Accurate chromosome segregation is a fundamental process in cell biology. During mitosis, chromosomes are segregated into daughter cells through interactions between centromeres and microtubules in the mitotic spindle.
KEYWORDS: chromosome segregation, centromere transcription, ncRNA, RNA Pol II, cenRNA, R-loop, CENP-A, CENP-C, Aurora-B, CPC, centromere, mitosis
ABSTRACT
Accurate chromosome segregation is a fundamental process in cell biology. During mitosis, chromosomes are segregated into daughter cells through interactions between centromeres and microtubules in the mitotic spindle. Centromere domains have evolved to nucleate formation of the kinetochore, which is essential for establishing connections between chromosomal DNA and microtubules during mitosis. Centromeres are typically formed on highly repetitive DNA that is not conserved in sequence or size among organisms and can differ substantially between individuals within the same organism. However, transcription of repetitive DNA has emerged as a highly conserved property of the centromere. Recent work has shown that both the topological effect of transcription on chromatin and the nascent noncoding RNAs contribute to multiple aspects of centromere function. In this review, we discuss the fundamental aspects of centromere transcription, i.e., its dual role in chromatin remodeling/CENP-A deposition and kinetochore assembly during mitosis, from a cell cycle perspective.
INTRODUCTION
Accurate segregation of chromosomes into daughter cells is one of the main goals of cell division. Errors in chromosome segregation lead to aneuploidy, which is correlated with cancer and other diseases (1, 2). Centromeres are specialized chromosomal domains that nucleate the assembly of the kinetochore, a structure responsible for chromosome attachment to spindle microtubules and chromosome movement during cell division. Despite a high level of functional conservation, a universal centromere-specific DNA sequence has not been identified. Indeed, most eukaryotic centromeres are organized on short tandem repeats assembled into higher-order repeats (3), which differ dramatically among species and exhibit significant variation between individuals of the same species (Fig. 1). In fact, establishing the exact sequence of these repetitive regions represents a significant technical challenge, especially for organisms such as humans, in which centromeres can extend for several megabases (4). As new sequencing technologies have emerged, it has become possible to assemble the linear sequence of human centromeres. A recent work using a nanopore sequencing strategy reported the complete centromere sequence of human chromosome Y (5), which represents a key advance in deciphering these hidden genome pieces. However, the repetitive nature of centromeres and the lack of linear reference models for most centromeres will continue to challenge genomic studies of centromere DNA and centromere RNA (cenRNA). Importantly, despite the lack of sequence conservation between organisms, work during the last decade has unveiled a role for transcription as a conserved property of centromere regions.
FIG 1.
Comparative analysis of centromere DNA organization in S. cerevisiae, S. pombe, D. melanogaster, and Homo sapiens. Schemes representing the main components of centromere regions in the different organisms are indicated. Despite the lack of conservation in size, with lengths ranging from 125 bp in S. cerevisiae to megabases in humans, or in sequence, the epigenetic loading of CENP-A/Cse4/Cnp1/Cid nucleosomes is a common feature of centromere definition.
Centromeres are determined epigenetically in most species, which is in agreement with the absence of sequence conservation (6, 7). Briefly, work in many organisms has shown that no DNA sequence is either necessary or sufficient to confer centromere identity. The notable exception to this paradigm is Saccharomyces cerevisiae, whose centromeres are specified by a conserved 125-bp sequence (reviewed in reference 6). Instead, centromeres are defined by the deposition of the histone H3 variant centromeric protein A (CENP-A), which leads to interspersed H3 and CENP-A nucleosomes along these chromosome regions (8, 9). Importantly, different studies uncovered the existence of posttranslational modifications on histone H3 (H3K4me2 and H3K36me2) at centromere nucleosomes that resemble transcriptionally active domains found in housekeeping genes (10, 11). Consistent with these marks on centromeric H3, transcription arising from centromere DNA was detected at endogenous, artificial, and neocentromeres (12–15). Depletion of these marks causes a rapid loss of transcription at the centromere, inefficient CENP-A loading, and kinetochore inactivation, linking centromere transcription to centromere identity (11). A more recent work suggested that centromere transcription promoted H3K9 acetylation, which in turn prevented heterochromatin formation at these regions (16). Taken together, these observations indicate that specialized chromatin present at the centromere promotes both centromere transcription and centromere identity.
The existence of centromere transcription was first reported for mouse satellite DNA (17, 18). To date, it is widely assumed that transcription has evolved as a fundamental aspect of centromeres that is conserved among all eukaryotes. Interestingly, the core and the flanking pericentric regions, the two main centromere domains, are both transcribed. Indeed, small interfering RNAs (siRNAs) derived from pericentromeric expression are essential for defining and maintaining this region of heterochromatin in Schizosaccharomyces pombe, and long noncoding RNAs (lncRNAs) derived from pericentric repeats are an important feature of heterochromatin formation in human and mouse cells (19–23). In contrast, transcripts from the central core of the centromere do not generate heterochromatin but appear to interact with centromere proteins to stabilize the centromere/kinetochore. In this review, we focus on recent advances in understanding transcription at the centromere core domain. For an extensive review of pericentromeric domain expression, we recommend reading the work of Chan and Wong (24).
