See Lowe et al. (doi:10.1093/brain/awy226) for a scientific commentary on this article.
Alzheimer’s disease targets discrete neural systems, but the mechanisms underlying this regionally selective vulnerability remain unknown. Grothe et al. report that Alzheimer’s disease pathology specifically affects brain regions with certain molecular properties, and that the biological pathways underlying this vulnerability differ for amyloid accumulation versus neurodegeneration.
Keywords: gene expression, PET, MRI, amyloid, tau
Abstract
Amyloid deposition and neurofibrillary degeneration in Alzheimer’s disease specifically affect discrete neuronal systems, but the underlying mechanisms that render some brain regions more vulnerable to Alzheimer’s disease pathology than others remain largely unknown. Here we studied molecular properties underlying these distinct regional vulnerabilities by analysing Alzheimer’s disease-typical neuroimaging patterns of amyloid deposition and neurodegeneration in relation to regional gene expression profiles of the human brain. Graded patterns of brain-wide vulnerability to amyloid deposition and neurodegeneration in Alzheimer’s disease were estimated by contrasting multimodal amyloid-sensitive PET and structural MRI data between patients with Alzheimer’s disease dementia (n = 76) and healthy controls (n = 126) enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Regional gene expression profiles were derived from brain-wide microarray measurements provided by the Allen brain atlas of the adult human brain transcriptome. In a hypothesis-driven analysis focusing on the genes coding for the amyloid precursor (APP) and tau proteins (MAPT), regional expression levels of APP were positively correlated with the severity of regional amyloid deposition (r = 0.44, P = 0.009), but not neurodegeneration (r = 0.01, P = 0.96), whereas the opposite pattern was observed for MAPT (neurodegeneration: r = 0.46, P = 0.006; amyloid: r = 0.08, P = 0.65). Using explorative gene set enrichment analysis, amyloid-vulnerable regions were found to be characterized by relatively low expression levels of gene sets implicated in protein synthesis and mitochondrial respiration. By contrast, neurodegeneration-vulnerable regions were characterized by relatively high expression levels of gene sets broadly implicated in neural plasticity, with biological functions ranging from neurite outgrowth and synaptic contact over intracellular signalling cascades to proteoglycan metabolism. At the individual gene level this data-driven analysis further corroborated the association between neurodegeneration and MAPT expression, and additionally identified associations with known tau kinases (CDK5, MAPK1/ERK2) alongside components of their intracellular (Ras-ERK) activation pathways. Sensitivity analyses showed that these pathology-specific imaging-genetic associations were largely robust against changes in some of the methodological parameters, including variation in the brain donor sample used for estimating regional gene expression profiles, and local variations in the Alzheimer’s disease-typical imaging patterns when these were derived from an independent patient cohort (BioFINDER study). These findings highlight that the regionally selective vulnerability to Alzheimer’s disease pathology relates to specific molecular-functional properties of the affected neural systems, and that the implicated biochemical pathways largely differ for amyloid accumulation versus neurodegeneration. The data provide novel insights into the complex pathophysiological mechanisms of Alzheimer’s disease and point to pathology-specific treatment targets that warrant further exploration in independent studies.
See Lowe et al. (doi:10.1093/brain/awy226) for a scientific commentary on this article.
Introduction
Alzheimer’s disease is a progressive neurodegenerative brain disorder that is characterized pathologically by accumulation of fibrillar amyloid-β protein, probably starting 10–20 years before cognitive symptom onset, and formation of tau-containing neurofibrillary tangles, which are both spatially and temporally closely associated with neuronal degeneration and cognitive symptoms. A striking but little understood feature of both pathological alterations is that they affect discrete neural systems in a fairly consistent regional pattern while other brain regions are widely spared or only affected in very late stages of the disease (Braak and Braak, 1991; Thal et al., 2002). Multimodal neuroimaging techniques now allow imaging diverse aspects of Alzheimer’s disease pathology in the living human brain, and greatly facilitate the brain-wide regional quantification of the pathological alterations due to markedly increased sampling rates when compared to laborious histopathological examinations. Thus, studies using amyloid-sensitive PET ligands have revealed a highly reproducible brain-wide pattern of regionally varying severity of amyloid deposition that is most pronounced in specific neocortical association areas. This amyloid pattern clearly deviates from the typical pattern of Alzheimer’s disease-related neurodegeneration as detected by structural MRI, which is most pronounced in allocortical regions primarily affected by neurofibrillary tangles (Jack et al., 2008; Whitwell et al., 2008; La Joie et al., 2012; Grothe and Teipel, 2016).
Elucidating the underlying principles that govern these differential regional vulnerabilities to amyloid deposition and neurodegeneration is of critical research interest, as it may yield important clues toward understanding the mechanisms of formation and progression of Alzheimer’s disease pathology. A promising line of research towards this goal aims at characterizing the Alzheimer’s disease-typical vulnerability patterns in relation to specific functional or structural properties of the human brain as assessed in healthy control populations (Buckner et al., 2009; Seeley et al., 2009; Vlassenko et al., 2010; Raj et al., 2012; Zhou et al., 2012; Iturria-Medina et al., 2014; Shinohara et al., 2014; Fjell et al., 2015; Oh et al., 2016; Mutlu et al., 2017). These studies collectively suggest that the regional vulnerability to Alzheimer’s disease pathology may at least partly be determined by specific systemic limitations associated with the anatomo-functional properties of the affected neural systems (Jagust and Mormino, 2011; Jagust, 2013). However, the precise properties that convey this elevated vulnerability remain poorly defined, and the strikingly different patterns of amyloid deposition and neurodegeneration strongly suggest that both aspects of Alzheimer’s disease pathology are governed by diverging mechanisms.
Here we aimed to characterize the properties underlying regionally selective vulnerability to Alzheimer’s disease-typical amyloid deposition and neurodegeneration further, by studying the neuroimaging patterns of these alterations in relation to the transcriptional architecture of the human brain as revealed by brain-wide regional gene expression profiling (Hawrylycz et al., 2012, 2015). Regionally varying gene expression profiles reflect the molecular properties underlying inter-regional differences in anatomical (Goel et al., 2014; Whitaker et al., 2016; Shin et al., 2017) and functional (Goyal et al., 2014; Hawrylycz et al., 2015; Richiardi et al., 2015; Wang et al., 2015; Krienen et al., 2016) brain tissue characteristics, and may thus be ideally suited to study the shared features of Alzheimer’s disease-vulnerable neural systems on a molecular level. While most previous studies examining relations between functional/structural characteristics of the human brain and regional vulnerability to Alzheimer’s disease pathology have exclusively focused on either amyloid (Buckner et al., 2009; Vlassenko et al., 2010; Iturria-Medina et al., 2014; Shinohara et al., 2014; Oh et al., 2016) or neurodegeneration (Seeley et al., 2009; Raj et al., 2012; Zhou et al., 2012; Fjell et al., 2015), we conjointly estimated the respective patterns using multimodal amyloid-PET and structural MRI acquisitions, and studied the molecular properties underlying regional vulnerability to each type of pathological marker.
Based on previous neuropathological evidence for a regional correlation between amyloid deposition in Alzheimer’s disease and amyloid precursor protein (APP) levels in neurologically normal brains (Shinohara et al., 2014), we first examined whether this spatial association may be reproduced using our combined neuroimaging-gene expression approach, and whether an analogous association may be observed between the MRI-based neurodegeneration pattern and expression levels of the microtubule-associated protein tau (MAPT) gene. Going beyond hypothesis-driven analyses of select candidate genes, we used explorative gene set enrichment analysis (GSEA) of the full genome-wide expression profiles (Subramanian et al., 2005) to characterize the molecular properties and related biochemical pathways underlying the distinct regional vulnerability patterns in a more comprehensive manner.
Material and methods
Neuroimaging datasets
The primary characterization of Alzheimer’s disease-typical patterns of amyloid deposition and neurodegeneration used in this study was based on our previous analysis of combined 18F-florbetapir-PET and high-resolution structural MRI data of well-characterized samples of patients with Alzheimer’s disease dementia (n = 75) and healthy elderly controls (n = 126) enrolled in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study (Grothe and Teipel, 2016).
Although the overall imaging patterns of Alzheimer’s disease-typical amyloid deposition and neurodegeneration are very well described in the Alzheimer’s disease neuroimaging literature (Jack et al., 2008; Whitwell et al., 2008; La Joie et al., 2012; Grothe and Teipel, 2016), differences in patient characteristics, imaging acquisitions (e.g. choice of amyloid radiotracer, specific MRI sequence), and routines used for image processing and analysis may lead to some local variations in the derived patterns. Thus, in a complementary sensitivity analysis we derived Alzheimer’s disease-typical patterns of amyloid deposition and neurodegeneration using combined 18F-flutemetamol-PET and high-resolution structural MRI data from independent samples of patients with Alzheimer’s disease dementia (n = 33) and healthy controls (n = 28) enrolled in the Swedish BioFINDER study. Only patients with Alzheimer’s disease dementia with a typical late-onset amnestic clinical presentation were selected from the larger patient sample of the BioFINDER cohort, given that phenotypic variants of Alzheimer’s disease are known to associate with atypical neurodegeneration patterns (Ossenkoppele et al., 2015).
