Short abstract
cGMP-dependent kinase-I (cGKI) is known to regulate spinal pain processing. This enzyme consists of two isoforms (cGKIα and cGKIβ) that show distinct substrate specificity and tissue distribution. It has long been believed that the α isoform is exclusively expressed in the adult dorsal root ganglion. The aim of the present study was to reexamine the expression of cGKI isoforms in the adult mouse dorsal root ganglion using isoform-specific cGKI antibodies whose specificities had been validated in the previous studies. Immunoblot and immunohistochemical analyses revealed the presence of both isoforms in the dorsal root ganglion. Moreover, cGKIα was found to be mainly expressed within the cytoplasm of small- to medium-sized peptidergic and nonpeptidegic C-fibers, whereas cGKIβ was located within the nuclei of a wide range of dorsal root ganglion neurons. In addition, glutamine synthetase-positive satellite glial cells expressed both isoforms to varying degrees. Finally, using an experimental model for neuropathic pain produced by L5 spinal nerve transection, we found that cGKIα expression was downregulated in the injured, but not in the uninjured, dorsal root ganglion. In contrast, cGKIβ expression was upregulated in both the injured and uninjured dorsal root ganglions. Also, injury-induced cGKIβ upregulation was found to occur in small-to-medium-diameter dorsal root ganglion neurons. These data thus demonstrate the existence of two differently distributed cGKI isoforms in the dorsal root ganglion, and may provide insight into the cellular and molecular mechanisms of pain.
Keywords: Cyclic GMP-dependent kinase-I, dorsal root ganglion, satellite glial cell, peripheral nerve injury, gene expression
Background
cGMP (cyclic guanosine monophosphate) is a second messenger generated by soluble guanylyl cyclase (sGC) or membrane-bound guanylyl cyclase (mGC) in response to stimulation with nitric oxide (NO) or natriuretic peptides (NPs), respectively.1,2 cGMP is known to play a key role in pathological pain processing, at least in part, via activation of cGMP-dependent kinase-I (cGKI; also known as PKG-I), which is abundantly expressed in the primary afferent neurons and their terminals in the spinal dorsal horn (DH).3,4 Indeed, gene knockout and pharmacological inhibition of cGKI attenuate pain hypersensitivity after peripheral nerve injury and inflammation.5–7 Intriguingly, recent elegant studies with nociceptor-specific cGKI knockout mice have shown that cGKI expressed in nociceptive sensory neurons is a mediator of neuronal hyperactivity in the primary afferents, spinal DH, and pain-related brain regions, including thalamus and cortex, during pathological pain conditions.8,9 Also, these mutant mice show a lack of pain hypersensitivity after intrathecal injection of NO donor or NP,9 indicating that NO–sGC–cGMP and NP–mGC–cGMP pathways are upstream components of cGKI.
Alternative splicing of the Prkg1 gene produces two isoforms of cGKI (cGKIα and cGKIβ) that differ in the first ∼100 NH2-terminal amino acid sequence and show distinct sensitivities to cGMP.2 Their N-termini mediate protein interactions and therefore contribute to the distinct substrate specificity and subcellular localization of the cGKI isoforms.2,10 Moreover, isoform-specific antibodies raised against the N-terminus of these respective enzymes have revealed that cGKIα and cGKIβ are differently distributed in mouse brain and various tissues.11,12 Several studies have demonstrated that the embryonic dorsal root ganglion (DRG) exclusively expresses the α isoform, whose transcript appears at E10.5.13,14 Also, by using commercially available antibodies, Sung et al.15 have shown that cGKIα, but not cGKIβ, is expressed in the adult DRG. In the present study, we reexamined the cellular and subcellular distribution of the α and β isoforms in the adult mouse DRG using the previously well-characterized cGKI isoform-specific antibodies.12 Furthermore, we also assessed the effects of peripheral nerve injury on the expression levels of these cGKI isoforms in the DRG.
Methods
Animals
All animal experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by the animal experimentation committees of Kansai Medical University and Niigata University. Eight- to twelve-week-old male wild-type C57BL/6N mice were used. The neuropathic pain model was prepared by selective transection of the L5 spinal nerve, as described previously.16
Western blot analysis
Total protein (10 μg) extracted from mouse tissues was separated on sodium dodecyl sulfate-polyacrylamide gels (7.5%). cGKIα and cGKIβ were detected by rabbit polyclonal antibodies (1:250) raised against the amino terminus of the respective kinases.12 Also, mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:500; MAB374, Merck Millipore, Temecula, CA, USA) antibody was used. Detection was performed with ECL Prime Western Blotting Detection Reagents (GE Healthcare, Piscataway, NJ, USA).
