Abstract
AIM
To explore the relationship between dynamic expression of high mobility group box-3 (HMGB3) and malignant transformation of hepatocytes.
METHODS
Expression of HMGB family proteins were observed in rat hepatocarcinogenesis models induced with 2-acetylaminofluorene. Alterations of HMGB3 were analyzed at the mRNA level by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and at the protein level by immunohistochemistry or Western blotting. HMGB3 in human liver cancer tissues were evaluated using bioinformatics databases from GEO, TCGA, and Oncomine. A specific HMGB3-shRNA was used to knock down HMGB3 expression in order to investigate its effects on proliferation and cell cycle in vitro and in vivo.
RESULTS
Elevated HMGB3 levels were first reported in hepatocarcinogenesis, with increasing expression from normal liver to cancer. Bioinformatic databases showed that HMGB3 expression in hepatocellular carcinoma tissues was significantly higher than that in normal liver tissues. Higher HMGB3 expression was discovered in liver cancer cells compared with LO2 cells in vitro. According to gene set enrichment analysis, HMGB3 mRNA levels were correlated with cell cycle and DNA replication pathways. Knocking down HMGB3 by specific shRNA significantly inhibited proliferation of HepG2 cells by cell cycle arrest and downregulating DNA replication related genes (cyclin B1, FEN1, and PCNA) at the mRNA and protein level. Furthermore, silencing HMGB3 significantly inhibited xenograft tumor growth (measured by Ki67) in vivo.
CONCLUSION
HMGB3 is involved in malignant transformation of hepatocytes and could be a useful biomarker for diagnosis and a potential target for therapy of liver cancer.
Keywords: Liver cancer, HMGB-3, Hepatocarcinogenesis, Proliferation, Tumor growth
Core tip: High mobility group box (HMGB) family proteins were correlated with hepatocellular carcinoma (HCC) development and progression. This current study examined the effects of HMGB3 on HCC both in vitro and in vivo. Overexpression of HMGB3 was observed in hepatic malignant transformation and HCC tissues in bioinformatic databases. Knockdown of HMGB3 significantly inhibited proliferation, cell cycle, and tumor growth of HCC cells, providing a novel insight for HCC research.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common and highly aggressive cancers worldwide, with the third most common cause of cancer related death[1]. Various factors can cause HCC, including cirrhosis, infections with Hepatitis B virus (HBV) or HCV, nonalcoholic fatty liver disease (NFALD), diabetes, aflatoxin B1, tobacco, and excessive alcohol consumption. Among them, chronic infections with HBV and HCV account for more than 60% of total HCC cases[2,3]. In past decades, although obvious improvement has been observed in therapeutic approaches, the prognosis of HCC remains poor because of aggressive invasiveness, frequent metastasis, and multi-drug resistance (MDR). Given that multiple genes and signaling pathways play crucial roles in the occurrence and development of HCC, target therapy is a promising approach for HCC treatment[4]. The Food and Drug Administration (FDA) has approved Sorafenib, a multi-kinase inhibitor, as a first-line treatment for advanced HCC. However, the overall effects are partially unsatisfying due to its low response rate and high frequency of adverse events[5,6]. Thus, it is of great importance to identify novel biomarkers for early diagnosis and potential targets against the progression of HCC.
The high mobility group (HMG)-box (HMGB) family belongs to the HMG protein superfamily[7] (HMGA[8], HMGB[9], and HMGN[10]). HMGB consists of four members (HMGB1, HMGB2, HMGB3, and HMGB4) with similar physiology and pathology features. It encodes proteins containing one or more DNA-binding motifs and participates in multiple cellular processes including cell differentiation, migration, and inflammatory-related activities. The HMGB family plays a complex role in carcinogenesis due to its diverse tumorigenic bioactivities in tumors. HBV functionally binds to HMG protein and activates it[11]. Mitochondrial biogenesis mediated by hypoxia promotes HCC growth through interaction between HMGB1 and Toll-like receptor 9[12]. HMGB1 secretion could be stimulated by HBX protein and subsequently enhance HCC metastasis[13]. HMGB1 signaling is also regulated by specific long noncoding RNA[14] or microRNA[15] showing pro- or anti- effects on invasion and metastasis of HCC[16]. Moreover, overexpression of HMGB2 is associated with aggressiveness and prognosis of HCC[17]. However, until now, there is rather less known about the HMGB3 or HMGB4 expression in HCC progression.
HMGB3 is a multifunctional protein with various roles in different cellular compartments and localized in the nucleus, chromosomes, and cytoplasm. It contributes to the balance between self-renewal and differentiation of hematopoietic stem cells, and enhances DNA flexibility to activate gene promoters[18]. Recently, abnormal HMGB3 has been characterized as pro-carcinogenic by promoting tumor growth, proliferation, invasion, and metastasis in several tumors including gastric[19], lung[20], esophageal[21], breast[22], colorectal[23], and urinary bladder[24]. However, the current knowledge concerning the positive and negative effects of HMGB3 on HCC development is not explicit. The aims of this study were to investigate the dynamic HMGB3 expression in hepatocarcinogenesis, bioinformatics databases, HCC cell lines, and a xenograft model, as well as to validate HMGB3 as a diagnostic marker or novel target gene for HCC.
MATERIALS AND METHODS
Rat hepatocarcinogenesis model
Forty Sprague-Dawley rats (4-6 wk old) were provided by the Experimental Animal Center of Nantong University. Living conditions included a clean environment, 12 h light/dark cycle, and 55% humidity as previously described[25]. The control group (n = 10) were fed a normal diet, and the hepatocarcinogenesis group (n = 30) were fed a diet with 0.05% 2-acetylaminofluorene (2-AAF, Sigma, United States). The rats were sacrificed at different times depending on their condition. Rat livers were used for pathology, RNA extraction, and quantitative analysis of HMGB expression. Following the determination of morphological changes in the rat livers and hematoxylin and eosin (H&E) staining, the hepatocarcinogenesis group was divided into three sub-groups: degeneration (n = 6), precancerous (n = 6), and HCC (n = 6). All procedures in vivo were performed according to the guidelines of Animal Care and Use Committee of Nantong University, China.