Detailed diagnostic procedures as well as inclusion and exclusion criteria for the diagnostic categories in the ADNI and BioFINDER cohorts have been reported previously (Grothe and Teipel, 2016; Hansson et al., 2017) and are detailed in the Supplementary material. Only clinically diagnosed patients with Alzheimer’s disease dementia with (PET or CSF) biomarker evidence of cerebral amyloidosis were included in the patient samples, and cognitively normal controls with biomarker evidence of cerebral amyloidosis were excluded from the control samples. Demographic and clinical characteristics of the ADNI and BioFINDER samples are summarized in Table 1. Written informed consent was obtained from all study participants according to the Declaration of Helsinki, and ethical approval for data collection and sharing was given by the institutional review boards of the participating institutions in the ADNI and BioFINDER studies.
Table 1.
Characteristics of ADNI and BioFINDER samples
| ADNI | BioFINDER | |||
|---|---|---|---|---|
| CN | AD | CN | AD | |
| n | 126 | 75 | 28 | 33 |
| Age, years | 72.7 ± 6.4 | 75.0 ± 8.5 | 75.0 ± 5.6 | 74.8 ± 5.3 |
| Sex (M/F) | 65/61 | 40/35 | 17/11 | 22/11 |
| Education, years | 16.8 ± 2.5 | 15.6 ± 2.8 | 12.5 ± 3.5 | 12.7 ± 3.8 |
| % APOE4a | 22% | 79% | 24% | 73% |
| CDR (0/0.5/1/2/3) | 126/0/0/0/0 | 0/32/42/1/0 | 28/0/0/0/0 | 0/10/18/4/1 |
| CDR-SOB | – | 4.5 ± 1.5 | – | 5.8 ± 3.6 |
| MMSE (a.u.) | 29.1 ± 1.2 | 22.9 ± 2.1 | 29.0 ± 1.1 | 22.1 ± 5.1 |
Average values are reported as mean ± SD.
AD = Alzheimer’s disease dementia patients; CDR-SOB = Clinical Dementia Rating – Sum of Boxes; CN = cognitively normal controls; F = female; M = male; MMSE = Mini-Mental State Examination; n = sample size.
aPersons with at least one APOE4 allele; APOE genotype was not available for eight ADNI participants (three cognitively normal and five with Alzheimer’s disease).
Neuroimaging acquisition and preprocessing
Details on image acquisition and preprocessing are provided in the Supplementary material. Briefly, all structural MRI data were acquired at 3T using high-resolution 3D T1-weighted imaging sequences. Preprocessing of these data followed a standard voxel-based morphometry approach within SPM software (http://www.fil.ion.ucl.ac.uk/spm/). Amyloid-PET data were acquired using 18F-florbetapir (ADNI) or 18F-flutemetamol (BioFINDER) radiotracers and PET scans were spatially normalized using registration parameters from co-registered MRIs and converted to voxel-wise maps of standard uptake value ratios (SUVRs) by scaling to signal in study-specific reference regions. Brain-wide patterns of Alzheimer’s disease-typical amyloid deposition and neurodegeneration were finally estimated using voxel-wise Z-scores of preprocessed maps of amyloid-PET SUVR and MRI-derived grey matter volume, respectively. These Z-score maps represent in each voxel the mean difference between patients with Alzheimer’s disease dementia and healthy controls scaled by the standard deviation of the control group (Grothe and Teipel, 2016; Hansson et al., 2017).
Gene expression dataset
Regional gene expression profiles were assessed using microarray-based measurements of regional gene expression levels in the adult human brain that have been made publicly available by the Allen Brain Institute (http://human.brain-map.org/; RRID: SCR_007416). This worldwide unique data source constitutes the most comprehensive assessment of the transcriptional architecture of the human brain to date, and includes ∼62 000 microarray probes collected from 3700 regional brain tissue samples in autopsy data of six adult individuals (24–57 years) who had no known history of neuropsychiatric or neurological conditions (Hawrylycz et al., 2012, 2015). In the dataset each regional sample is associated with an anatomical label as well as with a coordinate representing its approximate location in standard stereotactic space (MNI space). In the present study we used a previous anatomic mapping and inter-donor averaging of the regional expression values within the widely used anatomical parcellation scheme of the Desikan-Killiany cortical atlas (French and Paus, 2015). The final dataset includes median cortical expression profiles for 20 737 protein-coding genes of the human genome expressed as log2 expression values in each of the 34 Desikan-Killiany regions. Given that analyses of the bi-hemispheric data from the first two brain donors of the Allen brain atlas could not identify any interhemispheric asymmetries in gene expression (Hawrylycz et al., 2012), brain tissue collection in the subsequent brain donors was limited to the left hemisphere only (Hawrylycz et al., 2015). Thus, we restricted all our analyses to the more robust estimates of cortical gene expression profiles from the left hemisphere (Rittman et al., 2016; Romme et al., 2017; Shin et al., 2017).
Statistical analysis
In a first hypothesis-driven analysis, we aimed to quantify the spatial correspondence between Alzheimer’s disease-typical imaging signatures of amyloid deposition and neurodegeneration and regional expression levels of APP and MAPT genes. For this, the imaging patterns were first mapped from voxel-level to the cortical parcellation scheme of the Desikan-Killiany atlas (French and Paus, 2015), and spatial associations with the gene expression profiles were then assessed using Spearman correlations across the 34 left-hemispheric cortical regions (Krienen et al., 2016). Given that regional gene expression values vary to some degree across the six individual brain donors contributing to the Allen brain atlas (French and Paus, 2015), we further assessed the robustness of these imaging-genetic associations against variations in the donor sample used for calculating the median gene expression profiles. Thus, we used a Jackknife resampling technique to calculate median gene expression profiles for APP and MAPT using systematically varying subsamples of the data that only included four of the six brain donors (15 possible donor combinations). Spatial correlations with the Alzheimer’s disease-typical imaging patterns were then calculated for each of the median gene expression profiles and averaged using Fisher weighted means.
We next explored genome-wide regional gene expression profiles associated with the respective pathological imaging patterns using a data-driven approach in combination with GSEA (Subramanian et al., 2005). In contrast to the hypothesis-testing analyses of APP and MAPT candidate genes, the principal aim of this approach was to generate new hypotheses about genes and pathways involved in regional vulnerability to Alzheimer’s disease pathology. For each imaging pattern, we calculated spatial (Spearman) correlations for all 20 737 protein-coding genes in the dataset, and ranked the genes according to their spatial correlation values. The top (positive correlation) and bottom parts (negative correlation) of this ranked list contain the genes of interest, expression values of which increase or decrease, respectively, in relation to the regional vulnerability to Alzheimer’s disease pathology. GSEA is a statistical approach explicitly designed to extract meaningful and interpretable information from high-throughput microarray data. Rather than selecting arbitrary sets of differentially expressed genes from the top or bottom of a ranked list based on statistical significance or effect size thresholds, GSEA uses the whole information inherent in the list to statistically assess whether prespecified gene sets, as a group, are significantly over- or under-represented (enriched) at the extremes of the list. A non-random distribution of a gene set across the ranked list is quantified by the normalized enrichment score, which also accounts for the differing sizes of the examined functional gene sets. Statistical significance of this score is assessed using permutation tests, and P-values are corrected for the independent testing of multiple gene sets using the false discovery rate (FDR). The threshold for statistical significance is set to PFDR < 0.05. The gene sets are defined by the common implication of the included genes in particular biological states or processes and are retrieved from a curated database (the Molecular Signatures Database; http://software.broadinstitute.org/gsea/msigdb/index.jsp). In the present study, we explored 1533 gene sets, including 497 curated and peer-reviewed gene sets of functional pathways derived from the Reactome database (http://reactome.org/), and 1036 gene sets that group genes annotated by the same gene ontology (GO) term (http://www.geneontology.org/).
To account for partial redundancy of the analysed gene sets and to help identifying the principal biological pathways revealed by the GSEA approach, the identified gene sets were organized into a network structure using the Cytoscape plug-in ‘Enrichment Map’ (Merico et al., 2011). Here, each gene set is plotted as a node and edges represent gene overlap between sets, resulting in an automated network layout that groups related gene sets into network clusters representing a common underlying pathway.
Data availability
ADNI data is openly shared and was downloaded online at https://ida.loni.usc.edu. Processed ADNI data as well as BioFINDER data used in this study are not publicly available for download, but may be retrieved from the corresponding author upon request. Allen human brain atlas regional gene expression profiles mapped to the Desikan-Killiany cortical atlas were downloaded online at: https://figshare.com/articles/A_FreeSurfer_view_of_the_cortical_transcriptome_generated_from_the_Allen_Human_Brain_Atlas/1439749, and the original donor-specific microarray measurements are openly available at: http://human.brain-map.org/static/download.