Immunohistochemistry
Immunohistochemistry was performed on 10-μm sections of L4-6 DRG fixed by transcardial perfusion with 4% paraformaldehyde. Before immunolabeling, antigen unmasking was performed by microwave treatment for 10 min in 10-mM citrate buffer (pH 6.0). The sections were treated with 50% and then 100% methanol for 5 min for each. After having been washed with 0.1% Triton X-100 in PBS (PBST), the sections were incubated at room temperature for 1 h with blocking buffer containing 10% normal donkey serum in PBST; and then they were reacted overnight at 4°C with primary antibodies. Regarding isolectin B4 (IB4) labeling, sections were preincubated for 20 min at room temperature in PBST supplemented with 0.1 mM CaCl2, 0.1 mM MgCl2, and 0.1 mM MnCl2, and then treated overnight at 4°C with 50 μg/ml of fluorescein isothiocyanate-conjugated IB4 (L2895, Sigma-Aldrich, St. Louis, MO, USA) in the preincubation buffer, as described previously.17 We used the following primary antibodies: rabbit anti-cGKIα (1:25),12 rabbit anti-cGKIβ (1:25),12 mouse anti-NeuN conjugated to Alexa Fluor 488 (1:100; MAB377X, Merck Millipore), guinea pig anti-calcitonin gene-related peptide (CGRP; 1:200; CGRP-Gp-Af280, Frontier Institute Co., Ltd., Hokkaido, Japan), mouse anti-neurofilament 200 (1:500; N0142, Sigma-Aldrich), and guinea pig anti-glutamine synthetase (GS; 1:107)18 antibodies. Also, Alexa Fluor 488 anti-mouse, Alexa Fluor 488 anti-guinea pig, and Alexa Fluor 594 anti-rabbit (1:300; Life Technologies, Eugene, OR, USA) were used as secondary antibodies. The fluorescent images were acquired with an OLYMPUS laser scanning confocal microscope (FV1200; OLYMPUS, Tokyo, Japan). For quantification, the DRG sections from 3–4 mice were assessed. DRG neurons with nuclei were analyzed, and the cell size was manually measured using NIH ImageJ software. Also, the average fluorescence intensities within the cell body (both cytoplasm and nucleus) and nucleus were assessed to determine the subcellular distribution of cGKI isoforms. To calculate the percentage of cGKI isoform-labeled neurons in the DRG, we divided the number of cGKI isoform-positive neurons (10-times higher than background signal) by the total number of marker-positive neurons.
Quantitative real-time PCR
RNA was extracted with TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and reverse transcribed with the Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Quantitative real-time polymerase chain reaction (PCR) was performed with a Rotor-Gene Q (Qiagen, Hilden, Germany) and Rotor-Gene SYBR Green PCR Kit (Qiagen, Hilden, Germany). cGKIα and cGKIβ were detected using isoform-specific forward primers (cGKIα: 5′-ATCCGAGAGGTCGAAGGATCT-3′; cGKIβ: 5′-TTCTACCCCAAGAGCCCACA-3′) and a common reverse primer (5′-ATTCCACGGGGTACATACAGT-3′). The primer sequences for GAPDH were 5′-GCCATCAATGACCCCTTCATT-3′ (forward) and 5′-TCTCGCTCCTGGAAGATGG-3′ (reverse). GAPDH was used as an internal control for normalization.
Statistical analysis
The data were analyzed with Student’s t test. The criterion of significance was set at p < 0.05. All results were expressed as the means ± standard error of the mean (SEM).
Results
Distribution of cGKIα-positive immunoreactivity in the adult mouse DRG
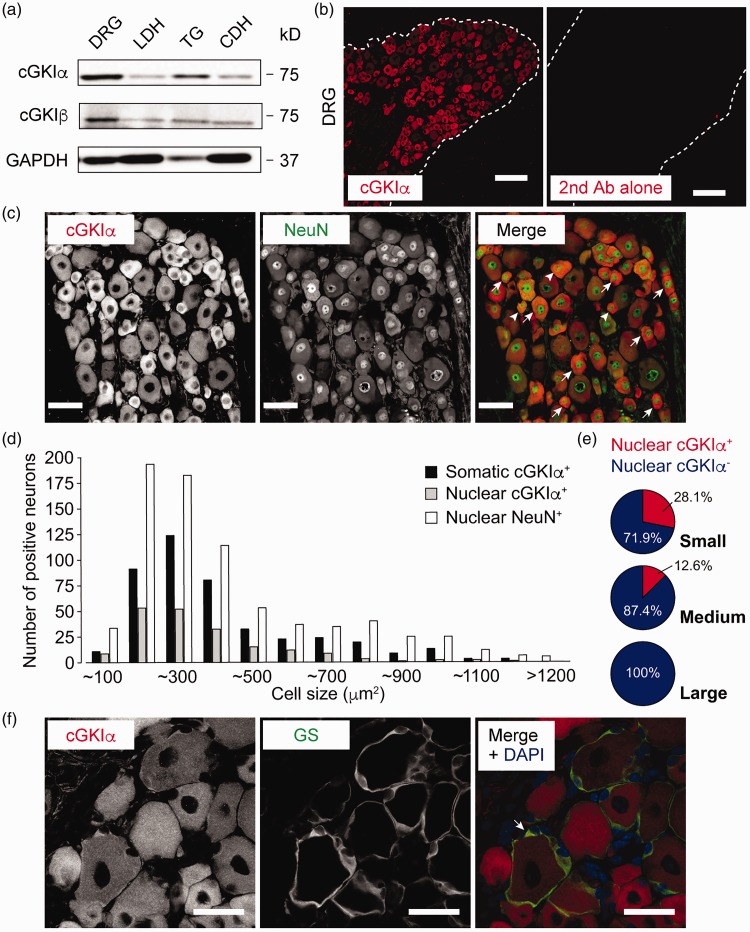
To evaluate the protein expression of cGKI isoforms in adult mouse sensory ganglia and spinal cord, we used the isoform-specific antibodies whose specificities had been earlier validated by immunoblotting and immunohistochemistry with recombinant proteins and/or tissues obtained from wild-type and cGKI-deficient animals.11,12,19 Western blot analysis with the cGKIα antibody revealed a protein band of the predicted size (75 kDa) in the homogenates of DRG, trigeminal ganglion (TG), and lumbar and cervical spinal DHs (Figure 1(a)). Likewise, the 75-kDa cGKIβ-immunoreactive bands were detected in those for the same tissues (Figure 1(a)). DRG and TG tended to express higher levels of cGKI isoforms than their corresponding spinal DHs (Figure 1(a)).