CENTROMERE TRANSCRIPTION
Centromere formation by deposition of CENP-A, also termed CID in Drosophila melanogaster, Cse4 in S. cerevisiae, and Cnp1 in S. pombe, is conserved among organisms. Using S. cerevisiae and S. pombe as yeast models, several studies have shown that centromere transcription is crucial for ensuring chromosome stability and accurate loading of Cse4 and Cnp1, respectively (25, 26). Analogously, studies of plants revealed that transcription at the centromere is essential for Cenp-A assembly and centromere formation and maintenance (27–29). In flies, transcription of repeat satellite III (SATIII), which spans megabases on the X-, 2nd-, and 3rd-chromosome centromeres of D. melanogaster, is required for accurate mitosis by ensuring the correct localization of CENP-A and centromere protein C (CENP-C) on all chromosomes (30). Finally, studies with human cells and Xenopus egg extracts demonstrated that transcriptional activity at centromeres is required to ensure correct CENP-C levels and CENP-A loading and accurate chromosome segregation (31–33).
In summary, centromere transcription has evolved as an essential feature of centromeric chromosome domains. The observation that centromeres are transcribed has suggested that the act of transcription or the product of transcription (lncRNA or cenRNA) may play a role in centromere function. When we consider possible roles for centromere transcription, it is also important to consider the cell cycle timing of transcription and how this is linked to various stages of centromere and kinetochore assembly. We outline the evidence that both process and product likely regulate aspects of centromere formation and function and discuss the fact that transcription may regulate different aspects of centromere function at different cell cycle stages.
TRANSCRIPTIONAL MACHINERY AT THE CENTROMERE
RNA polymerase II (RNA Pol II) is responsible for the bulk of centromere transcription, as demonstrated by the effects of specific drugs that differentially inhibit RNA Pol I, II, or III (31, 34). Importantly, a classic work in budding yeast demonstrated that high levels of transcription across a centromere lead to centromere inactivation (35). Moreover, a recent study using human artificial chromosomes also showed that only moderate levels of transcription are compatible with correct centromere function, which demonstrates the existence of tight control over this process (15). The analysis of lncRNAs (cenRNAs) generated from fission yeast centromere transcription revealed the existence of a 5′ cap and a poly(A) tail, supporting their RNA Pol II origin (36). In addition, previous studies using mutants of specific subunits of RNA Pol II provided evidence that Rbp7 is a key component promoting centromere transcription in fission yeast (37). More recently, immunofluorescence (IF) assays showed the presence of RNA Pol II phosphorylated at Ser2 of the C-terminal domain (CTD) at all human, fly, and frog centromeres, indicating active transcription elongation through the action of RNA Pol II (16, 30, 31, 38) during mitosis. RNA Pol III also orchestrates transcription of tRNA gene clusters located at the boundaries between the pericentric and core centromere domains in S. pombe, which functions to establish a barrier between the core domain and pericentric heterochromatin (39, 40). However, tRNA transcription is not likely to play a direct role in centromere assembly, as RNA Pol III inhibition does not affect centromere transcription (34).
Despite our knowledge of RNA polymerases acting at the centromere, little is known about the promoter and transcription factors (TFs) recruiting this machinery to those chromosome regions. Studies of budding yeast have revealed some of the molecular players driving centromere transcription. Cbf1 encodes a transcription factor that binds the centromere core domain of S. cerevisiae and is required for the production of centromere transcripts (26). Two other transcription factors, encoded by Ste12 and Dig1, regulate Cbf1 activity, and more importantly, loss of any of these factors causes defects in centromere function (26, 41). Analogously, the transcription factor encoded by Ams2 binds GATA core sequences located at the centromere of S. pombe, facilitating CENP-A loading and chromosome segregation (25). As yet, the characterization of TFs involved in centromere transcription in mammals is not as clear as that for yeast. To date, only the general CTDP1 factor (RNA Pol II subunit A C-terminal domain phosphatase), which promotes the Ser2-to-Ser5 phospho-transition and processivity of RNA Pol II, has been identified at the centromere of human cells (31). Due to the highly repetitive nature of higher eukaryotic centromeres, it has been difficult to identify sequences that might function as promoter regions and to define the full-length sequences of transcripts originating from the centromere. Studies of maize and tammar wallaby (28, 42) suggested that promoters for centromere transcription may reside within retroviral elements in the centromere. However, whether this is true for all centromere transcription or if transcription factors bind to retroelements within the centromere is unknown. Additionally, the recent full-length sequence of the human Y chromosome centromere revealed a lack of transposon sequences in the CENP-A-binding domain (5). It will thus be important to determine if transposons act as promoter regions within the centromere or if there are cryptic promoter elements present within the repeat sequences. Interestingly, work using a neocentromere that lacks α-satellite sequences and a structurally dicentric chromosome that contains two α-satellite arrays demonstrated that RNA Pol II localizes to active sites of kinetochore formation during mitosis (31), suggesting that mitotic centromere transcription is linked to the active kinetochore. At this point, we do not know if centromere proteins also promote transcription during interphase or if centromere transcription is a fundamental property of repetitive DNA sequences. Taken together, the data show that the TFs and binding domains involved in RNA Pol II recruitment in most organisms are still unknown.