Results
Multimodal neuroimaging patterns of amyloid deposition and neurodegeneration in Alzheimer’s disease
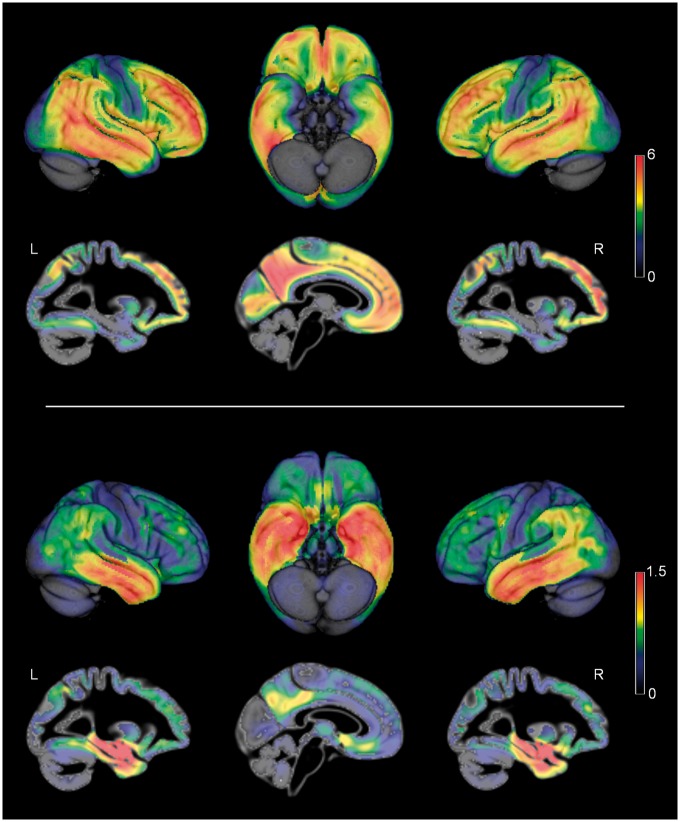
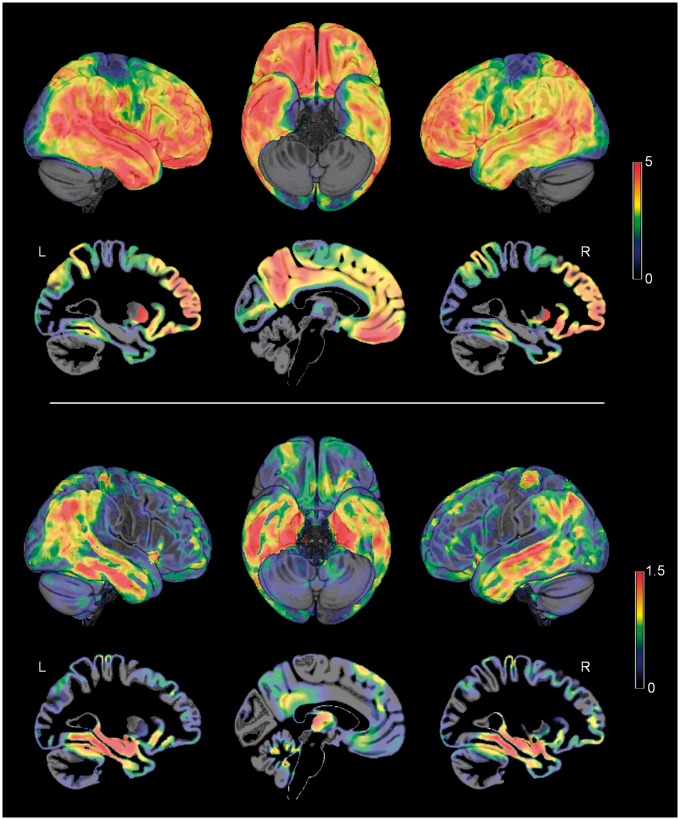
Figure 1 illustrates the brain-wide patterns of amyloid deposition and neurodegeneration in Alzheimer’s disease as estimated by contrasting amyloid-sensitive 18F-florbetapir-PET and high-resolution structural MRI data of patients with Alzheimer’s disease dementia and healthy controls from the ADNI cohort (Table 1). Regions of highest amyloid load correspond to distinct heteromodal association areas of the frontal, parietal, and lateral temporal lobes, whereas primary sensory-motor areas and the medial temporal lobe show the lowest vulnerability to amyloid deposition. By contrast, neurodegeneration is most pronounced in the medial temporal lobe, extending into lateral temporoparietal areas as well as anterior and posterior parts of the cingulate cortex. Primary sensory-motor areas, but also large parts of the frontal lobe appear to be relatively preserved from neurodegeneration.
Figure 1.
Alzheimer’s disease-typical neuroimaging patterns of amyloid deposition and neurodegeneration. Alzheimer’s disease-typical patterns of amyloid deposition (top) and neurodegeneration (bottom) as assessed by 18F-florbetapir-PET and structural MRI, respectively. Colour code reflects effect size (average z-score) of increased amyloid signal and decreased grey matter volume in patients with Alzheimer’s disease as compared to healthy controls (ADNI cohort). L/R = left/right hemisphere.
Associations of pathological imaging patterns with regional expression levels of APP and MAPT genes
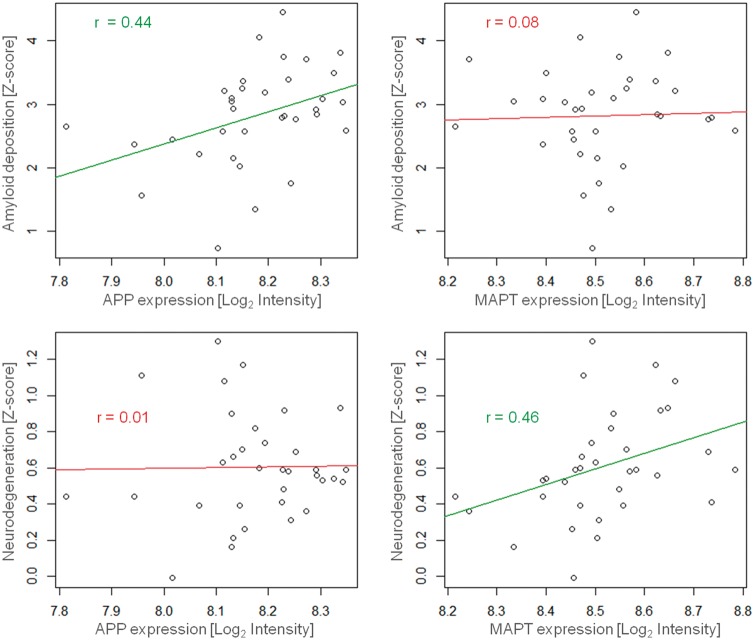
In a first hypothesis-driven analysis we assessed spatial associations of the Alzheimer’s disease-typical patterns of amyloid deposition and neurodegeneration with regional expression levels of APP and MAPT, respectively. As illustrated in Fig. 2, regional expression levels of APP were positively correlated with the severity of regional amyloid deposition (r = 0.44, P = 0.009), but not neurodegeneration (r = 0.01, P = 0.96). For MAPT expression levels the opposite pattern was observed, such that the severity of regional neurodegeneration in Alzheimer’s disease (r = 0.46, P = 0.006), but not severity of amyloid deposition (r = 0.08, P = 0.65), increased with increasing levels of regional MAPT expression. A sensitivity analysis across gene expression profiles estimated from varying subsamples of the data fully reproduced these pathology-specific associations with regional APP and MAPT expression (average correlations: APP–amyloid, r = 0.38, P = 0.03; APP–neurodegeneration, r = 0.15, P = 0.40; MAPT–neurodegeneration, r = 0.40, P = 0.02; MAPT–amyloid, r = 0.08, P = 0.65), indicating that these differential spatial associations are not primarily driven by the gene expression data of any particular brain donor(s).
Figure 2.
Associations of Alzheimer’s disease-typical patterns of amyloid deposition and neurodegeneration with regional expression levels of APP and MAPT genes. Regional severities of amyloid deposition (top) and neurodegeneration (bottom) are plotted against regional expression levels of APP (left) and MAPT (right) genes across 34 left-hemispheric cortical regions. Linear trends are indicated by regression lines and r indicates Spearman correlation coefficient. Green (top left and bottom right) and red (top right and bottom left) colours indicate statistical significance or non-significance, respectively, at a threshold of P < 0.05.
Genome-wide expression profiles associated with vulnerability to amyloid deposition and neurodegeneration in Alzheimer’s disease
Following up on the analysis based on APP and MAPT candidate genes, we aimed to determine gene expression profiles associated with vulnerability to Alzheimer’s disease-typical amyloid deposition and neurodegeneration in a more comprehensive manner using a data-driven approach.
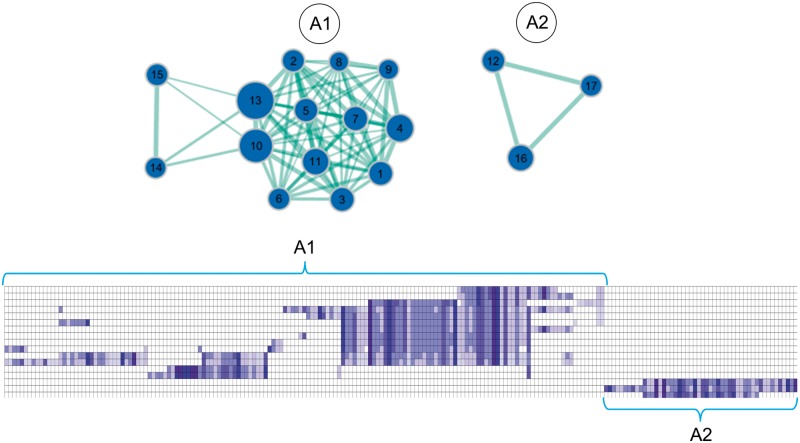
For the amyloid deposition pattern, GSEA revealed 18 gene sets to be negatively enriched and one gene set to be positively enriched in amyloid-vulnerable brain regions (Table 2). Visualization of the enrichment network showed that all but one of the underexpressed gene sets belonged to two distinct clusters, each grouping gene sets with overlapping gene members indicating a common underlying pathway (Fig. 3). The biggest cluster (A1) mainly contained gene sets implicated in different aspects of protein synthesis, but also gene sets related to response to viral infection and immunity (cross-presentation). The other cluster (A2) contained three overlapping gene sets representing pathways of mitochondrial respiration, including the citric acid cycle and oxidative phosphorylation.
