Abstract
Background and Aims
An expanding number of monogenic defects have been identified as causative of severe forms of very early-onset inflammatory bowel diseases [VEO-IBD]. The present study aimed at defining how next-generation sequencing [NGS] methods can be used to improve identification of known molecular diagnosis and to adapt treatment.
Methods
A total of 207 children were recruited in 45 paediatric centres through an international collaborative network [ESPGHAN GENIUS working group] with a clinical presentation of severe VEO-IBD [n = 185] or an anamnesis suggestive of a monogenic disorder [n = 22]. Patients were divided at inclusion into three phenotypic subsets: predominantly small bowel inflammation, colitis with perianal lesions, and colitis only. Methods to obtain molecular diagnosis included functional tests followed by specific Sanger sequencing, custom-made targeted NGS, and in selected cases whole exome sequencing [WES] of parents-child trios. Genetic findings were validated clinically and/or functionally.
Results
Molecular diagnosis was achieved in 66/207 children [32%]: 61% with small bowel inflammation, 39% with colitis and perianal lesions, and 18% with colitis only. Targeted NGS pinpointed gene mutations causative of atypical presentations, and identified large exonic copy number variations previously missed by WES.
Conclusions
Our results lead us to propose an optimised diagnostic strategy to identify known monogenic causes of severe IBD.
Keywords: Genetics and molecular epidemiology, paediatrics, VEO-IBD, monogenic disorders, TNGS
1. Introduction
Very early-onset inflammatory bowel diseases [VEO-IBD] are defined by the development of chronic intestinal inflammation before the age of 6 years. A subgroup of patients suffer from a rapid and severe disease evolution, which raises difficult diagnostic and therapeutic issues.1 In the latter patients, disease-causing mutations are identified in an increasing number of genes expressed in haematopoietic immune cells, in epithelial cells, or in both. Early molecular diagnosis is crucial in order to reduce mortality and morbidity, notably by defining whether haematopoietic stem cell transplantation [HSCT] is indicated.
Molecular diagnosis in VEO-IBD patients is classically based on precise phenotyping followed by selected functional tests and Sanger sequencing of candidate genes.2,3 Onset of severe bloody diarrhoea and perianal lesions a few weeks after birth suggests the existence of mutations inactivating the interleukin 10 [IL-10] receptor or IL10,4–7 whereas exudative diarrhoea in the context of autoimmunity—and notably type I diabetes—is highly suggestive of IPEX syndrome [Immune dysregulation, Polyendocrinopathy, Enteropathy, X-linked] due to mutations in Forkhead box P3 [FOXP3] gene.8
However, such approach to molecular diagnosis has limitations given the increasing number of genes in which mutations have been identified and given recurrent evidence of lack of strict phenotype-genotype correlations.9 Furthermore, in patients with overlapping clinical presentations, different pathophysiological defects require distinct therapeutic approaches.10 Therefore, precise molecular diagnosis is a prerequisite for offering these patients access to optimal care with the best possible outcome.
These issues stress the need to establish time- and cost-effective clinical diagnostic protocols.3,11 Next-generation DNA sequencing [NGS] methods allow efficient genotype-based diagnosis. Whole genome [WGS] and whole exome [WES] sequencing provide unbiased approaches, but they remain relatively expensive, and can raise difficult challenges for analysis and interpretation, which limits their use for diagnostic purposes. Targeted NGS [TNGS] —also called targeted gene panel sequencing—emerges as a powerful tool for fast and simple screening of genes of interest. Since 2014, TNGS was proven to be valuable in the identification of primary immunodeficiencies, notably in patients with atypical presentations.9,12,13 More recently, TNGS could identify causative mutations in small cohorts of VEO-IBD patients.1,14
The present study aimed at defining how next-generation sequencing [NGS] methods can be used to improve identification of known molecular defects and to adapt treatment accordingly. Based on our results, we propose an optimised diagnostic strategy to identify known monogenic causes of severe IBD depending on the phenotype of the patients.
2. Methods
2.1. Patients
A working group from ESPGHAN [European Society for Paediatric GastroEnterology, Hepatology and Nutrition] called GENIUS [GENetically and/or ImmUne-mediated enteropathieS] was created in 2009 in order to improve diagnosis of monogenic intestinal disorders and to optimise patients’ care. GENIUS [www.genius-group.org] gathers paediatric gastroenterologists from 45 centres [see Appendix in Supplementary data, available at ECCO-JCC online].
Inclusion criteria were chronic diarrhoea developed before the age of 6 years, with severe disease course requiring immunosuppressive treatments (including steroids, azathioprine, or anti-tumour necrosis factor [TNF]), surgery [partial or subtotal colectomy], and/or parenteral nutrition [185 patients]. Additional patients were included despite disease onset after the age of 6 years in the presence of a familial history suggestive of Mendelian inheritance [nine patients with affected siblings] or of a disease course refractory to treatment [13 patients]. Overall, 42 patients were from consanguineous families [20%]. Based on the clinical phenotype, patients were divided at inclusion into three subsets: 1] predominantly inflammatory small bowel inflammation with massive lymphocytic infiltrate and villous atrophy of the small bowel associated with various signs of autoimmunity; 2] inflammatory colitis associated with severe perianal lesions such as fistula or abscess; 3] severe isolated colitis. Patients were included in each centre after obtaining informed written consent for functional and genetic studies from a parent or legal guardian. The study adhered to the tenets of the revised Declaration of Helsinki and was approved by an institutional review board [CPP Ile de France II, 2014.01.04 AF].