Cell culture and transfection
Human HCC cells HepG2, SMMC7721, HCCLM3, Huh7, BEL7404 and normal hepatocyte L02 were purchased from Cell Bank of Chinese Academy of Science (Shanghai, China). All cell lines were maintained in Dulbecco modified Eagle medium or RPMI1640 medium supplemented with 10% fetal bovine serum and antibiotics at 37 °C in a humidified incubator with 5% CO2. Three candidate shRNAs were designed by Genema (Shanghai, China). Cell transfection was conducted according to manufacturer’s instructions. Briefly, once cells reached 80% confluence, plasmids were gently transfected into cells using a transfection regent kit. After incubation for 12 h, cells were treated with fresh complete medium. The transfection efficiency was observed using a fluorescence microscope after 24 h. shRNA sequences were as follows: shRNA-1, 5’-GGAAAGTTTGATGGTGCAAAG-3’; shRNA-2, 5’-CGATCATATTGTAGTCTCTCA-3’; shRNA-3, 5’-CCTCCCTATAAATGTGGTAGC-3’; and NC-shRNA, 5’-GGAAGACGATGTCCGGGAAAG-3’.
Histopathological examination
Sections of the formalin-fixed paraffin-embedded (FFPE) tissues were deparaffinized in xylene and rehydrated with a series of graded ethanol. After incubation in hematoxylin solution for 15 min, sections were counterstained in eosin solution using the Hematoxylin and Eosin (H&E) Staining Kit (Solarbio, China) according to the manufacturer’s instructions. Samples were viewed under a light microscope.
Bioinformatics analysis
In order to analyze the mRNA expression of HMGB3 in more HCC samples, gene expression profiling data from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (GSE-14520, GSE-5364, GSE-77314, and GSE-50579, United States), The Cancer Genome Atlas (TCGA) database, and Oncomine database (United States) were incorporated in this study. All data extracted from bioinformatics databases were presented as log2 value.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA v2.2) was performed to discover the differences in biological processes and signaling pathways in transcript levels between high and low HMGB3 expression in GSE-14520 and TCGA. Gene sets were obtained from the Molecular Signatures Database (MSigDB). Enrichment scores (ES) were calculated to estimate genes from a pre-defined gene set. The positive enrichment score indicated that the gene set was considered upregulated while negative score meant downregulated. The number of permutations was set to 1000, and P < 0.05 was considered significantly enriched.
MTT assay
Cell proliferation was detected with the MTT assays. HepG2 cells transfected with shRNA-1 and NC-shRNA were seeded in the 96-well plate at a concentration of 3000 cells/well. Then, MTT solution (0.5 mg/ml) was added to the appropriate wells for the 1st ,2nd, 3rd, and 4th d. Following a four hour incubation, DMSO was added. Then, the absorbance was detected at a wavelength of 490 nm.
Cell cycle
Cells were collected by trypsin and fixed in 70% methanol for 30 min. Then cells were resuspended in PBS containing 50 μg/mL propidium iodide (Invitrogen, United States) for one hour at room temperature. After that, samples were analyzed by a flow cytometer (BD Biosciences, United States). Percentage of each cycle phase was calculated by Modfit software.
Immunohistochemistry
Immunohistochemical analysis was performed as previously described[25]. In brief, tissue samples were fixed with formalin, embedded in paraffin, and cut into 4 μm thick sections. Following incubation at 70 °C for one hour, slides were deparaffinized in xylene and rehydrated with gradient ethanol. Antigen retrieval was conducted using EDTA solution at pH 8.0. After being blocked for one hour, slides were incubated with primary antibody overnight at 4 °C and then with the secondary antibody for two hours at room temperature. After that, slides were visualized by DAB, and counterstained by hematoxylin.
Western blotting
Total protein was extracted from cell lysates using RIPA solution according to the manufacturers’ instructions and separated by 10% SDS-PAGE. Then the samples were transferred onto a PVDF membrane. After blocking with PBS with 5% BSA, the membranes were incubated with primary antibodies at 1:1000 dilution (HMGB3, R&D, United States; Cyclin B1, FEN1, and PCNA, Abcam, United States; GAPDH, CST, United States) at 4 °C overnight. After secondary antibody incubation, the samples were detected using the ECL detection system (Bio-Rad, United States).
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted from tissues using Trizol (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized by using a reverse transcription kit (Invitrogen, CA, United States). Quantitative polymerase chain reaction (qPCR) was conducted by using SYBR Premix Ex Taq kit (Takara, Japan) according to the manufacturer’s instructions. The relative mRNA expression (normalized to GAPDH) was assessed using the 2-ΔΔCt (ΔΔCt=ΔCt[target gene]-ΔCt[GAPDH]) analysis method. The primers in this study were presented in Table 1.
Table 1.
Primers for real-time polymerase chain reaction
| Gene symbol | Species | Primer sequence (5’-3’) | Location |
| HMGB1 | Rat | F: GCTGACAAGGCTCGTTATGAA | 186-205 |
| R: CCTTTGATTTTGGGGCGGTA | 381-361 | ||
| HMGB2 | Rat | F: CGGGGCAAAATGTCCTCGTA | 28-47 |
| R: ATGGTCTTCCATCTCTCGGAG | 155-135 | ||
| HMGB3 | Rat | F: AGGTGACCCCAAGAAACCAAA | 9-29 |
| R: TCAGCAAAATTGACGGGAACC | 119-99 | ||
| HMGB4 | Rat | F: AGACCAGCTAAGGCCCAAG | 12-30 |
| R: CCTTTTCGTGCTTTGAGATGGAT | 172-150 | ||
| HMGB3 | Human | F: CCAAAGGGCAAGATGTCCG | 25-43 |
| R: TTGACAGGGACCTCTGGGTTT | 110-90 | ||
| CCNB1 | Human | F: AATAAGGCGAAGATCAACATGGC | 43-65 |
| R: TTTGTTACCAATGTCCCCAAGAG | 153-131 | ||
| FEN1 | Human | F: ATGACATCAAGAGCTACTTTGGC | 62-84 |
| R: GGCGAACAGCAATCAGGAACT | 142-122 | ||
| PCNA | Human | F: CCTGCTGGGATATTAGCTCCA | 77-97 |
| R: CAGCGGTAGGTGTCGAAGC | 185-167 |
HMGB: High mobility group box; FEN1: Flap structure-specific endonuclease 1; PCNA: Proliferating cell nuclear antigen; F: Forward primer; R: Reverse primer.