In addition to TFs, several studies have identified transcription-associated chromatin remodeling factors and RNA Pol II-associated proteins present at the centromere. FACT (facilitates chromatin transcription) and the Mediator complex are major regulators of transcriptional activity mediated by RNA Pol II in eukaryotes. FACT, a highly conserved heterodimer that consists of SSRP1 and SUPT16H subunits, promotes RNA Pol II elongation (43). Interestingly, both subunits interact with the CENP-A protein in different organisms (44, 45) and collaborate in its transcription-coupled loading at centromere domains by interacting with CAL1 (chromosome alignment defect 1), the CENP-A loading factor in Drosophila (32). In addition, FACT seems to play a role in correcting ectopic CENP-A loading by triggering its proteasome-mediated degradation in S. cerevisiae (46). The main function of Mediator, a large modular complex, is to serve as a bridge between regulatory factors (i.e., enhancers) and the general RNA Pol II machinery (47). Interestingly, three components that form a submodule of the complex (Med8, Med18, and Med20) regulate centromere function in S. pombe by moderating RNA Pol II-mediated transcription to ensure precise CENP-A loading (48). Whether Mediator has a conserved role at the centromere in higher eukaryotes needs further study.
DUAL ROLE FOR CENTROMERE TRANSCRIPTION
Role of transcription-coupled chromatin remodeling and cenRNA.
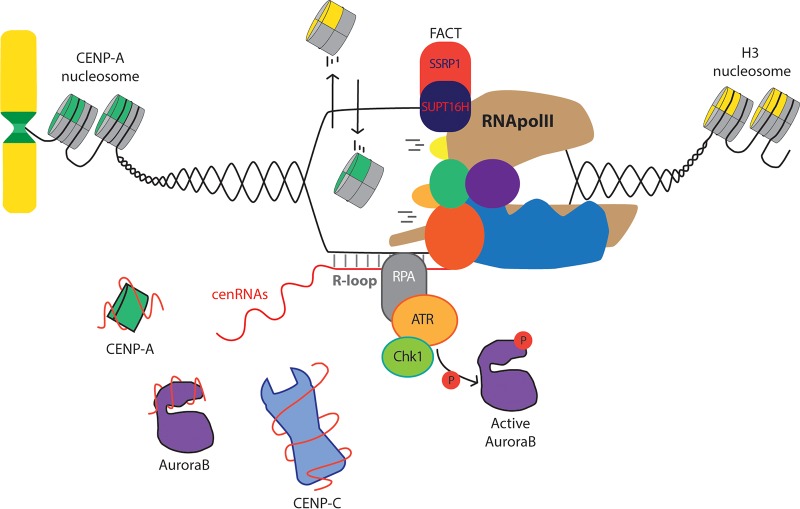
As noted above, the act of centromere transcription may affect centromere assembly in at least two distinct ways: by triggering a chromatin remodeling event mediated by the passage of RNA Pol II and by producing a functional cenRNA (Fig. 2). This dual effect plays a crucial role at centromeres, where chromatin is defined by CENP-A deposition and, as a consequence of its repetitive nature, noncoding RNAs are the only product of transcription. Although the specific loading of CENP-A at centromeres has been associated extensively with the transcription process in many organisms (47, 48), recent reports using strategies to separate the two outputs of transcription are starting to unveil the precise underlying molecular mechanism. Indeed, a study using ectopic centromeres in Drosophila revealed that the CENP-A loading factor CAL1 recruits FACT and RNA Pol II, which is essential for de novo CENP-A deposition (32). By employing human artificial chromosomes, Molina and colleagues (16) proposed a model in which RNA Pol II passage generates the appropriate epigenetic landscape required to destabilize H3 nucleosomes in order to promote CENP-A loading. A more recent study developed a novel method using tissue culture cells from Drosophila to understand the recruitment and chromatin incorporation of CENP-A. Indeed, a two-step model was proposed for CENP-A incorporation at the centromeres. In this model, CAL1-interacting CENP-A is recruited to centromere domains in a manner that is independent of transcription, and it remains loosely associated with chromatin until RNA Pol II passage facilitates exchange of H3 nucleosomes for stable incorporation of CENP-A into chromatin (49). From all these studies, it appears clear that the impact of transcription on chromatin, likely promoted by the FACT complex, is necessary for the stable incorporation of newly synthesized CENP-A into chromatin. Additionally, studies with Drosophila and human cells have shown that destruction of cenRNAs without affecting the process of transcription also results in defects in CENP-A and CENP-C deposition at centromeres (30, 34), suggesting that the transcriptional process and cenRNA product are both important for maintenance of centromeric chromatin. Based on this evidence, it seems likely that centromere transcription is temporally coupled to deposition of newly synthesized CENP-A.
FIG 2.
The process and product of centromeric transcription regulate key aspects of centromere biology. (Top) The topological effect generated by RNA Pol II passage favors the incorporation of CENP-A nucleosomes at centromere regions. (Bottom) As a consequence of transcription, nascent centromeric RNAs (cenRNAs) and DNA/RNA hybrids (R-loops) modulate distinct fundamental aspects of a plethora of centromere proteins.
Centromere RNAs.