2.2. DNA preparation
Genomic DNA was extracted from peripheral blood mononuclear cells [PBMC] isolated by Ficoll HyPaque Plus [GE Healthcare, Velizy-Villacoublay, France] from umbilical cord tissue [patient 25] or from whole blood lysed by BD lysis buffer [BD Biosciences, Le Pont de Claix, France] using the QIAamp® DNA Blood Mini Kit [Qiagen, Courtaboeuf, France].
2.3. Whole exome and targeted next-generation sequencing
A custom-made TNGS panel was designed and gathered 66 genes in which mutations can cause VEO-IBD or congenital diarrhoeal disorders [Supplementary Table 1, available as Supplementary data, available at ECCO-JCC online]. The choice of VEO-IBD associated genes was mostly guided by the list suggested by Uhlig in 201411 and Canani et al. in 2015.15 Some genes of common variable immunodeficiency were included as well, as they can lead to severe intestinal inflammation.16–22 Genes of severe combined immunodeficiencies, in which diarrhoea is usual but with a different clinical context, and genes associated with diseases with very specific and primarily extra-intestinal symptoms such as bullous skin diseases or albinism, were excluded as affected patients are unlikely to be diagnosed in paediatric gastroenterology centres.
Genomic DNA libraries were generated from DNA [3 µg for WES and 1 µg for TNGS] sheared with a Covaris S2 Ultrasonicator using SureSelectXT Library PrepKit [Agilent, Garches, France] on the Genomic Platform at the Imagine Institute. Capture by hybridisation was performed using either Agilent Sure Select All Exon V5 [Agilent, Les Ulis, France] for WES or biotinylated complementary 120-bp RNA baits designed with SureSelect SureDesign software [H. sapiens, hg19, GRCh37, February 2009] that cover the 1130 regions of interest [ROI] encompassing all exons of the 66 selected genes.
Targeted exons or ROI were pulled out with magnetic streptavidin beads, polymerase chain reaction [PCR]-amplified using indexing primers and sequenced on an Illumina HiSeq2500 HT system [Illumina, San Diego, USA]. Data analysis was performed with Paris Descartes University/Imagine Institute’s Bioinformatics core facilities. Paired-end sequences were mapped on the human genome reference [NCBI build37/hg19 version] using the Burrows-Wheeler Aligner. Downstream processing was carried out with the Genome Analysis Toolkit [GATK], SAMtools,23 and Picard, according to documented best practices [http://www.broadinstitute.org/gatk/guide/topic?name = best-practices]. Variant calls were made with the GATK Unified Genotyper based on the 72nd version of ENSEMBL database. Genome variations were defined using the in-house software PolyQuery for WES or PolyDiag for TNGS, which filters out irrelevant and common polymorphisms based on frequencies extracted from public databases: US National Center for Biotechnology Information database of single nucleotide polymorphisms [SNP] [dbSNP24], 1000 genomes,25 Exome Variant Server [EVS, http://evs.gs.washington.edu/EVS/], and Exome Aggregation Consortium [ExAC, http://exac.broadinstitute.org].26 Consequences of mutations on protein function were predicted using three algorithms: Polyphen2 [http://genetics.bwh.harvard.edu/pph2/],27 SIFT [Sorting Intolerant From Tolerant, J. Craig Venter Institute],28 and Mutation Taster [www.mutationtaster.org].29 Mutations were then ranked on the basis of the predicted impact of each variant by combined annotation-dependent depletion [CADD],30 and compared with the mutation significance cut-off [MSC], a gene-level specific cut-off for CADD scores [http://pec630.rockefeller.edu:8080/MSC/].31
2.4. Quality control of TNGS
DNA samples from all patients were sequenced between August 2015 and April 2016 in three consecutive runs. Mean coverage of the panel was 682 reads, and varied depending on the number of samples sequenced by run with medians of 826, 629, and 593 reads in runs 1 [29 samples], 2 [51 samples], and 3 [90 samples], respectively. ROI with the smallest coverage were the first exons of DGAT1 [50 to 200 reads] and MALT1 [10 to 100 reads], likely due to their high GC content which impairs capture, and exon 7 of ITCH [20 to 200 reads] and exon 1 of APOB [20 to 160 reads], which both contain repeated regions. Overall, all ROI were well covered, including NCF1, IKBKG, and NEUROG3, known to be difficult to capture. Due to the presence of pseudogenes, careful consideration was, however, required to attribute variants to IKBKG and NCF1. The sensitivity of TNGS was assessed by comparing against Sanger sequencing in a validation sample of 12 patients with established molecular diagnoses. All mutations were identified by TNGS, including a complex compound heterozygous defect in IL10RB [VS12] consisting of a large deletion of exon 3 on the maternal allele and a duplication of exon 6 on the paternal allele [Charbit-Henrion et al., in submission].
2.5. Gene validation
All mutations identified by WES or TNGS, and not previously described as disease-causing in the literature, were validated as follows.