Xenograft assay
Male BALB/c nude mice (4-6 wk old) were subcutaneously injected with 2 × 106 HepG2 cells transfected with shRNA-1 or NC-shRNA. Tumor size was measured every four days and calculated according to the formula (Volume= length × width2 × 1/2). Mice were sacrificed at the 30th d after injection. Tumors were weighed and fixed for further immunohistochemistry of HMGB3 and Ki67 (1:50, Abcam, United States). All procedures were approved by Animal care committee of Nantong University.
Statistics
The data in this study are presented as means ± standard deviation (SD) of at least three experiments. Comparisons between groups were performed using Two-tailed Student’s t-test. P values less than 0.05 were considered statistically significant.
RESULTS
Upregulating expression of HMGB3 in hepatocarcinogenesis
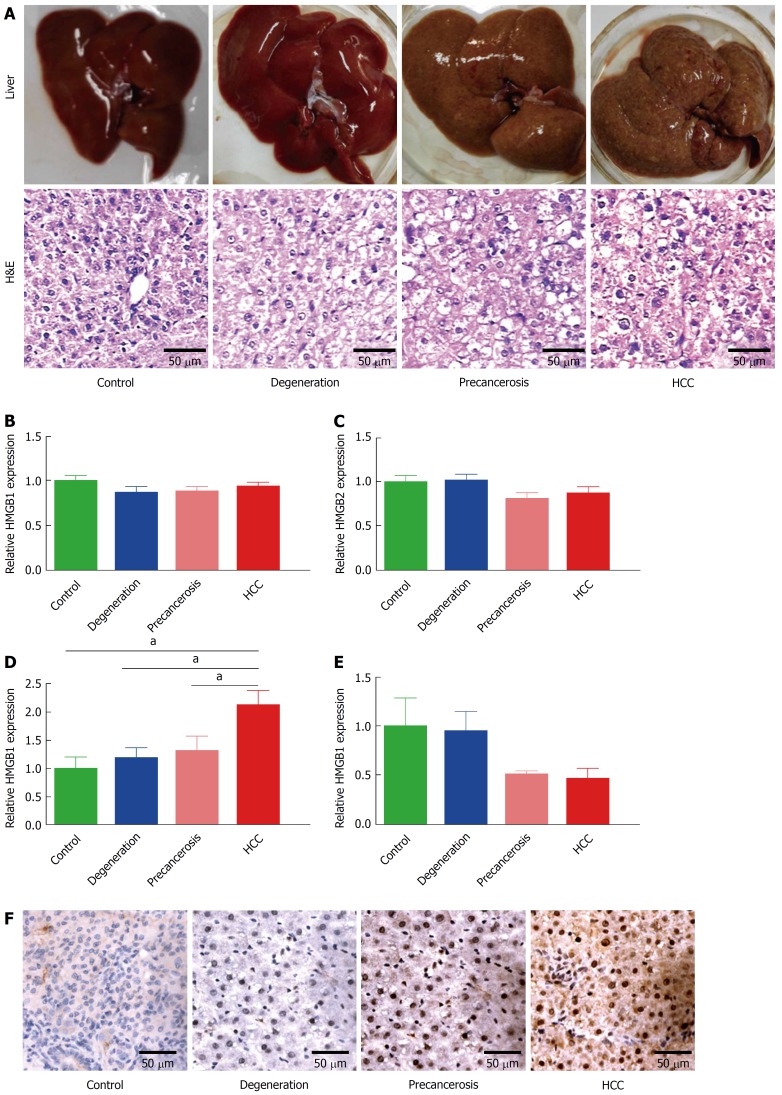
According to the morphological alteration and H&E staining, rats were divided into four groups: normal, degeneration, precancerous, and cancerous. The degeneration group was characterized as the granule-like degeneration in the cytoplasm. The precancerous group was characterized as dense nuclear chromatin and high ratio of nucleus to cytoplasm. The cancerous group showed denser nuclear chromatin, upper ratio of nucleus to cytoplasm, and loss of hepatic structure (Figure 1A). Then expression of the HMGB family (HMGB1, HMGB2, HMGB3, and HMGB4) were detected in liver tissues of each group above by RT-qPCR. No statistically significant changes of HMGB1 (Figure 1B) or HMGB2 (Figure 1C) were found among the four groups. Notably, hepatic HMGB3 expression had a significant increase during the transformation from normal hepatocytes to HCC (Figure 1D). HMGB4 expression was downregulated through the progression to HCC (Figure 1E). The upregulation of hepatic HMGB3 at protein level from normal to cancerous group (Figure 1F) were confirmed by IHC staining.
Figure 1.
Dynamic upregulation of HMGB3 in rat hepatocarcinogenesis. Rat hepatocarcinogenesis models were successfully made by consistent 2-AAF intake. A: The dynamic alterations of liver morphology (upper panel) and H&E staining (lower panel) of liver tissues in rat hepatocarcinogenesis. The livers of the rat model, according to the results of rat liver H&E staining, were divided into normal, degeneration, precancerous, and HCC group. B-E: the dynamic alterations of the HMGB family at the mRNA level in models were detected by RT-qPCR. B: HMGB1 mRNA. C: HMGB2 mRNA. D: HMGB3 mRNA. E: HMGB4 mRNA. Each band was presented as a relative value normalized to normal controls (n = 6). F: the immunohistochemical staining of rat HMGB3 expression in different groups. aP < 0.05. 2-AAF: 2-acetylaminofluorene; H&E: Hematoxylin and eosin; HMGB: High mobility group-box; RT-qPCR: Reverse transcription-quantitative polymerase chain reaction.