RNAs from centromeres are generated from the two main centromere domains: the pericentromeric area and the core region. Interestingly, depending on the origin, transcripts are processed through different pathways and have distinct molecular functions. Briefly, in S. pombe, transcripts derived from pericentromeric transcription are processed into siRNAs that induce chromatin silencing through recruitment of the Clr4 histone methyltransferase, and therefore they function to maintain the heterochromatic nature of these regions (31). In vertebrates, pericentric RNAs associate directly with Suv39 histone methyltransferases to promote heterochromatin formation (22, 23). Our knowledge about the structure and processing of RNAs generated from the core centromere (cenRNAs) is more limited. As mentioned previously, centromere DNA is based on short tandem repeats that extend for lengths of up to megabases in humans. Intriguingly, studies have revealed highly variable cenRNA lengths, ranging from 40 nucleotides (nt) in maize and tammar wallaby (42, 50) to several thousand nucleotides in flies or humans (30, 34, 51). In addition, studies have demonstrated that cenRNAs undergo posttranscriptional processing. In mouse cells, two long RNA precursors, of 2,000 nt and 4,000 nt, are processed into 120-nt cenRNAs by an unknown mechanism (51). Grenfell and colleagues (33) recently revealed that splicing is required for correct spindle and kinetochore assembly in Xenopus egg extracts. In that study, the authors show that the splicing machinery interacts with and promotes the processing of the frog centromere repeat 1 (fcr1) cenRNA precursor, which in turn is required for the accurate localization of CENP-C, Aurora-B, and MCAK (mitotic centromere-associated kinesin), key factors in orchestrating kinetochore function. In human cells, cenRNAs from chromosomes containing functionally distinct α-satellite arrays (34) show differences in stability depending on their origin. Interestingly, α-satellite transcripts from arrays that assemble an active centromere are more stable than those generated from arrays that do not assemble a centromere. Additionally, α-satellite transcripts produced from the functional centromere array physically associate with CENP-A and CENP-C, while α-satellite transcripts from inactive arrays interact with CENPB (34). In summary, the structural properties of cenRNAs and the posttranscriptional players regulating their metabolism are poorly understood. Deciphering which factors may impact cenRNA processing and stability and how their role is modulated across the cell cycle constitutes a challenging future goal.
cis versus trans.
One of the most intriguing phenomena surrounding cenRNA function is whether cenRNAs function in cis or in trans. Centromere RNA may act only near the site of transcription, possibly as a nascent transcript, or centromere RNAs may be processed and diffuse from the site of transcription to influence the function of all centromeres, possibly through interaction with mobile centromere factors. At this point, there is evidence to support both cis and trans roles for centromere RNAs. Studies of human cells have revealed several different mechanisms by which cenRNAs act in cis. McNulty and colleagues (34) demonstrated the existence of array-specific noncoding α-satellite RNAs in each single chromosomal centromere. Importantly, destruction of RNAs generated from one specific active centromere array affects the loading and localization of CENP-A and CENP-C proteins predominantly at that single centromere, which in turn causes the activation of a cell cycle checkpoint and arrest in S and G2. In a second study, Kabeche and colleagues (52) unveiled the existence of DNA/RNA hybrids (R-loops) (reviewed in reference 53) at all centromeres, generated as a consequence of RNA Pol II-mediated transcription. These R-loops, which are more abundant in mitosis, recruit replication protein A (RPA), a single-stranded-DNA-binding factor related to DNA damage sensing, to centromere regions. As a consequence, RPA promotes the recruitment and activation of ATR kinase, which activates Chk1, leading to stimulation of Aurora-B activity. In the reported study, R-loops were present at all centromeres, and it is not clear if the ATR signal was able to diffuse away from the centromere to act on other centromeres. It will be important to develop methods to manipulate R-loops at specific centromeres to determine if this pathway acts in cis or in trans. In summary, both reports have provided elegant evidence for two different cis-based modes of action for human cenRNAs.
In addition to works supporting a cis-acting role for cenRNAs, several studies support the action of cenRNAs in trans. Work in Drosophila showed that the SATIII RNA, generated from transcription of the X chromosome, may act in trans, as this transcript was also found at the centromeres of the other two major autosomes, chromosomes 2 and 3 (30). However, a recent study showed that SATIII RNA is also transcribed from arrays on chromosomes 2 and 3 (49). Moreover, although SATIII RNA depletion increased the frequency of lagging chromosomes in anaphase, it was not clear if SATIII RNA was required for the proper segregation of the 4th chromosome, which does not contain SATIII DNA. It will thus be essential to determine whether SATIII RNA acts in cis or in trans and, in the case of acting in trans, to understand how SATIII RNA is localized to centromeres distant from the site of transcription. In another work, using Xenopus as a model, our group reported an analogous trans-acting mechanism for cenRNAs (38). While the fcr1 centromeric repeat is present at approximately half of Xenopus centromeres (54), codetection of fcr1 RNA and DNA showed that fcr1 RNA is present at centromeres in a manner independent of the existence of the fcr1 DNA sequence. The fcr1 cenRNA interacts with Aurora-B, promoting its localization to the centromeres of all chromosomes, which supports the notion of fcr1 acting in trans (38). Again, the precise molecular mechanism mediating the localization of fcr1 RNA at the centromeres of chromosomes, distinct from its sites of transcription, remains unknown.
ROLE OF CENTROMERE TRANSCRIPTION DURING THE CELL CYCLE
Centromere transcription has been proposed to affect several aspects of centromere function, from CENP-A assembly to mitotic kinetochore signaling. To understand how centromere transcription may affect all of these processes, it is worthwhile to examine the timing of centromere transcription in relation to these various proposed functions (Fig. 3). To date, however, an extensive and detailed study measuring centromere transcription levels during the entire cell cycle and addressing how this process is regulated is lacking for most organisms. In addition, the fact that transcription-mediated chromatin remodeling and nascent cenRNAs can play roles simultaneously or at different cell cycle stages should be considered in interpreting results. In this section, we evaluate the most recent advances concerning the role of centromere transcription from a cell cycle perspective.
