See Contreras and Hippenmeyer (doi:10.1093/brain/awy218) for a scientific commentary on this article.
Autism spectrum disorders (ASDs) are complex conditions with diverse aetiologies. Szczurkowska et al. demonstrate that two ASD-related molecules – FGFR2 and Negr1 – physically interact to act on the same downstream pathway, and regulate cortical development and ASD-relevant behaviours in mice. Identifying common mechanisms in ASDs may reveal targets for pharmacological intervention.
Keywords: autism, development, cell adhesion, in utero electroporation, FGFR2 signaling
Abstract
Autism spectrum disorders are neurodevelopmental conditions with diverse aetiologies, all characterized by common core symptoms such as impaired social skills and communication, as well as repetitive behaviour. Cell adhesion molecules, receptor tyrosine kinases and associated downstream signalling have been strongly implicated in both neurodevelopment and autism spectrum disorders. We found that downregulation of the cell adhesion molecule NEGR1 or the receptor tyrosine kinase fibroblast growth factor receptor 2 (FGFR2) similarly affects neuronal migration and spine density during mouse cortical development in vivo and results in impaired core behaviours related to autism spectrum disorders. Mechanistically, NEGR1 physically interacts with FGFR2 and modulates FGFR2-dependent extracellular signal-regulated kinase (ERK) and protein kinase B (AKT) signalling by decreasing FGFR2 degradation from the plasma membrane. Accordingly, FGFR2 overexpression rescues all defects due to Negr1 knockdown in vivo. Negr1 knockout mice present phenotypes similar to Negr1-downregulated animals. These data indicate that NEGR1 and FGFR2 cooperatively regulate cortical development and suggest a role for defective NEGR1-FGFR2 complex and convergent downstream ERK and AKT signalling in autism spectrum disorders.
See Contreras and Hippenmeyer (doi:10.1093/brain/awy218) for a scientific commentary on this article.
Introduction
Autism spectrum disorders (ASDs) are a group of medical conditions with different aetiologies that originate during neurodevelopment. Although hundreds of diverse gene variants have been implicated in the pathogenesis of ASD, all ASDs are characterized by common core symptoms, such as early disruption in social skills, communication disabilities and repetitive behaviour, with most patients also suffering comorbidities (e.g. somatosensory perception impairment; Varghese et al., 2017; Yenkoyan et al., 2017). Consistent with the genetic cause and early onset of symptoms, defective brain development has been implicated in ASD. This includes cortical layering and migration defects as well as impaired dendritic spine density both in human patients and in mouse models (Hutsler et al., 2007; Hutsler and Zhang, 2010; Stoner et al., 2014; Reiner et al., 2016; Stouffer et al., 2016; Martinez-Cerdeno, 2017; Varghese et al., 2017). Thus, in recent years, researchers have repeatedly attempted to find a convergent pathway that may explain how the wide variety of ASD genetic variants converge into a core group of impaired processes during brain development and in behavioural phenotypes (Voineagu et al., 2011; Gokoolparsadh et al., 2016).
Among the numerous genes associated with ASD and key regulators of physiological as well as pathological brain development, a large body of literature indicates cell adhesion molecules (CAMs) and receptor tyrosine kinases (RTKs), extracellular signal-regulated kinase (ERK) and protein kinase B (AKT) signalling. In particular, both CAMs and RTKs are essential for proper neural migration and morphological maturation (Sobeih and Corfas, 2002; Rieger et al., 2009; Jossin and Cooper, 2011). Moreover, ERK and AKT are downstream of many RTK pathways and can be also activated by a large number of CAMs, positioning them as a possible convergent signalling pathway in neurodevelopment and ASD. Accordingly, growing evidence based on data from patients with ASD and animal models has indicated ERK and AKT as hubs for the convergence of genes and cellular pathways that are dysregulated in ASD (Ye et al., 2010; Kalkman, 2012; Pinto et al., 2014; Pucilowska et al., 2015; Wen et al., 2016). Interestingly, our laboratory has recently demonstrated a role for the immunoglobulin superfamily CAM neuronal growth regulator 1 (NEGR1; Funatsu et al., 1999; Schafer et al., 2005) in regulating neuronal morphological maturation by a functional interaction with the RTK fibroblast growth factor receptor 2 (FGFR2) through modulation of the FGFR2-ERK signalling pathway in vitro (Pischedda and Piccoli, 2015). Moreover, independent genetic studies have suggested a link among both NEGR1 and FGFR2 and brain disorders, including autism (Marshall et al., 2008; Pinto et al., 2010; Hussman et al., 2011; Casey et al., 2012; Michaelson et al., 2012; Neale et al., 2012; Genovese et al., 2015).
Here, we provide evidence in mice that the CAM NEGR1 and the RTK FGFR2 together regulate cortical development as well as core behaviours related to ASD by impinging on ERK and AKT signalling.
Materials and methods
All care of animals and experimental procedures were conducted in accordance with IIT and CIBIO licensing as well as the Italian Ministry of Health.
In vitro experiments
Cell lines, primary neuronal cultures and drug treatments
N2A (ATCC CCL-131) and HEK293 (ATCC CRL-1573) were grown in Dulbecco’s modified Eagle medium with 10% foetal bovine serum, 1% l-glutamine and 1% penicillin/streptomycin. Stable clones were isolated upon selection in G418 (1 mg/ml). Cultures were treated with 5 mM NH4Cl or 150 nM MG132 for 4, 6, 18, or 24 h. For the co-clustering assay, N2A wild-type and NEGR1 stable cells were co-transfected with FGFR2-GFP, dissociated in a solution not permissive for cell-to-cell aggregation (2 mM EDTA in PBS) and collected in Eppendorf tubes. Then, cells were counted and resuspended in cell-to-cell adhesion permissive medium (Hank’s Balanced Salt Solution, 2 mM MgCl2, 2 mM CaCl2) at a concentration of 4 × 106 cells/ml. Fifty thousand cells were then seeded and incubated for 30 min at 37°C in a 24-well plate with coverslips. Soluble FLAG-NEGR1 recombinant protein was purified as previously described (Pischedda and Piccoli, 2015). For the overlay experiments, HEK293 wild-type and FGFR2-myc stable clones were treated with 1 μg/ml of FLAG-Negr1 recombinant protein for 1 h at 4°C. Cells were subsequently washed twice with cold PBS, put in complete media supplemented with 20 ng/ml FGFb and then incubated at 37°C for 2 h. To monitor FGFR2 trafficking, HEK293 wild-type and NEGR1 stable cells were co-transfected with the indicated reporter plasmids and treated 48 h after with FGFb (10 ng/ml) and 5 mM NH4Cl for 4 h at 37°C to induce FGFR2-GFP activation and internalization.
Cortical neuron cultures were prepared from embryonic Day (E)17.5–18.5 mouse embryos (C57BL/6). Medium-density (150–200 cells/mm2) neuronal cultures were plated and grown on 12-well plastic tissue culture plates (Iwaki; Bibby Sterilin) as previously described (Corti et al., 2008; Pischedda et al., 2014). Neurons were treated with 10 ng/ml FGFb, 20 ng/ml FGF7/KGF, 5 ng/ml EGF (all from Peprotech) for 10 min alone or in combination with Dynasore (150 μM, Sigma-Aldrich), monensin (10 μM, 30 min Sigma-Aldrich), 5 mM NH4Cl or 150 nM MG132 for 4, 6, 18, or 24 h.
Plasmids and transfection
All cultures were transfected with Lipofectamine™ 2000 following the manufacturer’s protocol (Life Technologies). Strep-FLAG NEGR1 was described in Pischedda and Piccoli (2015), hFGFR2-GFP in Schuller et al. (2008), and FGFR2-myc in Mansukhani et al. (2000). RAB5A-mCherry (Addgene 27679), RAB7-mCherry (Addgene 55127) and LAMP1-mcherry (Addgene 45147) were purchased. The siRNA-resistant Strep-FLAG NEGR1 construct was generated by site-directed mutagenesis to introduce three silent mutations within the siRNA target sequence using the forward primer: 5′-GCCGTGGACAACATGCTCGTCAGGAAAGGTGACACAGCG-3′.
Lentiviral vectors expressing a siRNA cassette and GFP reporter were originally described in Wiznerowicz and Trono (2003). The mouse Negr1 and mouse Fgfr2 silencing lentiviral vectors used in biochemical assays (siNegr1 and siFGFR2) and in utero electroporation experiments (siNegr1) are described in Pischedda and Piccoli (2015). The FGFR2 silencing vectors used in in utero electroporation experiments (siRNA 2A: 5′-GCACACACTTACAGAGCACAA-3′ targeting mouse Fgfr2 3′UTR and siRNA 2B: 5′-CCTCTCTACGTCATAGTTGAA-3′ targeting mouse Fgfr2 exon 11) were originally described in Tarkkonen et al. (2012). The silencing cassettes, including a U6 promoter, and silencing sequences 2A and 2B were also cloned into a pDsRed-expression vector (Clontech).
Immunoblotting
All cultures and brain samples were washed in PBS and lysed in RIPA buffer (150 mM NaCl, 50 mM HEPES, 0.5% NP40, 1% sodium deoxycholate). After 1 h under mild agitation, lysate was clarified by centrifugation for 20 min at 16 000g. All procedures were performed at 4°C. To perform streptavidin pull-down, streptactin Superflow beads (IBA, 50% in ethanol) were diluted 1:1 with wash buffer containing 150 mM NaCl and 50 mM HEPES and centrifuged for 2 min at 2000g to purify the resin from the ethanol. The lysate was added to the beads and put under mild agitation for 1 h at 4°C. To perform co-immunoprecipitation assays, protein-G sepharose beads (Abcam, 20% in ethanol) were diluted 1:1 with wash buffer containing 150 mM NaCl and 50 mM HEPES, and centrifuged for 2 min at 2000g to purify the resin from the ethanol. The lysate was incubated with 1 µg goat anti-Negr1 antibody for 1 h at 4°C, then added to the beads and put under mild agitation for 1 h at 4°C. After incubation, the resin was washed twice with a wash buffer containing 150 mM NaCl, 50 mM HEPES, 0.1% Triton™ X-100. A brief centrifugation at 2000g followed each wash. Finally, the resin was resuspended in 60 μl of 2× Laemmli Buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue and 0.125 M Tris HCl, pH 6.8). To collect a crude membrane preparation, neuronal cultures were dissociated in an appropriate buffer (4 mM HEPES, 0.32 M sucrose, pH 7.4) and homogenized in a glass potter (10–15 strokes). Samples were centrifuged at 600g at 4°C for 10 min to remove the nuclear fraction. The resulting supernatant was ultracentrifuged at 100 000g for 1 h at 4°C, and the membrane fraction in the pellet was then lysed in RIPA buffer (150 mM NaCl, 50 mM HEPES, 0.5% NP40, 1% sodium deoxycholate). Protein samples were measured via standard Bradford assay (Bio-Rad). For protein identification and relative quantification by western blotting, a proper volume of sample containing an equal amount of proteins was diluted with 0.25% 5× Laemmli buffer and loaded onto 10% SDS-PAGE gels; the proteins were transferred onto nitrocellulose membrane (Sigma-Aldrich) at 80 V for 120 min at 4°C. The primary antibodies were applied overnight in a blocking buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20, and 5% non-fat dried milk); primary antibodies included rabbit anti-FGFR2 1:200 (Santa Cruz Biotechnology), rabbit anti-FGFR1 1:200 (Santa Cruz Biotechnology), goat anti-Negr1 1:1000 (R&D), mouse anti-Na+K+ ATPase (Abcam), rabbit anti-GFP 1:5000 (Life Technologies), rabbit anti-S6rp 1:2000, rabbit anti-p42/44 (pERK), rabbit anti-42/44 (ERK; Cell Signaling), rabbit anti-pAKT (Cell Signaling), rabbit anti-NSF (Cell Signaling), mouse anti-AKT pan (Cell Signaling), mouse anti-FLAG (Merck) and mouse anti-β-tubulin (Merck). The secondary antibodies (HRP-conjugated anti-mouse, anti-rabbit, or anti-goat; Jackson ImmunoResearch) were used at a dilution of 1:8000. The signal was detected using an ECL detection system (GE Healthcare). Images were acquired by the ChemiDoc™ Touch image system (Bio-Rad), and protein quantification was performed by measuring the optical density of the specific bands with ImageJ software (NIH). Unless otherwise stated, all other chemicals were purchased from Applichem.
Cell culture histology, immunostaining, image acquisition and analysis
Cells were fixed in 4% paraformaldehyde and 4% sucrose at room temperature. Rabbit anti-FLAG (Sigma, 1:200) antibodies were applied in GDB buffer (30 mM phosphate buffer pH 7.4, containing 0.2% gelatin, 0.5% Triton™ X-100, and 0.8 M NaCl) overnight at 4°C. Cy3- or Cy5-coupled secondary antibodies and 4’,6’-diamidino- 2-phenylindole (DAPI; Life Technologies) were diluted 1:1000 in GDB buffer and applied for 1 h. Positive cells were randomly chosen for quantification. Cover slips were mounted with ProLong® reagent (Life Technologies) and observed with an AxioObserver Z1 microscope equipped with an Apotome module (Zeiss) using a plan-Apochromat 63×/1.40 oil objective, pixel size 0.102 mm × 0.102 mm. The obtained images provided an axial resolution comparable to confocal microscopy (Schaefer et al., 2004; Garini et al., 2005). Co-localization studies were performed on the single plane generated by optical sectioning elaborated by the Apotome module. The determination of M1 coefficients (Manders et al., 1992) were performed using ImageJ (https://imagej.net/Welcome) and is indicative of GFP-FGFR2/mCherry reporter localization.
In vivo and ex vivo experiments
In utero electroporation
In utero electroporation was performed as previously described (Szczurkowska et al., 2016). Briefly, E15.5 timed-pregnant CD1 mice (Charles River SRL) were anaesthetized with isoflurane (induction, 4.0%; surgery, 2.0%), and the uterine horns were exposed by laparotomy. The DNA (scramble for control, Negr1 siRNA, Fgfr2 siRNA, Fgfr2 cDNA or Negr1 siRNA with Fgfr2 cDNA, 1.0–2.0 µg/µl in water) together with the dye Fast Green (0.3 mg/ml; Sigma) was injected (3–4 µl) through the uterine wall into one of the lateral ventricles of each embryo by a 30G needle (Pic Indolor). For uniform visualization of transfected neurons and to ensure balance of the total plasmid mass in each electroporation, we co-electroporated each plasmid together with td-Tomato or EGFP. The embryo’s head was carefully held between tweezer-type circular electrodes (3 mm diameter for standard electroporation or 10 mm diameter for tripolar electroporation; Nepa Gene) across the uterine wall. The third electrode was used for electroporation of the visual, prefrontal or motor cortices (3 mm × 5 mm diameter). For the electroporation, six electrical pulses (amplitude, 30 V; duration, 50 ms; intervals, 1) were delivered with a square-wave electroporation generator (CUY21EDIT; Nepa Gene). Then, the uterine horns were returned to the abdominal cavity, and embryos were allowed to continue their normal development.
