among the DEDs of caspase-8, -10, and FADD by FRET. (A) 293T cells were transfected with indicated constructs and harvested 16 h later for FRET analysis. Black line indicates the baseline for FRET by the YFP fusion proteins alone. The base line for CFP proteins are similar to YFP proteins (data not shown), and the red shows the intensity of FRET between CFP and YFP fusion proteins. (B) Jurkat I9.2 cells were transfected with the indicated CFP and YFP fusion proteins. Cells were then treated with anti-Fas (anti-APO1.3; Kamiya Biomedical), and FRET was measured by flow cytometry.
We also fused full-length caspase-8 and -10 with CFP and YFP, respectively. If caspase-8 and -10 go into the same complex, caspase-8-CFP and caspase-10-YFP will be in close proximity and FRET will be induced, whereas the separate recruitment of either caspase should produce no signal. We found that Fas-crosslinking induced FRET in Jurkat cells expressing caspase-8-CFP and caspase-10-YFP (Fig. 4B Right), which was comparable to FRET observed in cells expressing caspase-8-CFP plus caspase-8-YFP (Fig. 4B Center). FRET was not observed in cells expressing caspase-8-CFP and a control noninteracting fusion protein (TRAF1-YFP; Fig. 4B Left). Moreover, the relative lack of FRET before Fas-crosslinking indicates that overexpression of caspase-8 and -10 alone is not sufficient to cause random interactions of sufficient proximity for efficient energy transfer. Taken together, these data suggest that caspase-8 and -10 can be recruited likely within 50 Å, which would constitute the same Fas signaling complex.
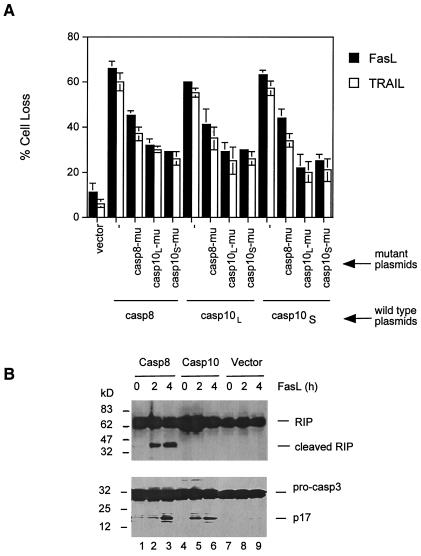
Caspase-8 and -10 Are the Major Mediators of Fas-Mediated Apoptosis but Have Distinct Cleavage Specificities.
Because the DEDs of caspase-8 and -10 can both interact with FADD DEDs, it is possible they may regulate each other by competing for the binding to FADD DED in the Fas signaling complex. To test this idea, we cotransfected mutant caspase-8 and -10 molecules with the inactive protease domains to determine whether they could dominantly interfere with apoptosis induced by DRs in I9.2 cells. We found that either mutated caspase-8 or -10 suppressed the effects of wild-type caspase-8 or -10 isoforms in mediating Fas- and TRAIL receptor-dependent apoptosis (Fig. 5A). Both mutant caspase-10S or -10L isoforms suppressed Fas- and TRAIL receptor-mediated apoptosis (Fig. 5A). Caspase-8 and -10 could therefore compete with each other in mediating apoptosis from death receptors. These data do not distinguish competition for FADD within the cytoplasm or at the Fas signaling complex; however, the former is unlikely because FADD and caspase-8 do not typically associate until both are recruited into a DR signaling complex (4).
Figure 5.
Requirement of both caspase-8 and -10 for Fas signaling. (A) I9.2 cells were transfected with 12 μg of caspase-8, caspase-10L, or caspase-10S plus 12 μg of vector control, mutant caspase-8, mutant caspase-10L, or mutant caspase-10S by electroporation. Four micrograms of GFP plasmid was included in each transfection. The cells were incubated with 30 μg/ml of FLAG-tagged FasL or TRAIL plus 1 μg/ml of anti-FLAG Ab (Alexis). After culture at 37°C for 24 h, GFP+ cells negative for annexin V staining were quantitated by flow cytometry, and the percentage of cell loss was calculated (13). Less than 5% transfected cells were positive for annexin V staining without FasL or TRAIL treatment. (B) I9.2 cells were transfected with caspase-8, -10, or control vector plus GFP. Between 55–65%, cells were GFP+ by flow cytometry analysis (data not shown). The cells were treated with 0.1 μg/ml of FLAG-tagged FasL plus 1 μg/ml of anti-FLAG Ab (Alexis), and cell lysates were used for Western blot analysis of RIP (Upper) and caspase-3 (Lower).
Caspase-8 is readily suppressed by caspase inhibitors such as CrmA and zVAD, whereas caspase-10 is more resistant (ref. 6; unpublished data). Hence, caspase-8 and -10 could have different substrate specificities or they may cleave the same substrates with different efficiency. We found that I9.2 cells reconstituted with caspase-8 underwent Fas-mediated cleavage of RIP, a known cellular substrate for caspase-8 (ref. 19; Fig. 5B), but found no RIP cleavage in I9.2 cells overexpressing caspase-10 induced by Fas (Fig. 5B Upper, lanes 4–6). A similar level of caspase-3 processing was observed in I9.2 cells overexpressing caspase-8 or -10 (Fig. 5B Lower, lanes 1–6). Therefore, RIP is preferentially recognized by caspase-8 but not caspase-10, implying biochemically distinct cleavage and inhibition specificities.