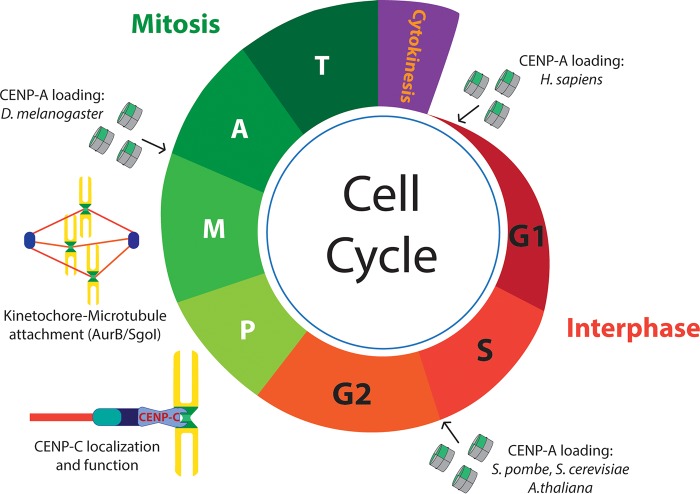
FIG 3.
Cell cycle perspective on centromere transcription. CENP-A loading occurs only once during the cell cycle, but at different stages depending on the species, i.e., in early G1 in humans, in the S/G2 transition in yeast or Arabidopsis, and in metaphase-anaphase (M/A) in Drosophila. The existence of centromere transcription during these stages facilitates the incorporation of CENP-A at these chromosome regions. Centromere transcription uncoupled from CENP-A loading also guarantees other centromere aspects, such as sensing the kinetochore-microtubule attachment tension status in metaphase by regulating the localization and function of Aurora-B (AurB) or shugoshin I (SgoI). In addition, transcription ensures the proper localization and function of CENP-C, a key RNA-binding factor linking CENP-A nucleosomes to the Mis12 complex during mitosis.
Role of centromere transcription and cenRNAs in CENP-A loading.
CENP-A deposition constitutes a key process in the assembly and propagation of centromeres and requires the precise incorporation of newly synthesized CENP-A molecules. Not surprisingly, CENP-A loading is a tightly controlled process that occurs just once during the cell cycle. Interestingly, the time frame in which new CENP-A is deposited differs among species (55). CENP-A is deposited in early G1 in vertebrates (56), during metaphase-anaphase in Drosophila (57, 58), and in S/G2 in S. cerevisiae, S. pombe, and Arabidopsis thaliana (29, 59, 60). Importantly, transcription has been observed at the centromere during all those cell cycle stages, which supports its role in remodeling chromatin to favor CENP-A loading. In S. pombe, RNA Pol II and Mediator show a peak of occupancy at the core centromere in S phase (48). During the rest of the cell cycle, both factors showed more moderate levels of occupancy, indicating that higher levels of centromere transcription are required at the time of CENP-A loading in fission yeast (48). For Drosophila tissue culture cells, analysis of an Rbp3 (RNA Pol II subunit)-green fluorescent protein (GFP) fusion protein across the cell cycle showed evident centromere localization of RNA Pol II, starting in mitosis and ending in G1. In addition, the RNA Pol II localization pattern correlated with the dynamics of 5-ethynyl uridine (EU)-labeled nascent RNA at centromere domains, again suggesting that centromere transcription is coupled with CENP-A incorporation (49). For humans, IF analysis showed actively elongating RNA Pol II enriched on the centromere during mitosis, before CENP-A deposition in G1 (31). In addition, one recent study showed that elongating RNA Pol II is present at centromeres, at high levels, until kinetochores have achieved stable microtubule attachment (61). Since CENP-A loading occurs in G1 in human cells, it seems unlikely that transcription during mitosis is required to facilitate CENP-A deposition into chromatin, but it may play a role in regulating other centromere proteins during this cell cycle stage.
Many studies have reported analyses of centromere transcript levels during different stages of the cell cycle. The quantification of minor satellite RNAs in mouse cells showed that levels vary greatly across the cell cycle, from barely detectable at G1 to a moderate peak at G2/M (62). In Drosophila, the levels of EU-labeled nascent RNAs at the centromere also experienced high variation during the cell cycle, being more abundant during mitosis and early G1 (49). In contrast, a recent work measuring the levels of α-satellite transcripts originating from specific centromeres in human cells revealed stable RNA levels during the entire cell cycle (34). Although they are informative, we must strictly consider those levels to be an indirect measure of transcription, since they reflect the rates of both synthesis and degradation of RNAs. In the future, it will be important to measure RNA Pol II centromere association by use of chromatin immunoprecipitation (ChIP) to more precisely pinpoint the timing of centromere transcription. Taken together, these studies have revealed the existence of variable dynamics for centromere transcripts across the cell cycle that differ among species. It is likely that one of the main functions of centromere transcription is to ensure the accurate and timely loading of CENP-A. Importantly, the presence of active RNA Pol II and cenRNAs outside the CENP-A loading time window suggests a role for transcription in the regulation of other functional aspects of centromeres during specific cell cycle stages.