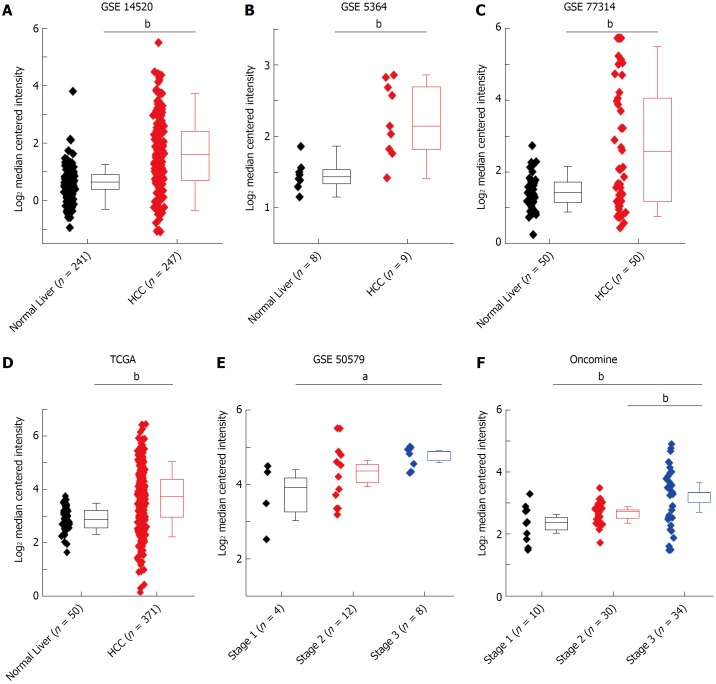
Validation of HMGB3 mRNA by bioinformatics databases
To further verify the HMGB3 expression in human HCC tissues, data from several bioinformatics databases (GEO, TCGA, and Oncomine) of normal livers (n = 359) and HCC tissues(n = 765) were analyzed. Compared with normal livers, HMGB3 had higher expression in HCC tissues according to the GSE14520 (fold change = 1.896, t = 11.270, P < 0.001, Figure 2A), GSE5364 (fold change = 1.720, t = 4.161, P = 0.002, Figure 2B), GSE77314 (fold change = 2.204, t = 4.473, P < 0.001, Figure 2C), and TCGA database (fold change = 1.709, t = 9.125, P < 0.001, Figure 2D). Meanwhile, GSE50597 presented upregulated HMGB3 expression at advance stage of HCC in comparison with that at early stage (fold change = 2.054, t = 3.046, P = 0.012, Figure 2E). Besides, Oncomine database elucidated that higher HMGB3 expression was detected in HCC tissues rather than liver cancer precursor (fold change = 1.469, t = 2.948, P = 0.005) or normal livers (fold change = 1.795, t = 3.380, P = 0.003, Figure 2F). Given the observation above, the data suggested that the upregulation of liver HMGB3 mRNA expression might be related to HCC progression.
Figure 2.
HMGB3 mRNA expression related to hepatocellular carcinoma by bioinformatic databases. Comparative analysis of human normal livers (n = 359) and HCC (n = 765) tissues from bioinformatics databases suggested that the upregulation of hepatic HMGB3 mRNA might be involved in HCC progression. The data of HMGB3 mRNA in HCC or normal liver were extracted from A: GSE-14520, B: GSE-5364, C: GSE-77314, D: the HMGB3 mRNA in TCGA database, E: GSE-50579, and F: the HMGB3 mRNA in Oncomine database. Values were presented as Log2 median centered intensity. aP < 0.05; bP < 0.01.GSE and GEO Series; TCGA: The Cancer Genome Atlas; HCC: Hepatocellular carcinoma.
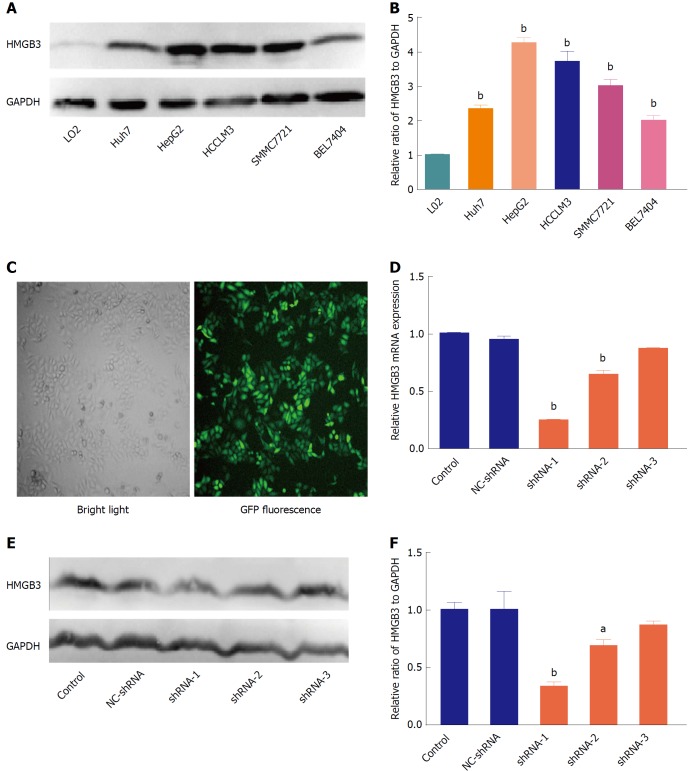
Knockdown of HMGB3 in HCC cell lines
To further determine the role of HMGB3 in HCC progression, HMGB3 expression was detected and silenced in HCC cell lines. In contrast to normal hepatocyte L02, HMGB3 was overexpressed in HCC cell lines Huh7, HepG2, HCCLM3, SMMC7721, and BEL7404 (Figure 3A and 3B). HepG2 had the highest expression of HMGB3 and was chosen to conduct RNAi using three specific shRNAs with GFP labeling (Figure 3C). Compared with the control and NC-shRNA group, HMGB3 expression was significantly (P < 0.001) downregulated with shRNA-1 at the mRNA (Figure 3D) and protein levels (Figure 3E and F).
Figure 3.