Figure 1.
cGKIα is mainly expressed in the cytoplasm of small- to medium-sized DRG neurons. (a) Western blot showing the protein expression of cGKI isoforms in adult mouse DRG, LDH, TG, and CDH. (b) cGKIα immunolabeling in the DRG. The sections exposed only to secondary antibody had no signals. (c–e) cGKIα is expressed in both the cytoplasm and nuclei of NeuN-positive DRG neurons. Among nuclear NeuN-positive cells, cGKIα-positive cell bodies (arrow) and cells containing nuclear cGKIα (arrowhead) are indicated (c), and their size distributions are shown (d). Pie graphs indicate the percentages of cGKIα-positive and -negative nuclei in small- (n = 615), medium- (n = 143), and large-sized (n = 5) neurons (e). (f) Some of the GS-positive satellite glial cells are also positive for cGKIα. Scale bars: (b) = 100 μm; (c) = 50 μm; (f) = 30 μm.
CDH: cervical dorsal horn; DAPI: 4′,6-diamidino-2-phenylindole; DH: dorsal horn; DRG: dorsal root ganglion; GS: glutamine synthetase; LDH: lumbar dorsal horn; NeuN: neuronal nuclei; cGKI: cyclic GMP-dependent kinase-I.
Immunohistochemical analysis with the cGKIα antibody detected intense signals in the DRG, whereas the secondary antibody used alone gave only background ones (Figure 1(b)). In order to characterize the cellular and subcellular distribution of the cGKIα isoform, we then carried out double immunolabeling for cGKIα and NeuN (neuronal marker) or GS (a marker for satellite glial cells). cGKIα immunoreactivity was found in both the cytoplasm and nuclei of NeuN-positive DRG and TG neurons (Figure 1(c) and data not shown). Quantitative analysis revealed that approximately 56.6% (432 of 763) and 25.0% (191 of 763) of the NeuN-positive DRG neurons were also positive for cGKIα in their cell bodies and nuclei, respectively. Furthermore, size-frequency analysis revealed that small- (<600 mm2 in area) and medium- (600–1200 mm2 in area), but not large- (>1200 mm2 in area) diameter DRG neurons contained cGKIα within their cell bodies as well as, but to a lesser extent, in their nuclei (Figure 1(d) and (e)), indicating that the α isoform was mainly expressed in the cytoplasm. On the other hand, at high magnification, we observed cytoplasmic cGKIα signals in a small number of GS-positive satellite glial cells in the DRG (Figure 1(f)).
cGKIα is mainly expressed in peptidergic and nonpeptidergic C-fiber neurons
Small-diameter DRG neurons can be classified into peptidergic and nonpeptidergic C-fibers that are marked by the expression of CGRP and IB4, respectively. We detected the colocalization of cGKIα and CGRP or IB4 in the DRG (Figure 2(a) and (b)). Quantitative analysis showed that approximately 59.7% of the CGRP-labeled neurons and 63.4% of the IB4-labeled ones were positive for cGKIα in their cell bodies. In addition, cGKIα immunoreactivities were also found in the nuclei of 18.3% and 27.2% of CGRP- and IB4-labeled DRG neurons, respectively. Furthermore, we also observed cGKIα expression in NF200-labeled medium- and large-sized A-fibers (Figure 2(c)). Within the NF200-positive population, approximately 44.7% and 17.9% of these neurons were positive for somatic and nuclear GKIα, respectively. Collectively, these findings suggest that cGKIα was mainly localized within the cytoplasm of peptidergic and nonpeptidergic C-fibers, being consistent with the data from the size-frequency analysis.
Figure 2.
Colocalization of cGKIα with CGRP, IB4, or NF200 in the DRG. (a–c) Double immunolabeling for cGKIα and CGRP (a), IB4 (b), or NF200 (c). Among marker-positive cells, cGKIα-positive cell bodies (arrow) and cells containing nuclear cGKIα (arrowhead) are indicated. Scale bars = 50 μm.