INFLUENCE OF CENTROMERE TRANSCRIPTION ON CENTROMERE PROTEIN FUNCTION
In addition to considering the timing of centromere transcription, it is important to consider the centromere proteins that have been reported to interact with cenRNAs or are impacted by the process of centromere transcription. Through analysis of the protein targets of cenRNA, we can gain insight into the timing and function of centromere transcription.
Modulation of CENP-C localization and function.
CENP-C was one of the first centromere proteins identified in humans (63). Homologs of this nonhistone centromere protein have been identified in most eukaryotic organisms (64). CENP-C plays a crucial role in stabilizing CENP-A nucleosomes and linking CENP-A chromatin with the microtubule-binding complexes of the kinetochore. Interestingly, several studies have reported a conserved RNA-binding activity for CENP-C (13, 30, 33, 34, 50). Indeed, the physical interaction of CENP-C with transcripts derived from centromere transcription is required for stable localization of CENP-C to the centromere in several different organisms (13, 30, 31, 50). However, how centromere transcription and the interaction with cenRNA promote stable CENP-C localization is not well understood. A report on maize showed that the DNA-binding activity of CENP-C is promoted by its interaction with single-stranded RNAs in vitro. Interestingly, deletion of the CENP-C RNA-binding domain results in CENP-C mislocalization in vivo. However, the fact that both RNA- and DNA-binding activities reside on the same protein domain needs to be considered in interpreting these results (50). Importantly, CENP-A loading depends on CENP-C, and vice versa (65, 66). It is tempting to speculate about centromere transcription reinforcing CENP-A loading by stabilizing CENP-C levels through the action of nascent centromeric RNAs. In the future, it will be important to determine how CENP-C interacts with cenRNA and how RNA binding by CENP-C affects interaction of this protein with CENP-A nucleosomes and microtubule-binding complexes at the kinetochore. Additionally, it will be important to determine when CENP-C interacts with cenRNA and if the RNA interaction is modulated during the cell cycle.
Role of centromere transcription as an Aurora-B activator and chromosome attachment sensor.
Aurora-B, which is a component of the chromosome passenger complex (CPC), is a member of the Aurora family of serine/threonine (S/T) kinases with a central role during cell division (67). Early in mitosis, Aurora-B is recruited to the chromosome arms and inner centromere region, where it regulates histone phosphorylation, cohesin release, and accurate spindle microtubule attachment to kinetochores (68). Interestingly, many recent reports have provided evidence that Aurora-B localization and activation are both regulated by centromere transcription and cenRNAs. Aurora-B was initially identified as a protein interacting with minor satellite RNAs in mouse cells (51) and was subsequently found to interact with cenRNAs in both human cells and Xenopus (38, 69). Our group has shown that Aurora-B interacts with many types of RNAs, including cenRNAs, in Xenopus egg extracts (70). Work in both humans and Xenopus has shown that centromere transcription and cenRNAs are required for normal Aurora-B localization and activation (38, 69). Moreover, Grenfell et al. (33) revealed that cotranscriptional recruitment of the RNA processing machinery to the centromere is also required for both localization and activation of Aurora-B in Xenopus. Work from our lab has shown that the CPC interacts directly with RNA through binding sites in Aurora-B and Borealin both in vitro and in vivo (38, 70). Consistent with a role for cenRNA in the normal activation of Aurora-B, blocking centromere transcription led to an increase in syntelic kinetochore attachments in Xenopus. This result suggests that there may be a relationship between centromere transcription during mitosis and the mechanism that senses kinetochore microtubule attachment. A recent report describing the presence of R-loops at the centromere during mitosis may suggest a plausible mechanism by which tension can be transmitted to Aurora-B (52). Kabeche et al. demonstrated that R-loops are present at centromeres specifically during mitosis and are required for normal Aurora-B activation. During mitosis, RPA binds to centromeric R-loops, promoting the centromere localization and further activation of ATR kinase in mitosis, which triggers Chk1-mediated Aurora-B stimulation (71) to preserve correct chromosome segregation (52). During mitosis, Aurora-B localizes to the inner centromere region, where it functions to correct improper kinetochore-microtubule attachments (72). It is currently unclear how centromere transcription or the presence of R-loops is involved in this process. It is tempting to speculate that centromere transcription may be responsive to the kinetochore-microtubule attachment status and that this information is transmitted to Aurora-B through centromere RNAs or the ATR pathway. Kabeche et al. showed that ATR interacts with CENP-F to promote localization of ATR to the kinetochore. However, recent work in human cells showed that CENP-F deletion has no effect on cell viability or mitotic chromosome segregation (73). It will be important to determine if CENP-F is required for ATR localization to the kinetochore in all cell types or if there are alternative pathways for ATR kinetochore localization. In the future, it will also be interesting to determine if the presence of centromeric R-loops is influenced by kinetochore-microtubule attachment and how this information is transmitted to Aurora-B. Additionally, it will be important to understand how RNA binding to the CPC influences the kinase activity of this complex. In summary, many reports support the notion of an additional role for centromere transcription, independent of its participation in CENP-A loading, potentially as a tension/attachment sensor during mitosis to ensure accurate chromosome segregation. At this point, it is most clear that the product of transcription is important during mitosis, but it will be critical to determine if the chromatin remodeling activity is also required during mitosis.