Novel mutations in SKIV2L and TTC37 were confirmed by reassessment of the clinical phenotype. The patients presented the canonical signs of syndromic diarrhoea within a tricho-hepato-enteric syndrome32: congenital diarrhoea [4/4], hair abnormalities with trichorrhexis nodosa [4/4], intrauterine growth retardation [3/4], oligohydramnios [2/4], facial dysmorphia [2/4], and immune defect [3/4]. Novel mutations in MYO5B and EPCAM were confirmed by histology (haemotoxylin and eosin [HE], periodic acid-Schiff [PAS] staining for MYO5B) and immunochemistry [EPCAM staining] performed on formalin-fixed intestinal biopsies. EPCAM was revealed with clone 323/A3 [Acris Antibodies GmbH, Montluçon, France] as described.33 Novel FOXP3 mutations were validated by demonstrating a reduced frequency of CD4+ CD25+ CD127+ FOXP3+ regulatory T cells or abnormal suppressive function.34,35 IL-10 receptor functional test was performed as described.6 Novel XIAP mutations were authenticated by showing impaired IL-8 production by PBMC stimulated with N-acetyl-muramyl-M-alanyl-D-isoglutamine hydrate [Muramyl-dipeptide/MDP, Sigma, St. Quentin Fallavier, France] or lipopolysaccharide [LPS, Sigma] as described previously,36 and protein expression was assessed by XIAP intracellular staining performed on 2 × 106 fixed-permeabilised PBMC using the Intraprep Permeabilization Reagent kit [Beckman Coulter, Villepinte, France] and incubated with anti-XIAP antibody [clone 28/hILP, BD] or isotype control antibody [mouse IgG1, Sony Biotechnologies, Weybridge, UK] and with secondary PE-goat anti-mouse IgG1 antibody. For extracellular staining, cells were incubated with anti-CD3-Bv510, anti-CD14-Pe/Cy7, anti-CD11c-APC, anti-CD86-Bv421 [Sony Biotechnologies], and anti-CD19-Bv711 [BD Biosciences]. LRBA and NCF1 mutations were confirmed by absent protein expression on western blot performed on cell lysates according to standard protocols. Proteins were detected with monoclonal antibodies against human NCF1 [sc-14015 phox-p47, Santa Cruz Biotechnology, Dallas, USA], LRBA [clone HPA023597, Sigma], horseradish peroxidase-linked secondary antibodies or horseradish peroxidase-conjugated GAPDH rabbit antibody [Ozyme, Montigny-le-Bretonneux, France], and ECL Prime Western Blotting Detection reagent [GE Healthcare].
3. Results
3.1. Description of the cohort
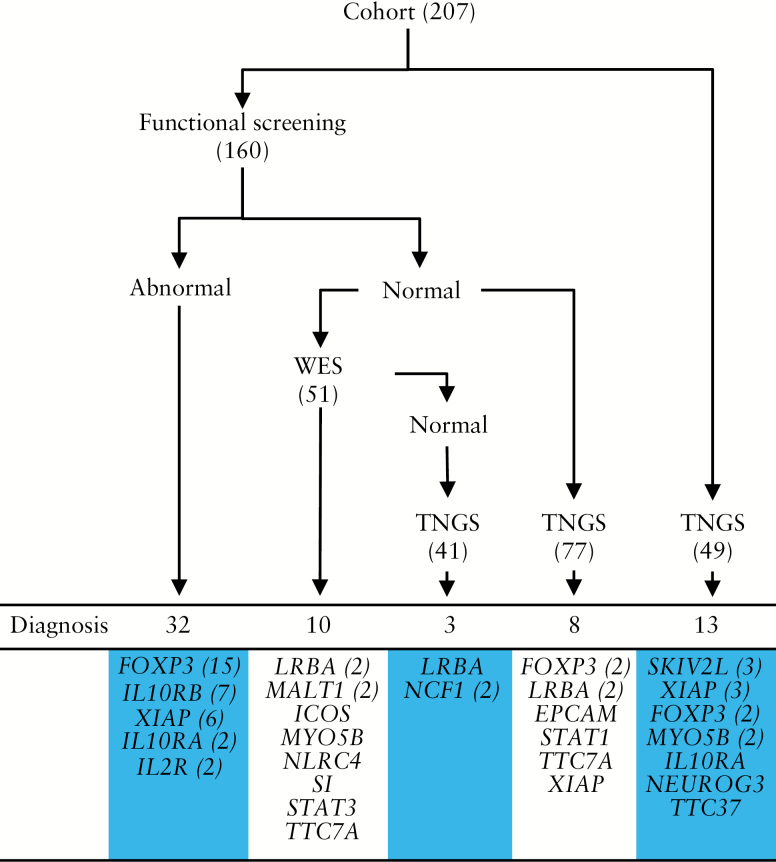
Between August 2009 and August 2015, 207 consecutive patients with severe chronic diarrhoea were recruited; 144 patients had started their disease before the age of 2 years [70%] and 22 patients after the age of 6 years [Table 1]. As described in Methods, and shown in Table 2, patients were divided into three groups based on phenotype: 51 patients with small bowel inflammation and autoimmune features; 33 with colitis and perianal lesions; and 123 with colitis only. As shown in Figure 1, 160 patients benefited from functional tests that were orientated by the clinical presentation: numbers and phenotype of regulatory T cells were determined in patients with small bowel inflammation, and signalling downstream IL-10 receptor or XIAP was assessed in patients with colitis with or without perianal lesions. In 32 patients with abnormal results, Sanger sequencing was used to confirm mutations in FOXP3, IL2RA, IL10RA, IL10RB, or XIAP. Between 2013 and 2015, WES was performed on DNA trios [DNA from child and both parents] in 51 cases with normal functional tests and very severe phenotypes. A known monogenic disorder was identified in 10 patients [20%]. In 2015, TNGS was introduced and performed in all patients previously investigated but left undiagnosed and