CGRP: calcitonin gene-related peptide; IB4: isolectin B4; cGKI: cyclic GMP-dependent kinase-I.
Distribution of cGKIβ-positive immunoreactivity in the adult mouse DRG
Next, we examined the cellular and subcellular distribution of the cGKIβ isoform in the DRG. cGKIβ was localized in both cytoplasm and nuclei of NeuN-positive DRG and TG neurons (Figure 3(a) and data not shown). Approximately, 43.7% (296 of 677) and 69.3% (469 of 677) of NeuN-labeled DRG neurons were positive for cGKIβ in their cell bodies and nuclei, respectively. Furthermore, the size-frequency analysis indicated that about 92.2% of the cGKIβ-positive cell bodies were small, 7.4% were medium, and 0.3% were large (Figure 3(b)). In contrast, a large proportion of adult DRG neurons, regardless of cell size, contained cGKIβ within their nuclei (Figure 3(b) and (c)), indicating that the β isoform was mainly located within the nuclei. Additionally, we found that cGKIβ was strongly expressed in the cytoplasm of GS-positive satellite glial cells (Figure 3(d)).
Figure 3.
cGKIβ is mainly expressed in the nuclei of a wide range of DRG neurons. (a–c) cGKIβ is expressed in both the cytoplasm and nucleus of NeuN-positive DRG neurons. Among nuclear NeuN-positive cells, cGKIβ-positive cell bodies (arrow) and cells containing nuclear cGKIβ (arrowhead) are indicated (a), and their size distributions are shown (b). Pie graphs indicate the percentages of cGKIβ-positive and -negative nuclei in small- (n = 544), medium- (n = 121), and large-sized (n = 12) neurons (c). (d) cGKIβ is expressed in GS-positive satellite glial cells. Scale bars: (a) = 50 μm; (d) = 30 μm.
DAPI: 4′,6-diamidino-2-phenylindole; GS: glutamine synthetase; NeuN: neuronal nuclei; cGKI: cyclic GMP-dependent kinase-I.
cGKIβ is mainly expressed in the nuclei of peptidergic and nonpeptidergic C-fibers as well as in those of A-fibers
Using double immunolabeling, we detected colocalization of cGKIβ and cell type–specific markers, such as CGRP, IB4, and NF200 (Figure 4(a) to (c)). In agreement with data from the size-frequency analysis, quantitative analysis showed that about 59.0% of the CGRP-labeled neurons, 80.0% of IB4-labeled neurons, and 65.2% of NF200-labeled neurons were positive for nuclear cGKIβ. On the other hand, somatic cGKIβ was expressed in 37.4% of CGRP-labeled neurons, 79.6% of IB4-labeled neurons, and 28.4% of NF200-labeled neurons. Taken together, these observations suggest that cGKIβ was mainly located within the nuclei of a wide range of DRG neurons.
Figure 4.
Colocalization of cGKIβ with CGRP, IB4, or NF200 in the DRG. (a–c) Double immunolabeling for cGKIβ and CGRP (a), IB4 (b), or NF200 (c). Among marker-positive cells, cGKIβ-positive cell bodies (arrow) and cells containing nuclear cGKIβ (arrowhead) are indicated. Scale bars = 50 μm.
CGRP: calcitonin gene-related peptide; IB4: isolectin B4; cGKI: cyclic GMP-dependent kinase-I.
Expression levels of cGKI isoforms are differently regulated by nerve injury
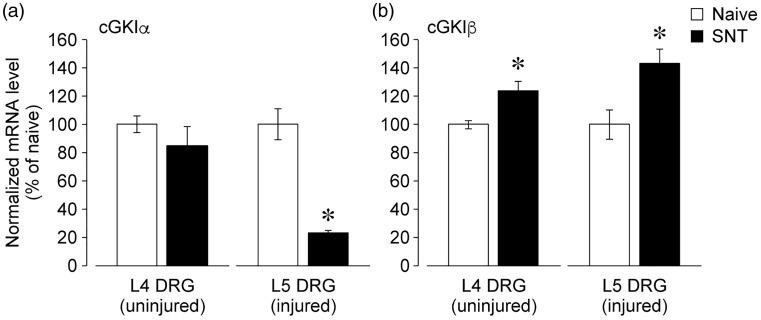
We next assessed whether peripheral nerve injury could affect the expression of cGKI isoforms in the DRG. Seven days after L5 spinal nerve transection, the injured L5 and adjacent uninjured L4 DRG were separately collected to determine the messenger RNA (mRNA) levels in them. Real-time PCR analysis showed that cGKIα expression was reduced in the injured, but not in the uninjured, DRG (Figure 5(a)). In contrast, cGKIβ expression was significantly elevated in both the injured and uninjured DRGs (Figure 5(b)).
Figure 5.
Injury-induced decrease in cGKIα expression and increase in cGKIβ expression in the DRG. (a and b) Real-time PCR analysis of cGKIα (a) and cGKIβ (b) mRNA expression in the injured L5 and uninjured L4 DRG at day 7 after L5 spinal nerve transection. Data are expressed as means ± SEM from experiments using 3–4 mice. *p < 0.05, versus naive mice.