Slice histology, immunostaining, image acquisition and analysis
E18 brains were dissected and fixed for 24 h in 4% paraformaldehyde (PFA) in PBS, whereas postnatal day (P)7 and P20–25 brains were fixed by transcardial perfusion with PFA solution. Then brains were cryopreserved in 30% sucrose and sectioned coronally in 80-µm thick slices with a microtome-refrigerator (Microm HM 450 Sliding Microtome equipped with Freezing Unit Microm KS34, Thermo Scientific). Slices were counterstained with Hoechst (2.5 µg/µl; Sigma). All slices were mounted in Vectashield Mounting Medium (Vector Laboratories). For analysis of migration, dendritic arborization and spine density, image acquisition and analysis was performed on GFP or Tomato fluorescence from transfected cells. For migration analysis, images of randomly chosen cortical slices at the level of the somatosensory cortex were acquired on a Neurolucida epifluorescence microscope equipped with the software Neurolucida (MicroBrightField) and a 20× objective (NA 0.7). The contrast of the images was adjusted to enhance the fluorescence of cell bodies while attenuating the signal from neuronal processes to facilitate cell counting. One image was acquired per animal. For dendrite morphology analysis, pyramidal neurons were acquired using a confocal laser-scanning microscope (TCS SP5; Leica Microsystems) equipped with a 63× immersion objective (NA 1.4). To allow a better visualization of neuronal processes, the contrast of the images was adjusted to decrease the fluorescence of cell bodies. Reconstructions of representative pyramidal neurons were drawn with Adobe Photoshop (Adobe System). Analysis of total branch number and length, and Sholl analysis were performed with ImageJ software (NeuronJ and Sholl Analysis plugins). For spine counting, confocal images of transfected neurons were acquired using a confocal laser-scanning microscope (TCS SP5; Leica Microsystems) equipped with a 63× immersion objective (NA 1.4) with 1.5× digital zoom (1-µm thick z-stacks) and projected on a 2D image. On each image, the apical dendrite of a neuron was visually identified, and spines were counted on the whole visible length of one to three collateral dendrites for each image and divided by this length. When the apical dendrite was not clearly visible, spines on one to three randomly-chosen dendrites were counted on the whole visible length and divided by this length. The densities of spines for each dendrite were averaged per each image. One image for one to three different slices were acquired per animal and quantifications for each slice were averaged together.
For immunostaining, free-floating slices were permeabilized and blocked with PBS containing 0.3% Triton™ X-100, 10% normal goat serum (NGS) and 0.2% bovine serum albumin (BSA). Primary antibodies were incubated in PBS containing 0.3% Triton™ X-100, 5% NGS and 0.1% BSA [rat anti-CTIP2, 1:100 (Abcam), rabbit anti-Cux1, 1:300 (Santa Cruz), rabbit anti-FGFR2 1:200 (Abcam)]. Immunostaining was detected using Alexa Fluor® 543 or Alexa Fluor® 647 fluorescent secondary antibody (Invitrogen), 1:600, incubated in PBS containing 0.3% Triton™ X-100, 5% NGS and 0.1% BSA. Slices were counterstained with Hoechst (2.5 µg/µl; Sigma). Stained slices were acquired using a confocal laser-scanning microscope (TCS SP5; Leica Microsystems) equipped with a 20× immersion objective (1-µm thick z-stacks) and projected on a 2D image. One representative slice per condition was acquired.
Golgi-Cox staining, image acquisition and analysis
Animals were perfused transcardially with 0.9% saline. Whole brains were dissected and immersed in the Golgi-Cox solution (5% potassium dichromate, 5% mercuric chloride and 5% potassium chromate) for 35 days. Then, brains were transferred to a 30% sucrose solution and stored in the dark at 4°C. Two-hundred-micrometre thick coronal slices were cut at room temperature with a microtome (Microm HM 450 Sliding Microtome) and transferred onto 1% gelatin-coated slides to initiate the staining process in humidified chambers. Ammonium hydroxide was applied for 10 min. Next, slices were treated with an increasing grade of ethanol (50%, 75%, 95% and 100%), treated with xylene for 30 min, and then mounted in DPX mounting medium (Sigma-Aldrich). A stack of images (1-µm thick z-stacks) from randomly chosen cortical slices at the level of the somatosensory cortex were acquired on a Neurolucida microscope equipped with the software Neurolucida (MicroBrightField) and a 100× objective (NA 1.3). One stack of 80–150 images (1 μm step) was acquired per cell for a total of 10 cells per animal. On each image stack, the apical dendrite of a neuron was visually identified, and spines were counted on the whole visible length of one to three collateral dendrites for each image and divided by this length. The densities of spines for each dendrite were averaged per each image.
In situ hibridization
Negr1 mRNA was localized by in situ hybridization with digoxigenin (DIG)-labelled cRNA probes, as described previously (Heinrich et al., 2006). Sense and antisense probes were generated against full-length murine Negr1 cDNA (Schafer et al., 2005) and diluted in hybridization buffer at a final concentration of 100 ng/ml. Microscopy was performed with an AxioImager 2 (ZEN software; Zeiss). Images were taken with a digital camera (AxioCam MRc5 and a 10-fold objective, Plan Apochromat, NA 0.45).
All images in all experiments were acquired in a random order and, when possible, in a single session to minimize errors caused by fluctuation in laser output and degradation of fluorescence.
All image acquisitions and data analysis were performed by an experimenter blind to the experimental conditions.
Behavioural experiments
All animals were housed in a climate-controlled animal facility (22 ± 2°C) and maintained on a 12 h light/dark cycle with ad libitum access to food and water. For animals (CD1 mice) electroporated with the experimental siRNA (Negr1 siRNA or Fgfr2 siRNA), littermates electroporated with control plasmids were used in the same session as controls. Negr1 siRNA-electroporated littermates were used as controls for the Negr1 siRNA/FGFR2cDNA animals. For Negr1−/− animals, wild-type littermates were used as controls. The Negr1−/− animals are in a C57BL6/J background. In all experiments we used males and females.
Ultrasonic vocalization analysis
Each pup was separated at P4 from the mother and littermates and placed in an empty container (diameter, 5 cm; height, 3 cm), located in a sound-attenuating Styrofoam box (diameter, 30 cm; height, 40 cm). Calls were recorded for 5 min by an ultrasound microphone sensitive to frequencies of 10–180 kHz (Avisoft UltraSoundGate condenser microphone capsule CM16, Avisoft Bioacoustics) and Avisoft Recorder software (Version 3.2). Data analysis was performed using Avisoft SASLab Pro (Version 4.40). The total number of calls during the 5 min recording session was quantified.
Hot plate test
Response to an acute thermal stimulus was measured in pups at P9 using an adapted hot plate test from a previously described protocol (Gioiosa et al., 2008; DeLorey et al., 2011). Since animals used for in utero electroporation and Negr1−/− mice are in different backgrounds, the hot plate temperature was adjusted accordingly for each strain. In particular, the experimenter held the pup between the thumb and forefinger in an upright position and gently placed the hind paws of the mouse on the surface of the hot plate kept at constant temperature of 50°C (CD1 mice, in utero electroporation experiments) or 55°C (Negr1−/− mice in C57BL6/J background). The latency to withdraw the paw from the hot plate was measured. To prevent any heat injury to pups, a cut-off latency of 30 s was applied.
Social interaction/self-grooming test
Animals were tested for reciprocal social interactions and self-grooming behaviour as previously described (Silverman et al., 2010). Briefly, animals at P20–25 were placed one-by-one in a Plexiglas® box (30 cm × 30 cm) with a non-littermate peer of the same age. A 10-min interaction period was recorded in ambient light using a Handycam (SONY HDR-XR155) placed above the box with the aid of a tripod. Parameters considered for subsequent analysis of the videos included total social sniffing time by the experimental animal (electroporated or Negr1−/−) on the wild-type peer and self-grooming time of the experimental animal.
Statistical analysis
Statistical analysis was performed with Student’s t-test or one- or two-way ANOVA and Holm-Sidak or Bonferroni post hoc comparison. For heteroskedastic datasets and/or for datasets of non-normal distribution, Mann-Whitney test or Kruskal-Wallis and Dunn’s post hoc comparison were used. For the datasets in Supplementary Figs 3B, 5E and 10B, the chi-squared test was performed on the probability distribution of the datasets. For statistical purposes, the last layer (G) was removed from dataset in Supplementary Fig. 3B and the last three layers (E, F and G) were removed from dataset in Supplementary Fig. 10D. To better appreciate the different distributions, we showed the cumulative distribution in the figure. Outliers were excluded only from the final pool of data by a Grubb’s test iteratively until no outliers were found. All boxplots represent 10–90 percentiles.
Data availability
All data from this work are available on the free repository OFS and can be accessed on: https://osf.io/5a673/?view_only=35ccdcaf059f48efa581eab210abbc73
Results
Negr1 or Fgfr2 downregulation similarly impairs the radial migration and spine density of pyramidal neurons in the somatosensory cortex in vivo
To investigate the roles of NEGR1 and FGFR2 during neuronal development in vivo, we first addressed their expression in the mouse brain cortex at different developmental stages. In particular, in situ hybridization in the perspective somatosensory cortex with specific antisense probes for Negr1 at E15 revealed Negr1 mRNA expression in the subventricular zone, cortical plate and marginal zone (Supplementary Fig. 1A). Postnatally (P7) and in the adult, Negr1 appeared to be progressively more confined to the upper layers of the somatosensory and visual cortex, with strong expression in layer IV (Supplementary Fig. 1 A and B). Conversely, Negr1 mRNA expression was low in the motor cortex and prefrontal cortex at all ages (Supplementary Fig. 1A–C). For FGFR2, immunohistochemistry with specific antibodies against the protein revealed that its expression was widespread across the whole cortex at all ages (Supplementary Fig. 1D).
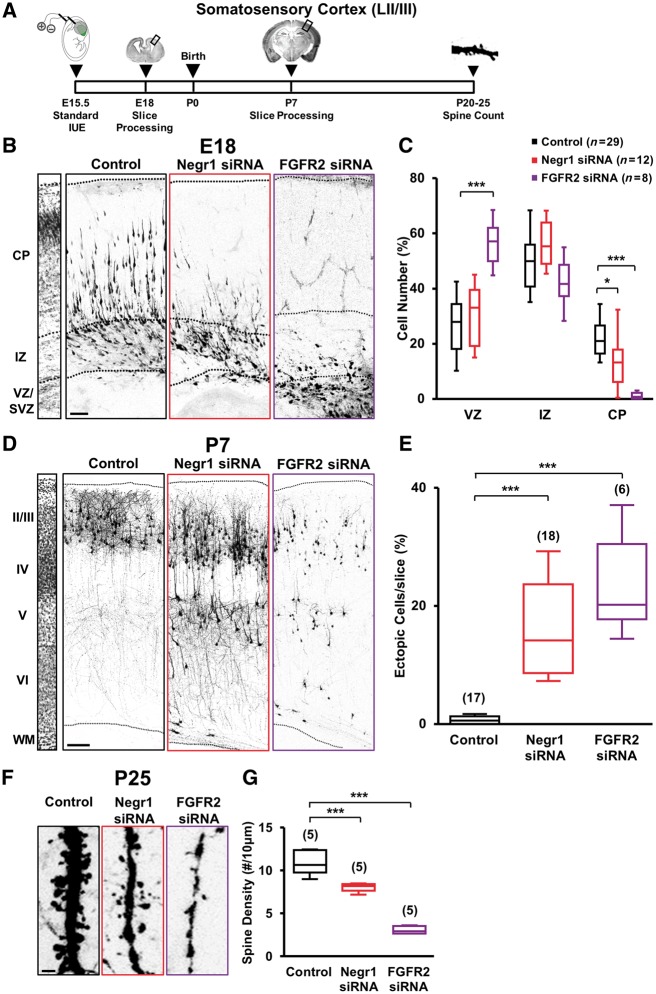
Given the strong expression of Negr1 mRNA and FGFR2 protein in the subventricular zone of the developing somatosensory cortex at a time and location when newborn neurons that are committed to layer II/III are generated, we investigated whether NEGR1 and FGFR2 have a role in the development of these neurons. We took advantage of in utero electroporation to transfect E15.5 subventricular zone neural progenitor cells with vectors expressing EGFP (to visualize transfected cells) together with small interfering RNA (siRNA) to target endogenous Negr1 or Fgfr2, or scrambled control siRNA (Pischedda et al., 2014; Pischedda and Piccoli, 2015). After allowing in vivo development, we examined the distribution of EGFP-positive neurons at different postnatal stages (Fig. 1A). We found strikingly similar phenotypes for Negr1 and Fgfr2 siRNA-transfected animals. In particular, we quantified the number of neurons expressing scrambled siRNA, Negr1 siRNA or Fgfr2 siRNA residing at the ventricular zone, intermediate zone, or cortical plate at E18 and normalized this number to the total number of transfected cells. Control scrambled siRNA cells were distributed across the ventricular zone, intermediate zone, and cortical plate. Interestingly, NEGR1 or FGFR2 knockdown neurons were mostly located in the ventricular zone and intermediate zone, with only a small percentage reaching the cortical plate (Fig. 1B and C). By P7, almost all siRNA control-transfected cells reached layer II/III as expected, whereas a large fraction of Negr1 or Fgfr2 siRNA-expressing cells were arrested in layer V and did not cross the border between layers IV and V (Fig. 1D and E). However, these cells retained their upper-layer identity as they were immunopositive for the LII-IV marker CUX1 and immunonegative for the deep-layer marker CTIP2 (Supplementary Fig. 2). Therefore, we defined these cells as ‘ectopic’ neurons. In addition to the ectopic positioning of NEGR1- or FGFR2-defective neurons in the deep cortical layers, we also found a subtle defect in the positioning of siRNA-positive cells within layers II/III. Quantification of NEGR1- and FGFR2-depleted neurons in seven subregions of upper cortical layers (layers II–IV) revealed indeed that the siRNA-transfected neurons accumulated to lower subregions compared to controls (Supplementary Fig. 3). Finally, when we allowed transfected animals to develop until P35, we still found NEGR1- or FGFR2-downregulated cells ectopically arrested in layer V, indicating that the neuronal migration defects were long-lasting (Supplementary Fig. 4). To further investigate the effect of NEGR1 or FGFR2 downregulation in the cortex, we turned our attention to the posterior part of the brain. As the expression of NEGR1 is high in the visual cortex (Supplementary Fig. 1B), we electroporated Negr1 or Fgfr2, or scrambled control siRNA in the visual cortex with the aid of the tripolar configuration for in utero electroporation (dal Maschio et al., 2012; Szczurkowska et al., 2016). The results were consistent with the somatosensory cortex, with a number of ectopic cells and a defect in the positioning of siRNA-positive cells within layer II/III at P7 (Supplementary Fig. 5A–E).