CONCLUSIONS AND FUTURE DIRECTIONS
The current evidence strongly supports a conserved role for centromere transcription in several aspects of centromere function. However, we have very little understanding of the molecular players in this process, and many outstanding questions remain. How centromere/chromosome arm expression is regulated during the cell cycle constitutes one of the most interesting aspects. This fact is especially important during mitosis, when most transcription from chromosome arms is depleted (74, 75), while centromeres seem to escape from this general inhibition. In addition, several studies support a trans-acting mode of action for cenRNAs, with execution of their role in centromeres different from their place of origin. The molecular players driving the transport of centromere transcripts are unknown, and whether cenRNAs impact any aspect of chromosome regions different from centromeres is also uncharacterized. Moreover, whether a transcript generated from chromosome arm expression plays a role at the centromere needs to be elucidated. Finally, although increased expression of repetitive sequences has been described for many cancer types (76, 77), understanding the effect of erroneous/ectopic centromere expression on the appearance and progression of cancer represents a clear direction for future studies.
ACKNOWLEDGMENTS
Work on centromere transcription in the lab of M.D.B. is supported by the NIH (grant 1R01 GM122893).
We acknowledge Judith Sharp for helpful comments on the manuscript.
Biographies

Carlos Perea-Resa obtained a Ph.D. in molecular biology and genetics from the Complutense University of Madrid in 2015, and most of his research career was developed at the Centro de Investigaciones Biologicas (CIB) of the Spanish Research Council (CSIC), studying posttranscriptional gene expression regulation in response to stress. He currently works as a research fellow in Michael Blower's laboratory, associated with the Molecular Biology Department of the Massachusetts General Hospital (MGH-HMS), which he joined in April 2016. During his first 2 years as a postdoc, he is focusing on centromere biology and aiming to understand different functional aspects of centromere expression in human cells.

Michael D. Blower is an associate professor of genetics at Harvard Medical School and Massachusetts General Hospital. He received his Ph.D. from the University of California, San Diego, where he worked with Gary Karpen on the centromere-specific histone CID. He performed postdoctoral research at the University of California, Berkeley, where he worked with Rebecca Heald and Karsten Weis on mitotic spindle assembly. Since 2006, he has been a professor at Harvard Medical School, where his lab has focused on the regulation of the transcriptome during mitosis.
REFERENCES
- 1.Gordon DJ, Resio B, Pellman D. 2012. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 2.Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Cell Biol 16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 3.Aldrup-McDonald ME, Sullivan BA. 2014. The past, present, and future of human centromere genomics. Genes (Basel) 5:33–50. doi: 10.3390/genes5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miga KH. 2015. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res 23:421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- 5.Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazei KV, Paten B, Haussier D, Willard HF, Akeson M, Miga KH. 2018. Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 36:321–323. doi: 10.1038/nbt.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpen GH, Allshire RC. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet 13:489–496. doi: 10.1016/S0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan BA, Blower MD, Karpen GH. 2001. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet 2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 8.Blower MD, Sullivan BA, Karpen GH. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330. doi: 10.1016/S1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodor DL, Mata JF, Sergeev M, David AF, Saliminian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. 2014. The quantitative architecture of centromeric chromatin. Elife 3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan BA, Karpen GH. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann JH, Rodríguez MG, Martins NM, Kimura DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH. 2003. Transcription within a functional human centromere. Mol Cell 12:509–516. doi: 10.1016/S1097-2765(03)00279-X. [DOI] [PubMed] [Google Scholar]
- 13.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH. 2007. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. 2009. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. 2008. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC. 2016. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 7:13334. doi: 10.1038/ncomms13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harel J, Hanania N, Tapiero H, Harel L. 1968. RNA replication by nuclear satellite DNA in different mouse cells. Biochem Biophys Res Commun 33:696–701. doi: 10.1016/0006-291X(68)90352-5. [DOI] [PubMed] [Google Scholar]
- 18.Cohen AK, Huh TY, Helleiner CW. 1973. Transcription of satellite DNA in mouse L-cells. Can J Biochem 51:529–532. [DOI] [PubMed] [Google Scholar]
- 19.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 20.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 21.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martiense RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 22.Velazquez Camacho O, Galan C, Swist-Rosowska K, Ching R, Gamalinda M, Karabiber F, De la Rosa-Velazquez I, Engist B, Koschorz B, Shukeir N, Onishi-Seebacher M, van de Nobelen S, Jenuwein T. 2017. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife 6:e25293. doi: 10.7554/eLife.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O'Neill RJ, Sullivan BA, Straight AF. 2017. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife 6:e25299. doi: 10.7554/eLife.25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan FL, Wong LH. 2012. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res 40:11178–11188. doi: 10.1093/nar/gks921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ES, Saitoh S, Yanagida M, Takahashi K. 2003. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11:175–187. doi: 10.1016/S1097-2765(03)00011-X. [DOI] [PubMed] [Google Scholar]