Table 2.
Differentially-expressed gene sets in amyloid-vulnerable brain regions
| Cluster | Gene set name | Gene set number | Size | NES | FDR P-value | Repl. NES | Repl. FDR P-value |
|---|---|---|---|---|---|---|---|
| Negatively enriched | |||||||
| A1 | 3 UTR MEDIATED TRANSLATIONAL REGULATION (REACTOME) | 1 | 94 | −2.66 | <0.001 | −2.52 | <0.001 |
| A1 | PEPTIDE CHAIN ELONGATION (REACTOME) | 2 | 76 | −2.58 | <0.001 | −2.64 | <0.001 |
| A1 | NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX (REACTOME) | 3 | 94 | −2.52 | <0.001 | −2.46 | <0.001 |
| A1 | TRANSLATION (REACTOME) | 4 | 134 | −2.48 | <0.001 | −2.21 | <0.001 |
| A1 | INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION (REACTOME) | 5 | 90 | −2.44 | <0.001 | −2.43 | <0.001 |
| A1 | STRUCTURAL CONSTITUENT OF RIBOSOME (GO) | 6 | 68 | −2.42 | <0.001 | −2.43 | <0.001 |
| A1 | SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE (REACTOME) | 7 | 98 | −2.40 | <0.001 | −2.28 | <0.001 |
| A1 | FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX (REACTOME) | 8 | 44 | −2.31 | <0.001 | −2.12 | <0.001 |
| A1 | ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S (REACTOME) | 9 | 51 | −2.30 | <0.001 | −2.11 | <0.001 |
| A1 | METABOLISM OF MRNA (REACTOME) | 10 | 199 | −2.29 | <0.001 | −1.93 | <0.001 |
| A1 | INFLUENZA LIFE CYCLE (REACTOME) | 11 | 124 | −2.27 | <0.001 | −2.24 | <0.001 |
| A1 | METABOLISM OF RNA (REACTOME) | 13 | 242 | −2.15 | <0.001 | −1.84 | <0.001 |
| A1 | ER PHAGOSOME PATHWAY (REACTOME) | 14 | 59 | −2.12 | 0.002 | −1.67 | 0.001 |
| A1 | ANTIGEN PROCESSING CROSS PRESENTATION (REACTOME) | 15 | 72 | −2.03 | 0.006 | −1.58 | 0.011 |
| A2 | RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS (REACTOME) | 12 | 81 | −2.15 | 0.009 | −1.90 | <0.001 |
| A2 | TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT (REACTOME) | 16 | 116 | −2.02 | 0.007 | −1.81 | <0.001 |
| A2 | RESPIRATORY ELECTRON TRANSPORT (REACTOME) | 17 | 65 | −2.01 | 0.008 | −1.81 | <0.001 |
| - | SMOOTH MUSCLE CONTRACTION (REACTOME) | 18 | 22 | −2.00 | 0.007 | −1.62 | 0.022 |
| Positively enriched | |||||||
| - | OLFACTORY SIGNALING PATHWAY (REACTOME) | 19 | 307 | 2.25 | 0.006 | 2.13 | <0.001 |
NES = normalized enrichment score; Repl. = Replication values using neuroimaging patterns derived from the BioFINDER cohort.
Figure 3.
Network structure of negatively enriched gene sets in amyloid-vulnerable brain regions. Negatively enriched gene sets (blue circles) are plotted in a graph structure, where the overlap in gene members between different gene sets is indicated by the width of cyan lines. Gene sets are numbered according to their order in Table 2 and circle diameters reflect the size of each gene set. The matrix plot below shows the cluster structure of the leading-edge genes of the enriched gene sets (hierarchical clustering with average linkage). Each column corresponds to a leading-edge gene as detailed in Supplementary Table 1. Rows 1–14 contain the leading-edge genes of gene sets in cluster A1, rows 15–17 those of gene sets in cluster A2.
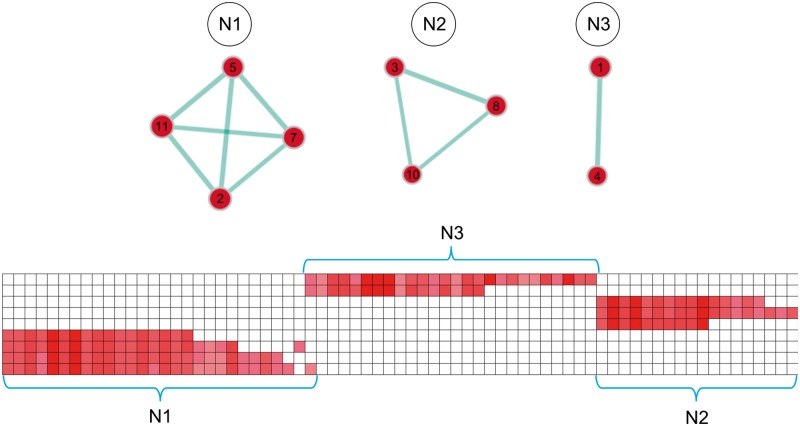
For the neurodegeneration pattern, GSEA revealed 11 gene sets to be positively enriched in neurodegeneration-vulnerable brain regions, but no significantly negatively enriched gene sets (Table 3). Again, visualization of the enrichment network structure showed that several of the overexpressed gene sets overlapped in their gene members, resulting in three different clusters that contained gene sets related to ‘cellular differentiation and neurite formation’ (N1, four gene sets), ‘extracellular signal-regulated kinase (MAPK/ERK) pathways’ (N2, three gene sets), and ‘proteoglycan metabolism’ (N3, two gene sets) (Fig. 4).
Table 3.
Differentially-expressed gene sets in neurodegeneration-vulnerable brain regions
| Cluster | Gene set name | Gene set number | Size | NES | FDR P-value | Repl. NES | Repl. FDR P-value |
|---|---|---|---|---|---|---|---|
| Positively enriched | |||||||
| N1 | NEURITE_DEVELOPMENT (GO) | 2 | 52 | 2.15 | 0.004 | 1.87 | <0.001 |
| N1 | AXONOGENESIS (GO) | 5 | 42 | 2.09 | 0.005 | 1.97 | <0.001 |
| N1 | CELLULAR MORPHOGENESIS DURING DIFFERENTIATION (GO) | 7 | 48 | 2.07 | 0.005 | 1.97 | <0.001 |
| N1 | NEURON_DEVELOPMENT (GO) | 11 | 60 | 2.01 | 0.009 | 1.66 | 0.01 |
| N2 | SIGNALLING TO RAS (REACTOME) | 3 | 26 | 2.10 | 0.007 | 1.76 | 0.01 |
| N2 | SIGNALLING TO ERKS (REACTOME) | 8 | 35 | 2.04 | 0.007 | 1.53 | 0.013 |
| N2 | SHC RELATED EVENTS (REACTOME) | 10 | 16 | 2.02 | 0.009 | 1.91 | 0.003 |
| N3 | CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM (REACTOME) | 1 | 45 | 2.16 | 0.009 | 1.96 | <0.001 |
| N3 | A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS (REACTOME) | 4 | 24 | 2.09 | 0.005 | 2.04 | <0.001 |
| - | OXIDOREDUCTASE ACTIVITY ACTING ON THE ALDEHYDE OR OXO GROUP OF DONORSNAD OR NADP AS ACCEPTOR (GO) | 6 | 16 | 2.08 | 0.005 | 1.87 | 0.003 |
| - | RAP1 SIGNALLING (REACTOME) | 9 | 15 | 2.03 | 0.009 | 1.57 | 0.031 |
NES = normalized enrichment score; Repl. = Replication values using neuroimaging patterns derived from the BioFINDER cohort.
Figure 4.
Network structure of positively enriched gene sets in neurodegeneration-vulnerable brain regions. Positively enriched gene sets (red circles) are plotted in a graph structure, where the overlap in gene members between different gene sets is indicated by the width of cyan lines. Gene sets are numbered according to their order in Table 3 and circle diameters reflect the size of each gene set. The matrix plot below shows the cluster structure of the leading-edge genes of the enriched gene sets (hierarchical clustering with average linkage). Each column corresponds to a leading-edge gene as detailed in Supplementary Table 2. Rows 1 and 2 contain the leading-edge genes of gene sets in cluster N3, rows 3–5 those of gene sets in cluster N2, and rows 6–10 those of gene sets in cluster N1.
Core differentially expressed genes within the identified pathways
Because statistical inference in GSEA is based on functional gene sets as a whole, not all of the genes included in a significant gene set need to be differentially expressed. The principal genes that account for a gene set’s enrichment signal are called the leading-edge subset, and a leading-edge analysis aims to find commonalities among the most relevant genes of the identified pathways by clustering the respective leading-edge subsets (Subramanian et al., 2005). Detailing on the structure of the enrichment networks, this analysis revealed that the enrichment signals of the clustered gene sets were driven by highly overlapping subsets of differentially expressed genes (Figs 3, 4 and Supplementary Tables 1 and 2).
Thus, for cluster A1 of the amyloid-specific gene sets (protein synthesis) these included primarily genes coding for ribosomal proteins (RPS, RPL), which were the main contributors to the enrichment signal in 12 of 14 gene sets of this cluster, including those related to viral infection (Supplementary Table 1). The two gene sets related to immunity/cross-presentation were linked to cluster A1 by overlap of several leading-edge genes coding for proteins belonging to the proteasome (PSM) family. Enrichment signal of gene sets in cluster A2 (mitochondrial respiration) was driven by genes coding for several key enzymes of the citric acid cycle, as well as components of all complexes of the electron transport chain and ATP synthase.