DRG: dorsal root ganglion; mRNA: messenger ribonucleic acid; SNT: spinal nerve transection; cGKI: cyclic GMP-dependent kinase-I.
Nerve injury–induced upregulation of cGKIβ protein expression occurs in small- to medium-sized DRG neurons
To further evaluate the change of expression pattern of cGKIβ protein at day 7 after nerve injury, we assessed cGKIβ immunoreactivity in the naive and injured L5 DRG by performing double immunolabeling experiments. Nerve injury was found to cause an elevation of cGKIβ immunoreactivity in NeuN-positive DRG neurons (Figure 6(a)). Moreover, size-frequency analysis showed that injury-induced cGKIβ upregulation occurred in both cell bodies and nuclei of small- to medium-sized NeuN-positive DRG neurons (n = 666 and 487 cells, respectively, for naive and nerve-injured mice; four mice per group (Figure 6(b)). In contrast, these changes were absent in large-sized NeuN-positive DRG neurons (Figure 6(b)). On the other hand, there were no obvious changes in cGKIβ immunoreactivity in GS-positive satellite glial cells after injury (Figure 6(c)).
Figure 6.
Injury-induced cGKIβ upregulation in the small-to-medium-diameter DRG neurons. The naive and injured L5 DRGs at day 7 after L5 spinal nerve transection were analyzed. (a) Double immunolabeling showing injury-induced cGKIβ upregulation in NeuN-positive cells. (b) Quantification of signal intensity for somatic and nuclear cGKIβ in the small-, medium- and large-sized NeuN-positive DRG neurons. Data are expressed as means ± SEM from experiments using four mice. *p < 0.05, versus naive mice. (c) Double immunolabeling showing no changes of cGKIβ expression level in GS-positive cells after injury. Arrow indicates cGKIβ-positive and GS-positive cell. Scale bars = 50 μm.
DRG: dorsal root ganglion; GS: glutamine synthetase; NeuN: neuronal nuclei; SNT: spinal nerve transection; cGKI: cyclic GMP-dependent kinase-I.
Discussion
To our knowledge, this is the first study reexamining cGKIα and cGKIβ expression in the adult mouse DRG using the isoform-specific cGKI antibodies that had been used earlier to define the spatial expression patterns of cGKI isoforms in mouse brain and tissues.11,12,19 In contrast to the previous studies showing the absence of cGKIβ protein expression in the adult rat DRG,15,20 we here showed that cGKIβ, besides cGKIα, was expressed in the adult mouse DRG and TG, as shown by the results of immunoblot and immunohistochemical analyses. Although the reasons for these discrepancies remain unclear, they may include the difference in antibodies and method used, such as our introduction of microwave treatment for antigen retrieval. Another possible explanation could be the obvious species difference between rat and mouse.
Among DRG neurons, the small- to medium-sized peptidergic and nonpeptidergic C-fibers and NF200-positive A-fibers expressed varying levels of cGKIα and cGKIβ protein in their cytoplasm. Also, both the β and, to a lesser extent, α isoforms were located within the nuclei of small- to medium-sized cells. One obvious difference between the two isoforms was that cGKIβ, but not cGKIα, was expressed in the nuclei of a large portion of the large-sized DRG neurons. Nuclear localization of cGKI has also been observed in many types of cells, including neurons, smooth muscle cells, osteoblasts, and neutrophils.21–24 cGMP binding to cGKI allosteric sites triggers a conformational change in the enzyme, resulting in the exposure of a functional nuclear localization signal.25 Indeed, cGMP causes nuclear translocation of cGKI isoforms.26–28 The subcellular distribution of cGKI isoforms can be affected by interactions with partner proteins. For instance, the inositol receptor–associated cGKI substrate, an endoplasmic reticulum membrane protein, interacts with cGKIβ, but not with cGKIα, to negatively regulate nuclear translocation of the enzyme.27 Thus, it is conceivable that the difference in subcellular localization of cGKI isoforms among sensory fibers could be attributed to the presence of distinct anchoring partners.