Silencing HMGB3 in hepatocellular carcinoma cell lines. A: The protein expression of HMGB3 was detected in different HCC cell lines (Huh7, HepG2, HCCLM3, SMMC7721, and BEL7404), and normal hepatocytes L02 using western blotting. GAPDH was used as an internal reference. B: Each bar represents the corresponding intensity in A normalized to GAPDH. C: Representative morphology of HepG2 cells transfected with GFP-labeling shRNA in bright light and fluorescence. D: RT-qPCR was performed to detect HMGB3 mRNA levels in HepG2 cells transfected with different shRNAs and control. Relative value of HMGB3 was calculated according to the 2−ΔΔCt method. E: Western blotting was conducted to analyze the HMGB3 protein expression in cells of the shRNA-transfected group and control group. F: Each bar represents the corresponding intensity in E normalized to GAPDH. aP < 0.05; bP < 0.01.
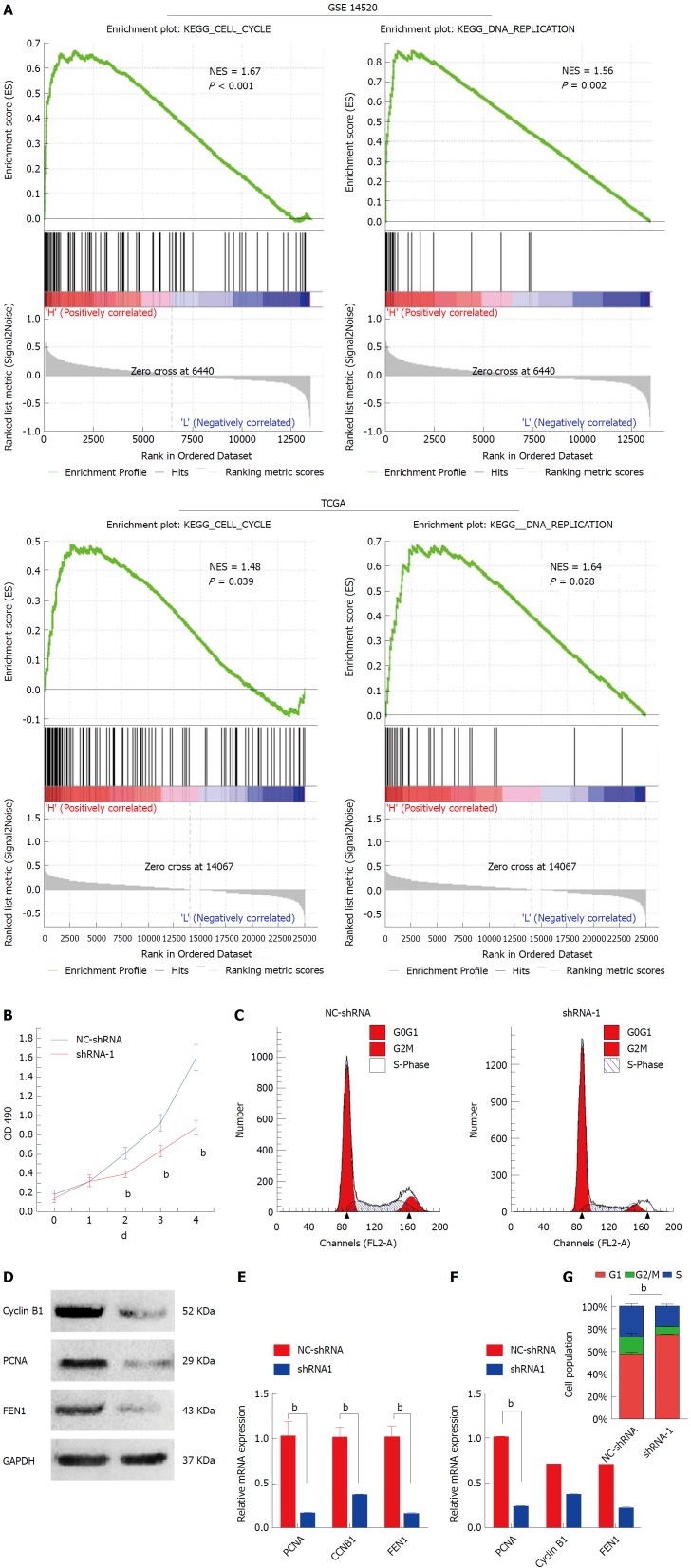
Silencing HMGB3 inhibited proliferation and regulated cell cycle of HCC cells
Gene set enrichment analysis was performed to sort the pathways enriched in distinct phenotype labels according to HMGB3 levels. In both of GSE14520 and TCGA, high expression of HMGB3 was correlated with cell cycle and DNA replication (Figure 4A). Thus, the proliferation activity and cell cycle were studied in HMGB3-knockdown HepG2 cells. As shown in Figure 4B, knockdown of HMGB3 using shRNA-1 significantly inhibited the proliferation of HepG2 cells. Furthermore, silencing HMGB3 could lead to an obvious arrest of HepG2 cells in the G1 phase (Figure 4C and G). In addition, consistent with the results of GSEA, HMGB3 knockdown also inhibited the expression of cell cycle and DNA replication related genes Cyclin B1, proliferating cell nuclear antigen (PCNA) and flap structure-specific endonuclease 1 (FEN1), indicating that HMGB3 might promote the proliferation of HCC cells by regulating cell cycle and DNA replication pathway (Figure 4D-F).
Figure 4.
Knockdown HMGB3 inhibited cell cycle and proliferation of hepatocellular carcinoma cells. A: Gene set enrichment analysis (GSEA) was conducted to sort the pathways according to HMGB3 expression in GSE14520 and TCGA. Cell cycle and DNA replication pathways were found to be significantly correlated with HMGB3 expression. B: Proliferation of HepG2 cells transfected with shRNA-1 and NC-shRNA was detected using the MTT method. C: Cell cycle of HepG2 cells was analyzed after transfection with shRNA using flow cytometry. D: According to the GSEA analysis, three genes (CCNB1, PCNA, and FEN1) involved in cell cycle and DNA replication were detected after shRNA transfection using RT-qPCR. E: Western blotting was conducted to observe the protein expression of cyclin B1, PCNA, and FEN1 in the NC-shRNA and shRNA-1 group. F: Each bar represents the corresponding intensity in E normalized to GAPDH. G: Percentage columns represent the distribution of the cell cycle in corresponding groups. bP < 0.01.