On the other hand, overlapping leading-edge subsets of the neurodegeneration-specific cluster N1 (cellular differentiation/neurite formation) comprised genes coding for diverse classes of molecules jointly involved in developmental processes such as neurite outgrowth, axonal guidance, cytoskeletal flexibility, and synaptic contact (Supplementary Table 2). Some prominent examples include neurexin (NRXN1), neuropilin (NRP2), semaphorin (SEMA4F) and roundabout guidance (ROBO1, ROBO2) receptors, apolipoprotein E (APOE), microtubule-associated protein tau (MAPT), as well as cyclin dependent kinase 5 (CDK5) and its activator p35/25 (CDK5R1). Leading-edge genes of cluster N2 (MAPK/ERK signalling) comprised members from all steps of the MAPK/ERK signalling cascade, such as genes coding for adaptor/docking proteins (e.g. GBR2), Ras proteins (HRAS, KRAS, NRAS), and mitogen-activated protein kinases (e.g. MAPK1). Cluster N3 primarily included leading-edge genes coding for the glypican (GPC) and syndecan (SDC) families of heparan sulfate proteoglycans.
Sensitivity analysis using Alzheimer’s disease-typical neuroimaging patterns derived from an independent cohort
To assess the robustness of our findings against study-related variations in the Alzheimer’s disease-typical neuroimaging patterns, we examined whether the identified associations with gene expression profiles could be replicated when using regional vulnerability patterns derived from multimodal imaging data of an independent cohort of patients with Alzheimer’s disease dementia and healthy controls (BioFINDER cohort, Table 1). Although some regional differences are evident when compared to the ADNI data, overall very similar brain-wide patterns of Alzheimer’s disease-typical amyloid deposition and neurodegeneration were revealed in this cohort (Fig. 5), resulting in high spatial correlations between the cohort-specific imaging patterns of r = 0.80 for amyloid deposition and r = 0.73 for neurodegeneration (both P < 0.001). In accordance with our initial findings, the amyloid deposition pattern was spatially correlated with APP expression (r = 0.36, P = 0.03), but not MAPT expression (r = 0.12, P = 0.50). Similarly, the neurodegeneration pattern showed a higher spatial correlation with MAPT expression (r = 0.29, P = 0.10) than with APP expression (r = 0.08, P = 0.66), although the MAPT-neurodegeneration association only reached trend-level statistical significance here. Importantly, all of the differentially expressed gene sets identified by the GSEA approach in our primary analysis were also significantly enriched with respect to the vulnerability patterns defined by this independent cohort (including all of the MAPT-containing gene sets of cluster N1) (Tables 2 and 3).
Figure 5.
Alzheimer’s disease-typical neuroimaging patterns of amyloid deposition and neurodegeneration in replication cohort. Alzheimer’s disease-typical patterns of amyloid deposition (top) and neurodegeneration (bottom) as assessed by 18F-flutemetamol-PET and structural MRI, respectively, in the replication cohort (BioFINDER cohort; Table 1). Colour code reflects effect size (average z-score) of increased amyloid signal and decreased grey matter volume in patients with Alzheimer’s disease as compared to healthy controls. L/R = left/right hemisphere.
Discussion
Understanding the regionally heterogeneous distribution of pathological alterations in Alzheimer’s disease and the shared characteristics of those neural systems that are more vulnerable to Alzheimer’s disease pathology than others is critical for a deeper understanding of Alzheimer’s disease pathogenesis. By studying brain-wide patterns of Alzheimer’s disease-typical neuroimaging abnormalities in relation to the transcriptional architecture of the human brain, this study shows that regional vulnerability to Alzheimer’s disease pathology is linked to distinct molecular characteristics of the affected brain regions. Moreover, the deviating regional vulnerabilities to amyloid deposition and neurodegeneration were found to be associated with largely differing molecular profiles corresponding to distinct biochemical pathways of cellular functioning.
Regional correlations of Alzheimer’s disease-typical amyloid and neurodegeneration patterns with expression levels of APP and MAPT
Our initial hypothesis-driven analysis showed that regional expression levels of APP in the human brain are predictive of the regional severity of amyloid deposition in Alzheimer’s disease as measured by amyloid-sensitive PET. This indicates that a high regional content of amyloid precursor protein, which probably relates to specific characteristics of synaptic functioning in the local neuronal tissue (Cirrito et al., 2005; Hick et al., 2015; Klevanski et al., 2015), may increase a brain region’s vulnerability to the accumulation of amyloid pathology in later life. Such a scenario would be supported by findings in trisomy 21, where the presence of an additional copy of the APP gene leads to an increased age-related amyloid accumulation in individuals with Down syndrome (Lao et al., 2016; Doran et al., 2017). A regional correlation between immunohistologically-determined APP levels in neurologically normal controls and accumulation of amyloid pathology in Alzheimer’s disease had been shown previously based on neuropathological assessments across 12 different brain regions (Shinohara et al., 2014). While the neuroimaging approach used in the present study only provides an indirect surrogate marker for the extent of amyloid pathology, it enables considerably bigger sample sizes and higher regional sampling rates for a robust and spatially comprehensive estimation of the brain-wide pattern of amyloid deposition.
In addition to the regional correlation between APP expression and amyloid deposition, our analyses also demonstrated an analogous spatial association between MAPT expression levels and severity of neurodegeneration in Alzheimer’s disease. The Alzheimer’s disease-typical neurodegeneration pattern as measured by structural MRI closely resembles long-standing neuropathological estimates of the regional distribution of neurofibrillary tangle pathology in Alzheimer’s disease (Braak and Braak, 1991; Whitwell et al., 2008, 2012). Most recent findings from tau-sensitive PET imaging data largely confirm the spatial correspondence between accumulation of tau pathology and MRI-measured neurodegeneration in Alzheimer’s disease (Cho et al., 2016b; Xia et al., 2017), and further highlight marked differences between the typical allocortical-predominant pattern of tau/neurodegeneration and the neocortical-predominant pattern of amyloid deposition (Cho et al., 2016a; Sepulcre et al., 2017). Our finding of a regional correlation between MAPT expression levels and Alzheimer’s disease-typical neurodegeneration agrees with findings from a recent study that examined differences in gene expression data from the Allen brain atlas between brain regions with high versus low vulnerability to tau pathology as defined by the Braak staging scheme (Freer et al., 2016). The study demonstrated differential expression of a set of candidate genes coding for proteins known to be involved in pathological protein aggregation in Alzheimer’s disease, which on an individual gene level also included high MAPT expression levels in tau-vulnerable regions. These data collectively suggest that neural systems with a physiologically high content of tau may also be more vulnerable to pathological alterations of tau processing and subsequent neurofibrillary degeneration as the brain ages. Similarly, increased MAPT expression caused by a recently detected rare microduplication of the MAPT locus was found to lead to prominent neurofibrillary tangle pathology and an Alzheimer’s disease dementia phenotype in the absence of notable amyloid pathology (Le Guennec et al., 2017). However, although the previous study by Freer et al. (2016) focused exclusively on tau-vulnerable regions, the analysed set of candidate genes did not distinguish between proteins involved in tau or amyloid aggregation, and thus it remains unknown whether the differential expression of this group of genes was primarily driven by tau-related proteins. An important methodological difference to our current study is the way in which vulnerable brain regions are defined and used to analyse the gene expression data. Freer et al. (2016) manually selected dichotomous sets of tau-vulnerable and tau-resistant regions from the Allen brain atlas based on correspondence to previous descriptions of tau vulnerability in the literature [i.e. the Braak staging scheme (Braak and Braak, 1991)]. By contrast, in the current study we used MRI data of well-characterized samples of patients with Alzheimer’s disease and healthy controls to estimate a graded regional pattern of brain-wide vulnerability to Alzheimer’s disease-typical neurodegeneration. This procedure allowed us to exploit the exceptionally dense regional gene expression information provided by the Allen brain atlas (Hawrylycz et al., 2012) in a spatial correlation approach.
Together, the distinct spatial associations of Alzheimer’s disease-typical amyloid deposition and neurodegeneration with APP and MAPT expression levels support the hypothesis that the differential regional vulnerabilities to these pathological hallmarks are at least partly determined by regional differences in the neurotypical expression levels of the precursor proteins underlying the respective proteinopathic alteration. Using GSEA to more fully characterize the genome-wide expression profiles associated with vulnerability to amyloid deposition and neurodegeneration revealed that these different aspects of Alzheimer’s disease pathology are generally related to markedly distinct biochemical pathways.
Biochemical pathways underlying vulnerability to amyloid deposition
Amyloid-vulnerable brain regions were characterized by relatively low expression levels of gene sets implicated in protein synthesis and mitochondrial respiration. Besides genes of the ribosomal complex itself, the negatively enriched gene sets related to protein synthesis also included several underexpressed genes implicated in protein folding, degradation, and quality control mechanisms, including genes coding for proteasome proteins (PSM family), endoplasmatic chaperones (e.g. HSPA1B, CALR and PDIA3), and those implicated in non-sense mediated mRNA decay (e.g. CASC3). Neural systems with physiologically low expression levels of these genes might be more vulnerable to age-related dysfunctions in translation, protein folding and quality control mechanisms. While there is ample evidence implicating disturbed post-translational control mechanisms in amyloid aggregation and Alzheimer’s disease (Muchowski, 2002; Malgaroli et al., 2006), relatively less data are available for the role of ribosomal protein synthesis. Although it has long been known that Alzheimer’s disease brain tissue is characterized by a severely decreased ribosomal protein synthesis, this has usually been interpreted as a consequence of pathology accumulation rather than its cause (Sajdel-Sulkowska and Marotta, 1984; Langstrom et al., 1989). However, a more recent study indicated that ribosome dysfunction may be among the earliest neurochemical alterations in Alzheimer’s disease, although its relation to amyloid accumulation was not assessed directly (Ding et al., 2005). Potential mechanisms that could link ribosome dysfunction to amyloid accumulation include translational errors, such as ribosomal frameshifting (van Leeuwen et al., 2006; Wills and Atkins, 2006), or a decreased ribosomal protein folding capability (Pathak et al., 2017).