also systematically in the newly included patients [n = 49]. TNGS identified causative mutations in 24/167 patients [14%]. As expected, diagnosis yield varied depending on whether TNGS was used to complete previous diagnostic approaches or as the first method. Thus, TNGS reached eight molecular diagnoses [10%] in the 77 functionally tested patients who had not benefited from WES [because DNA was not available from both parents: n = 45; or because of less severe phenotypes]. However, when used as first screening [n = 49], diagnosis yield of TNGS was much higher, with 13 molecular defects identified [26.5%]. Importantly, in our study, TNGS had a superior diagnostic yield compared with WES. Indeed, it allowed diagnosis in three patients who had been already investigated by WES but harboured copy number variations hardly detectable by this method [one girl with large LRBA deletion, two siblings with large NCF1 deletion]. Overall, a known monogenic disorder was identified in 66/207 patients [32%].
Table 1.
Diagnosis yield according to age at disease onset.
| Small bowel inflammation | Colitis + perianal | Colitis only | Total | |
|---|---|---|---|---|
| Whole cohort | ||||
| Number of patients | 51 | 33 | 123 | 207 |
| Number of molecular diagnoses | 31 | 13 | 22 | 66 |
| 61% | 39% | 18% | 32% | |
| With disease onset before age 2 years | ||||
| Number of patients | 45 | 22 | 77 | 144 |
| Number of molecular diagnoses | 30 | 12 | 17 | 59 |
| 67% | 55% | 22% | 41% | |
| With disease onset after age 6 years | ||||
| Number of patients | 2a | 7b | 13c | 22 |
| Number of molecular diagnoses | 1 | 1 | 2 | 4 |
| 50% | 14% | 15% | 18% | |
aTwo patients from two multiplex families.
bFour patients from four multiplex families and three patients with refractory disease.
cThree patients from three multiplex families and 10 patients with refractory disease.
Table 2.
Phenotype based genetic characterisation of VEO-IBD patients.
| Small bowel inflammation | Colitis + perianal | Colitis only | |
|---|---|---|---|
| Number of patients | 51 | 33 | 123 |
| % of boys | 78% | 72% | 58% |
| Number of diagnosed patients | 31 | 13 | 22 |
| % of molecular diagnoses | 61% | 39% | 18% |
| % of diagnoses done by | |||
| Functional tests | 55% | 92% | 14% |
| WES | 19.5% | 0% | 18% |
| TNGS | 26% | 8% | 68% |
| FOXP3 [19] | IL10RA [3] | EPCAM [1] | |
| IL2RA [2] | IL10RB [7] | ICOS [1] | |
| LRBA [5] | XIAP [3] | MYO5B [2] | |
| MALT1 [2] | NCF1 [2] | ||
| MYO5B [1] | NLRC4 [1] | ||
| NEUROG3 [1] | SI [1] | ||
| STAT3 [1] | SKIV2L [3] | ||
| STAT1 [1] | |||
| TTC37 [1] | |||
| TTC7A [2] | |||
| XIAP [7] |
List of genes and number of patients for each gene in brackets.
WES, whole exome sequencing; TNGS, targeted next-generation sequencing.
Figure 1.
Cohort screening for gene identification. Number of diagnoses obtained by each method. Number of patients for each gene defect is reported in brackets. WES: whole exome sequencing; TNGS: targeted next-generation sequencing.
3.2. Yield of molecular diagnoses according to the clinical phenotype
By combining selected functional tests and next-generation sequencing, approximately 30% of all the patients included in the present cohort received a molecular diagnosis. Yet, as mentioned above, clinical presentation differed between patients and functional tests were only available to validate a few gene defects. To optimise diagnostic work-up, we then analysed whether the frequency of identified molecular defects differed between the three groups of patients and, within each group of patients, when and how functional testing and TNGS had been most useful for diagnosis [Table 2].