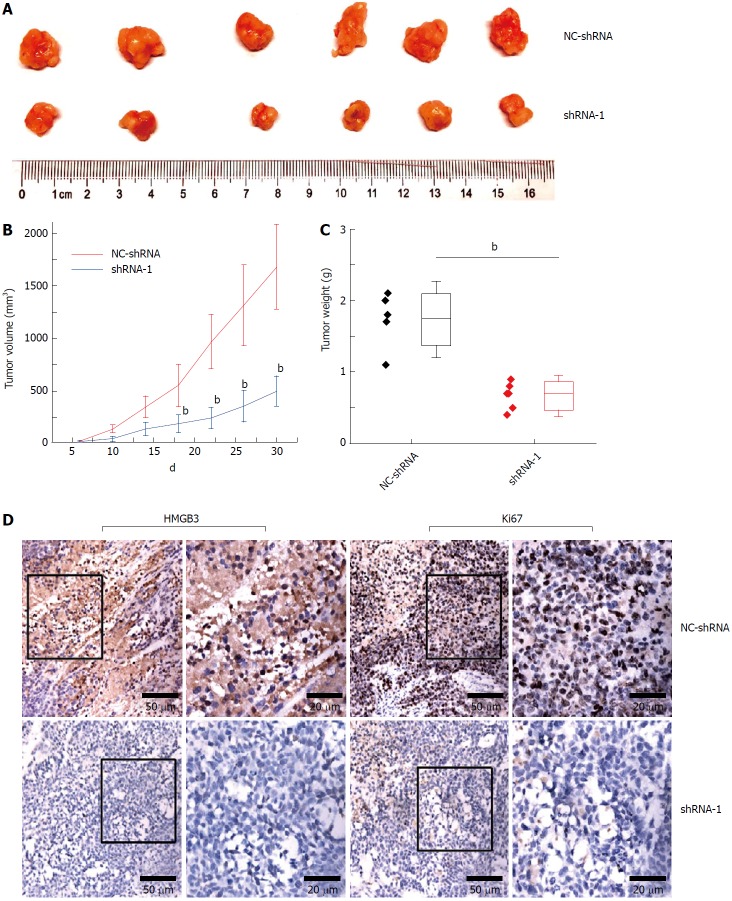
Silencing HMGB3 inhibited tumor growth
Compared with the NC-shRNA group, significantly decreased tumor volume and weight (P < 0.001) was observed in the shRNA-1 group at the 30th d after subcutaneous injection (Figure 5A and C). In addition, silencing HMGB3 by shRNA-1 significantly impeded the growth of xenograft tumors according to the growth curves (Figure 5B). Furthermore, IHC results showed that Ki67 expression in tumor tissues of the shRNA-1 group was significantly lower (P < 0.01) than that of the NC-shRNA group, indicating that HMGB3 might contribute to the proliferation of HCC cells in vivo (Figure 5D).
Figure 5.
Silencing HMGB3 suppressed growth of xenograft tumor. HepG2 cells transfected with NC-shRNA and shRNA-1 were subcutaneously injected into mice. A: The morphology of the xenograft tumor in the NC-shRNA or shRNA-1 group at 30th d. B: The growth curves of tumors derived from HepG2 cells in the NC-shRNA or shRNA-1 group. C: The weight of xenograft tumors in the NC-shRNA or shRNA-1 group. D: The immunochemical staining of HMGB3 and Ki67 in xenograft tumor tissues in the NC-shRNA or shRNA-1 group. bP < 0.01.
DISCUSSION
The HMGB family has been recognized as an important regulator in tumor progression[18]. Although the HMGB family with various physiological and pathological functions was previously associated with liver cancer, neither HMGB1 nor HMGB2 was reported to exhibit dynamic expression in tumorigenesis[26]. In this study, the rat hepatocarcinogenesis model was conducted to analyze the expression characteristics from HMGB1, HMGB2, HMGB3, and HMGB4. No statistical differences in HMGB1 or HMGB2 expression were observed. However, HMGB3 and HMGB4 expression were upregulated and downregulated, respectively through the malignant transformation in vivo. To further verify the HMGB3 expression in HCC, several HCC-related bioinformatics databases were assessed. Interestingly, consistent with the results of HCC model, analyses of normalized log2 transformed microarray expression data sets clearly confirmed the significant upregulation of HMGB3 mRNA in human HCC tissues, and especially in advanced HCC tissues, indicating that HMGB3 might be an oncogenic protein involved in the malignant transformation of hepatocytes.
HMGB proteins can assist in either activating or repressing transcription[27]. In adult vertebrates, HMGB1 is found in all cell types, whereas HMGB3 mRNA was reported to be absent in most adult tissues[7,28]. Indeed, overexpression of HMGB3 has been discovered in several cancer types and correlated with clinical features of cancer patients, including advanced tumor-node-metastasis (TNM) stage, serosal invasion, and overall survival[23,28]. The rat hepatocarcinogenesis model also indicated the potential role of HMGB3 in HCC progression. However, the expression features of HMGB3 and its roles in HCC are still unclear. Thus, the current study further investigated HMGB3 expression in HCC cell lines. Interestingly, overexpression of HMGB3 was observed in HCC cells rather than normal hepatocytes, which was consistent with the hepatocarcinogenesis model and bioinformatic analysis.