Figure 1.
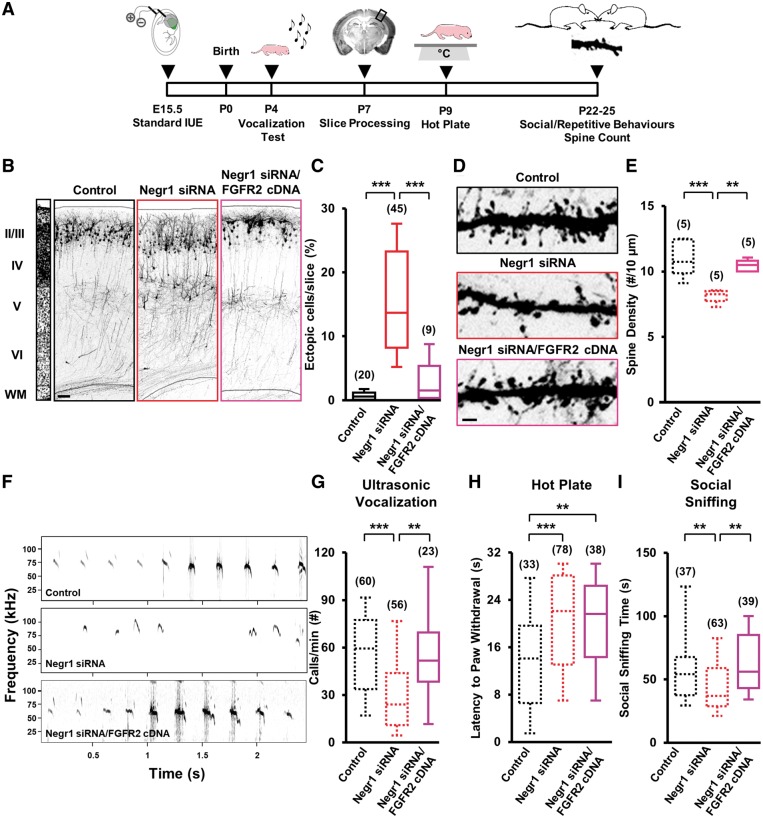
Negr1 or FGFR2 downregulation affects late-born neuron migration and spine density in vivo. (A) Experimental protocol with standard in utero electroporation for targeting LII/III of the mouse somatosensory cortex. (B) Confocal images of GFP fluorescence in coronal sections of somatosensory cortices at E18 transfected in utero (at E15.5) with scrambled siRNA (control), Negr1 siRNA or Fgfr2 siRNA. Slices were counterstained with nuclear staining DAPI for visualization of cortical layers (left). Scale bar = 100 µm. Dotted lines indicate cortical zone borders. VZ = ventricular zone; IZ = intermediate zone; CP = cortical plate. (C) Quantification of the number of transfected neurons residing at the ventricular zone, intermediate zone, or cortical plate in experiments as in B. Data are expressed as a percentage of the total number of fluorescent cells in the section. Asterisks indicate statistically significant difference (two-way ANOVA against control; post hoc Holm-Sidak: *P < 0.05, ***P < 0.001). Numbers in parentheses indicate total number of processed animals (1 slice/animal). (D) GFP fluorescence in coronal sections of somatosensory cortices of transfected animals at P7. Left: DAPI counterstaining. Scale bar = 100 µm. Dotted lines indicate slice border. WM = white matter. (E) Quantification of the number of transfected neurons that did not complete their migration in experiments as in D. Data are expressed as average percentage of the total number of fluorescent cells in the section. Asterisks indicate statistically significant difference (Kruskal-Wallis test against control; Dunn’s post hoc: ***P < 0.001). Numbers in parenthesis indicate total number of processed animals (1 slice/animal). (F) GFP fluorescence in dendrites of somatosensory cortical neurons of P25 transfected mice. Scale bar = 5 μm. (G) Quantification of the spine density in experiments as in F (P20–25 mice). Data are expressed as average. Asterisks indicate statistically significant difference (one-way ANOVA against control; Holm-Sidak post hoc: ***P < 0.001. Numbers in parentheses indicate total number of processed animals (1–3 dendrites/slice, 1–3 slices/animal).
Since Negr1 expression appeared to be stronger in the somatosensory and visual cortices in comparison to other cortical areas (Supplementary Fig. 1), we next investigated whether the effect of siRNA-mediated downregulation of Negr1 was only restricted to these areas or was common to other cortical regions. We first focused on the motor cortex, which can be visualized on the same coronal section of the somatosensory cortex, allowing a direct comparison. Negr1 mRNA expression was clearly stronger and localized in layer IV in the somatosensory cortex, whereas it progressively faded when entering the motor cortex (Supplementary Fig. 6A–C). When we downregulated Negr1 in the motor and somatosensory cortices by tripolar in utero electroporation (Supplementary Fig. 6A), we found that the defect in migration (i.e. neurons arrested in layer V) was mostly present in the somatosensory cortex and faded (as Negr1 mRNA expression did) with progression to the motor cortex (Supplementary Fig. 5C and 6B). These results on Negr1 siRNA animals were paralleled by those of a similar analysis in the prefrontal cortex (Supplementary Fig. 6D and E), which also lacks Negr1 mRNA expression in layer IV (Supplementary Fig. 1A–C). Downregulation of Fgfr2 in the prefrontal cortex did not lead to any major phenotype of neuronal migration as well, with only a few ectopic cells detectable (Supplementary Fig. 6E).
NEGR1 and FGFR modulate neurite outgrowth in different systems in vitro (Schafer et al., 2005; Cavallaro and Dejana, 2011; Pischedda et al., 2014; Pischedda and Piccoli, 2015; Sanz et al., 2015). To compare the possible effects of NEGR1 or FGFR2 depletion on neuronal morphology in vivo, we performed GFP fluorescence-based reconstructions of neurons located in layer II/III and ectopic neurons located in layer V in the somatosensory cortex of Negr1 or Fgfr2 siRNA animals at P7 (Supplementary Fig. 7A). By analysing the number of branches and total branch length as well as by Sholl analysis, we found that knockdown of Negr1 or Fgfr2 severely impaired neuronal morphology. Interestingly, each genetic manipulation affected dendrite formation in a different manner (Supplementary Fig. 7).
Previous studies have described the impact of both FGFR2 and NEGR1 on dendritic spines in vitro (Umemori and Sanes, 2008; Pischedda et al., 2014). Thus, we investigated the dendritic spine density in Negr1 or Fgfr2 siRNA animals at P20–25 by GFP fluorescence-based reconstructions. Cells transfected with Negr1 siRNA or Fgfr2 siRNA in somatosensory as well as visual cortex similarly showed a significant decrease in spine density compared to controls (Fig. 1F, G, Supplementary Fig. 5F and G).
Finally, since the use of siRNA can be hampered by off-target effects, we transfected siRNA-resistant cDNA constructs (Supplementary Fig. 8A–D) together with Negr1 or Fgfr2 siRNA by in utero electroporation and examined the neuronal migration and spine density of transfected neurons in the somatosensory cortex. We found that overexpression of the siRNA-resistant variants of Negr1 and Fgfr2 rescued both phenotypes in Negr1- or Fgfr2-downregulated neurons at P7 (Supplementary Fig. 8E–I).
These data suggest that NEGR1 and FGFR2 play a major role in cortical development in vivo. Because of the similarity of phenotypes in late-born pyramidal-neuron migration and spine density of NEGR1 and FGFR2-defective neurons, these two molecules possibly interact with converging functions in these processes. Conversely, the effects on dendritic arborization are, in fact, specific for each molecule.
Negr1 or Fgfr2 downregulation impairs core behaviours related to autism spectrum disorder
Patients with ASD and animal models may display an abnormal laminar cytoarchitecture and cortical disorganization of neurons as well as abnormalities in dendritic spines (Stoner et al., 2014; Reiner et al., 2016; Hori and Hoshino, 2017; Martinez-Cerdeno, 2017; Varghese et al., 2017; Dang et al., 2018). Moreover, many ASD-associated genes are mostly expressed in pyramidal neurons located in the upper layers of the cortex, as we found here for Negr1 (Parikshak et al., 2013). Thus, we next investigated whether Negr1 and Fgfr2 downregulation in mice would cause core symptoms and sensory abnormalities relevant to ASD.
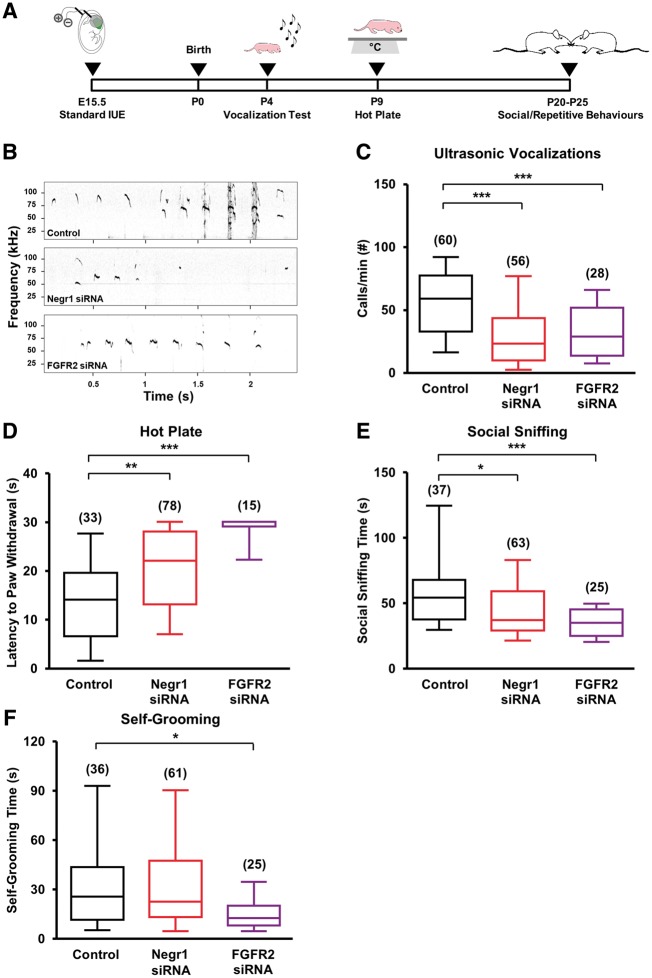
First, we investigated the consequences of Negr1 or Fgfr2 downregulation in the somatosensory cortex on ultrasound vocalizations emitted by pups upon separation from their mother and littermates. Indeed, the somatosensory cortex in mice is believed to be analogous to the language area in the human cortex (Sia et al., 2013), and ultrasonic vocalizations have been widely used to test communication/social skills in mouse models of ASD (Scattoni et al., 2009; Wohr and Scattoni, 2013). We electroporated E15.5 embryos from different dams with Negr1 siRNA or Fgfr2 siRNA. In each litter, we also electroporated some embryos with scrambled siRNA as intra-litter controls (Fig. 2A). We tested the animals at P4 for ultrasonic vocalizations and found that pups transfected with Negr1 or Fgfr2 siRNA vocalized significantly less in comparison to control littermates (Fig. 2B and C). By contrast, downregulation of Negr1 or Fgfr2 in the prefrontal cortex did not cause vocalization impairments (Supplementary Fig. 6D–F).
Figure 2.
FGFR2 or NEGR1 downregulation in the somatosensory cortex results in alterations in core behaviours related to ASD. (A) Experimental protocol. (B) Example of a fraction of the recordings of ultrasonic vocalizations emitted upon isolation from dam and littermates by P4 pups transfected in utero. (C) Quantification of the number of ultrasonic vocalizations during the entire isolation in experiments as in B. Data are expressed as the average of the total number of emitted calls/min. Asterisks indicate statistically significant difference (one-way ANOVA against control, post hoc Holm-Sidak: ***P < 0.001). (D) Quantification of the latency to paw withdrawal of transfected P9 pups after placement on a hot plate. Data are expressed as the average time spent on the hot plate until first pain reaction. Asterisks indicate statistically significant difference (Kruskal-Wallis test against control, post hoc Dunn’s: **P < 0.01, ***P < 0.001). (E) Quantification of the time spent performing social sniffing by transfected animals at P20–25 during juvenile play with a stranger peer from a different litter. Data are expressed as the average total time that a transfected animal spent at sniffing during the entire trial. Asterisks indicate statistically significant difference (Kruskal-Wallis test against control, post hoc Dunn’s: *P < 0.05, ***P < 0.001). (F) Quantification of the average total time spent by transfected animals on self-grooming behaviour at P20–25, during 10 min of juvenile play with a stranger mouse from a different litter. Asterisks indicate statistically significant difference (Kruskal-Wallis test against control, post hoc Dunn’s: *P < 0.05). For all panels, numbers in parentheses indicate total number of analysed animals, 3–13 litters per experimental case.
Second, we investigated whether Negr1 or Fgfr2 downregulation in the somatosensory cortex caused sensory deficits using a hot plate test at P9 (Silverman et al., 2010). We found that pups transfected with Negr1 siRNA or Fgfr2 siRNA showed a longer latency to respond to the acute thermal stimulus when placed on the heated plate in comparison to control littermates (Fig. 2D).
Finally, we examined whether Negr1 or Fgfr2 downregulation compromised social and repetitive behaviours later in life. In particular, we recorded the social sniffing time in Negr1 siRNA or Fgfr2 siRNA mice at P20–25 during juvenile play with a non-transfected stranger peer from another litter (McFarlane et al., 2008). We found that Negr1- or Fgfr2-downregulated mice engaged in less social interactions than control animals (Fig. 2E). During the same recording session, we examined Negr1- and Fgfr2-downregulated animals for self-grooming, a model of complex repetitive, self-directed behaviour (Kalueff et al., 2016). While the grooming time of Negr1 siRNA animals was not significantly changed compared to controls, it was reduced in Fgfr2 siRNA animals (Fig. 2F).
These data show that Negr1 or Fgfr2 downregulation in neurons destined for layer II/III of the somatosensory cortex causes behavioural changes reminiscent of ASD-relevant phenotypes.