Among 51 patients [37 boys, 72.5%] who presented with small bowel inflammation, a molecular diagnosis was obtained in 31/51 patients [61%]. In 19/31 patients, number or phenotype of Tregs were abnormal. In 17 of these 19 patients, Sanger sequencing identified mutations in FOXP3 [n = 15] and in IL2RA [n = 2]. Other mutations were identified either by WES in six patients or TNGS in eight patients. Mutations identified by WES were: a homozygous MALT1 mutation in two siblings with decreased number of Tregs,34 a homozygous LRBA mutation in two other siblings,37 a de novo gain-of-function mutation of STAT3 in one girl, and a homozygous MYO5B mutation in one boy in whom marked intestinal inflammation had erroneously led to ascribe the severe diarrhoea to an immune-mediated process. Mutations identified by TNGS included FOXP3 mutations in four boys, for three of whom only DNA was available and in one case normal number and phenotype of regulatory T cells but with abnormal suppressive function; a homozygous NEUROG3 mutation in one boy with chronic diarrhoea since the first months of life and type 1 diabetes since the age of one38; compound heterozygous LRBA mutations in three patients, one boy with two missense mutation, and two siblings with one missense mutation and a large deletion [from exon 3 to exon 48]. As indicated above, this large deletion in LRBA had been missed by WES performed before TNGS.
The group with colitis and perianal lesions included 33 patients of whom 73% were boys. Among the 33 patients, 13 [40%] were diagnosed with mutations in IL10RA [n = 4], IL10RB [n = 6], or XIAP [n = 3]. Based on their phenotype highly suggestive of defective IL-10 or XIAP signalling, all patients except one benefited from functional tests.6,36 Overall, diagnosis was oriented in 12 patients by functional tests, which showed either lack of responsiveness of peripheral monocytes to the inhibitory effect of exogenous IL-10 [n = 9] or lack of stimulation of the NOD2-XIAP pathway by muramyl-dipeptide [n = 3]. Sanger sequencing confirmed mutations in IL10RA [n = 3], IL10RB [n = 6], or XIAP [n = 3]. In one patient for whom only DNA was available, TNGS identified a homozygous IL10RA mutation.
The group of patients with only colitis comprised 123 patients of whom 59% were male. As defined above, they displayed neither autoimmune symptoms nor perianal lesions. Only 22 of them reached a molecular diagnosis [18%]. Three boys [3/22, 13.5%] were diagnosed with XIAP mutations after abnormal functional screening and targeted Sanger sequencing of XIAP. WES allowed diagnosis in four additional patients [4/22, 18%] harbouring mutations in four different genes: NLRC4, SI, TTC7A, and ICOS. However, most molecular diagnoses in this group [15/22, 68%] were obtained by TNGS. TNGS pinpointed mutations in EPCAM [n = 1], MYO5B [n = 2], NCF1 [in two siblings with a large homozygous deletion which was missed by WES], SKIV2L [n = 3], STAT1 [n = 1], TTC37 [n = 1], TTC7A [n = 1], and XIAP in four patients: in three boys for whom only DNA was available and in a 17-year-old girl with severe and refractory VEO-IBD since the age of 6 months. Because of X-linked inheritance, XIAP deficiency had not been considered. Following identification of a premature stop codon by TNGS, abnormal signalling downstream XIAP and reduced protein expression in 50% of PBMC were demonstrated. The same mutation was found in her sister, who had also developed very severe IBD. Of note, in three additional patients, rare variants identified by TNGS were ruled out, either because they were incompatible with the clinical phenotype [a rare heterozygous variant in PLCG2 was highly unlikely to be causative since the patient did not display any auto-inflammatory symptoms] or through functional testing [two variants in MYO5B in a patient with normal PAS staining of duodenal sections; two rare variants in IL21R but normal STAT3 phosphorylation of PBMC in response to IL-21].
3.3. Yield of molecular diagnoses depending on age at disease onset
As mentioned above, diagnostic yield on the whole cohort was 32%; 70% of the patients started their disease before the age of two years [144/207, Table 1]. Among the latter patients, diagnostic yield was slightly higher [41%]. Further analysis shows that, in patients with small bowel inflammation or colitis with perianal lesions, the vast majority of molecular diagnoses were identified in children with disease onset before the age of two years. In patients included despite later age at disease onset [n = 22], a monogenic defect was identified in four cases [18%].
4. Discussion
An expanding number of monogenic defects have been identified as causative of severe forms of VEO-IBD. These diseases are individually rare but, collectively, they raise difficult diagnostic and therapeutic issues due to their severity and considerable genetic heterogeneity. Paediatric gastroenterologists from ESPGHAN created the GENIUS working group in 2009 with the goals of fostering clinical knowledge of these diseases and facilitating development of therapeutic strategies based on precise molecular diagnosis. The resulting cooperation between 45 paediatric centres allowed us to comprehensively investigate the potential monogenic causes of severe VEO-IBD and to propose guidelines for the diagnosis of known molecular defects.
The 207 patients described in this study represent the largest European cohort to date. Overall, 32% of patients obtained a molecular diagnosis. Diagnosis yield, however, varied when patients were stratified by their phenotypes. Molecular diagnosis was obtained in two-thirds of children presenting with small bowel inflammation suggestive of ‘IPEX’ syndrome, and in almost half of patients having colitis with severe perianal lesions. In contrast, diagnosis yield in the ‘Colitis only’ subset was relatively low [less than 20%]. These differences remain to be explained. A possible hypothesis is that not all cases of early-onset colitis have a monogenic cause, notably in patients with disease onset beyond the first 2 years of life and in absence of familial history.
