Abstract
Aging contributes to cellular stress and neurodegeneration. Our understanding is limited regarding the tissue-restricted mechanisms providing protection in postmitotic cells throughout life. Here, we show that spinal cord motoneurons exhibit a high abundance of asymmetric dimethyl arginines (ADMAs) and the presence of this posttranslational modification provides protection against environmental stress. We identify protein arginine methyltransferase 8 (PRMT8) as a tissue-restricted enzyme responsible for proper ADMA level in postmitotic neurons. Male PRMT8 knock-out mice display decreased muscle strength with aging due to premature destabilization of neuromuscular junctions. Mechanistically, inhibition of methyltransferase activity or loss of PRMT8 results in accumulation of unrepaired DNA double-stranded breaks and decrease in the cAMP response-element-binding protein 1 (CREB1) level. As a consequence, the expression of CREB1-mediated prosurvival and regeneration-associated immediate early genes is dysregulated in aging PRMT8 knock-out mice. The uncovered role of PRMT8 represents a novel mechanism of stress tolerance in long-lived postmitotic neurons and identifies PRMT8 as a tissue-specific therapeutic target in the prevention of motoneuron degeneration.
SIGNIFICANCE STATEMENT Although most of the cells in our body have a very short lifespan, postmitotic neurons must survive for many decades. Longevity of a cell within the organism depends on its ability to properly regulate signaling pathways that counteract perturbations, such as DNA damage, oxidative stress, or protein misfolding. Here, we provide evidence that tissue-specific regulators of stress tolerance exist in postmitotic neurons. Specifically, we identify protein arginine methyltransferase 8 (PRMT8) as a cell-type-restricted arginine methyltransferase in spinal cord motoneurons (MNs). PRMT8-dependent arginine methylation is required for neuroprotection against age-related increased of cellular stress. Tissue-restricted expression and the enzymatic activity of PRMT8 make it an attractive target for drug development to delay the onset of neurodegenerative disorders.
Keywords: ADMA, aging, CREB1, motoneuron, neurodegeneration, PRMT8
Introduction
The rate of cellular aging and the appearance of age-related pathologies are modulated by highly conserved stress response and repair pathways (Haigis and Yankner, 2010). Accumulating evidence indicates that the longevity of a cell or organism depends on its ability to properly regulate signaling pathways that counteract perturbations, such as protein misfolding, DNA damage, or oxidative stress (Kourtis and Tavernarakis, 2011). However, stress responses must fundamentally differ between proliferating cells, which can be easily replaced, and cells that are terminally differentiated and nonreplaceable, such as postmitotic neurons. In proliferating cells, the risk of propagating cellular defects is great, which can be potentially threatening the survival of the entire organism. Therefore, elimination of damaged proliferating cells is the most straightforward solution. In contrast, in long-lived neurons, this could damage the integrity of the complex neural network, so alternative mechanisms need to be used to properly maintain the homeostasis of these cells (Herrup and Yang, 2007; Kole et al., 2013). Identifying and understanding molecular mechanisms regulating cellular stress resistance in postmitotic neurons is important for defining novel therapeutic targets in late-onset neurodegenerative diseases.
Posttranslational modifications (PTMs), such as methylation, acetylation, and phosphorylation are crucial for balanced functions of stress response pathways and dysregulated PTMs can potentially lead to a pathological state (Lake and Bedford, 2007; Dantuma and van Attikum, 2016). Arginine is a positively charged residue that is often found in protein motifs, including the RGG/RG motifs (Thandapani et al., 2013). The RGG sequences within these motifs are substrate recognition sites for type I and type II protein arginine methyltransferases (PRMTs) catalyzing monomethylation and asymmetric or symmetric dimethylation of arginine residues, respectively (Bedford and Clarke, 2009; Blanc and Richard, 2017). In the last decade, hundreds of methylarginine proteins have been detected by mass spectrometry and other proteomic and molecular techniques (Hart-Smith et al., 2012; Bremang et al., 2013; Guo et al., 2014; Auburger et al., 2016). These methyltransferase substrate proteins are involved in transcriptional regulation (TAF15, CRTC2), pre-mRNA splicing (FUS, G3BP1, hnRNP A1, DDX3X, SAM68), mRNA translation (EIF3, EIF4), DNA-damage response (MRE11, 53BP1, FUS), and regulation of apoptosis (PABP1). Prior in vitro studies established that asymmetric arginine dimethylation of amyotrophic lateral sclerosis (ALS)-related FUS, TAF15, hnRNPA1, and methylation of many others is required for adequate stress response (Liu and Dreyfuss, 1995; Tradewell et al., 2012; Yamaguchi and Kitajo, 2012; Shorter and Taylor, 2013). Other studies implicated the involvement of arginine methylation in DNA damage response through the methylation of MRE11 and 53BP1 (Boisvert et al., 2005a,b). Together, these results strongly indicate a regulatory role for asymmetric arginine methylation in the regulation of stress response of neurons and the pathogenesis of neurodegenerative diseases.