- 26.Ohkuni K, Kitagawa K. 2012. Role of transcription at centromeres in budding yeast. Transcription 3:193–197. doi: 10.4161/trns.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, Birchler JA, Parrott WA, Dawe RK. 2003. A molecular view of plant centromeres. Trends Plant Sci 8:570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Topp CN, Zhong CX, Dawe RK. 2004. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci U S A 101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. 2006. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosic S, Kohler F, Erhardt S. 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207:335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Chong KH, Wong LH. 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A 109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CC, Bower S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O'Neill RJ, Mellone BG. 2015. Establishment of centromeric chromatin by the article establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev Cell 34:73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grenfell AW, Heald R, Strzelecka M. 2016. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol 214:783. doi: 10.1083/jcb.20160402908222016c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNulty SM, Sullivan LL, Sullivan BA. 2017. Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell 42:226.e6–240.e6. doi: 10.1016/j.devcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill A, Bloom K. 1987. Genetic manipulation of centromere function. Mol Cell Biol 7:2397–2405. doi: 10.1128/MCB.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. 2011. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem 286:23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 1:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blower MD. 2016. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep 15:1624–1633. doi: 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott KC, Merrett SL, Willard HF, Carolina N. 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 24:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 40.Scott KC, White CV, Willard HF. 2007. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One 2:e1099. doi: 10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemmerich P, Stoyan T, Wieland G, Koch M, Lechner J, Diekmann S. 2000. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc Natl Acad Sci U S A 97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carone DM, Longo MS, Ferreri GC, Harris M, Shook N, Bulazei KV, Carone BR, Obergfell C, O'Neill MJ, O'Neill RJ. 2009. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 43.Formosa T. 2012. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foltz DR, Jansen LE, Black BE, Bailley AO, Yates JR III, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 45.Barth TK, Schade GO, Schmidt A, Vetter I, Wirth M, Heun P, Thomae AW, Imhof A. 2014. Identification of novel Drosophila centromere-associated proteins. Proteomics 14:2167–2178. doi: 10.1002/pmic.201400052. [DOI] [PubMed] [Google Scholar]
- 46.Deyter GMR, Biggins S. 2014. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev 28:1815–1826. doi: 10.1101/gad.243113.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutourina J. 2018. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19:262–274. doi: 10.1038/nrm.2017.115. [DOI] [PubMed] [Google Scholar]
- 48.Carlsten JO, Szilagyi Z, Liu B, Lopez MD, Szaszi E, Djupedal I, Nystrom T, Ekwall K, Gustafsson CM, Zhu X. 2012. Mediator promotes CENP-A incorporation at fission yeast. Mol Cell Biol 32:4035–4043. doi: 10.1128/MCB.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bobkov GOM, Gilbert N, Heun P. 2018. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J Cell Biol 217:1957–1972. doi: 10.1083/jcb.201611087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Y, Topp CN, Dawe RK. 2010. DNA binding of centromere protein C (CENP-C) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouzinba-Segard H, Guais A, Francastel C. 2006. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A 103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabeche L, Nguyen HD, Buisson R, Zou L. 2018. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359:108–114. doi: 10.1126/science.aan6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santos-Pereira JM, Aguilera A. 2015. R loops: new modulators of genome dynamics and function. Nat Rev Genet 16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 54.Edwards NS, Murray AW. 2005. Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell 16:1800–1810. doi: 10.1091/mbc.e04-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nechemia-Arbely Y, Fachinetti D, Cleveland DW. 2012. Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp Cell Res 318:1353–1360. doi: 10.1016/j.yexcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jansen LET, Black BE, Foltz DR, Cleveland DW. 2007. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuh M, Lehner CF, Heidmann S. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17:237–243. [DOI] [PubMed] [Google Scholar]
- 58.Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I. 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet 7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. 2004. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol 14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 60.Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. 2008. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell 19:682–690. doi: 10.1091/mbc.e07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Qu Q, Rice A. 2015. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol Cell 59:426–436. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 62.Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C. 2009. Non-coding murine centromeric transcripts associate with and potentiate Aurora-B kinase. Nucleic Acids Res 37:5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Earnshaw WC, Rothfield N. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 64.Talbert PB, Bryson TD, Henikoff S. 2004. Adaptive evolution of centromere proteins in plants and animals. J Biol 3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland A, Desai A, Jansen LET, Cleveland DW. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol 183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lampson MA, Cheeseman IM. 2011. Sensing centromere tension: Aurora-B and the regulation of kinetochore function. Trends Cell Biol 21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ideue T, Cho Y, Nishimura K, Tani T. 2014. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19:528–538. doi: 10.1111/gtc.12149. [DOI] [PubMed] [Google Scholar]
- 70.Jambhekar A, Emerman AB, Schweidenback CTH, Blower MD. 2014. RNA stimulates Aurora-B kinase activity during mitosis. PLoS One 9:e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petsalaki E, Akoumianaki T, Black EJ, Gillespie DA, Zachos G. 2011. Phosphorylation at serine 331 is required for Aurora-B activation. J Cell Biol 195:449–466. doi: 10.1083/jcb.201104023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 73.McKinley KL, Cheeseman IM. 2017. Large-scale analysis of CRISPR/Cas9 cell-cycle reveals the diversity of p53-dependent responses to cell-cycle defects. Dev Cell 40:405–420. doi: 10.1016/j.devcel.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prescott DM, Bender MA. 1962. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- 75.Liang K, Woodfin AR, Slaughter BD, Unruh JR, Box AC, Rickels RA, Gao X, Haug JS, Jaspersen SL, Shilatifard A. 2015. Mitotic transcriptional activation: clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol Cell 60:435–445. doi: 10.1016/j.molcel.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, Haber DA. 2011. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 4:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu Q, Hoong N, Aslanian A, Hara T, Benner C, Heinz S, Miga KH, Ke E, Verma S, Soroczynski J, Yates JR III, Hunter T, Verma IM. 2018. Heterochromatin-encoded satellite RNAs induce breast cancer. Mol Cell 70:842–853. doi: 10.1016/j.molcel.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]