In our settings, Sanger sequencing orientated by abnormal functional tests, WES and TNGS presented comparable diagnostic yields. The first method allowed molecular diagnosis in 32 out of 160 tested patients [20%]; WES identified a known monogenic disorder in 10 patients out of 51 [20%]. It may seem that TNGS was less efficient, with a global diagnostic yield of only 14% [24/167]. Nonetheless, one needs to analyse separately patients who benefited from TNGS as a first screening test [13 molecular diagnoses out of 49 patients, 26.5%] and patients who were functionally tested [eight molecular diagnoses out of 77 patients, 10%]. Altogether our findings confirm and extend results obtained in two previous studies in smaller cohorts of VEO-IBD patients, in which TNGS could identify causative mutations in 4/251 and in 5/71 patients,14 respectively. Furthermore, in patients with higher genetic heterogeneity such as those with isolated colitis, TNGS is more efficient [68% of monogenic defects were identified by TNGS in this group] as it can reveal diagnosis in patients with atypical clinical presentation, as exemplified in the present study in one girl with XIAP deficiency.
Of note, a molecular diagnosis was identified in four out of 22 patients who had started their disease after 6 years. These four patients [with defects in LRBA, NCF1, and XIAP] would have remained undiagnosed with a cut-off for inclusion at 6 years. Given increasing evidence of de novo mutations, of incomplete penetrance, and of delayed disease onset in patients with mutations in notably XIAP, CTLA4, LRBA, STAT3, or NLRC4,17–22,36,37,39 we thus recommend that access to TNGS screening should be considered not only in all severe VEO-IBD patients, but also in older children or teenagers with very severe intestinal diseases refractory to medical treatment. The modest cost per patient renders this option economically affordable and may prove instrumental to guide therapeutic strategy. This is exemplified in our cohort by a case of XIAP deficiency diagnosed in a young adult, who had already undergone colectomy, thus too late to propose treating the colitis by HSCT.40
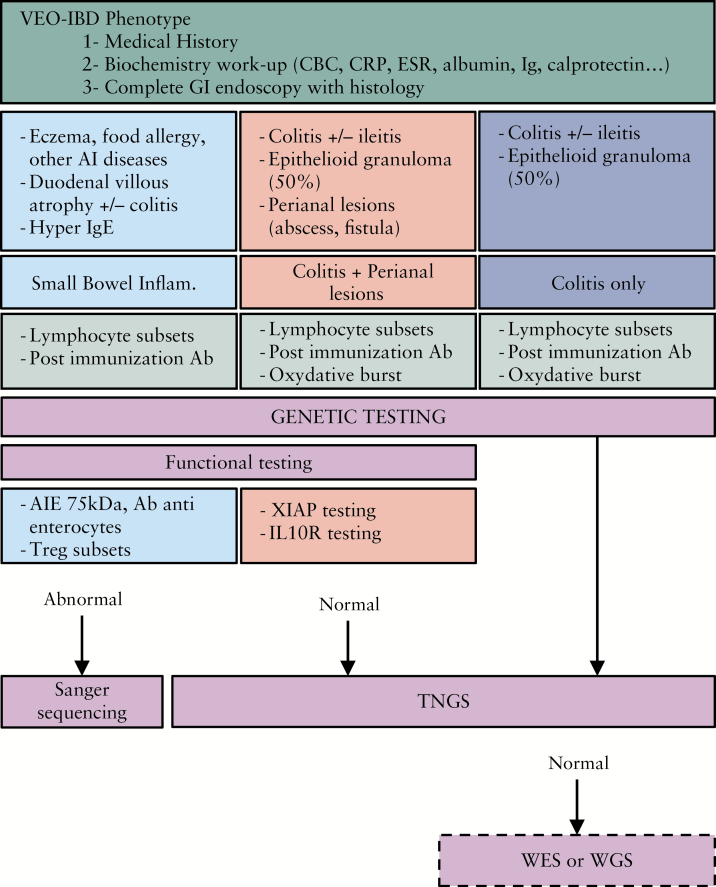
Based on the retrospective analysis of this large multicentre cohort of severe VEO-IBD patients, we propose an algorithm to obtain a molecular diagnosis in an optimised manner [Figure 2]. It has to be validated prospectively in an independent cohort of patients. In addition to severe chronic diarrhoea starting early in life, some atypical clinical features may orientate towards specific monogenic defects [hair abnormalities, dysmorphia, autoimmunity, eczema, allergy, immune deficiency, etc]. Combined with laboratory tests and endoscopic and histology findings, this precise phenotyping allows division of patients into three groups as described above. Since many monogenic forms of VEO-IBD are primary immunodeficiencies, standard immunological work-up is recommended [see Figure 2]. When pathognomonic symptom associations evoke a specific monogenic disorder, corresponding functional tests should be considered as they may quickly orientate molecular diagnosis within only a few days.
Figure 2.
Proposed algorithm for molecular diagnosis in VEO-IBD. VEO-IBD: very early-onset inflammatory bowel disease; CBC: cell blood count; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; Ig: immunoglobulin; GI: gastro-intestinal; AI: autoimmune; Ab: antibodies; Treg: regulatory T cells; small bowel inflam: small bowel inflammation; TNGS: targeted next-generation sequencing; WES: whole exome sequencing; WGS: whole genome sequencing.