Thus far, 11 members of this enzyme family have been identified (Bedford and Clarke, 2009; Blanc and Richard, 2017). PRMT8 is a unique member of the family because it shows highly tissue-specific expression by being restricted to the CNS (Taneda et al., 2007; Kousaka et al., 2009). PRMT8 has been shown to act as a posttranslational modifier of various proteins (Kim et al., 2008; Pahlich et al., 2008). Similar to PRMT1, its closest paralog, PRMT8, is involved in the epigenetic control of gene expression and normal function of neurons (Simandi et al., 2015). However, the in vivo biological role of PRMT8 in the CNS and the mechanisms resulting in neural defects remain largely unknown except for very recent studies that described the role of PRMT8 in Purkinje cells (Kim et al., 2015) and excitatory synaptic function (Penney et al., 2017).
Here, we show that asymmetric dimethyl arginine (ADMA) level declines during embryonic development in the mouse. Strikingly, choline acetyltransferase (ChAT)+ MNs selectively maintain high ADMA level in the adult spinal cord. Fused in sarcoma (FUS), a prominent arginine methyltransferase target, shows similar cell-type-restricted expression. Inhibition of methyltransferases results in accumulation of DNA double-stranded breaks (DSBs), altered FUS kinetics at DNA DSBs, and overall a more vulnerable cellular state and decreased cellular viability. We show that, among the asymmetric arginine methyltransferases, PRMT8 is selectively expressed in the spinal cord MNs. Loss of PRMT8 results in a progressive decrease in muscle strength due to the dysfunction and gradual loss of MNs in aging animals. The persistent stress in the absence of methyltransferase activity leads to decreased cAMP response-element-binding protein 1 (CREB1) level and insufficient activation of prosurvival and regeneration gene network in response to aging-related oxidative and ER stress. This work is a proof-of-concept showing that ADMA has a nonproliferation-associated, cell-type-restricted role and is required for proper stress response in postmitotic neurons. Based on our findings, PRMT8 should be considered as a novel component of neural stress resistance and a target for future drug discovery in neurodegenerative diseases.
Materials and Methods
Cell culture.
Undifferentiated NSC34 cells were maintained in proliferation media (DMEM, 10% FBS, 1% antibiotics). For differentiation, cells were seeded at a concentration of 5000 cells/cm2 and, 24 h after plating, medium was changed to differentiation medium (1:1 DMEM/F12, 1% FBS, 1% nonessential amino acid, and 1 μm all-trans retinoic acid). Cells were allowed to differentiate up to 4 d and differentiation medium was changed on the second day. Differentiated cells were treated with stress inducers for 3 h before RNA isolation and 12 h for immunoblot analysis. Endoplasmic Reticulum (ER) stress was induced by thapsigargin (Millipore) in a 0.2 μm (cell viability assay) or 1 μm (gene expression, protein analysis) final concentration. Sodium arsenite (Sigma-Aldrich) was used in a 0.1 μm (cell viability assay) or 10 μm concentration (gene expression, protein analysis) to induce oxidative stress. ADOX (Sigma-Aldrich) was used in a 40 μm final concentration. Human induced pluripotent stem cells (iPSCs) were differentiated to motoneurons (MNs) stress as described previously (Das et al., 2007).
Gene-silencing assays.
siRNA constructs were purchased from Sigma-Aldrich for targeting the mouse Creb1 (Mm02_00289365) and Prmt8 (Mm01_00128129). MISSION siRNA fluorescent universal negative control #1 (Sigma-Aldrich) was used as a control. NSC34 cells were plated and differentiated 2 d before transfection. Transfection of NSC34 cells was performed using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer's instructions.
Mice.
The PRMT8-null mouse line was generated by the European Conditional Mouse Mutagenesis Program and Knockout Mouse Project (EUCOMM/KOMP) and Sanger Mouse Genetics Project (EPD0105_1_A03) and was purchased from the European Mouse Mutant Archive (EM:04479). For genotyping of PRMT8 knock-out mice, tail DNA was extracted and multiplex allele-specific PCR was performed using the following PCR primers: forward (P1), 5′-CCTGGCACTTTGAGGTGTTG-3′, and reverse (P2), 5′-GTCTGATGGAATGGGCCTG-3′, which generated a 380 bp product for the Prmt8 wild-type allele; and reverse (P3), 5′-TCGTGGTATCGTTATGCGCC-3′, which generated a 252 bp product for the Prmt8 knock-out allele. Male mice were used for the experiments.
Onset ages of the disease in G93A SOD1-transgenic mice were determined as the time when the animals reached peak body weight and the end stage of the disease were determined when the animals could not right themselves within 10 s when placed on their side, typically ∼12–15 weeks of age.
Animals that underwent surgical maneuvers (both recovery and terminal surgeries) were anesthetized with ketamine–xylazine combined intramuscular anesthesia as follows: ketamine hydcrochloride (Ketavet; Pharmacia & Upjohn) at 110 mg/kg body weight and xylazine (Rompun; Bayer) at 12 mg/kg body weight.
Grip strength, Rotarod, and limb clasping.