The finding that amyloid-vulnerable brain regions are characterized by comparably low expression levels of mitochondrial respiration genes is particularly intriguing as it coincides with a previous report of unexpectedly low rates of oxidative phosphorylation (when compared to overall glucose metabolism) in these brain regions; a phenomenon referred to as ‘aerobic glycolysis’ (Vlassenko et al., 2010). Based on observations of activity-dependent increases in synaptic amyloid levels in animal models (Cirrito et al., 2005), regional vulnerability to amyloid deposition has been linked to high functional loads of the affected brain regions (Bero et al., 2011; Jagust and Mormino, 2011; Ovsepian and O’Leary, 2016). Accordingly, in humans it could be demonstrated that the severity of amyloid deposition is spatially associated with the regional distribution of diverse markers of heightened neuronal activity in neurologically normal controls, including rates of 18F-fluorodeoxyglucose (FDG)-PET measured glucose metabolism (Oh et al., 2016), activation and connectivity patterns in functional MRI data (Buckner et al., 2009; Sperling et al., 2009), as well as immunohistological levels of synaptic marker proteins (Shinohara et al., 2014). However, notwithstanding these overall brain-wide associations, comparably high neuronal activity is also characteristic for some brain regions that are largely spared from amyloid deposition (e.g. the primary visual cortex), indicating that vulnerability to amyloid deposition may relate to some specific characteristic of neuronal/synaptic activity, rather than high activity levels per se (Oh et al., 2016; Ovsepian and O’Leary, 2016). The highly interconnected heteromodal association areas that are primarily affected by amyloid deposition are characterized by a range of neuronal properties that allow them to support large-scale synchronous activity, such as optimized biophysical properties for maintaining spike bursts (Buckner et al., 2009; Ovsepian and O’Leary, 2016). This functional characteristic might put these brain regions at increased need for rapid energy generation through the use of aerobic glycolysis (Vlassenko et al., 2010; Jagust and Mormino, 2011), thus possibly providing the link to the low expression of mitochondrial respiration genes observed here. Notably, experimental manipulation of mitochondrial function in animal models could provide initial evidence for a causal influence of decreased mitochondrial energy production on increased amyloid deposition through disrupted clearance mechanisms (Scheffler et al., 2012; Kukreja et al., 2014). Although a previous study in humans could link genetic variants within oxidative phosphorylation genes to an increased Alzheimer’s disease risk, no effects on CSF markers of amyloid pathology were observed in this study (Biffi et al., 2014). The relation between mitochondrial energy metabolism and amyloid pathology in the natural human disease course remains to be elucidated in more detail.
Interestingly, APP was not included among the genes identified in the genome-wide enrichment analysis. This may indicate that APP only plays a secondary role for determining regional amyloid load in sporadic Alzheimer’s disease when compared to other pathways, consistent with previous findings (Shinohara et al., 2014). While an alternative explanation would be that APP and its processing pathways are not well represented in the analysed gene set databases, additional analyses on APP-related gene sets argue against such an analytic confound (Supplementary material).
Biochemical pathways underlying vulnerability to Alzheimer’s disease-typical neurodegeneration
In contrast to the low expression levels of specific gene sets in amyloid-vulnerable areas, brain regions vulnerable to Alzheimer’s disease-typical neurodegeneration were characterized by comparably high expression levels of genes implicated in biological functions ranging from neurite outgrowth and synaptic contact over intracellular signalling cascades to proteoglycan metabolism. Interestingly, at the individual gene level this data-driven analysis corroborated the association with MAPT expression, and additionally identified some of the best described tau kinases (CDK5, MAPK1/ERK2) alongside components of their Ras-ERK activation pathways (Ferrer et al., 2001; Cruz et al., 2003; Mazanetz and Fischer, 2007).
While at first sight the overexpressed gene sets appear to represent separate biochemical pathways, a common denominator of the related functions may be a high capacity for neuroplastic change. Thus, the physiological functions of tau protein and its associated kinases have been implicated in cytoskeletal flexibility, dendritic spine formation, and also more functional aspects of synaptic plasticity such as long-term depression (Kimura et al., 2014; Mita et al., 2016; Wang and Mandelkow, 2016; Huang et al., 2017). Tau function is physiologically regulated by its phosphorylation state, and even transient hyperphosphorylation of tau can be observed under specific physiological conditions, where it has been associated with adaptive neuroprotective changes of the synapse (Arendt et al., 2003; Wang and Liu, 2008). Furthermore, both signalling in the MAPK/ERK pathway and proteoglycan metabolism have been implicated in diverse aspects of neural plasticity. Specifically, Ras-mediated activation of downstream MAPK/ERK kinases has been shown to regulate plastic mechanisms of synaptic function (Stornetta and Zhu, 2011), whereas heparan sulfate proteoglycans play relevant roles in activity-dependent synaptic reorganization and neurogenesis (Matsumoto-Miyai et al., 2009; Lugert et al., 2017; Minge et al., 2017). Similarly, the overexpression of APOE within the molecular profile related to neurodegeneration vulnerability might be explained by its physiological function in synaptic plasticity (Kim et al., 2014), although allelic variants of this Alzheimer’s disease risk gene have been more closely linked to amyloid deposition than to tau/neurodegeneration (Kim et al., 2009; Gonneaud et al., 2016; Grothe et al., 2017) (see Supplementary material for an extended discussion of this rather unexpected spatial association).
This post hoc interpretation of the neurodegeneration-related molecular profile to be reflective of a high plastic potential would be consistent with previous reports highlighting the disproportionately high capacity for plastic change in tau-vulnerable brain regions (Arendt, 2004; Walhovd et al., 2016). This coincidence is particularly striking in the medial temporal allocortex, which attracts by far the highest amount of neurofibrillary pathology (Braak and Braak, 1991) and at the same time is one of the most plastic neural systems in the human brain, characterized by high rates of experience-dependent synaptic reorganization and the selective capability of adult neurogenesis (Banks et al., 2014; Goncalves et al., 2016). Based on diverse lines of evidence it has been hypothesized that a high potential for lifelong adaptive reorganization, while instrumental for supporting memory formation and other higher cognitive functions, may become a risk factor for maladaptive processes resulting in pathological tau hyperphosphorylation and associated neurodegeneration as the brain ages (Neill, 1995; Mesulam, 1999; Arendt, 2004). In this context it is notable that a dual role in neural plasticity and neurofibrillary degeneration not only characterizes tau and its phosphorylation pathways (Wang and Liu, 2008; Wang and Mandelkow, 2016), but also applies to heparan sulfate proteoglycans, which in addition to their role in neural plasticity (Matsumoto-Miyai et al., 2009; Minge et al., 2017) have long been identified as key factors in the formation of neurofibrillary tangle pathology in Alzheimer’s disease (Goedert et al., 1996; van Horssen et al., 2003; Holmes et al., 2013). Thus, in neuropathological studies heparan sulfate proteoglycans were found to co-localize with hyperphosphorylated tau at the earliest stages of neurofibrillary tangle pathology, and in vitro experiments revealed that they can prevent tau from binding to microtubules and induce its aggregation into paired helical filaments (Goedert et al., 1996). More recent work could further show that heparan sulfate proteoglycans promote transcellular tau propagation by binding extracellular tau fibrils on the cell surface (Holmes et al., 2013). Together, the distinct molecular profile associated with vulnerability to Alzheimer’s disease-typical neurodegeneration identified in the present study is largely consistent with the notion of a selective vulnerability caused by a high potential for plastic change.
Limitations
In this study we used a cortical parcellation scheme of 34 anatomically defined brain regions that is widely used in the neuroimaging literature (Desikan-Killiany cortical atlas; French and Paus, 2015). However, when compared to the spatially detailed parcellations revealed by recent multi-modal mappings of human brain organization (Glasser et al., 2016), this anatomical parcellation likely involves averaging gene expression data across structurally and functionally heterogeneous brain areas. Although the used gene expression data from the Allen brain atlas provide the anatomically most comprehensive transcriptome data for the human brain available to date (Keil et al., 2018), the spatial coverage would probably not be sufficient to accurately estimate gene expression profiles at much higher spatial resolutions. Even for the relatively coarse anatomical parcellation into 34 cortical regions, some of the regions only contain a limited number of tissue samples stemming from a subset of available brain donors (French and Paus, 2015). Thus, at higher regional granularities the decreasing number of tissue samples per region would markedly increase the influence of noise from single microarray measurements and interindividual variance on the estimated gene expression profiles. Similarly, the limited availability of right hemispheric gene expression measurements in the Allen brain atlas has prevented us from studying potential effects of brain asymmetries on the imaging-genetic spatial associations. However, interhemispheric symmetry is evident in the Alzheimer’s disease-typical imaging patterns (Figs 1 and 5) and can also be reasonably assumed for the gene expression profiles (Hawrylycz et al., 2012; Pletikos et al., 2014).