Previous studies have demonstrated that cGKI is widely expressed in the DRG neurons;5,9,29 however, to our knowledge, cGKI expression in the satellite glial cells has not been characterized until now. As satellite glial cells form a thin envelope around DRG neurons, besides their characteristic morphology, immunohistochemical staining in combination with marker proteins, such as GS, is required to assess colocalization. In the present study, using high-magnification observation, we obtained evidence that both cGKIβ and, to a lesser extent, cGKIα were expressed in the cytoplasm of GS-positive satellite glial cells. In the DRG, NO mediates signaling from neurons to satellite glial cells.30,31 Indeed, satellite glial cells show an elevation of their cGMP levels in response to NO and nerve trauma.30,32 Accordingly, sGC is expressed in the satellite glial cells, but not in neurons;33 however, there is controversial evidence for neuronal sGC expression.9,34 It is important to note that satellite glial cells are morphologically and functionally heterogeneous35 and that not all satellite glial cells express sGC and synthesize cGMP.32,33,36 Given that sGC is not colocalized with cGKI in the DRG,33 one of the important questions is how cGKI can be activated independently of sGC/cGMP signaling in the satellite glial cells. In this context, oxidants can induce a disulfide-bond formation between two cysteine residues (Cys42) of cGKIα homodimer, thereby causing an activation of the kinase in a cGMP-independent manner.37 Further, the disulfide dimerization of cGKIα is increased in the DRG after nerve injury, and knock-in mice carrying the C42S version of cGKIα show a reduction in neuropathic pain.5 However, a recent paper by Kalyanaraman et al.38 has argued against oxidation-induced cGKIα activation. Another possibility is an activation of cGKI by cyclic adenosine monophosphate39,40 or cyclic cytidine 3′,5′-monophosphate.41 These hypotheses will need to be tested by future studies. On the other hand, in the DRG neurons, cGMP production most likely occurs via NP/mGC signaling.4 Moreover, the activation of mGC-coupled NP receptors type A and B is functionally coupled with cGKI in the DRG neurons.42,43
Previous study has demonstrated that nerve injury reduces cGKIα expression in the DRG.5 Further, we here showed that injury-induced cGKIα downregulation occurred in the injured, but not in the uninjured, DRG. Since the cGKIα downregulation does not occur in the C42S cGKIα knock-in mice, oxidants supposedly regulate cGKIα expression in the DRG after injury.5 As opposed to the α isoform, cGKIβ expression was found to be significantly elevated in both the injured and uninjured DRGs. Intriguingly, injury-induced cGKIβ upregulation occurred in NeuN-positive neurons, but not in GS-positive satellite glial cells. Furthermore, we showed that injury caused an increase in cGKIβ protein expression in small- to medium-sized DRG neurons, but not in large-sized neurons. These observations may imply a possible role of neuronal cGKIβ in neuropathic pain. Unfortunately, however, the genetic and pharmacological tools that specifically target the cGKIβ isoform are still lacking. Interestingly, injury causes cGKI activation and its retrograde transport to the DRG somata.15 cGKI is known to target nuclear proteins, including histone and transcriptional factors, and to regulate gene expression.2 For instance, cGKIβ, but not cGKIα, interacts with transcriptional regulator TFII-I and stimulates TFII-I-mediated transactivation in a cGMP-dependent manner.26 Therefore, it is tempting to hypothesize that nuclear cGKI isoforms could regulate gene expression in the DRG after injury, thereby contributing to neuropathic pain. To elucidate roles of cGKI isoforms in neuropathic pain, cell type- and isoform-specific cGKI knockout mice will be needed in future studies.
Conclusions
The present study demonstrated that the two isoforms of cGKI were differently expressed in the adult mouse DRG and that the expression level of the β isoform was elevated in both the injured and uninjured DRGs. Elucidation of pathological roles of cGKI isoforms and their upstream regulators and downstream effectors will promote a better understanding of the cellular and molecular mechanisms of chronic pain.
Authors’ contributions
HU and SI designed the study. HU, SM, and TK performed the biochemical and animal experiments. HU analyzed the data. MW and JS generated antibodies used in the study. HU and SI wrote the manuscript. All authors read and approved the final manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants-in-aid for scientific research (22390063 to SI, 11J09224 and 17K14959 to HU, and 16K09001 to TK) from the Japan Society for the Promotion of Science.
References
- 1.Schlossmann J, Schinner E. cGMP becomes a drug target. Naunyn Schmiedebergs Arch Pharmacol 2012; 385: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francis SH, Busch JL, Corbin JD, Sibley D. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev 2010; 62: 525–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidtko A, Tegeder I, Geisslinger G. No NO, no pain? The role of nitric oxide and cGMP in spinal pain processing. Trends Neurosci 2009; 32: 339–346. [DOI] [PubMed] [Google Scholar]