HMGB3 has been reported to play crucial roles in tumor progression by contributing to malignant behaviors and regulating oncogenic pathways. For instance, HMGB3 could enhance the migration and growth of gastric cancer cells via activation of the Wnt pathway[23]. It also could increase the proliferation and invasion of breast cancer cells as a target gene of miRNA-205[29]. However, for HCC, the malignant behaviors and mechanism mediated by HMGB3 still remain unclear. Thus, bioinformatic analysis was conducted to explore the underlying roles. Notably, GSEA based on GEO database and TCGA jointly indicated that overexpression of HMGB3 might be associated with cell cycle and DNA replication pathways. As expected, corresponding in vitro studies showed that silencing HMGB3 could significantly inhibit proliferation and induce cell cycle arrest in HCC cells. To further confirm potential mechanisms responsible for the anti-proliferation effects, we explored expression of Cyclin B1, PCNA, and FEN1, which were cell cycle and DNA replication related genes highly enriched in high-HMGB3-mediated pathways in GSEA. Consistent with this prediction, knockdown of HMGB3 obviously downregulated the three genes at the mRNA and protein levels, which have been reported to promote tumor progression[30-32]. Collectively, HMGB3 might promote proliferation of HCC cells by regulating cell cycle and DNA replication pathways.
Although HMGB3 has been correlated with proliferation, chemoresistance, and migration of cancer cells in previous studies[23,33,34], as far as we know, there was no xenograft assays to evaluate HMGB3 as a regulator of tumor growth in vivo. Given the interesting results from the rat model and in vitro study, our current study further evaluated HMGB3 as an important regulator of tumor growth in vivo. Xenograft tumors derived from HMGB3-silenced HCC cells grew slower than the control tumors, with an obvious reduction in the proliferation marker Ki67. It suggested that HMGB3 might contribute to the tumor growth in vivo via regulating proliferation of HCC cells.
In conclusion, to the best of our knowledge, this is the first report to investigate HMGB3 expression and indicate that it may be a novel diagnostic marker or therapeutic target for HCC. Here, the findings are promising, and the initial evidence confirmed that HMGB3 is one of the key molecules in HCC progression. Future studies should clarify the molecular mechanisms of the upregulation of HMGB3 expression and its important role in hepatocarcinogenesis to elucidate how HMGB3 might promote proliferation of HCC cells and tumor growth by regulating cell cycle and DNA replication pathways.
ARTICLE HIGHLIGHTS
Research background
Hepatocellular carcinoma (HCC) is one of the most common and fatal malignancies worldwide with a multi-factorial, multistep, complex process, and poor prognosis. Early diagnosis of HCC at an early stage is of the utmost importance. This found a new molecular biomarker to monitor the malignant transformation of hepatocytes.
Research motivation
Although serum alpha fetoprotein (AFP) level is a useful tumor marker for the detection and monitoring of HCC, the false-negative rate with AFP level alone may be as high as 40% for patients with early stage HCC. Even in patients with advanced HCC, the AFP levels may remain normal in 15%-30% of the patients. New specific markers, such as circulating HS-GGT, HS-AFP or AFP-L3, miRNA, GPC-3, and GP73, have been developed to improve the sensitivity, specificity, early detection, and prediction of prognosis. However, the overall results have been unsatisfactory.
Research objectives
The most urgent needs are to find sensitive markers for early diagnosis or monitor postoperative recurrence, and to give adequate treatment for HCC. It has many characteristics, such as fast infiltrating growth, metastasis in early stage, high-grade malignancy, and poorly therapeutic efficacy, thus the prognosis is poor and early detection is of the utmost importance. The present study focused on exploring the relationship between dynamic expression of HMGB-3 and malignant transformation of hepatocytes.
Research methods
Dynamic models of rat hepatocarcinogenesis were made to investigate the expression of the high mobility group box (HMGB) family. HMGB3 expression was measured at the protein level by immunohistochemistry or Western blotting and at the mRNA level by real time PCR. Human HMGB3 expression was evaluated using bioinformatics databases and its mechanisms were analyzed in vitro. Xenograft growth was also measured.
Research results
HMGB3 expression was upregulated through the malignant transformation of liver cells in vivo.
Research conclusions
The upregulation of liver HMGB3 expression was found by dynamic model of hepatocytes malignant transformation with the alterations of rat liver histopathology. This was confirmed by mining HMGB3 expression in human HCC tissues in bioinformatic databases. Further studies elucidating the role HMGB3 plays in regulating HCC progression, suggests that HMGB3 could be a novel marker for early diagnosis or a molecular therapy target.
Research perspectives
HMGB3 has been confirmed as one of the key molecules in the HMGB family with HCC development. However, the molecular mechanisms of the upregulation of HMGB3 expression and its important role in hepatocarcinogenesis should be clarified in promoting proliferation of HCC cells and tumor growth and regulating cell cycle and DNA replication pathways in the future.
ACKNOWLEDGMENTS
The authors thank Dr. FitzGibbon T for comments on earlier drafts of the manuscript.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional animal care and use committee statement: The study protocol was approved by the Animal Medical Ethics Committee of Affiliated Hospital of Nantong University.
Conflict-of-interest statement: The authors declare no conflicts of interest.
Data sharing statement: No additional unpublished data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 19, 2018
First decision: June 6, 2018
Article in press: June 27, 2018
P- Reviewer: Hann HW, Namisaki T S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Yin SY
Contributor Information
Wen-Jie Zheng, Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Min Yao, Department of Immunology, Medical School of Nantong University, Nantong 226001, Jiangsu Province, China.
Miao Fang, Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Li Wang, Department of Medical Informatics, Medical School of Nantong University, Nantong 226001, Jiangsu Province, China.