When functional tests fail to orientate toward a molecular defect, NGS methods need to be considered, given the genetic heterogeneity and important phenotype-genotype overlaps. TNGS, WES, and WGS are non-hypothesis driven assays that allow molecular diagnosis in patients with atypical clinical presentations. In a routine clinical setting, TNGS is the method to prefer as it is cost- and time-effective while providing optimal coverage of the genes of interest [see Supplementary Table 2, available as Supplementary data, available at ECCO-JCC online]. Regularly updating the panel of genes to be targeted upon identification of novel disease-causing genes should progressively increase sensitivity and diagnostic yield. WES could be performed as a second-line genetic assay when TNGS has failed to reveal a molecular diagnosis. WES—and furthermore WGS—will allow identification of novel causative genes, but they require an important investment in terms of cost, sequencing delay, analysis, and validation, which restricts their use to research protocols only. In our cohort, WES allowed us to ascribe VEO-IBD to biallelic mutations in DUOX2,41 and more recently to biallelic mutations in intestinal alkaline phosphatase.42 Furthermore, novel candidate genes are currently under validation in three distinct families.
In conclusion, development of accurate and cost-effective diagnosis tools to screen a putative monogenic VEO-IBD population is indispensable to obtain molecular diagnosis and optimise patients’ care. Further genomic analyses including WES and, likely in the near future, WGS, should help in identifying novel candidate genes. Overall, genetic characterisation of VEO-IBD will help broaden knowledge of the phenotypic spectrum of inherited intestinal disorders. Identifying key genes for gut homeostasis may also help in designing new therapeutic targets for patients suffering from chronic intestinal diseases.
Funding
This work was supported by ERC-2013-AdG-339407-IMMUNOBIOTA, Investissement d’Avenir ANR-10-IAHU-01, Fondation des Maladies Rares, and Association François Aupetit. FC-H was supported by a fellowship from INSERM. JN received a fellowship from the Polish National Science Centre [2015/16/T/NZ5/00168].
Conflict of Interest
The authors do not declare any conflict of interest.
Author Contributions
FCH, FR, and NCB designed the study; FCH and SH were in charge of designing the TNGS; OA and CF performed TNGS; FCH, MP, RDL, and JN analysed TNGS data; BB and SR extracted cells and DNA and performed ELISA experiments; JB performed histological analysis; FR, MCS, FM, and EL performed analysis of regulatory T cells and LRBA protein; SL and CL analysed XIAP expression; FCH, NCB, MP FR, AF, CP, FRL, and SL reviewed and analysed data; FCH, FR, MP, and NCB wrote the manuscript that was reviewed by all authors. Members of GENIUS group were in charge of the patients and acquired clinical data.
Supplementary Material
Acknowledgments
The authors acknowledge the use of the bioresources of the Necker Imagine DNA biobank [BB-033-00065]. They are grateful to the members of the Department of Clinical Research of the Imagine Institute, headed by Elisabeth Hullier-Ammar, for their guidance with ethical procedures, and to Pauline Touche for her help with the Immunobiota protocol.
References
- 1. Kammermeier J, Drury S, James CT, et al. . Targeted gene panel sequencing in children with very early onset inflammatory bowel disease – evaluation and prospective analysis. J Med Genet 2014;51:748–55. [DOI] [PubMed] [Google Scholar]
- 2. Picard C, Fischer A. Contribution of high-throughput DNA sequencing to the study of primary immunodeficiencies. Eur J Immunol 2014;44:2854–61. [DOI] [PubMed] [Google Scholar]
- 3. Uhlig HH. Monogenic diseases associated with intestinal inflammation: implications for the understanding of inflammatory bowel disease. Gut 2013;62:1795–805. [DOI] [PubMed] [Google Scholar]
- 4. Glocker EO, Kotlarz D, Boztug K, et al. . Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009;361:2033–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glocker EO, Frede N, Perro M, et al. . Infant colitis – it’s in the genes. Lancet 2010;376:1272. [DOI] [PubMed] [Google Scholar]
- 6. Begue B, Verdier J, Rieux-Laucat F, et al. . Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol 2011;106:1544–55. [DOI] [PubMed] [Google Scholar]
- 7. Pigneur B, Escher J, Elawad M, et al. . Phenotypic characterization of very early-onset IBD due to mutations in the IL10, IL10 receptor alpha or beta gene: a survey of the Genius Working Group. Inflamm Bowel Dis 2013;19:2820–8. [DOI] [PubMed] [Google Scholar]
- 8. Verbsky JW, Chatila TA. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked [IPEX] and IPEX-related disorders: an evolving web of heritable autoimmune diseases. Curr Opin Pediatr 2013;25:708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoddard JL, Niemela JE, Fleisher TA, Rosenzweig SD. Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Front Immunol 2014;5:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lo B, Zhang K, Lu W, et al. . AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science 2015;349:436–40. [DOI] [PubMed] [Google Scholar]