Finally, we note that our explorative GSEA approach explicitly focused on the identification of pathways that were most robustly associated with each imaging pattern separately, and may thus not be sensitive for the detection of interacting pathways associated with both amyloid and neurodegeneration. A more focused investigation of interacting gene networks that may potentially link both types of Alzheimer’s disease pathology is a promising avenue for future research.
Conclusion
This study shows that the regional vulnerability to Alzheimer’s disease pathology relates to specific molecular properties of the affected neural systems, and that the biochemical pathways underlying this vulnerability largely differ for amyloid accumulation versus neurodegeneration. While these molecular properties may equip the respective neural systems with specific functional characteristics necessary for efficient information processing in the context of higher cognitive functions, they may come with the downside of an elevated vulnerability to maladaptive cellular processes and pathological alterations during brain ageing. The identification of distinct molecular properties underlying regional vulnerability to amyloid deposition and neurodegeneration provides novel insights into the complex pathophysiological mechanisms of Alzheimer’s disease and points to pathology-specific treatment targets that warrant further exploration in independent studies.
Funding
The present study was funded by a grant from the Alzheimer Forschung Initiative e.V. to M.J.G. (grant number #16037), and NIH grants 1K23EB019023 and R01AG052653-01A1 to J.S. M.S. is supported by the Knut and Alice Wallenberg Foundation, the Wallenberg Centre for Molecular and Translational Medicine, and the Swedish Research Council.
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.usc.edu) and from the Swedish BioFINDER study. A complete list of BioFINDER members can be found at http://biofinder.se/the_biofinder_study_group/. The BioFinder study was supported by the European Research Council, the Swedish Research Council, the Swedish Brain Foundation, the Swedish Alzheimer Association, the Marianne and Marcus Wallenberg Foundation, the Skåne University Hospital Foundation, and the Swedish federal government under the ALF agreement. ADNI data collection and sharing for this project was funded by ADNI grants [(National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012)]. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuroimaging at the University of Southern California.
Competing interests
O.H. has acquired research support (for the institution) from Roche, GE Healthcare, Biogen, AVID Radiopharmaceuticals, Fujirebio, and Euroimmun. In the past 2 years, he has received consultancy/speaker fees (paid to the institution) from Lilly, Roche, and Fujirebio.
Supplementary Material
Glossary
Abbreviations
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- GSEA
gene set enrichment analysis
- MAPK/ERK
mitogen-activated protein kinases/extracellular signal-regulated kinases
References
- Arendt T. Neurodegeneration and plasticity. Int J Dev Neurosci 2004; 22: 507–14. [DOI] [PubMed] [Google Scholar]
- Arendt T, Stieler J, Strijkstra AM, Hut RA, Rudiger J, Van der Zee EA et al. . Reversible paired helical filament-like phosphorylation of tau is an adaptive process associated with neuronal plasticity in hibernating animals. J Neurosci 2003; 23: 6972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks PJ, Warburton EC, Brown MW, Bashir ZI. Mechanisms of synaptic plasticity and recognition memory in the perirhinal cortex. Prog Mol Biol Transl Sci 2014; 122: 193–209. [DOI] [PubMed] [Google Scholar]
- Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME et al. . Neuronal activity regulates the regional vulnerability to amyloid-beta deposition. Nat Neurosci 2011; 14: 750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Sabuncu MR, Desikan RS, Schmansky N, Salat DH, Rosand J et al. . Genetic variation of oxidative phosphorylation genes in stroke and Alzheimer’s disease. Neurobiol Aging 2014; 35: 1956.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–59. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T et al. . Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci 2009; 29: 1860–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Choi JY, Hwang MS, Kim YJ, Lee HM, Lee HS et al. . In vivo cortical spreading pattern of tau and amyloid in the Alzheimer disease spectrum. Ann Neurol 2016a; 80: 247–58. [DOI] [PubMed] [Google Scholar]
- Cho H, Choi JY, Hwang MS, Lee JH, Kim YJ, Lee HM et al. . Tau PET in Alzheimer disease and mild cognitive impairment. Neurology 2016b; 87: 375–83. [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Yamada KA, Finn MB, Sloviter RS, Bales KR, May PC et al. . Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron 2005; 48: 913–22. [DOI] [PubMed] [Google Scholar]
- Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 2003; 40: 471–83. [DOI] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci 2005; 25: 9171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran E, Keator D, Head E, Phelan MJ, Kim R, Totoiu M et al. . Down syndrome, partial trisomy 21, and absence of Alzheimer’s disease: the role of APP. J Alzheimers Dis 2017; 56: 459–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer I, Blanco R, Carmona M, Ribera R, Goutan E, Puig B et al. . Phosphorylated map kinase (ERK1, ERK2) expression is associated with early tau deposition in neurones and glial cells, but not with increased nuclear DNA vulnerability and cell death, in Alzheimer disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Brain Pathol 2001; 11: 144–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Amlien IK, Sneve MH, Grydeland H, Tamnes CK, Chaplin TA et al. . The roots of Alzheimer’s disease: are high-expanding cortical areas preferentially targeted? Cereb Cortex 2015; 25: 2556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer R, Sormanni P, Vecchi G, Ciryam P, Dobson CM, Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer’s disease. Sci Adv 2016; 2: e1600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Paus T. A FreeSurfer view of the cortical transcriptome generated from the Allen Human Brain Atlas. Front Neurosci 2015; 9: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E et al. . A multi-modal parcellation of human cerebral cortex. Nature 2016; 536: 171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 1996; 383: 550–3. [DOI] [PubMed] [Google Scholar]
- Goel P, Kuceyeski A, LoCastro E, Raj A. Spatial patterns of genome-wide expression profiles reflect anatomic and fiber connectivity architecture of healthy human brain. Hum Brain Mapp 2014; 35: 4204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves JT, Schafer ST, Gage FH. Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell 2016; 167: 897–914. [DOI] [PubMed] [Google Scholar]
- Gonneaud J, Arenaza-Urquijo EM, Fouquet M, Perrotin A, Fradin S, de La Sayette V et al. . Relative effect of APOE epsilon4 on neuroimaging biomarker changes across the lifespan. Neurology 2016; 87: 1696–703. [DOI] [PubMed] [Google Scholar]
- Goyal MS, Hawrylycz M, Miller JA, Snyder AZ, Raichle ME. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab 2014; 19: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe MJ, Teipel SJ. Spatial patterns of atrophy, hypometabolism, and amyloid deposition in Alzheimer’s disease correspond to dissociable functional brain networks. Hum Brain Mapp 2016; 37: 35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe MJ, Villeneuve S, Dyrba M, Bartres-Faz D, Wirth M. Multimodal characterization of older APOE2 carriers reveals selective reduction of amyloid load. Neurology 2017; 88: 569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson O, Grothe MJ, Strandberg TO, Ohlsson T, Hagerstrom D, Jogi J et al. . Tau pathology distribution in Alzheimer’s disease corresponds differentially to cognition-relevant functional brain networks. Front Neurosci 2017; 11: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL et al. . Canonical genetic signatures of the adult human brain. Nat Neurosci 2015; 18: 1832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA et al. . An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hick M, Herrmann U, Weyer SW, Mallm JP, Tschape JA, Borgers M et al. . Acute function of secreted amyloid precursor protein fragment APPsalpha in synaptic plasticity. Acta Neuropathol 2015; 129: 21–37. [DOI] [PubMed] [Google Scholar]
- Holmes BB, DeVos SL, Kfoury N, Li M, Jacks R, Yanamandra K et al. . Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci USA 2013; 110: E3138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Lin X, Liang Z, Zhao T, Du S, Loy MMT et al. . Cdk5-dependent phosphorylation of liprinalpha1 mediates neuronal activity-dependent synapse development. Proc Natl Acad Sci USA 2017; 114: E6992–7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Toussaint PJ, Evans AC. Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput Biol 2014; 10: e1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM et al. . 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain 2008; 131 (Pt 3): 665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust W. Vulnerable neural systems and the borderland of brain aging and neurodegeneration. Neuron 2013; 77: 219–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Mormino EC. Lifespan brain activity, beta-amyloid, and Alzheimer’s disease. Trends Cogn Sci 2011; 15: 520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil JM, Qalieh A, Kwan KY. Brain transcriptome databases: a user’s guide. J Neurosci 2018, in press. doi: 10.1523/JNEUROSCI.1930-17.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron 2009; 63: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Basak J. Apolipoprotein E in synaptic plasticity and Alzheimer’s disease: potential cellular and molecular mechanisms. Mol Cells 2014; 37: 767–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Whitcomb DJ, Jo J, Regan P, Piers T, Heo S et al. . Microtubule-associated protein tau is essential for long-term depression in the hippocampus. Philos Trans R Soc Lond B Biol Sci 2014; 369: 20130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevanski M, Herrmann U, Weyer SW, Fol R, Cartier N, Wolfer DP et al. . The APP intracellular domain is required for normal synaptic morphology, synaptic plasticity, and hippocampus-dependent behavior. J Neurosci 2015; 35: 16018–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci USA 2016; 113: E469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukreja L, Kujoth GC, Prolla TA, Van Leuven F, Vassar R. Increased mtDNA mutations with aging promotes amyloid accumulation and brain atrophy in the APP/Ld transgenic mouse model of Alzheimer’s disease. Mol Neurodegener 2014; 9: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie R, Perrotin A, Barre L, Hommet C, Mezenge F, Ibazizene M et al. . Region-specific hierarchy between atrophy, hypometabolism, and beta-amyloid (Abeta) load in Alzheimer’s disease dementia. J Neurosc 2012; 32: 16265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langstrom NS, Anderson JP, Lindroos HG, Winblad B, Wallace WC. Alzheimer’s disease-associated reduction of polysomal mRNA translation. Brain Res Mol Brain Res 1989; 5: 259–69. [DOI] [PubMed] [Google Scholar]