Zhi-Zhen Dong, Department of Diagnostics, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Deng-Fu Yao, Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China. yaodf@ahnmc.com.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 2.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM, et al. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–1337. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yi SW, Choi JS, Yi JJ, Lee YH, Han KJ. Risk factors for hepatocellular carcinoma by age, sex, and liver disorder status: A prospective cohort study in Korea. Cancer. 2018;124:2748–2757. doi: 10.1002/cncr.31406. [DOI] [PubMed] [Google Scholar]
- 4.Eso Y, Marusawa H. Novel approaches for molecular targeted therapy against hepatocellular carcinoma. Hepatol Res. 2018;48:597–607. doi: 10.1111/hepr.13181. [DOI] [PubMed] [Google Scholar]
- 5.Trojan J, Waidmann O. Role of regorafenib as second-line therapy and landscape of investigational treatment options in advanced hepatocellular carcinoma. J Hepatocell Carcinoma. 2016;3:31–36. doi: 10.2147/JHC.S112537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst) 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Gerlitz G. HMGNs, DNA repair and cancer. Biochim Biophys Acta. 2010;1799:80–85. doi: 10.1016/j.bbagrm.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajino K, Yamamoto T, Hayashi J, Umeda T, Takahara T, Hino O. Recombination hot spot of hepatitis B virus genome binds to members of the HMG domain protein family and the Y box binding protein family; implication of these proteins in genomic instability. Intervirology. 2001;44:311–316. doi: 10.1159/000050063. [DOI] [PubMed] [Google Scholar]
- 12.Tohme S, Yazdani HO, Liu Y, Loughran P, van der Windt DJ, Huang H, Simmons RL, Shiva S, Tai S, Tsung A. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth through HMGB1 and TLR9 interaction. Hepatology. 2017;66:182–197. doi: 10.1002/hep.29184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, Dong Z, Yang P, Wang X, Jin G, Yu H, Chen L, Li L, Tang L, Bai S, et al. Hepatitis B virus X protein stimulates high mobility group box 1 secretion and enhances hepatocellular carcinoma metastasis. Cancer Lett. 2017;394:22–32. doi: 10.1016/j.canlet.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D, Cao J, Zhong Q, Zeng L, Cai C, Lei L, Zhang W, Liu F. Long noncoding RNA PCAT-1 promotes invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway in hepatocellular carcinoma. Biomed Pharmacother. 2017;95:1187–1193. doi: 10.1016/j.biopha.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 15.Lv G, Wu M, Wang M, Jiang X, Du J, Zhang K, Li D, Ma N, Peng Y, Wang L, et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int. 2017;37:1354–1364. doi: 10.1111/liv.13424. [DOI] [PubMed] [Google Scholar]
- 16.Lu L, Qiu C, Li D, Bai G, Liang J, Yang Q. MicroRNA-505 suppresses proliferation and invasion in hepatoma cells by directly targeting high-mobility group box 1. Life Sci. 2016;157:12–18. doi: 10.1016/j.lfs.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 17.Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, Park SY, Choi YJ, Do IG, Joh JW, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16:5511–5521. doi: 10.1158/1078-0432.CCR-10-0825. [DOI] [PubMed] [Google Scholar]
- 18.Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochim Biophys Acta. 2010;1799:114–118. doi: 10.1016/j.bbagrm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Gong Y, Cao Y, Song L, Zhou J, Wang C, Wu B. HMGB3 characterization in gastric cancer. Genet Mol Res. 2013;12:6032–6039. doi: 10.4238/2013.December.2.1. [DOI] [PubMed] [Google Scholar]
- 20.Song N, Liu B, Wu JL, Zhang RF, Duan L, He WS, Zhang CM. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34:2599–2603. doi: 10.1007/s13277-013-0807-y. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang Y, Luo X, Liu C, Xie J, Cai C. Increased expression of HMGB3: a novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:345–352. [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Wu Y, Liu A, Tang X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477:768–773. doi: 10.1016/j.bbrc.2016.06.133. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L, Hu Y, Zhang F, Li X, Zhang W, Li X. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/β-catenin pathway. PLoS One. 2017;12:e0179741. doi: 10.1371/journal.pone.0179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Cai Y, Zhao H, Xu Z, Sun Q, Luo M, Gu L, Meng M, Han X, Sun H. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol. 2015;36:4785–4792. doi: 10.1007/s13277-015-3130-y. [DOI] [PubMed] [Google Scholar]
- 25.Yan XD, Yao M, Wang L, Zhang HJ, Yan MJ, Gu X, Shi Y, Chen J, Dong ZZ, Yao DF. Overexpression of insulin-like growth factor-I receptor as a pertinent biomarker for hepatocytes malignant transformation. World J Gastroenterol. 2013;19:6084–6092. doi: 10.3748/wjg.v19.i36.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afrin R, Arumugam S, Rahman A, Wahed MI, Karuppagounder V, Harima M, Suzuki H, Miyashita S, Suzuki K, Yoneyama H, et al. Curcumin ameliorates liver damage and progression of NASH in NASH-HCC mouse model possibly by modulating HMGB1-NF-κB translocation. Int Immunopharmacol. 2017;44:174–182. doi: 10.1016/j.intimp.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Yanai H, Ban T, Taniguchi T. High-mobility group box family of proteins: ligand and sensor for innate immunity. Trends Immunol. 2012;33:633–640. doi: 10.1016/j.it.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Tang HR, Luo XQ, Xu G, Wang Y, Feng ZJ, Xu H, Shi YW, Zhang Q, Wu LG, Xue CQ, et al. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol. 2012;18:7319–7326. doi: 10.3748/wjg.v18.i48.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgamal OA, Park JK, Gusev Y, Azevedo-Pouly AC, Jiang J, Roopra A, Schmittgen TD. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One. 2013;8:e76402. doi: 10.1371/journal.pone.0076402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 32.He L, Luo L, Zhu H, Yang H, Zhang Y, Wu H, Sun H, Jiang F, Kathera CS, Liu L, et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol Oncol. 2017;11:640–654. doi: 10.1002/1878-0261.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo S, Wang Y, Gao Y, Zhang Y, Chen M, Xu M, Hu L, Jing Y, Jing F, Li C, et al. Knockdown of High Mobility Group-Box 3 (HMGB3) Expression Inhibits Proliferation, Reduces Migration, and Affects Chemosensitivity in Gastric Cancer Cells. Med Sci Monit. 2016;22:3951–3960. doi: 10.12659/MSM.900880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Nishikawa R, Kato M, Okato A, Arai T, Kojima S, Yamazaki K, Naya Y, Ichikawa T, Seki N. Regulation of HMGB3 by antitumor miR-205-5p inhibits cancer cell aggressiveness and is involved in prostate cancer pathogenesis. J Hum Genet. 2018;63:195–205. doi: 10.1038/s10038-017-0371-1. [DOI] [PubMed] [Google Scholar]