NEGR1 physically interacts with FGFR2
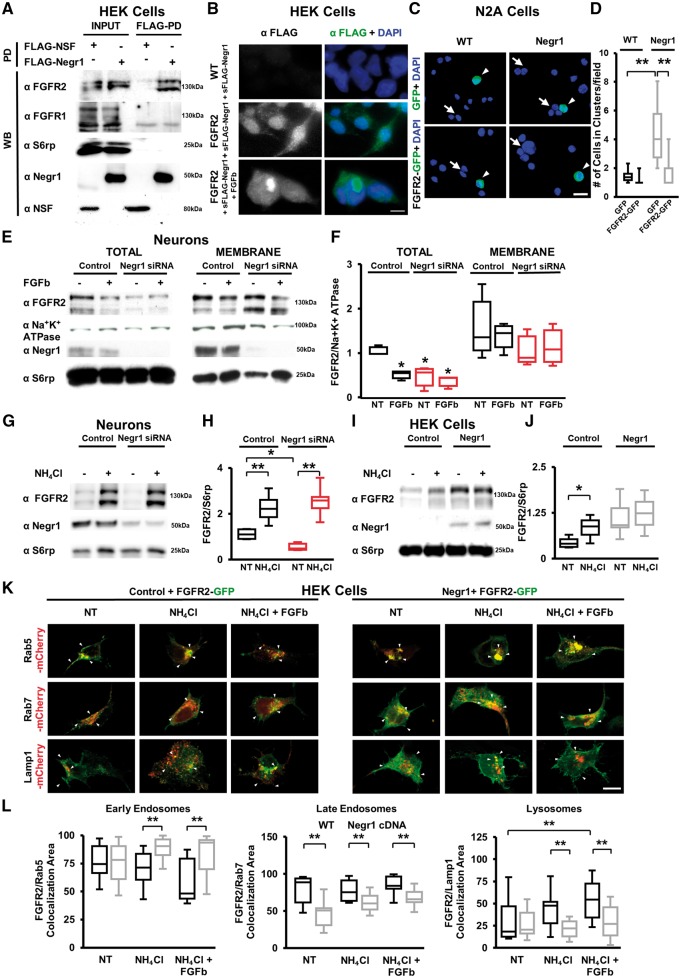
Our data on neuronal migration, spine density and social behaviour suggest that NEGR1 and FGFR2 may converge into a unique signalling pathway to regulate certain aspects of cortical development and some behaviours related to ASD. Since other IgLON family members interact physically with RTKs to exert their functions (McKie et al., 2012), we first examined whether the IgLON NEGR1 and the RTK FGFR2 interact physically using co-immunoprecipitation assays. We subjected HEK cells transfected with FLAG-NEGR1 or FLAG-N-ethylmaleimide sensitive factor (NSF, an intracellular protein used as a control) to FLAG pull-down (FLAG-PD) upon chemical cross-linking. FGFR2 specifically co-immunoprecipitated with NEGR1, but not with FGFR1 or the control ribosomal protein S6rp (Fig. 3A). We confirmed these results by immunoprecipitation of endogenous NEGR1 and FGFR2 proteins in brain lysates prepared from adult Negr1−/− and wild-type littermates (Supplementary Fig. 9A). Moreover, we verified that FGFR2–NEGR1 interaction was also present during development in wild-type pups (Supplementary Fig. 9B).
Figure 3.
NEGR1 influences FGFR2 intracellular trafficking. (A) Representative immunoblots on protein extracts from lysates of HEK293 cells expressing control FLAG-NSF or FLAG-NEGR1, chemically cross-linked, solubilized and processed for FLAG-immunoprecipitation (pull down, FLAG-PD). NEGR1 interacts with FGFR2 but not with FGFR1, S6 ribosomial protein (S6rp) or ubiquitously expressed ATPase N-ethylmaleimide sensitive factor (NSF). INPUT, 10% of total lysate. Similar experiments were repeated four times. (B) Representative fluorescence images of FLAG and DAPI immunostainings in wild-type or FGFR2-expressing HEK293 cells (FGFR2) exposed to purified soluble FLAG-NEGR1 (sFLAG-Negr1) or first to sFLAG-Negr1 and subsequently to the pan-FGFRs activator FGFb. Scale bar = 20 μm. (C) Representative images of GFP fluorescence and DAPI staining in wild-type or N2A cells stably expressing FLAG-NEGR1 assayed for cluster-formation when transfected with GFP alone or FGFR2-GFP. Arrows indicate clustered cells; arrowheads indicate GFP-expressing cells. Scale bar = 20 μm. (D) Quantification of the number of cells organized in clusters in experiments as in C. Data are expressed as average number of cells organized in cluster. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Bonferroni test: ***P < 0.001, n = 5 independent experiments). (E) Representative immunoblots on protein extracts from lysates of DIV14 cortical neurons infected at DIV4 with control scramble siRNA (Control) or Negr1 siRNA, treated at DIV 14 with FGFb or vehicle (NT), and then processed to isolate total and membrane-bound protein fractions. Membrane-bound protein enrichment was appreciated by staining with anti Na+/K+ ATPase antibody, while cytosolic contamination was monitored using an S6rp antibody. (F) Quantification of FGFR2 protein level in the two fractions, expressed as optical density and normalized versus Na+/K+ ATPase amount. Data are expressed as average of FGFR2/Na+-K+ ATPase ratio in vehicle-treated (NT) or drug-treated neurons. Asterisks indicate statistically significant difference versus control (one-way ANOVA, post hoc Tukey: ***P < 0.001, n = 5 independent experiments). (G) Representative immunoblots on protein extracts from lysates of DIV14 cortical neurons infected at DIV4 with control scramble siRNA (Control) or Negr1 siRNA and treated (or not, NT) at DIV 14 with NH4Cl. (H) Quantification of FGFR2 protein level expressed as optical density normalized versus S6rp amount. Data are expressed as average of FGFR2/S6rp ratio in vehicle-treated (NT) or drug-treated neurons. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Bonferroni: *P < 0.05; ***P < 0.001, n = 6 independent experiments). (I) Representative immunoblots on protein extracts from lysates of wild-type or FLAG-NEGR1-expressing HEK293 cells treated with NH4Cl. S6rp was used as an internal standard. (J) Quantification of FGFR2 protein level expressed as optical density normalized versus S6rp amount. Data are expressed as average of the FGFR2/S6rp ratio in vehicle-treated (NT) or drug -treated cells. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Bonferroni: *P < 0.05, n = 6 independent experiments). (K) Representative images of mCherry (red) and GFP fluorescence (green) in wild-type (control) or HEK293 cells stably expressing FLAG-Negr1 and transfected with FGFR2-GFP together with mCherry-Rab5 (marker of early endosomes), mCherry-Rab7 (marker of late endosomes) or mCherry-LAMP1 (marker of lysosomes)-expressing vectors. Arrowheads indicate co-localization of FGFR2-GFP and mCherry. Scale bar = 20μm. (L) Quantification of co-localization area between GFP-FGFR2 and the indicated reporter. Data are expressed as percentage of GFP-FGFR2 signal co-localizing (M1 coefficient) with mCherry signal. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Bonferroni: *P < 0.05, **P < 0.01, n = 5 independent experiments).
NEGR1 is prone to proteolytic cleavage and it can exist in a soluble form (Pischedda and Piccoli, 2015; Sanz et al., 2015). Thus, we investigated whether soluble NEGR1 added to media binds to FGFR2 expressed on a cellular membrane. We incubated living wild-type and FGFR2-overexpressing HEK293 cells (FGFR2) with recombinant FLAG-NEGR1 protein (1 μg/ml, 1 h at 4°C) and processed them for anti-FLAG immunocytochemistry. FLAG-NEGR1 bound exclusively to FGFR2-overexpressing cells, indicating that NEGR1 and FGFR2 interact in their native state (Fig. 3B). Interestingly, upon treatment with the pan-FGFR agonist FGFb (20 ng/ml, 2 h at 37°C), which induces FGFR internalization (Auciello et al., 2013), we identified FLAG-positive aggregates in the perinuclear region (Fig. 3B), suggesting that NEGR1 binding to FGFR2 is followed by co-internalization into endocytic vesicles upon stimulation.
IgLON family members organize homo- and heterodimers in trans (Gil et al., 2002; Sanz et al., 2015). Thus, we next assayed a stable N2A cell line expressing FLAG-NEGR1 (NEGR1) for its propensity to aggregate in vitro. We considered the formation of cell clusters upon dissociation and incubation in adhesion-permissive conditions as a readout of trans-dimerization. In agreement with previous findings (Lee et al., 2012), we found that NEGR1-expressing cells formed a higher number of multicellular aggregates compared to control wild-type cells (Fig. 3C and D). Interestingly, co-expression of FGFR2 reduced the formation of NEGR1-positive cell aggregates (Fig. 3C and D) without perturbing NEGR1 expression (Supplementary Fig. 9C and D).
These data suggest that NEGR1 and FGFR2 may physically interact both in cis and trans.
NEGR1 regulates FGFR2 intracellular fate
Upon agonist binding, RTKs are internalized and redistributed towards endosome-mediated recycling or degradation (Maxfield and McGraw, 2004; Auciello et al., 2013). Interestingly, the IgLON family member OPCML modulates the RTK FGFR1 subcellular distribution (McKie et al., 2012). Thus, we investigated whether the physical interaction of NEGR1 with FGFR2 alters FGFR2 intracellular trafficking. We isolated the total and membrane-bound protein fraction from in vitro Day (DIV)14 cortical neurons infected at DIV4 with viral Negr1 siRNA constructs or control scrambled siRNA. Moreover, we stimulated cultured neurons with the pan-FGFR agonist FGFb (10 ng/ml, 10 min) to trigger receptor internalization (Auciello et al., 2013). Negr1 silencing induced a significant reduction in the total FGFR2 protein level independent of FGFb-mediated FGFR2 internalization (Fig. 3E, F and Supplementary Fig. 9E). Conversely, the FGFR2 protein levels in the membrane fraction were not significantly affected (Fig. 3E and F), suggesting that endogenous NEGR1 controls the intracellular fate of FGFR2 in cortical neurons.
To examine the possible role of NEGR1 in the intracellular fate of FGFR2, we first analysed FGFR2 protein turnover. We assessed the FGFR2 protein levels in neuronal cultures treated at DIV14 with NH4Cl (5 mM), a blocker of lysosomal activity, or MG132 (150 nM), a proteasome inhibitor, as a control for cytosolic protein degradation. As expected for a membrane protein, we found that FGFR2 was actively, rapidly and selectively degraded by the lysosomal (but not proteasomal) compartment, as demonstrated by the significantly increased levels of FGFR2 expression that only appeared upon NH4Cl2 treatment (Fig. 3G, H and Supplementary Fig. 9F–I). Thus, we next investigated whether NEGR1 influences this lysosome-mediated FGFR2 degradation by monitoring the FGFR2 levels upon lysosomal inhibition in DIV14 cortical cultures infected at DIV4 with Negr1 siRNA. Negr1 silencing reduced the FGFR2 basal levels and increased FGFR2 susceptibility to lysosomal degradation (Fig. 3G and H). To complement such evidence, we monitored the FGFR2 levels in control or FLAG-NEGR1-overexpressing HEK293 cells in the presence (or not) of NH4Cl (5 mM, 6 h). In control cells, NH4Cl treatment induced a robust increase of the FGFR2 levels (with no effect on NEGR1 levels; Supplementary Fig. 9H and I); conversely, NEGR1-overexpressing cells did not accumulate FGFR2 upon lysosomal blockage (Fig. 3I and J) in agreement with the experiment in neuronal cultures. Altogether, these data show that NEGR1 may prevent depletion of intracellular FGFR2.
To understand whether Negr1 modulates the fate of FGFR2 further, we tracked the FGFR2 intracellular route. Upon endocytosis, RTKs merge with early/recycling (Rab5+) endosomes. Next, internalized RTKs can be redirected towards the membrane or to (Rab7+) late endosomes that will ultimately fuse with (LAMP1+) lysosomes (Maxfield and McGraw, 2004). Thus, in control or NEGR1-expressing HEK293 cells, we overexpressed GFP-FGFR2 together with mCherry-Rab5, mCherry-Rab7 or mCherry-LAMP1 to label early endosomal compartments, late endosomal compartments or lysosomes, respectively. We treated cultures with NH4Cl to prevent receptor degradation and with FGFb to stimulate FGFR2 endocytosis (Fig. 3K, L and Supplementary Fig. 10). By evaluating the co-localization of GFP fluorescence from GFP-FGFR2 with diverse mCherry reporters, we found that NEGR1 expression increased FGFR2 sorting to (Rab5+) early endosomes and reduced its shuffling to (Rab7+) late endosome or (LAMP1+) lysosomes. In conclusion, our data suggest that NEGR1 spares FGFR2 from lysosomal degradation and directs the receptor towards the recycling compartment.
NEGR1 enhances FGFR2 signalling
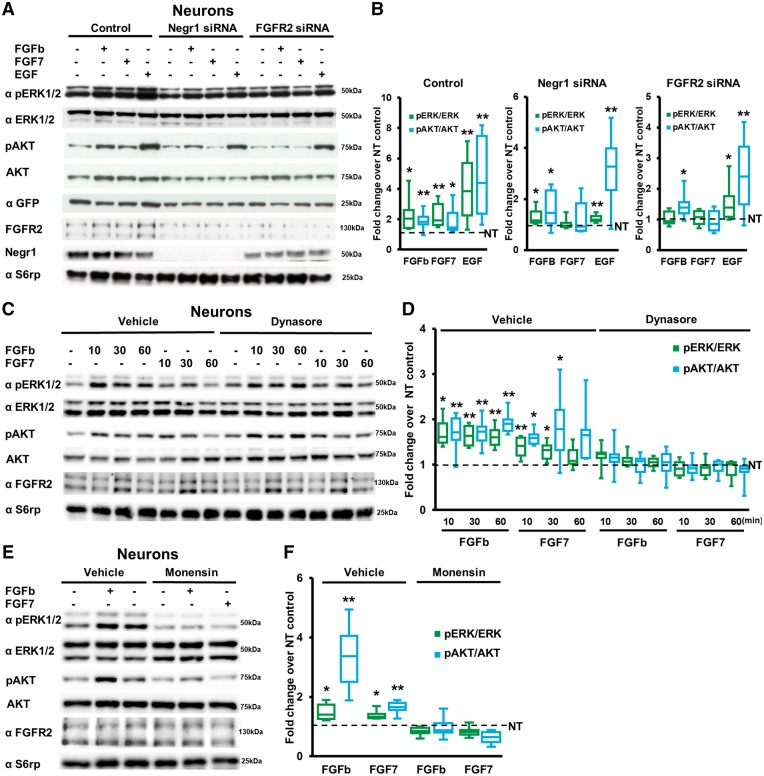
To address whether the Negr1 physical interaction with FGFR2 and regulation of its degradation eventually affects its signalling, we focused on two downstream effectors of the FGFR2 pathway relevant to ASD (i.e. ERK1/2 and AKT; Ornitz and Itoh, 2015). In particular, we infected neuronal cultures at DIV4 with viruses expressing control scrambled siRNA, Negr1 siRNA or Fgfr2 siRNA. We treated them with vehicle (PBS, 10 min), FGFR pan-agonist FGFb (10 ng/ml, 10 min), specific FGFR2 agonist FGF7 (20 ng/ml, 10 min) or specific EGFR agonist EGF (5 ng/ml, 10 min) and evaluated ERK1/2 and AKT phosphorylation. As expected, we found that in control-infected cultures, all three agonists induced a significant increase in ERK and AKT phosphorylation. Notably, Negr1 silencing impaired FGF7-induced increases in phosphorylated-ERK and phosphorylated-AKT, similar to FGFR2 downregulation (Fig. 4A and B).