- 4.Tegeder I, Scheving R, Wittig I, Geisslinger G. SNO-ing at the nociceptive synapse? Pharmacol Rev 2011; 63: 366–389. [DOI] [PubMed] [Google Scholar]
- 5.Lorenz JE, Kallenborn-Gerhardt W, Lu R, Syhr KM, Eaton P, Geisslinger G, Schmidtko A. Oxidant-induced activation of cGMP-dependent protein kinase Ialpha mediates neuropathic pain after peripheral nerve injury. Antioxid Redox Signal 2014; 21: 1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegeder I, Del Turco D, Schmidtko A, Sausbier M, Feil R, Hofmann F, Deller T, Ruth P, Geisslinger G. Reduced inflammatory hyperalgesia with preservation of acute thermal nociception in mice lacking cGMP-dependent protein kinase I. Proc Natl Acad Sci USA 2004; 101: 3253–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidtko A, Ruth P, Geisslinger G, Tegeder I. Inhibition of cyclic guanosine 5′-monophosphate-dependent protein kinase I (PKG-I) in lumbar spinal cord reduces formalin-induced hyperalgesia and PKG upregulation. Nitric Oxide 2003; 8: 89–94. [DOI] [PubMed] [Google Scholar]
- 8.Gangadharan V, Wang X, Luo C. Cyclic GMP-dependent protein kinase-I localized in nociceptors modulates nociceptive cortical neuronal activity and pain hypersensitivity. Mol Pain 2017; 13: 1744806917701743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo C, Gangadharan V, Bali KK, Xie RG, Agarwal N, Kurejova M, Tappe-Theodor A, Tegeder I, Feil S, Lewin G, Polgar E, Todd AJ, Schlossmann J, Hofmann F, Liu DL, Hu SJ, Feil R, Kuner T, Kuner R. Presynaptically localized cyclic GMP-dependent protein kinase 1 is a key determinant of spinal synaptic potentiation and pain hypersensitivity. PLoS Biol 2012; 10: e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann F, Ammendola A, Schlossmann J. Rising behind NO: cGMP-dependent protein kinases. J Cell Sci 2000; 113(Pt 10): 1671–1676. [DOI] [PubMed] [Google Scholar]
- 11.Feil S, Zimmermann P, Knorn A, Brummer S, Schlossmann J, Hofmann F, Feil R. Distribution of cGMP-dependent protein kinase type I and its isoforms in the mouse brain and retina. Neuroscience 2005; 135: 863–868. [DOI] [PubMed] [Google Scholar]
- 12.Geiselhoringer A, Gaisa M, Hofmann F, Scholssmann J. Distribution of IRAG and cGKI-isoforms in murine tissues. FEBS Lett 2004; 575: 19–22. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt H, Werner M, Heppenstall PA, Henning M, Moré MI, Kühbandner S, Lewin GR, Hofmann F, Feil R, Rathjen FG. cGMP-mediated signaling via cGKIalpha is required for the guidance and connectivity of sensory axons. J Cell Biol 2002; 159: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z, Wang Z, Gu Y, Feil R, Hofmann F, Ma L. Regulate axon branching by the cyclic GMP pathway via inhibition of glycogen synthase kinase 3 in dorsal root ganglion sensory neurons. J Neurosci 2009; 29: 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung YJ, Chiu DT, Ambron RT. Activation and retrograde transport of protein kinase G in rat nociceptive neurons after nerve injury and inflammation. Neuroscience 2006; 141: 697–709. [DOI] [PubMed] [Google Scholar]
- 16.Uchida H, Matsumura S, Okada S, Suzuki T, Minami T, Ito S. RNA editing enzyme ADAR2 is a mediator of neuropathic pain after peripheral nerve injury. FASEB J 2017; 31: 1847–1855. [DOI] [PubMed] [Google Scholar]
- 17.Unezaki S, Sasaki A, Mabuchi T, Matsumura S, Katano T, Nakazawa T, Nishio N, Andoh T, Yamamoto T, Nakatsuka T, Kuraishi Y, Ito S. Involvement of Tyr1472 phosphorylation of NMDA receptor NR2B subunit in postherpetic neuralgia in model mice. Mol Pain 2012; 8: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takasaki C, Yamasaki M, Uchigashima M, Konno K, Yanagawa Y, Watanabe M. Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur J Neurosci 2010; 32: 1326–1336. [DOI] [PubMed] [Google Scholar]
- 19.Schinner E, Schramm A, Kees F, Hofmann F, Schlossmann J. The cyclic GMP-dependent protein kinase Ialpha suppresses kidney fibrosis. Kidney Int 2013; 84: 1198–1206. [DOI] [PubMed] [Google Scholar]
- 20.Tao YX, Hassan A, Haddad E, Johns RA. Expression and action of cyclic GMP-dependent protein kinase Ialpha in inflammatory hyperalgesia in rat spinal cord. Neuroscience 2000; 95: 525–533. [DOI] [PubMed] [Google Scholar]
- 21.Gudi T, Casteel DE, Vinson C, Boss GR, Pilz RB. NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene 2000; 19: 6324–6333. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Krämer A, Robinson PJ. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J 1999; 18: 4549–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broderick KE, Zhang T, Rangaswami H, Zeng Y, Zhao X, Boss GR, Pilz RB. Guanosine 3′,5′-cyclic monophosphate (cGMP)/cGMP-dependent protein kinase induce interleukin-6 transcription in osteoblasts. Mol Endocrinol 2007; 21: 1148–1162. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt TA, Lincoln TM, Pryzwansky KB. Vimentin is transiently co-localized with and phosphorylated by cyclic GMP-dependent protein kinase in formyl-peptide-stimulated neutrophils. J Biol Chem 1991; 266: 21274–21280. [PubMed] [Google Scholar]