- 11. Uhlig HH, Schwerd T, Koletzko S, et al. ; COLORS in IBD Study Group and NEOPICS. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 2014;147:990–1007.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nijman IJ, van Montfrans JM, Hoogstraat M, et al. . Targeted next-generation sequencing: a novel diagnostic tool for primary immunodeficiencies. J Allergy Clin Immunol 2014;133:529–34. [DOI] [PubMed] [Google Scholar]
- 13. Yu H, Zhang VW, Stray-Pedersen A, et al. . Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next-generation sequencing. J Allergy Clin Immunol 2016;138:1142–51.e2. [DOI] [PubMed] [Google Scholar]
- 14. Petersen BS, August D, Abt R, et al. . Targeted gene panel sequencing for early-onset inflammatory bowel disease and chronic diarrhea. Inflamm Bowel Dis 2017;23:2109–20. [DOI] [PubMed] [Google Scholar]
- 15. Canani RB, Castaldo G, Bacchetta R, Martín MG, Goulet O. Congenital diarrhoeal disorders: advances in this evolving web of inherited enteropathies. Nat Rev Gastroenterol Hepatol 2015;12:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malamut G, Verkarre V, Suarez F, et al. . The enteropathy associated with common variable immunodeficiency: the delineated frontiers with celiac disease. Am J Gastroenterol 2010;105:2262–75. [DOI] [PubMed] [Google Scholar]
- 17. Flanagan SE, Haapaniemi E, Russell MA, et al. . Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet 2014;46:812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haapaniemi EM, Kaustio M, Rajala HL, et al. . Autoimmunity, hypogammaglobulinemia, lymphoproliferation, and mycobacterial disease in patients with activating mutations in STAT3. Blood 2015;125:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Milner JD, Vogel TP, Forbes L, et al. . Early-onset lymphoproliferation and autoimmunity caused by germline STAT3 gain-of-function mutations. Blood 2015;125:591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuehn HS, Ouyang W, Lo B, et al. . Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science 2014;345:1623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schubert D, Bode C, Kenefeck R, et al. . Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat Med 2014;20:1410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alangari A, Alsultan A, Adly N, et al. . LPS-responsive beige-like anchor [LRBA] gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J Allergy Clin Immunol 2012;130:481–8.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherry ST, Ward MH, Kholodov M, et al. . dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adzhubei IA, Schmidt S, Peshkin L, et al. . A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073–81. [DOI] [PubMed] [Google Scholar]
- 29. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014;11:361–2. [DOI] [PubMed] [Google Scholar]
- 30. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Itan Y, Shang L, Boisson B, et al. . The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 2016;13:109–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fabre A, Breton A, Coste ME, et al. . Syndromic [phenotypic] diarrhoea of infancy/tricho-hepato-enteric syndrome. Arch Dis Child 2014;99:35–8. [DOI] [PubMed] [Google Scholar]
- 33. Salomon J, Goulet O, Canioni D, et al. . Genetic characterization of congenital tufting enteropathy: epcam associated phenotype and involvement of SPINT2 in the syndromic form. Hum Genet 2014;133:299–310. [DOI] [PubMed] [Google Scholar]
- 34. Charbit-Henrion F, Jeverica AK, Bègue B, et al. ; GENIUS Group. Deficiency in mucosa-associated lymphoid tissue lymphoma translocation 1: a novel cause of IPEX-like syndrome. J Pediatr Gastroenterol Nutr 2017;64:378–84. [DOI] [PubMed] [Google Scholar]
- 35. Moes N, Rieux-Laucat F, Begue B, et al. . Reduced expression of FOXP3 and regulatory T-cell function in severe forms of early-onset autoimmune enteropathy. Gastroenterology 2010;139:770–8. [DOI] [PubMed] [Google Scholar]
- 36. Aguilar C, Lenoir C, Lambert N, et al. . Characterization of Crohn disease in X-linked inhibitor of apoptosis-deficient male patients and female symptomatic carriers. J Allergy Clin Immunol 2014;134:1131–41.e9. [DOI] [PubMed] [Google Scholar]
- 37. Bakhtiar S, Ruemmele F, Charbit-Henrion F, et al. . Atypical manifestation of LPS-responsive beige-like anchor deficiency syndrome as an autoimmune endocrine disorder without enteropathy and immunodeficiency. Front Pediatr 2016;4:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Germán-Díaz M, Rodriguez-Gil Y, Cruz-Rojo J, et al. . A new case of congenital malabsorptive diarrhea and diabetes secondary to mutant neurogenin-3. Pediatrics 2017, Jul 19. doi: 10.1542/peds.2016-2210. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 39. Romberg N, Al Moussawi K, Nelson-Williams C, et al. . Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat Genet 2014;46:1135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coelho R, Peixoto A, Amil-Dias J, et al. . Refractory monogenic Crohn’s disease due to X-linked inhibitor of apoptosis deficiency. Int J Colorectal Dis 2016;31:1235–6. [DOI] [PubMed] [Google Scholar]
- 41. Parlato M, Charbit-Henrion F, Hayes P, et al. . First identification of biallelic inherited DUOX2 inactivating mutations as a cause of very early onset inflammatory bowel disease. Gastroenterology 2017;153:609–11.e3. [DOI] [PubMed] [Google Scholar]
- 42. Parlato M, Charbit-Henrion F, Pan J, et al. . Human ALPI deficiency causes inflammatory bowel disease and highlights a key mechanism of gut homeostasis. EMBO Mol Med 2018; 10. doi: 10.15252/emmm.201708483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.