Figure 4.
NEGR1 influences FGFR2 signalling. (A) Representative immunoblots on protein extracts from lysates of DIV14 cortical neurons infected at DIV4 with viruses bearing plasmids expressing GFP together with control siRNA (Control), Negr1 siRNA or Fgfr2 siRNA and treated at DIV 14 with vehicle, FGFb, FGF7 or EGF. Equal level of infection was monitored by GFP expression, whereas total protein amount was monitored by S6rp staining. (B) Quantification of the ratio between pERK and total ERK, or pAKT and total AKT protein expression. Data are expressed as average percentage of pERK/ERK or pAKT/AKT ratio in drug-treated neurons normalized to vehicle-treated neurons (NT, dotted line). Asterisks indicate statistically significant difference versus vehicle-treated neurons (one sample t-test versus hypothetical value 1: *P < 0.05, **P < 0.01, n = 8 independent experiments). (C) Representative immunoblots on protein extracts from lysates of DIV14 cortical neurons treated with vehicle or dynasore to prevent receptor internalization and then stimulated with FGFb or FGF7 for the time indicated. (D) Quantification of the ratio between pERK and total ERK or pAKT and total AKT protein expression in experiments as in C. Data are expressed as average percentage of pERK/ERK or pAKT/AKT ratio in drug-treated neurons normalized to control vehicle-treated neurons (NT, dotted line); Asterisks indicate statistically significant difference versus vehicle-treated neurons (one sample t-test versus hypothetical value 1: *P < 0.05, **P < 0.01, n = 8 independent experiments). (E) Representative immunoblots on protein extracts from lysates of DIV14 cortical neurons treated with vehicle or monensin to prevent receptor recycling and then stimulated with FGFb or FGF7 for 10 min. (F) Quantification of the ratio between pERK and total ERK or pAKT and total AKT protein expression. Data are expressed as average percentage of pERK/ERK or pAKT/AKT ratio in drug-treated neurons normalized to vehicle treated (NT, dotted line). Asterisks indicate statistically significant difference versus vehicle treated (one sample t-test versus hypothetical value 1: *P < 0.05, **P < 0.01, n = 8 independent experiments).
The above data suggest that Negr1 silencing impairs FGFR2-ERK/AKT signalling. However, our biochemical investigation indicates that NEGR1 affects the intracellular sorting of FGFR2 rather than membrane-exposed FGFR2. Thus, we investigated whether FGFR2 also signals along its intracellular route. FGFRs are internalized in a dynamin-dependent mechanism (Auciello et al., 2013) and only then sorted to recycling endosomes. Thus, we treated neuronal cortical cultures with vehicle (DMSO, 30 min), the dynamin inhibitor dynasore (150 µM, 30 min; Macia et al., 2006) or the receptor-recycling inhibitor monensin (10 µM, 30 min; Felder et al., 1990; Nishimura et al., 2015) and investigated ERK1/2 and AKT signalling upon stimulation with FGFb (10 ng/ml) or FGF7 (20 ng/ml). Both inhibition of dynamin-dependent endocytosis and receptor recycling abolished the increase in ERK1/2 and AKT phosphorylation upon short- or long-term FGFb and FGF7 treatments (Fig. 4C–F).
These data indicate that FGFR2-ERK/AKT signalling occurs along the intracellular route upon internalization from the cell surface. They further suggest that Negr1 downregulation influences FGFR2 signalling by perturbing receptor targeting towards the recycling compartment.
FGFR2 overexpression rescues migration, spine defects and core behaviours related to autism spectrum disorder by Negr1 downregulation
Given the in vitro data showing that Negr1 downregulation affects ERK and AKT signalling by directing FGFR2 to the degradation compartment, we reasoned that overexpression of FGFR2 may rescue the neurodevelopmental and behavioural deficits in Negr1-downregulated mice. To test this hypothesis, we overexpressed FGFR2 together with Negr1 siRNA in neurons migrating to layer II/III by in utero electroporation at E15.5 (Fig. 5A). At P7, we found that cells cotransfected with Negr1 siRNA and Fgfr2 cDNA were able to migrate properly to layer II/III (Fig. 5B and C). Moreover, overexpression of FGFR2 also significantly rescued the defect in positioning of Negr1 siRNA-positive cells within layer II/III (Supplementary Fig. 3). In addition, we found that FGFR2 overexpression prevented the reduction of spine density (but not dendrite morphology, Supplementary Fig. 7A–C and F) in Negr1-downregulated neurons (Fig. 5D and E). Notably, when we overexpressed FGFR2 in the somatosensory cortex, we did not find any effect on neuronal migration, spine density or ultrasonic vocalization (Supplementary Fig. 11).
Figure 5.
FGFR2 overexpression rescues migration, spine density and core behaviours related to ASD caused by Negr1 downregulation in pups in vivo. (A) Experimental protocol. (B) GFP fluorescence in coronal sections of somatosensory cortices at P7 after in utero transfection (at E15.5) with scramble siRNA (control), Negr1 siRNA or co-transfection of Negr1 siRNA together with Fgfr2 cDNA. Left: DAPI counterstaining. Scale bar = 100 μm. (C) Quantification of the number of transfected neurons that did not complete their migration in experiments as in B. Data are expressed as average percentage of the total number of fluorescent cells in the section. Asterisks indicate statistically significant difference (Kruskal-Wallis test; Dunn’s post hoc: ***P < 0.001). Numbers in parentheses indicate total number of processed animals (1 slice/animal). (D) GFP fluorescence in dendrites derived from somatosensory cortical neurons of P20–25 transfected mice. Arrows: dendritic spines. Scale bar = 5 μm. (E) Quantification of spine density in experiments as in D. Data are expressed as average of spine density. Data from control and Negr1 siRNA animals (dotted lines) are taken from Fig. 1G and reported here for comparison. Asterisks indicate statistically significant difference (one-way ANOVA; Holm-Sidak post hoc: **P < 0.01, ***P < 0.001). Numbers in parenthesis indicate total number of processed animals (1–3 slices/animal). (F) Example of a fraction of the recordings of ultrasonic vocalizations emitted by transfected pups upon isolation from their dam and littermates at P4. (G) Quantification of the number of ultrasonic vocalizations during the entire isolation in experiments as in F. Data are expressed as the average of the total number of emitted calls/min. Data from control and Negr1 siRNA animals (dotted lines) are taken from Fig. 2C and reported here for comparison. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Holm-Sidak: **P < 0.01, ***P < 0.001). (H) Latency to paw withdrawal of transfected pups at P9 after placement on a hot plate. Data are expressed as the average time spent on the hot plate until first pain reaction. Data from control and Negr1 siRNA animals (dotted lines) are taken from Fig. 2D and reported here for comparison. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Holm-Sidak: **P < 0.01, ***P < 0.001). (I) Quantification of the time spent performing social sniffing by transfected animals during juvenile play with a stranger peer from a different litter at P20–25. Data are expressed as the average total time that a transfected animal spent at sniffing during the entire trial. Data from control and Negr1 siRNA animals (dotted lines) are taken from Fig. 2E and reported here for comparison. Asterisks indicate statistically significant difference (one-way ANOVA, post hoc Holm-Sidak: **P < 0.01). For all behavioural data, numbers in parenthesis indicate total number of analysed animals, 3–15 litters per experimental case.
Next, we investigated the effect of FGFR2 overexpression on behavioural defects caused by Negr1 downregulation. We found that FGFR2 co-transfection rescued the effect of Negr1 silencing on ultrasonic vocalizations of P4 pups (Fig. 5F and G) and social sniffing of P20–25 mice (Fig. 5I). Conversely, deficits observed at P9 in the hot plate test upon Negr1 downregulation were not rescued by co-transfection with Fgfr2 cDNA (Fig. 5H).
Altogether, these data demonstrate that NEGR1 is required to maintain the function of FGFR2 signalling in regulating neuronal migration, spine density and social communication/interaction behaviours relevant to ASD.
Negr1 −/− mice display phenotypes similar to Negr1 siRNA-electroporated animals
In utero electroporation only allows targeting of a limited number of neurons at a specific developmental time and in a specific brain region. Interestingly, loss of FGFR2 in conditional knockout animals strongly alters cortical development (Ever et al., 2008; Stevens et al., 2010). However, mice with a full FGFR2 knockout die at ∼E10–11 (Xu et al., 1998), making it impossible to study the long-term effects of embryonal loss of FGFR2. Thus, we investigated how constitutive deletion of NEGR1 in Negr1−/− animals (Lee et al., 2012) affects cortical development and core behaviours related to ASD. First, we assessed Negr1−/− animals for neuronal migration deficits. To this aim, we tracked neurons committed to upper cortical layers via Cux1 staining in brain slices obtained from pups at P7. Consistent with the location of Negr1 siRNA neurons in the lower subregions of the upper cortical layers (Supplementary Fig. 3) and Cux1-positive neurons ectopically located at the border between layers IV and V in Negr1 siRNA animals (Fig. 1D and Supplementary Fig. 2), and in agreement with the perturbation in the cortical lamination of conditional ERK knockout mice (Pucilowska et al., 2012), we found that the distribution of Cux1-positive cells was shifted towards layer V in the somatosensory cortex of Negr1−/− animals in comparison to wild-type littermates (Fig. 6A). This resulted in a net increase of the Cux1-positive layer II–IV thickness in the cortex of the Negr1−/− animals (Fig. 6B). Moreover, we also found a significant percentage of Cux1-positive cells ectopically located in cortical layer V of Negr1−/− animals (Fig. 6A and B). Finally, again consistent with what we found in Negr1 siRNA neurons, we also observed a decrease in spine density in Negr1−/− animals (Fig. 6C and D).
Figure 6.
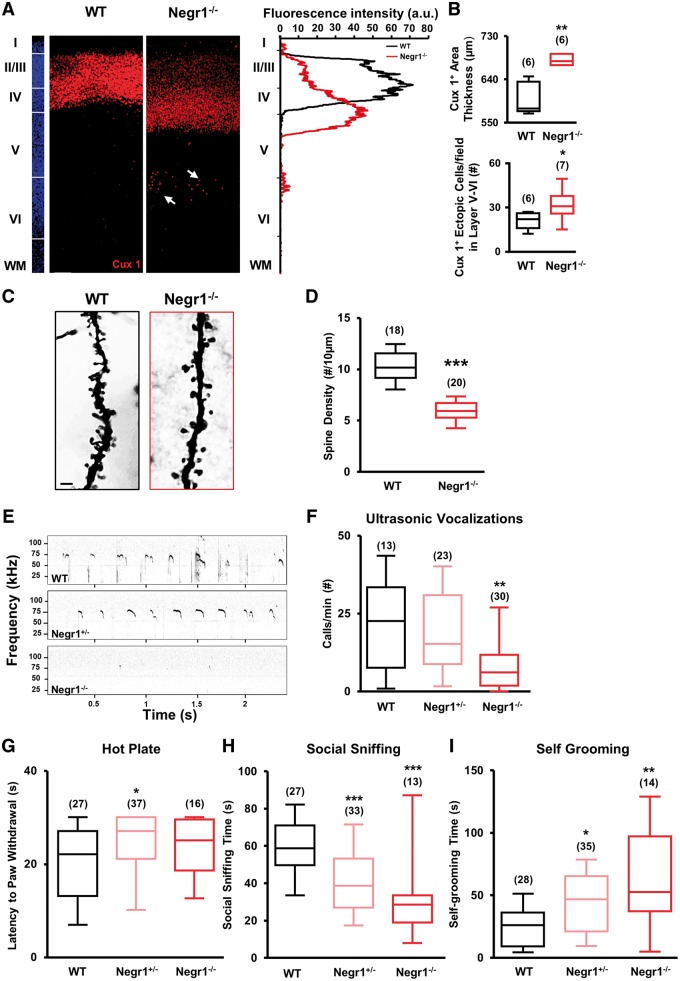
Negr1–/– animals display abnormalities in cortical upper layering and core behaviours related to ASD. (A) Left: Confocal images of Cux1 immunostaining (red) in the somatosensory cortex of wild-type and Negr1−/− littermates at P7. Arrows: ectopic cells. Slices were counterstained with DAPI (blue). Scale bar = 50 µm. Right: Quantification of the fluorescence intensity of Cux1 staining in the images from left, showing a shift towards layer IV in the Negr1−/− animal in comparison with wild-type. (B) Top: Quantification of the thickness of layers II–IV based on the distribution of Cux1-positive cells in wild-type and Negr1−/− animals. Data are expressed as average thickness of Cux1-stained area. Asterisks indicate statistically significant difference versus wild-type (Mann Whitney test: **P < 0.01). Bottom: Quantification of the number of Cux1-positive cells ectopically located outside layers II–IV in wild-type and Negr1−/− animals. Data are expressed as number of Cux1-positive cells in layer V–VI. Asterisks indicate statistically significant difference versus wild-type (unpaired Student’s t-test: *P < 0.05). Numbers in parenthesis indicate total number of processed animals (1 slice/animal). (C) Light-transmitted image of Golgi-Cox stained slices of the somatosensory cortex of wild-type and Negr1−/− littermates at P7. Scale bar = 2 µm. (D) Quantification of spine density in experiments as in C. Data are expressed as average of spine density. Asterisks indicate statistically significant difference (Student’s t-test: ***P < 0.001). Numbers in parenthesis indicate total number of processed cells from two different animals per genotype (3 slices/animal). (E) Example of a fraction of the recordings of ultrasonic vocalizations emitted by P4 pups upon isolation from their dam and littermates. (F) Quantification of the number of ultrasonic vocalizations during the entire isolation in experiments as in C. Data are expressed as the average of the total number of emitted calls/min. Asterisks indicate statistically significant difference versus wild-type (one-way ANOVA against wild-type, post hoc Holm-Sidak: **P < 0.01). (G) Latency to paw withdrawal of P9 pups after placement on a hot plate. Asterisks indicate statistically significant difference versus wild-type (one-way ANOVA against wild-type; post hoc Holm-Sidak: *P < 0.05). (H) Quantification of the time spent performing social sniffing by P20–25 animals during juvenile play with a stranger peer from a different litter. Asterisks indicate statistically significant difference versus wild-type (one-way ANOVA against wild-type; post hoc Holm-Sidak: ***P < 0.001). (I) Quantification of the average total time spent by P20–25 animals on self-grooming behaviour, during 10 min of juvenile play with a stranger mouse from a different litter. Asterisks indicate statistically significant difference versus wild-type (Kruskal-Wallis test against wild-type; post hoc Dunn’s: *P < 0.05, **P < 0.01). For all behavioural data, numbers in parenthesis indicate total number of analysed animals, 4–11 litters per experimental case.