Discussion
Decreased caspase-10 functions were associated with defective DR signaling in various immune cells in autoimmune lymphoproliferative syndrome, thereby suggesting a potentially important function of caspase-10 in both lymphocyte homeostasis and immunological tolerance (10). However, caspase-10 has not been identified in biochemical complexes formed in response to DR signaling in tumor lines (8, 9). We have now shown that caspase-10 is demonstrated in DR signaling complexes in nontransformed human T cells and that the level of caspase-10 protein expression varies in different cell types. Furthermore, we show by biochemical and biophysical studies that caspase-10 is activated by self-aggregation and that it associates with DR signaling molecules and restores Fas-induced apoptosis in Jurkat cells defective for caspase-8. In these respects, caspase-10 seems to function similarly to caspase-8, at least in transfected Jurkat cells.
T cells derived from patients harboring an inactivating mutation in caspase-8 are only partially deficient in Fas-dependent apoptosis (Fig. 6 and Decreased Apoptosis Caused by Caspase Deficiencies, which are published as supporting information on the PNAS web site, www.pnas.org). In these caspase- 8-deficient primary human T cells, caspase-10 can undergo Fas-mediated processing and activation (data not shown). A dominant interfering caspase-10 almost completely blocks Fas-mediated apoptosis in these caspase-8-deficient T cells (Fig. 6). Therefore, caspase-8 and -10 are apparently both required for normal Fas signaling in primary human T cells. We found that mutant caspase-8 and -10 could cross-inhibit one another (Fig. 5A) and can be found to enter the same complexes in response to Fas stimulation (Figs. 2B and 4B). These findings suggest that these caspases are intimately connected during DR signaling and explain why signaling deficits in multiple DRs were detected in patients with defective caspase-10 molecules but no apparent defects in caspase-8 (10). However, we found that caspase-10 differed from caspase-8. Caspase-10 did not mediate Fas-induced RIP cleavage in reconstituted Jurkat I9.2 cells. The genetic evidence from T cells of caspase-8- and -10-deficient patients therefore substantiates our observations with transfection studies of Jurkat cells. Taken together, our data suggest that caspase-10 is an apical initiator caspase in DR signaling but with potentially different substrate specificities or functions.
We speculate that caspase-8 may be inactivated by virally encoded inhibitors or the processes of malignant transformation to promote cell longevity, whereas caspase-10 could provide an alternative pathway for cell elimination by apoptosis. Accordingly, to become transformed, it might be necessary to down-regulate caspase-10. We found that caspase-10 is highly expressed in primary cells derived from the immune system; however, caspase-10 expression is low or absent in many transformed cell lines (Fig. 2 and data not shown). We also noted that virally induced immortalization in the case of EBV-transformed B cells can be associated with the absence of caspase-10. Hence, it is possible that decreased caspase-10 contributes to malignancy by causing apoptosis resistance.
Supplementary Material
Acknowledgments
We thank Dr. J. Blenis for I9.2 cells and Dr. M. Peter for polyclonal anti-Fas. J.W. is an investigator for the Cancer Research Institute.
Abbreviations
- FasL
Fas ligand
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- FRET
fluorescent resonance energy transfer
- DED
death-effector-domain
- DR
death receptor
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- CID
chemical inducer of dimerization
- FKBP
FK506 binding protein
- PHA
phytohemagglutinin
- DC
dendritic cell
- GFP
green fluorescent protein
- EBV
Epstein–Barr virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Locksley R M, Killeen N, Lenardo M J. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 3.Chinnaiyan A M, O'Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 4.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Chinnaiyan A M, Kischkel F C, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincenz C, Dixit V M. J Biol Chem. 1997;272:6578–6583. doi: 10.1074/jbc.272.10.6578. [DOI] [PubMed] [Google Scholar]
- 8.Kischkel F C, Lawrence D A, Chuntharapai A, Schow P, Kim K J, Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 9.Sprick M R, Weigand M A, Rieser E, Rauch C T, Juo P, Blenis J, Krammer P H, Walczak H. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Zheng L, Lobito A, Chan F K, Dale J, Sneller M, Yao X, Puck J M, Straus S E, Lenardo M J. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 11.Gronbaek K, Dalby T, Zeuthen J, Ralfkiaer E, Guidberg P. Blood. 2000;95:2184–2185. [PubMed] [Google Scholar]
- 12.Juo P, Kuo C J, Yuan J, Blenis J. Curr Biol. 1998;8:1001–1008. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Lobito A A, Shen F, Hornung F, Winoto A, Lenardo M J. Eur J Immunol. 2000;30:155–163. doi: 10.1002/1521-4141(200001)30:1<155::AID-IMMU155>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Siegel R M, Frederiksen J K, Zacharias D A, Chan F K, Johnson M, Lynch D, Tsien R Y, Lenardo M J. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 15.Chinnasamy D, Chinnasamy N, Enriquez M J, Otsu M, Morgan R A, Candotti F. Blood. 2000;96:1309–1316. [PubMed] [Google Scholar]
- 16.MacCorkle R A, Freeman K W, Spencer D M. Proc Natl Acad Sci USA. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvesen G S, Dixit V M. Proc Natl Acad Sci USA. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Devin A, Rodriguez Y, Liu Z G. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.