- Lao PJ, Betthauser TJ, Hillmer AT, Price JC, Klunk WE, Mihaila I et al. . The effects of normal aging on amyloid-beta deposition in nondemented adults with Down syndrome as imaged by carbon 11-labeled Pittsburgh compound B. Alzheimers Dement 2016; 12: 380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec K, Quenez O, Nicolas G, Wallon D, Rousseau S, Richard AC et al. . 17q21.31 duplication causes prominent tau-related dementia with increased MAPT expression. Mol Psychiatry 2017; 22: 1119–25. [DOI] [PubMed] [Google Scholar]
- Lugert S, Kremer T, Jagasia R, Herrmann A, Aigner S, Giachino C et al. . Glypican-2 levels in cerebrospinal fluid predict the status of adult hippocampal neurogenesis. Sci Rep 2017; 7: 46543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgaroli A, Vallar L, Zimarino V. Protein homeostasis in neurons and its pathological alterations. Curr Opin Neurobiol 2006; 16: 270–4. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Miyai K, Sokolowska E, Zurlinden A, Gee CE, Luscher D, Hettwer S et al. . Coincident pre- and postsynaptic activation induces dendritic filopodia via neurotrypsin-dependent agrin cleavage. Cell 2009; 136: 1161–71. [DOI] [PubMed] [Google Scholar]
- Mazanetz MP, Fischer PM. Untangling tau hyperphosphorylation in drug design for neurodegenerative diseases. Nat Rev Drug Discov 2007; 6: 464–79. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Bader GD. Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol Biol 2011; 781: 257–77. [DOI] [PubMed] [Google Scholar]
- Mesulam MM. Neuroplasticity failure in Alzheimer’s disease: bridging the gap between plaques and tangles. Neuron 1999; 24: 521–9. [DOI] [PubMed] [Google Scholar]
- Minge D, Senkov O, Kaushik R, Herde MK, Tikhobrazova O, Wulff AB et al. . Heparan sulfates support pyramidal cell excitability, synaptic plasticity, and context discrimination. Cereb Cortex 2017; 27: 903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita N, He X, Sasamoto K, Mishiba T, Ohshima T. Cyclin-dependent Kinase 5 regulates dendritic spine formation and maintenance of cortical neuron in the mouse brain. Cereb Cortex 2016; 26: 967–76. [DOI] [PubMed] [Google Scholar]
- Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron 2002; 35: 9–12. [DOI] [PubMed] [Google Scholar]
- Mutlu J, Landeau B, Gaubert M, de La Sayette V, Desgranges B, Chetelat G. Distinct influence of specific versus global connectivity on the different Alzheimer’s disease biomarkers. Brain 2017; 140: 3317–28. [DOI] [PubMed] [Google Scholar]
- Neill D. Alzheimer’s disease: maladaptive synaptoplasticity hypothesis. Neurodegeneration 1995; 4: 217–32. [DOI] [PubMed] [Google Scholar]
- Oh H, Madison C, Baker S, Rabinovici G, Jagust W. Dynamic relationships between age, amyloid-beta deposition, and glucose metabolism link to the regional vulnerability to Alzheimer’s disease. Brain 2016; 139 (Pt 8): 2275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Moller C, Lehmann M et al. . Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp 2015; 36: 4421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsepian SV, O’Leary VB. Neuronal activity and amyloid plaque pathology: an update. J Alzheimers Dis 2016; 49: 13–19. [DOI] [PubMed] [Google Scholar]
- Pathak BK, Mondal S, Banerjee S, Ghosh AN, Barat C. Sequestration of ribosome during protein aggregate formation: contribution of ribosomal RNA. Sci Rep 2017; 7: 42017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AM, Sedmak G, Meyer KA, Zhu Y, Cheng F et al. . Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 2014; 81: 321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron 2012; 73: 1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T et al. . BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 2015; 348: 1241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittman T, Rubinov M, Vertes PE, Patel AX, Ginestet CE, Ghosh BCP et al. . Regional expression of the MAPT gene is associated with loss of hubs in brain networks and cognitive impairment in Parkinson disease and progressive supranuclear palsy. Neurobiol Aging 2016; 48: 153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romme IA, de Reus MA, Ophoff RA, Kahn RS, van den Heuvel MP. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol Psychiatry 2017; 81: 495–502. [DOI] [PubMed] [Google Scholar]
- Sajdel-Sulkowska EM, Marotta CA. Alzheimer’s disease brain: alterations in RNA levels and in a ribonuclease-inhibitor complex. Science 1984; 225: 947–9. [DOI] [PubMed] [Google Scholar]
- Scheffler K, Krohn M, Dunkelmann T, Stenzel J, Miroux B, Ibrahim S et al. . Mitochondrial DNA polymorphisms specifically modify cerebral beta-amyloid proteostasis. Acta Neuropathol 2012; 124: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009; 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J, Grothe MJ, Sabuncu M, Chhatwal J, Schultz AP, Hanseeuw B et al. . Hierarchical organization of tau and amyloid deposits in the cerebral cortex. JAMA Neurol 2017; 74: 813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, French L, Xu T, Leonard G, Perron M, Pike GB et al. . Cell-specific gene-expression profiles and cortical thickness in the human brain. Cereb Cortex 2017: 1–11. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Fujioka S, Murray ME, Wojtas A, Baker M, Rovelet-Lecrux A et al. . Regional distribution of synaptic markers and APP correlate with distinct clinicopathological features in sporadic and familial Alzheimer’s disease. Brain 2014; 137 (Pt 5): 1533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Laviolette PS, O’Keefe K, O’Brien J, Rentz DM, Pihlajamaki M et al. . Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron 2009; 63: 178–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stornetta RL, Zhu JJ. Ras and Rap signaling in synaptic plasticity and mental disorders. Neuroscientist 2011; 17: 54–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Rub U, Orantes M, Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology 2002; 58: 1791–800. [DOI] [PubMed] [Google Scholar]
- van Horssen J, Wesseling P, van den Heuvel LP, de Waal RM, Verbeek MM. Heparan sulphate proteoglycans in Alzheimer’s disease and amyloid-related disorders. Lancet Neurol 2003; 2: 482–92. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, van Tijn P, Sonnemans MA, Hobo B, Mann DM, Van Broeckhoven C et al. . Frameshift proteins in autosomal dominant forms of Alzheimer disease and other tauopathies. Neurology 2006; 66 (2 Suppl 1): S86–92. [DOI] [PubMed] [Google Scholar]
- Vlassenko AG, Vaishnavi SN, Couture L, Sacco D, Shannon BJ, Mach RH et al. . Spatial correlation between brain aerobic glycolysis and amyloid-beta (Abeta) deposition. Proc Natl Acad Sci USA 2010; 107: 17763–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Westerhausen R, de Lange AM, Brathen AC, Grydeland H, Engvig A et al. . Premises of plasticity—and the loneliness of the medial temporal lobe. Neuroimage 2016; 131: 48–54. [DOI] [PubMed] [Google Scholar]
- Wang GZ, Belgard TG, Mao D, Chen L, Berto S, Preuss TM et al. . Correspondence between resting-state activity and brain gene expression. Neuron 2015; 88: 659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Liu F. Microtubule-associated protein tau in development, degeneration and protection of neurons. Prog Neurobiol 2008; 85: 148–75. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E. Tau in physiology and pathology. Nat Rev Neurosci 2016; 17: 5–21. [DOI] [PubMed] [Google Scholar]
- Whitaker KJ, Vertes PE, Romero-Garcia R, Vasa F, Moutoussis M, Prabhu G et al. . Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci USA 2016; 113: 9105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Dickson DW, Murray ME, Weigand SD, Tosakulwong N, Senjem ML et al. . Neuroimaging correlates of pathologically defined subtypes of Alzheimer’s disease: a case-control study. Lancet Neurol 2012; 11: 868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Josephs KA, Murray ME, Kantarci K, Przybelski SA, Weigand SD et al. . MRI correlates of neurofibrillary tangle pathology at autopsy: a voxel-based morphometry study. Neurology 2008; 71: 743–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Atkins JF. The potential role of ribosomal frameshifting in generating aberrant proteins implicated in neurodegenerative diseases. RNA 2006; 12: 1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Makaretz SJ, Caso C, McGinnis S, Gomperts SN, Sepulcre J et al. . Association of in vivo [18F]AV-1451 tau PET imaging results with cortical atrophy and symptoms in typical and atypical Alzheimer disease. JAMA Neurol 2017; 74: 427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 2012; 73: 1216–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADNI data is openly shared and was downloaded online at https://ida.loni.usc.edu. Processed ADNI data as well as BioFINDER data used in this study are not publicly available for download, but may be retrieved from the corresponding author upon request. Allen human brain atlas regional gene expression profiles mapped to the Desikan-Killiany cortical atlas were downloaded online at: https://figshare.com/articles/A_FreeSurfer_view_of_the_cortical_transcriptome_generated_from_the_Allen_Human_Brain_Atlas/1439749, and the original donor-specific microarray measurements are openly available at: http://human.brain-map.org/static/download.