- 25.Gudi T, Lohmann SM, Pilz RB. Regulation of gene expression by cyclic GMP-dependent protein kinase requires nuclear translocation of the kinase: identification of a nuclear localization signal. Mol Cell Biol 1997; 17: 5244–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casteel DE, Zhuang S, Gudi T, Tang J, Vuica M, Desiderio S, Pilz RB. cGMP-dependent protein kinase I beta physically and functionally interacts with the transcriptional regulator TFII-I. J Biol Chem 2002; 277: 32003–32014. [DOI] [PubMed] [Google Scholar]
- 27.Casteel DE, Zhang T, Zhuang S, Pilz RB. cGMP-dependent protein kinase anchoring by IRAG regulates its nuclear translocation and transcriptional activity. Cell Signal 2008; 20: 1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desch M, Sigl K, Hieke B, Salb K, Kees F, Bernhard D, Jochim A, Spiessberger B, Höcherl K, Feil R, Feil S, Lukowski R, Wegener JW, Hofmann F, Schlossmann J. IRAG determines nitric oxide- and atrial natriuretic peptide-mediated smooth muscle relaxation. Cardiovasc Res 2010; 86: 496–505. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y, Chao DS, Santillano DR, Cornwell TL, Nairn AC, Greengard P, Lincoln TM, Bredt DS. cGMP-dependent protein kinase in dorsal root ganglion: relationship with nitric oxide synthase and nociceptive neurons. J Neurosci 1996; 16: 3130–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thippeswamy T, Morris R. The roles of nitric oxide in dorsal root ganglion neurons. Ann NY Acad Sci 2002; 962: 103–110. [DOI] [PubMed] [Google Scholar]
- 31.Hanani M. Satellite glial cells in sensory ganglia: from form to function. Brain Res Brain Res Rev 2005; 48: 457–476. [DOI] [PubMed] [Google Scholar]
- 32.Shi TJ, Holmberg K, Xu ZQ, Steinbusch H, de Vente J, Hökfelt T. Effect of peripheral nerve injury on cGMP and nitric oxide synthase levels in rat dorsal root ganglia: time course and coexistence. Pain 1998; 78: 171–180. [DOI] [PubMed] [Google Scholar]
- 33.Schmidtko A, Gao W, Konig P, Heine S, Motterlini R, Ruth P, Schlossmann J, Koesling D, Niederberger E, Tegeder I, Friebe A, Geisslinger G. cGMP produced by NO-sensitive guanylyl cyclase essentially contributes to inflammatory and neuropathic pain by using targets different from cGMP-dependent protein kinase I. J Neurosci 2008; 28: 8568–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruscheweyh R, Goralczyk A, Wunderbaldinger G, Schober A, Sandkühler J. Possible sources and sites of action of the nitric oxide involved in synaptic plasticity at spinal lamina I projection neurons. Neuroscience 2006; 141: 977–988. [DOI] [PubMed] [Google Scholar]
- 35.Nascimento RS, Santiago MF, Marques SA, Allodi S, Martinez AM. Diversity among satellite glial cells in dorsal root ganglia of the rat. Braz J Med Biol Res 2008; 41: 1011–1017. [DOI] [PubMed] [Google Scholar]
- 36.Thippeswamy T, Morris R. Evidence that nitric oxide-induced synthesis of cGMP occurs in a paracrine but not an autocrine fashion and that the site of its release can be regulated: studies in dorsal root ganglia in vivo and in vitro. Nitric Oxide 2001; 5: 105–115. [DOI] [PubMed] [Google Scholar]
- 37.Burgoyne JR, Madhani M, Cuello F, Charles RL, Brennan JP, Schröder E, Browning DD, Eaton P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science 2007; 317: 1393–1397. [DOI] [PubMed] [Google Scholar]
- 38.Kalyanaraman H, Zhuang S, Pilz RB, Casteel DE. The activity of cGMP-dependent protein kinase Ialpha is not directly regulated by oxidation-induced disulfide formation at cysteine 43. J Biol Chem 2017; 292: 8262–8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White RE, Kryman JP, El-Mowafy AM, Han G, Carrier GO. cAMP-dependent vasodilators cross-activate the cGMP-dependent protein kinase to stimulate BK(Ca) channel activity in coronary artery smooth muscle cells. Circ Res 2000; 86: 897–905. [DOI] [PubMed] [Google Scholar]
- 40.Burnette JO, White RE. PGI2 opens potassium channels in retinal pericytes by cyclic AMP-stimulated, cross-activation of PKG. Exp Eye Res 2006; 83: 1359–1365. [DOI] [PubMed] [Google Scholar]
- 41.Desch M, Schinner E, Kees F, Hofmann F, Seifert R, Schlossmann J. Cyclic cytidine 3',5'-monophosphate (cCMP) signals via cGMP kinase I. FEBS Lett 2010; 584: 3979–3984. [DOI] [PubMed] [Google Scholar]
- 42.Zhang FX, Liu XJ, Gong LQ, Yao JR, Li KC, Li ZY, Lin LB, Lu YJ, Xiao HS, Bao L, Zhang XH, Zhang X. Inhibition of inflammatory pain by activating B-type natriuretic peptide signal pathway in nociceptive sensory neurons. J Neurosci 2010; 30: 10927–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishimoto I, Tokudome T, Horio T, Soeki T, Chusho H, Nakao K, Kangawa K. C-type natriuretic peptide is a Schwann cell-derived factor for development and function of sensory neurones. J Neuroendocrinol 2008; 20: 1213–1223. [DOI] [PubMed] [Google Scholar]