Next, we performed the ultrasonic vocalization test on Negr1+/− and Negr1−/− pups and wild-type littermates at P4. We found that Negr1−/− pups emitted a significantly lower number of ultrasonic vocalizations than wild-type and Negr1+/− pups (Fig. 6E and F), consistent with Negr1 siRNA animals. Furthermore, to assess the sensory deficits in Negr1+/− and Negr1−/−, we performed the hot plate test at P9 and found that, as with Negr1 siRNA animals, both genotypes showed a longer latency to respond to thermal stimuli than wild-type animals, although only Negr1+/− animals reached statistical significance (Fig. 6G). Next, we tested young animals for social and repetitive behaviours during juvenile play, scoring social sniffing and self-grooming time. Consistent with Negr1 siRNA animals, we found that both Negr1+/− and Negr1−/− animals spent less time sniffing the stranger mouse than wild-type animals (Fig. 6H). Moreover, both Negr1+/− and Negr1−/− animals performed self-grooming much more frequently than their wild-type littermates (Fig. 6I).
These data are highly consistent with the results from Negr1-siRNA animals and demonstrate that constitutive deletion of Negr1 is associated with cortical layering defects and impairment in social and sensory alterations relevant to ASD.
Discussion
NEGR1, FGFR2, and downstream ERK and AKT signalling in cortical development
The complex assembly of the six-layered structure of the mammalian cerebral cortex depends on a series of consequent events. These include radial migration of newborn pyramidal neurons from the ventricular zone/subventricular zone and development of their morphological properties. Among the numerous players implicated in these two processes, CAMs and RTK signalling are considered to be crucial (Sobeih and Corfas, 2002; Schmid and Anton, 2003; Rieger et al., 2009; Jossin and Cooper, 2011). Here, we propose a novel mechanism for the interaction between CAMs and RTKs to regulate key steps of cortical development in a cooperative fashion. We observed that in vivo downregulation of either the CAM NEGR1 or the RTK FGFR2 similarly impaired radial migration and dendritic spine densities of pyramidal neurons in the mouse somatosensory cortex. We further identified ERK and AKT as downstream effectors of this interaction, and provided evidence that NEGR1 and FGFR2 not only converge on the same signalling pathways but rather represent a unique molecular complex that possibly cooperatively regulates these downstream pathways.
This hypothesis is supported by the previous literature and our present findings. First, NEGR1 is a glycophosphatidylinositol-anchored cell surface protein, and these kinds of proteins often serve as organizing platforms to regulate RTKs signalling, such as FGF receptors (Citores et al., 1999). Second, CAMs can directly or indirectly bind to growth factor receptors and modulate their signalling (Cavallaro and Dejana, 2011). For example, NCAM interacts with FGFR1, thereby stabilizing it on the membrane and sustaining its intracellular signalling (Francavilla et al., 2009). In addition, OPCML—which, like NEGR1, belongs to the IgLON CAM subgroup—is able to increase FGFR1 degradation while hampering ERK and AKT signalling in cancer cells (McKie et al., 2012). Here, we demonstrate in neurons that NEGR1 favours the recycling of FGFR2 as opposed to its degradation. In particular, we found that NEGR1 downregulation favours FGFR2 lysosomal degradation, while NEGR1 overexpression drives the receptor toward recycling. Notably, NEGR1 downregulation did not affect the levels of FGFR2 exposed at the cell membrane. This suggests that NEGR1 modulates FGFR2 signalling by influencing its intracellular pool rather than the membrane-exposed fraction. While it is understood that membrane receptors signal when specific ligands bind at the plasma membrane, some receptors stay active and continue to signal also along the endocytic pathway (Sorkin and Von Zastrow, 2002). Intriguingly, we found that indeed the majority of FGFR2-related signalling occurs during FGFR2 recycling, and that NEGR1 sustains FGFR2 signalling via regulation of its intracellular trafficking. Moreover, our in vitro data link the NEGR1–FGFR2 interaction to ERK and AKT signalling, which are essential for proper neural migration and neuronal morphological maturation (Pucilowska et al., 2012; Cao et al., 2013; Itoh et al., 2016).
Also our in vivo findings are consistent with the hypothesis that NEGR1 and FGFR2 form a molecular complex that cooperatively regulates downstream signalling. Indeed, FGFR2 overexpression rescued the neuronal migration and spine density phenotypes that we described in NEGR1-downregulated animals. In these experiments, overexpression of Fgfr2 cDNA in utero possibly compensated the impaired FGFR2 signalling because of the increase of FGFR2 degradation by NEGR1 downregulation. On the other hand, overexpression of FGFR2 failed to rescue the dendrite arborization defects observed in NEGR1-knockdown animals. This suggests that NEGR1 and FGFR2 functionally cooperate to control the same signalling pathway (e.g. ERK and AKT activation as our in vitro data would indicate) to regulate neuronal migration and spine formation, but their interaction might be irrelevant for the control of neurite branching in vivo (Huang et al., 2017). This is consistent with our observation that NEGR1 downregulation and FGFR2 downregulation differently altered dendrite branching.
In addition to regulating intracellular signalling in a non-adhesive manner (e.g. in our case, through regulation of FGFR2 trafficking and its signalling), CAMs may also exert their functions by regulating cell-to-cell adhesion (Juliano, 2002) or by a combination of the two (Cavallaro and Dejana, 2011). Interestingly, IgLON family members take part in homophilic as well as heterophilic interactions in cis (Lodge et al., 2000; Reed et al., 2004) and trans (Schafer et al., 2005). In the complex environment of the developing brain—where membranes of different neurons come in close juxtaposition during migration—it is likely that NEGR1 interacts in trans homophilically with NEGR1 and/or in trans heterophilically with FGFR2 located on other neurons. Interestingly, while FGFR2 expression during development is ubiquitous, we found that NEGR1 is prominently expressed in the superior layers of the cortex (mainly in layer 4) at early postnatal stages. NEGR1 may thus be required in the somatosensory cortex to influence migration of neurons committed to upper layers while they move through lower cortical layers, especially through layer IV. Indeed, layer IV is characterized by a high cell density (Schuz and Palm, 1989) and may act as a physical barrier for migrating neurons. Accordingly, we showed that in brain areas devoid of a proper layer IV, such as the prefrontal or the motor cortices, NEGR1 action is dispensable. Our in vitro findings show that NEGR1 expression promotes the formation of cellular clusters, supporting the adhesive role of NEGR1 in trans. Interestingly, co-expression of FGFR2 almost abrogated NEGR1-induced cellular clustering, suggesting that the NEGR1-FGFR2 complex in cis weakens the homophilic Negr1 interaction in trans. We hypothesize that expression of both NEGR1 and FGFR2 on migrating neurons is necessary to allow proper migration: NEGR1 always sustains FGFR2-driven ERK1/2 and AKT signalling, while FGFR2, when forming cis heterophilic complexes with NEGR1, prevents NEGR1 itself from trans adhesive homophilic binding. Thus, the driving force fuelled by ERK1/2 and AKT signalling together with reduced cell-to-cell adhesion may assure that migrating neurons pass the layer IV block and eventually reach upper layers. However, FGFR2 can bind to a variety of other membrane molecules and therefore regulate a myriad of potentially interactive signalling pathways to generate diverse cellular responses. This is in line with our results where we showed that knocking-down FGFR2 elicits a stronger phenotype than knocking-down NEGR1 (e.g. morphological maturation phenotype). Indeed, upon knocking-down of FGFR2, we may not only affect its interaction with NEGR1, but also with other molecules involved in its downstream signalling (Williams et al., 1994; Lonic et al., 2013).
NEGR1, FGFR2, and downstream ERK and AKT signalling in neurodevelopmental disorders
In agreement with the role of CAMs and RTKs in cortical development, a wide variety of genetic variants of these molecules or alterations of their signalling have been associated with a number of neurodevelopmental disorders (Wang et al., 2017a), including schizophrenia (Zhang et al., 2015), Rett syndrome (Miyake et al., 2011; Mellios et al., 2018), attention deficit hyperactivity disorder (ADHD; Alasehirli et al., 2015), Tourette syndrome (Clarke et al., 2012), fragile X syndrome (Wang et al., 2012, 2017a; Kovacs et al., 2014) and ASD (Pinto et al., 2010; Stewart, 2015). In particular, NEGR1 has been recently listed among genes influencing intelligence in humans (Sniekers et al., 2017), schizophrenia (Karis et al., 2018), dyslexia (Veerappa et al., 2013) and ASD (Marshall et al., 2008; Michaelson et al., 2012; Pinto et al., 2014, 2010). Notably, a microdeletion of the NEGR1 gene was described in two siblings that presented cognitive disabilities, ADHD, speech problems and features of autism in one of them (Genovese et al., 2015). Moreover, gene-association studies have also implicated FGFR2 as a candidate gene in ASD (Wentz et al., 2014; Coci et al., 2017). Interestingly, FGFR2 mutations are causative of syndromic intellectual disabilities, such as Crouzon syndrome (Fernandes et al., 2016) and Apert syndrome whose patients may also suffer from ASD (Morey-Canellas et al., 2003; Koca, 2016). Finally, alterations in ERK and AKT signalling have been widely implicated in schizophrenia and intellectual disability (Hirayama-Kurogi et al., 2017; Wang et al., 2017a), and both an increase (Kalkman, 2012; Faridar et al., 2014; Pucilowska et al., 2015; Onore et al., 2017) as well as a decrease of ERK or AKT signalling (Satoh et al., 2011; Nicolini et al., 2015; Yufune et al., 2015) have been described in ASD. Accordingly, we described here the impact of NEGR1 and FGFR2 downregulation on brain development and behavioural abnormalities related to ASD. Moreover, our data show that NEGR1 and FGFR2 together impinge on ERK/AKT signalling, supporting the accumulating evidence indicating each of these two pathways as one possible converging downstream signalling among the thousands of genes (many encoding CAMs and RTKs) and molecular mechanisms involved in ASD aetiology (Pinto et al., 2010, 2014; Lanz et al., 2013; Subramanian et al., 2015; Wen et al., 2016; Magdalon et al., 2017). Nevertheless, whereas the majority of previous studies mostly provided in silico evidence, here, we provide in vivo experimental evidence in mice together with in vitro mechanistic insights regarding the possible involvement of the ERK and AKT pathways in ASD aetiology.
However, how can defective brain development relate to aberrant behavioural phenotypes? Aberrant migration and morphological maturation can result in cell death, ectopic positioning of neurons and/or abnormal synaptic connections which, eventually, can cause neuronal miswiring. Accordingly, in a number of neurodevelopmental disorders, such as intellectual disability (Allen et al., 1998), epilepsy (Penagarikano et al., 2011; Stouffer et al., 2016), schizophrenia (Owen et al., 2016) and ASD (Wegiel et al., 2010), defective radial migration and morphological maturation have been described. In particular, case studies show that individuals with ASD can display heterotopias and focal laminar disorganization (Bailey et al., 1998; Hutsler et al., 2007) with patches of abnormal laminar cortical cytoarchitecture (Stoner et al., 2014). Consistently, a large number of genetic or drug-induced animal models of ASD show defective neuronal migration (Kuwagata et al., 2009; Penagarikano et al., 2011; Hori and Hoshino, 2017; Dang et al., 2018). Interestingly, defects in dendritic arborization have also been reported in ASD patients with both stunted (Raymond et al., 1996) and increased dendritic branching (Copf, 2016), which is again reflected in animal studies (Snow et al., 2008; Pathania et al., 2014; Hullinger et al., 2016). Finally, defective dendritic spine formation has been extensively reported in both ASD patients and animal models (Hatanaka et al., 2017; Stephenson et al., 2017; Wang et al., 2017b). Specifically, a decreased number of mature dendritic spines, increased number of immature dendritic spines, and overall increase of spine numbers have been all observed in post-mortem tissues (Hutsler and Zhang, 2010; Martinez-Cerdeno, 2017) and ASD animal models (Varghese et al., 2017). Moreover mutations in genes that are key regulators of spine formation lead to ASD-like behaviours in mice (Monteiro and Feng, 2017; Stephenson et al., 2017). Here, we provide evidence that defective neuronal migration and morphological maturation in the somatosensory cortex are associated with deficits in core behaviours related to ASD in mice. Notably, we showed that FGFR2 overexpression is capable of rescuing neuronal migration, spine density and the behavioural phenotypes, but not dendritic arborization defects due to Negr1 silencing. This evidence strengthens the causality among defective migration, spine density and ASD behaviours and suggests that, at least in our model, correct dendritic arborization is dispensable. On the other hand, neurogenesis impairment has also been observed in both patients with ASD and animal models (Packer, 2016) and FGF signalling (including ERK and AKT signalling) has been implicated in cell proliferation (Ohkubo et al., 2004; Fournier et al., 2012; Kang and Hebert, 2015). In addition, LSAMP and NTM (two members of the IgLON superfamily) regulate cell proliferation and apoptosis (Singh et al., 2018a), and FGFR2 signalling is needed for cell survival during brain development (Saarimaki-Vire et al., 2007; Meier et al., 2014). Therefore, we cannot exclude that defective neurogenesis or neuronal survival may also contribute to the aberrant behaviours that we describe here in NEGR1- and FGFR2-downregulated mice.
Defective neuronal migration and morphological maturation may cause neuronal miswiring and eventually contribute to aberrant behavioural phenotypes in NEGR1- and FGFR2-downregulated animals. Accordingly, Negr1−/− animals have abnormal neuronal wiring in the entorhinal cortex and present social deficits (Singh et al., 2018b). Moreover, many genes associated with ASD display cellular- and laminar-specific enrichments in glutamatergic projection neurons located in superficial layers II–IV of the cortex, as we show here for NEGR1 (Parikshak et al., 2013; Willsey et al., 2013). Interestingly, disruption of cortical projection neurons has been hypothesized to lead to improper long-range connectivity, ultimately resulting in ASD phenotypes (Hutsler and Zhang, 2010; Kana et al., 2011). In this context, the aberrant communication, sociability and social recognition that we described in animals downregulated for NEGR1 or FGFR2 specifically in the somatosensory cortex can be explained. Indeed, whereas the reduction of the number of ultrasonic vocalizations could be directly ascribed to the defects in the somatosensory cortex, which is suggested to be the murine analogue of the cortical language area in humans (Sia et al., 2013), social behaviours in juvenile mice are not typically described as being dependent on somatosensory cortex activity. Nevertheless, cortical regions containing ectopic and morphologically defective cells can become part of aberrant neuronal circuits, as already described for mice with focal heterotopia due to downregulation of genes other than Negr1 and Fgfr2 (Ishii et al., 2015). Interestingly, these other mice exhibit cognitive deficits and increased seizure susceptibility, but different from NEGR1- and FGFR2-downregulated animals, they show no major defects in behavioural paradigms for ASD-related behaviours (Ishii et al., 2015). This may indicate that different mutations and/or different migration phenotypes may lead to dissimilar behavioural outcomes. Notably, patients with a range of psychiatric symptoms also often present aberrant neuronal migration in regions other than those allegedly responsible for the defective behaviours (Muraki and Tanigaki, 2015; Reiner et al., 2016).
Alternatively, besides circuit miswiring, alterations in sensory processing may contribute, at least in part, to abnormal social behaviour by hampering the interaction of the defective animals with their environment, as already suggested in autistic patients (Hilton et al., 2010; Suarez, 2012). Nevertheless, the fact that overexpression of FGFR2 was not able to rescue aberrant sensory perception (i.e. pain sensitivity), but was able to rescue the social interaction deficits in Negr1-siRNA animals does not support this idea. Moreover, the latter findings indicate that NEGR1 and FGFR2 probably do not converge on the pathways responsible for pain perception. On the other hand, our data on decreased pain sensitivity in Negr1- or FGFR2-downregulated animals are in agreement with reports describing painless self-injurious acts associated with somatosensory cortex defects in children with ASD (Duerden et al., 2014; Gu et al., 2018). Moreover, our results on pain sensitivity upon FGFR2 downregulation are consistent with the notion that spinal cord-derived FGF7 (a specific agonist of FGFR2) is necessary for nociceptive pain perception and that Fgf7−/− animals show decreased pain sensitivity (Liu et al., 2015).
Interestingly, both NEGR1-downregulated animals and Negr1−/− mice showed an increase in self-grooming behaviour, although in NEGR1-downregulated animals, the trend did not reach statistical significance. By contrast, FGFR2 downregulation decreased this behaviour. The latter effect is in agreement with recent findings showing hyperactive ERK signalling to be a cause of obsessive compulsive disorder-like (i.e. self-grooming) behaviours in mice (Ullrich et al., 2018). Nevertheless, since we tested self-grooming in a social context (Silverman et al., 2010), we cannot exclude that the behaviour that we observed also has a social meaning and is not entirely obsessive/repetitive in nature.
In summary, although a causative correlation between defective brain development and aberrant behavioural outcomes in neurodevelopmental disorders has been long inferred, experimental evidence in animal models is still not strong. In this context, our data showed that FGFR2 overexpression in Negr1-downregulated animals was able to rescue migration as well as spine density (but not dendrite morphology) together with communication as well as social (but not repetitive) behaviours: therefore, we experimentally demonstrated a link between specific brain defects and specific aberrant behaviours relevant to ASD.
Conclusion
Our study suggests a role for the interaction of CAMs and RTKs in brain development and neurodevelopmental disorders. In particular, in spectrum disorders, such as ASD, in which thousands of genes have been implicated and there is still no pharmacological intervention available, our results indicate the NEGR1–FGFR2 complex and downstream convergent ERK-AKT signalling pathways as potential targets for early pharmacological intervention.
Supplementary Material
Acknowledgements
We acknowledge Giacomo Pruzzo for technical support with the tripolar electrode for in utero electroporation and Monica Morini and all the animal facility staff for their support.
Glossary
Abbreviations
- AKT
protein kinase B
- ASD
autism spectrum disorder
- CAM
cell adhesion molecule
- ERK
extracellular signal-regulated kinase
- RTK
receptor tyrosine kinase
Funding
The support of Telethon-Italy (grants TCP15021 and GGP13187 to L.C., and TDPG00514 to G.P.), and CARIPLO foundation (grant 2013 0879 to L.C.) is acknowledged. This project has received partial funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 725563 to L.C.).
References
- Alasehirli B, Oguz E, Gokcen C, Erbagci AB, Orkmez M, Demiryurek AT. Relationship between soluble intercellular adhesion molecules and attention-deficit/hyperactivity disorder. Int J Psychiatry Med 2015; 50: 238–47. [DOI] [PubMed] [Google Scholar]
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA et al. . PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet 1998; 20: 25–30. [DOI] [PubMed] [Google Scholar]
- Auciello G, Cunningham DL, Tatar T, Heath JK, Rappoport JZ. Regulation of fibroblast growth factor receptor signalling and trafficking by Src and Eps8. J Cell Sci 2013; 126: 613–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M et al. . A clinicopathological study of autism. Brain 1998; 121 (Pt 5): 889–905. [DOI] [PubMed] [Google Scholar]
- Cao FJ, Zhang X, Liu T, Li XW, Malik M, Feng SQ. Up-regulation of Ras/Raf/ERK1/2 signaling in the spinal cord impairs neural cell migration, neurogenesis, synapse formation, and dendritic spine development. Chin Med J 2013; 126: 3879–85. [PubMed] [Google Scholar]
- Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R et al. . A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet 2012; 131: 565–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U, Dejana E. Adhesion molecule signalling: not always a sticky business. Nat Rev Mol Cell Biol 2011; 12: 189–97. [DOI] [PubMed] [Google Scholar]
- Citores L, Wesche J, Kolpakova E, Olsnes S. Uptake and intracellular transport of acidic fibroblast growth factor: evidence for free and cytoskeleton-anchored fibroblast growth factor receptors. Mol Biol Cell 1999; 10: 3835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RA, Lee S, Eapen V. Pathogenetic model for Tourette syndrome delineates overlap with related neurodevelopmental disorders including Autism. Transl Psychiatry 2012; 2: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coci EG, Auhuber A, Langenbach A, Mrasek K, Riedel J, Leenen A et al. . Novel unbalanced translocations affecting the long arms of chromosomes 10 and 22 cause complex syndromes with very severe neurodevelopmental delay, speech impairment, autistic behavior, and epilepsy. Cytogenet Genome Res 2017; 151: 171–8. [DOI] [PubMed] [Google Scholar]
- Copf T. Impairments in dendrite morphogenesis as etiology for neurodevelopmental disorders and implications for therapeutic treatments. Neurosci Biobehav Rev 2016; 68: 946–78. [DOI] [PubMed] [Google Scholar]
- Corti V, Sanchez-Ruiz Y, Piccoli G, Bergamaschi A, Cannistraci CV, Pattini L et al. . Protein fingerprints of cultured CA3-CA1 hippocampal neurons: comparative analysis of the distribution of synaptosomal and cytosolic proteins. BMC Neurosci 2008; 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dal Maschio M, Ghezzi D, Bony G, Alabastri A, Deidda G, Brondi M et al. . High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat Commun 2012; 3: 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang T, Duan WY, Yu B, Tong DL, Cheng C, Zhang YF et al. . Autism-associated Dyrk1a truncation mutants impair neuronal dendritic and spine growth and interfere with postnatal cortical development. Mol Psychiatry 2018; 23: 747–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorey TM, Sahbaie P, Hashemi E, Li WW, Salehi A, Clark DJ. Somatosensory and sensorimotor consequences associated with the heterozygous disruption of the autism candidate gene, Gabrb3. Behav Brain Res 2011; 216: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerden EG, Card D, Roberts SW, Mak-Fan KM, Chakravarty MM, Lerch JP et al. . Self-injurious behaviours are associated with alterations in the somatosensory system in children with autism spectrum disorder. Brain Struct Funct 2014; 219: 1251–61. [DOI] [PubMed] [Google Scholar]
- Ever L, Zhao RJ, Eswarakumar VP, Gaiano N. Fibroblast growth factor receptor 2 plays an essential role in telencephalic progenitors. Dev Neurosci 2008; 30: 306–18. [DOI] [PubMed] [Google Scholar]
- Faridar A, Jones-Davis D, Rider E, Li J, Gobius I, Morcom L et al. . Mapk/Erk activation in an animal model of social deficits shows a possible link to autism. Mol Autism 2014; 5: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S, Miller K, Moehren G, Ullrich A, Schlessinger J, Hopkins CR. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell 1990; 61: 623–34. [DOI] [PubMed] [Google Scholar]
- Fernandes MB, Maximino LP, Perosa GB, Abramides DV, Passos-Bueno MR, Yacubian-Fernandes A. Apert and Crouzon syndromes-cognitive development, brain abnormalities, and molecular aspects. Am J Med Genet A 2016; 170: 1532–7. [DOI] [PubMed] [Google Scholar]
- Fournier NM, Lee B, Banasr M, Elsayed M, Duman RS. Vascular endothelial growth factor regulates adult hippocampal cell proliferation through MEK/ERK- and PI3K/Akt-dependent signaling. Neuropharmacology 2012; 63: 642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla C, Cattaneo P, Berezin V, Bock E, Ami D, de Marco A et al. . The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J Cell Biol 2009; 187: 1101–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu N, Miyata S, Kumanogoh H, Shigeta M, Hamada K, Endo Y et al. . Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J Biol Chem 1999; 274: 8224–30. [DOI] [PubMed] [Google Scholar]
- Garini Y, Vermolen BJ, Young IT. From micro to nano: recent advances in high-resolution microscopy. Curr Opin Biotechnol 2005; 16: 3–12. [DOI] [PubMed] [Google Scholar]
- Genovese A, Cox DM, Butler MG. Partial deletion of chromosome 1p31.1 including only the neuronal growth regulator 1 gene in two siblings. J Pediatr Genet 2015; 4: 23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil OD, Zhang L, Chen S, Ren YQ, Pimenta A, Zanazzi G et al. . Complementary expression and heterophilic interactions between IgLON family members neurotrimin and LAMP. J Neurobiol 2002; 51: 190–204. [DOI] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Klanfer N, Bryant CD, Evans CJ et al. . Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm Behav 2008; 53: 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoolparsadh A, Sutton GJ, Charamko A, Green NF, Pardy CJ, Voineagu I. Searching for convergent pathways in autism spectrum disorders: insights from human brain transcriptome studies. Cell Mol Life Sci 2016; 73: 4517–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zhou TJ, Anagnostou E, Soorya L, Kolevzon A, Hof PR, Fan J. Heightened brain response to pain anticipation in high-functioning adults with autism spectrum disorder. Eur J Neurosci 2018; 47: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Kabuta T, Wada K. Disturbance in maternal environment leads to abnormal synaptic instability during neuronal circuitry development. Front Neurosci 2017; 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Nitta N, Flubacher A, Muller M, Fahrner A, Kirsch M et al. . Reelin deficiency and displacement of mature neurons, but not neurogenesis, underlie the formation of granule cell dispersion in the epileptic hippocampus. J Neurosci 2006; 26: 4701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton CL, Harper JD, Kueker RH, Lang AR, Abbacchi AM, Todorov A et al. . Sensory responsiveness as a predictor of social severity in children with high functioning autism spectrum disorders. J Autism Dev Disord 2010; 40: 937–45. [DOI] [PubMed] [Google Scholar]
- Hirayama-Kurogi M, Takizawa Y, Kunii Y, Matsumoto J, Wada A, Hino M et al. . Downregulation of GNA13-ERK network in prefrontal cortex of schizophrenia brain identified by combined focused and targeted quantitative proteomics. J Proteomics 2017; 158: 31–42. [DOI] [PubMed] [Google Scholar]
- Hori K, Hoshino M. Neuronal migration and AUTS2 syndrome. Brain Sci 2017; 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Lynn Miskus M, Lu HC. FGF-FGFR mediates the activity-dependent dendritogenesis of layer IV neurons during barrel formation. J Neurosci 2017; 37: 12094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullinger R, Li M, Wang J, Peng Y, Dowell JA, Bomba-Warczak E et al. . Increased expression of AT-1/SLC33A1 causes an autistic-like phenotype in mice by affecting dendritic branching and spine formation. J Exp Med 2016; 213: 1267–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussman JP, Chung RH, Griswold AJ, Jaworski JM, Salyakina D, Ma D et al. . A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism 2011; 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Love T, Zhang H. Histological and magnetic resonance imaging assessment of cortical layering and thickness in autism spectrum disorders. Biol Psychiatry 2007; 61: 449–57. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res 2010; 1309: 83–94. [DOI] [PubMed] [Google Scholar]
- Ishii K, Kubo K, Endo T, Yoshida K, Benner S, Ito Y et al. . Neuronal heterotopias affect the activities of distant brain areas and lead to behavioral deficits. J Neurosci 2015; 35: 12432–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Higuchi M, Oishi K, Kishi Y, Okazaki T, Sakai H et al. . PDK1-Akt pathway regulates radial neuronal migration and microtubules in the developing mouse neocortex. Proc Natl Acad Sci USA 2016; 113: E2955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci 2011; 14: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu Rev Pharmacol Toxicol 2002; 42: 283–23. [DOI] [PubMed] [Google Scholar]
- Kalkman HO. Potential opposite roles of the extracellular signal-regulated kinase (ERK) pathway in autism spectrum and bipolar disorders. Neurosci Biobehav Rev 2012; 36: 2206–13. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, Fentress JC. Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat Rev Neurosci 2016; 17: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana RK, Libero LE, Moore MS. Disrupted cortical connectivity theory as an explanatory model for autism spectrum disorders. Phys Life Rev 2011; 8: 410–37. [DOI] [PubMed] [Google Scholar]
- Kang W, Hebert JM. FGF signaling is necessary for neurogenesis in young mice and sufficient to reverse its decline in old mice. J Neurosci 2015; 35: 10217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karis K, Eskla KL, Kaare M, Taht K, Tuusov J, Visnapuu T et al. . Altered expression profile of IgLON family of neural cell adhesion molecules in the dorsolateral prefrontal cortex of schizophrenic patients. Front Mol Neurosci 2018; 11: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koca TT. Apert syndrome: a case report and review of the literature. North Clin Istanb 2016; 3: 135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs T, Bansagi B, Kelemen O, Keri S. Neuregulin 1-induced AKT and ERK phosphorylation in patients with fragile X syndrome (FXS) and intellectual disability associated with obstetric complications. J Mol Neurosci 2014; 54: 119–24. [DOI] [PubMed] [Google Scholar]
- Kuwagata M, Ogawa T, Shioda S, Nagata T. Observation of fetal brain in a rat valproate-induced autism model: a developmental neurotoxicity study. Int J Dev Neurosci 2009; 27: 399–405. [DOI] [PubMed] [Google Scholar]
- Lanz TA, Guilmette E, Gosink MM, Fischer JE, Fitzgerald LW, Stephenson DT et al. . Transcriptomic analysis of genetically defined autism candidate genes reveals common mechanisms of action. Mol Autism 2013; 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AW, Hengstler H, Schwald K, Berriel-Diaz M, Loreth D, Kirsch M et al. . Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One 2012; 7: e41537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wu QF, Li JY, Liu XJ, Li KC, Zhong YQ et al. . Fibroblast growth factor 7 is a nociceptive modulator secreted via large dense-core vesicles. J Mol Cell Biol 2015; 7: 466–75. [DOI] [PubMed] [Google Scholar]
- Lodge AP, Howard MR, McNamee CJ, Moss DJ. Co-localisation, heterophilic interactions and regulated expression of IgLON family proteins in the chick nervous system. Brain Res Mol Brain Res 2000; 82: 84–94. [DOI] [PubMed] [Google Scholar]
- Lonic A, Powell JA, Kong Y, Thomas D, Holien JK, Truong N et al. . Phosphorylation of serine 779 in fibroblast growth factor receptor 1 and 2 by protein kinase C(epsilon) regulates Ras/mitogen-activated protein kinase signaling and neuronal differentiation. J Biol Chem 2013; 288: 14874–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 2006; 10: 839–50. [DOI] [PubMed] [Google Scholar]
- Magdalon J, Sanchez-Sanchez SM, Griesi-Oliveira K, Sertie AL. Dysfunctional mTORC1 signaling: a convergent mechanism between syndromic and nonsyndromic forms of autism spectrum disorder? Int J Mol Sci 2017; 18: 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci 1992; 103 (Pt 3): 857–62. [DOI] [PubMed] [Google Scholar]
- Mansukhani A, Bellosta P, Sahni M, Basilico C. Signaling by fibroblast growth factors (FGF) and fibroblast growth factor receptor 2 (FGFR2)-activating mutations blocks mineralization and induces apoptosis in osteoblasts. J Cell Biol 2000; 149: 1297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J et al. . Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 2008; 82: 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V. Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev Neurobiol 2017; 77: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol 2004; 5: 121–32. [DOI] [PubMed] [Google Scholar]
- McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav 2008; 7: 152–63. [DOI] [PubMed] [Google Scholar]
- McKie AB, Vaughan S, Zanini E, Okon IS, Louis L, de Sousa C et al. . The OPCML tumor suppressor functions as a cell surface repressor-adaptor, negatively regulating receptor tyrosine kinases in epithelial ovarian cancer. Cancer Discov 2012; 2: 156–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Giesert F, Delic S, Faus-Kessler T, Matheus F, Simeone A et al. . FGF/FGFR2 signaling regulates the generation and correct positioning of Bergmann glia cells in the developing mouse cerebellum. PLoS One 2014; 9: e101124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK et al. . MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry 2018; 23: 1051–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X et al. . Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 2012; 151: 1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Hirasawa T, Soutome M, Itoh M, Goto Y, Endoh K et al. . The protocadherins, PCDHB1 and PCDH7, are regulated by MeCP2 in neuronal cells and brain tissues: implication for pathogenesis of Rett syndrome. BMC Neurosci 2011; 12: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci 2017; 18: 147–57. [DOI] [PubMed] [Google Scholar]
- Morey-Canellas J, Sivagamasundari U, Barton H. A case of autism in a child with Apert’s syndrome. Eur Child Adolesc Psychiatry 2003; 12: 100–2. [DOI] [PubMed] [Google Scholar]
- Muraki K, Tanigaki K. Neuronal migration abnormalities and its possible implications for schizophrenia. Front Neurosci 2015; 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A et al. . Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 2012; 485: 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolini C, Ahn Y, Michalski B, Rho JM, Fahnestock M. Decreased mTOR signaling pathway in human idiopathic autism and in rats exposed to valproic acid. Acta Neuropathol Commun 2015; 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Takiguchi S, Ito S, Itoh K. EGFstimulated AKT activation is mediated by EGFR recycling via an early endocytic pathway in a gefitinibresistant human lung cancer cell line. Int J Oncol 2015; 46: 1721–9. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Uchida AO, Shin D, Partanen J, Vaccarino FM. Fibroblast growth factor receptor 1 is required for the proliferation of hippocampal progenitor cells and for hippocampal growth in mouse. J Neurosci 2004; 24: 6057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onore C, Yang H, Van de Water J, Ashwood P. Dynamic Akt/mTOR signaling in children with autism spectrum disorder. Front Pediatr 2017; 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 2015; 4: 215–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet 2016; 388: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer A. Neocortical neurogenesis and the etiology of autism spectrum disorder. Neurosci Biobehav Rev 2016; 64: 185–95. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V et al. . Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013; 155: 1008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry 2014; 4: e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H et al. . Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 2011; 147: 235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L et al. . Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 2014; 94: 677–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R et al. . Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda F, Piccoli G. The IgLON family member Negr1 promotes neuronal arborization acting as soluble factor via FGFR2. Front Mol Neurosci 2015; 8: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischedda F, Szczurkowska J, Cirnaru MD, Giesert F, Vezzoli E, Ueffing M et al. . A cell surface biotinylation assay to reveal membrane-associated neuronal cues: Negr1 regulates dendritic arborization. Mol Cell Proteomics 2014; 13: 733–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucilowska J, Puzerey PA, Karlo JC, Galan RF, Landreth GE. Disrupted ERK signaling during cortical development leads to abnormal progenitor proliferation, neuronal and network excitability and behavior, modeling human neuro-cardio-facial-cutaneous and related syndromes. J Neurosci 2012; 32: 8663–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucilowska J, Vithayathil J, Tavares EJ, Kelly C, Karlo JC, Landreth GE. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J Neurosci 2015; 35: 3190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol 1996; 91: 117–19. [DOI] [PubMed] [Google Scholar]
- Reed J, McNamee C, Rackstraw S, Jenkins J, Moss D. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J Cell Sci 2004; 117: 3961–73. [DOI] [PubMed] [Google Scholar]
- Reiner O, Karzbrun E, Kshirsagar A, Kaibuchi K. Regulation of neuronal migration, an emerging topic in autism spectrum disorders. J Neurochem 2016; 136: 440–56. [DOI] [PubMed] [Google Scholar]
- Rieger S, Senghaas N, Walch A, Koster RW. Cadherin-2 controls directional chain migration of cerebellar granule neurons. PLoS Biol 2009; 7: e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimaki-Vire J, Peltopuro P, Lahti L, Naserke T, Blak AA, Vogt Weisenhorn DM et al. . Fibroblast growth factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci 2007; 27: 8581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz R, Ferraro GB, Fournier AE. IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J Biol Chem 2015; 290: 4330–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y, Endo S, Nakata T, Kobayashi Y, Yamada K, Ikeda T et al. . ERK2 contributes to the control of social behaviors in mice. J Neurosci 2011; 31: 11953–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Crawley J, Ricceri L. Ultrasonic vocalizations: a tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci Biobehav Rev 2009; 33: 508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer LH, Schuster D, Schaffer J. Structured illumination microscopy: artefact analysis and reduction utilizing a parameter optimization approach. J Microsc 2004; 216: 165–74. [DOI] [PubMed] [Google Scholar]
- Schafer M, Brauer AU, Savaskan NE, Rathjen FG, Brummendorf T. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol Cell Neurosci 2005; 29: 580–90. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Anton ES. Role of integrins in the development of the cerebral cortex. Cereb Cortex 2003; 13: 219–24. [DOI] [PubMed] [Google Scholar]
- Schuller AC, Ahmed Z, Levitt JA, Suen KM, Suhling K, Ladbury JE. Indirect recruitment of the signalling adaptor Shc to the fibroblast growth factor receptor 2 (FGFR2). Biochem J 2008; 416: 189–99. [DOI] [PubMed] [Google Scholar]
- Schuz A, Palm G. Density of neurons and synapses in the cerebral cortex of the mouse. J Comp Neurol 1989; 286: 442–55. [DOI] [PubMed] [Google Scholar]
- Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science 2013; 342: 987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci 2010; 11: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Lillevali K, Gilbert SF, Bregin A, Narvik J, Jayaram M et al. . The combined impact of IgLON family proteins Lsamp and Neurotrimin on developing neurons and behavioral profiles in mouse. Brain Res Bull 2018a; 140: 5–18. [DOI] [PubMed] [Google Scholar]
- Singh K, Loreth D, Pottker B, Hefti K, Innos J, Schwald K et al. . Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated Negr1 gene. Front Mol Neurosci 2018b; 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E et al. . Genome-wide association meta-analysis of 78 308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 2017; 49: 1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow WM, Hartle K, Ivanco TL. Altered morphology of motor cortex neurons in the VPA rat model of autism. Dev Psychobiol 2008; 50: 633–9. [DOI] [PubMed] [Google Scholar]
- Sobeih MM, Corfas G. Extracellular factors that regulate neuronal migration in the central nervous system. Int J Dev Neurosci 2002; 20: 349–57. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol 2002; 3: 600–14. [DOI] [PubMed] [Google Scholar]
- Stephenson JR, Wang X, Perfitt TL, Parrish WP, Shonesy BC, Marks CR et al. . A novel human CAMK2A mutation disrupts dendritic morphology and synaptic transmission, and causes ASD-related behaviors. J Neurosci 2017; 37: 2216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens HE, Smith KM, Maragnoli ME, Fagel D, Borok E, Shanabrough M et al. . Fgfr2 is required for the development of the medial prefrontal cortex and its connections with limbic circuits. J Neurosci 2010; 30: 5590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart LT. Cell adhesion proteins and the pathogenesis of autism spectrum disorders. J Neurophysiol 2015; 113: 1283–6. [DOI] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S et al. . Patches of disorganization in the neocortex of children with autism. N Engl J Med 2014; 370: 1209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer MA, Golden JA, Francis F. Neuronal migration disorders: focus on the cytoskeleton and epilepsy. Neurobiol Dis 2016; 92: 18–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MA. Sensory processing in children with autism spectrum disorders and impact on functioning. Pediatr Clin North Am 2012; 59: 203–14, xii–xiii. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Timmerman CK, Schwartz JL, Pham DL, Meffert MK. Characterizing autism spectrum disorders by key biochemical pathways. Front Neurosci 2015; 9: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurkowska J, Cwetsch AW, dal Maschio M, Ghezzi D, Ratto GM, Cancedda L. Targeted in vivo genetic manipulation of the mouse or rat brain by in utero electroporation with a triple-electrode probe. Nat Protoc 2016; 11: 399–412. [DOI] [PubMed] [Google Scholar]
- Tarkkonen KM, Nilsson EM, Kahkonen TE, Dey JH, Heikkila JE, Tuomela JM et al. . Differential roles of fibroblast growth factor receptors (FGFR) 1, 2 and 3 in the regulation of S115 breast cancer cell growth. PLoS One 2012; 7: e49970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich M, Weber M, Post AM, Popp S, Grein J, Zechner M et al. . OCD-like behavior is caused by dysfunction of thalamo-amygdala circuits and upregulated TrkB/ERK-MAPK signaling as a result of SPRED2 deficiency. Mol Psychiatry 2018; 23: 444–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemori H, Sanes JR. Signal regulatory proteins (SIRPS) are secreted presynaptic organizing molecules. J Biol Chem 2008; 283: 34053–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese M, Keshav N, Jacot-Descombes S, Warda T, Wicinski B, Dickstein DL et al. . Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol 2017; 134: 537–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerappa AM, Saldanha M, Padakannaya P, Ramachandra NB. Family-based genome-wide copy number scan identifies five new genes of dyslexia involved in dendritic spinal plasticity. J Hum Genet 2013; 58: 539–47. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S et al. . Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011; 474: 380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhou K, Fu Z, Yu D, Huang H, Zang X et al. . Brain development and Akt signaling: the crossroads of signaling pathway and neurodevelopmental diseases. J Mol Neurosci 2017a; 61: 379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li H, Takumi T, Qiu Z, Xu X, Yu X et al. . Distinct defects in spine formation or pruning in two gene duplication mouse models of autism. Neurosci Bull 2017b; 33: 143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Snape M, Klann E, Stone JG, Singh A, Petersen RB et al. . Activation of the extracellular signal-regulated kinase pathway contributes to the behavioral deficit of fragile x-syndrome. J Neurochem 2012; 121: 672–9. [DOI] [PubMed] [Google Scholar]
- Wegiel J, Kuchna I, Nowicki K, Imaki H, Marchi E, Ma SY et al. . The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol 2010; 119: 755–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Alshikho MJ, Herbert MR. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS One 2016; 11: e0153329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz E, Vujic M, Karrstedt EL, Erlandsson A, Gillberg C. A case report of two male siblings with autism and duplication of Xq13-q21, a region including three genes predisposing for autism. Eur Child Adolesc Psychiatry 2014; 23: 329–36. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Furness J, Walsh FS, Doherty P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 1994; 13: 583–94. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA et al. . Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 2013; 155: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 2003; 77: 8957–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Scattoni ML. Behavioural methods used in rodent models of autism spectrum disorders: current standards and new developments. Behav Brain Res 2013; 251: 5–17. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM et al. . Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 1998; 125: 753–65. [DOI] [PubMed] [Google Scholar]
- Ye H, Liu J, Wu JY. Cell adhesion molecules and their involvement in autism spectrum disorder. Neurosignals 2010; 18: 62–71. [DOI] [PubMed] [Google Scholar]
- Yenkoyan K, Grigoryan A, Fereshetyan K, Yepremyan D. Advances in understanding the pathophysiology of autism spectrum disorders. Behav Brain Res 2017; 331: 92–101. [DOI] [PubMed] [Google Scholar]
- Yufune S, Satoh Y, Takamatsu I, Ohta H, Kobayashi Y, Takaenoki Y et al. . Transient blockade of ERK phosphorylation in the critical period causes autistic phenotypes as an adult in mice. Sci Rep 2015; 5: 10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yu H, Jiang S, Liao J, Lu T, Wang L et al. . Evidence for association of cell adhesion molecules pathway and NLGN1 polymorphisms with schizophrenia in chinese han population. PLoS One 2015; 10: e0144719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from this work are available on the free repository OFS and can be accessed on: https://osf.io/5a673/?view_only=35ccdcaf059f48efa581eab210abbc73