Phenotyping characterization was performed in the Department of Behavioral Neurobiology (Institute of Experimental Medicine of the Hungarian Academy of Sciences, Budapest, Hungary). For grip strength measurement, animals were held by the base of the tail and allowed to grasp a steel grip gauge (2 mm) with their forepaws. Then, the mice were gently pulled away from the grip gauges in a steady fashion until the grip was released. The force exerted at the gauge at the time of grip release is determined by the mouse itself and is recorded as the grip force. At each time point, forelimb forces were the average of five measurements.
For the accelerating Rotarod test, the rotating rod apparatus (ITC Life Science) was used to measure the motor coordination and ability of mice to improve motor skill performance with training. Mice were placed on a rod and the rod was accelerated from 0 to 45 rpm in 2 min. The time periods that individual mice spent on the rod without falling were recorded. Three trials with a 30 min intertrial interval were performed every day for 3 d. Hindlimb position forelimb clasping was observed and scored as described by Guyenet et al. (2010).
Muscle tension recording.
Animals that were 3, 6, and 12 months old were randomly selected for tension recording (n = 5 in both the control and the knock-out groups) and anesthetized with ketamine–xylazine at the 3, 6, and 12 months survival period and the tibialis anterior (TA) and extensor digitorum longus (EDL) muscles of hindlimb were prepared for tension recording. The distal tendons were dissected free and attached to strain gauges and the exposed parts of the muscles were kept moist with Krebs' saline solution. Isometric contractions were then elicited from the muscles by stimulating the common peroneal nerve with bipolar electrodes. The length of each muscle was adjusted so as to produce the maximum twitch tension. Single twitch and tetanic (40–100 Hz) contractions were displayed and recorded on a computer; all the additional recording hardware and software were developed by Supertech (Kellényi system). Maximum tetanic tension was achieved at a stimulation frequency of ∼100 Hz. An estimate of the numbers of motor axons supplying the muscles was obtained by stimulating the common peroneal nerve with stimuli of increasing intensity and recording the stepwise increments of twitch contractions.
Force measurements in single myocyte preparations.
Force development during isometric contractions was measured in fast skeletal muscle fibers from EDL muscles of 3- and 12-month-old control and PRMT8-null mice. Permeabilized EDL myocytes were mounted between two thin needles with silicone adhesive and viewed under an inverted microscope. One needle was attached to a force transducer and the other to an electromagnetic motor. The measurements were performed at 15°C and the average sarcomere length was adjusted to 2.3 μm. The generation of total isometric force production was measured at saturating Ca2+ concentration (pCa 4.75). When a steady force level was reached, myocyte length was reduced by 20% within 2 ms and then quickly restretched. As a result, the force first dropped from the peak isometric level to zero (difference = maximal Ca2+-activated force) and then started to redevelop. After 8 s of force redevelopment, the myocyte was transferred into the relaxing solution. Maximal Ca2+-activated force was normalized to cross-sectional area.
Muscle histochemistry.
Motor end plates and their axons were visualized by using a combined silver impregnation and acetylcholine esterase histochemistry as described previously (Namba et al., 1967).
Retrograde labeling.
The operations were performed under deep ketamine–xylazine anesthesia as described above using sterile procedures. On the left side, the sciatic nerve was sectioned and the proximal stump of the nerve was covered with a few Fast Blue crystals. Five animals per group were used (from 6- and 12-month-old control and the knock-out groups). Four days after the application of this fluorescent dye, the animals were reanesthetized and perfused transcardially with 4% paraformaldehyde in 0.1 mol/L phosphate buffer, pH 7.4.
Quantification of motor fibers of the L4 ventral root.
Semithin sections were cut from L4 ventral root (n = 5 in each group). Remnants of fixative were carefully washed out from the root and the tissue was treated in 1% OsO4 (Agar Scientific) in PBS for 1 h, dehydrated in a graded ethanol series and propylene oxide, and then embedded in Durcupan (Fluka). Semithin sections (0.5 μm) were cut from the L4 ventral root on a Leica Ultracut-R ultramicrotome. Images of the whole cross-sectional area of the nerve were taken with an Olympus DP70 camera attached to an Olympus BX-50 microscope. Myelinated fibers were counted with the aid of ImageJ image analysis software. Myelinated axons displaying pathological alterations were not included in the axon counts.
Cell counts and quantification of neuromuscular junction (NMJ) pathological alterations.
The numbers of retrograde labeled cells and/or anti-vesicular acetylcholine transporter (VAChT)-positive cells were determined on 25-μm-thick serial cryostat sections. To avoid double counting of neurons present in consecutive sections, the retrograde labeled neurons and VAChT+ MNs were mapped with the aid of an Olympus drawing tube and their locations compared with those of the labeled neurons in the previous section (Nógrádi et al., 2007; Pintér et al., 2010).
To analyze the denervated NMJ structures, every third longitudinal section of TA and EDL muscles processed for a combined silver–acetylcholinesterase histochemistry were mounted onto gelatin-coated glass slides. Morphological characterization of NMJ changes was scored based according to recent studies (Bruneteau et al., 2015) and (Valdez et al., 2010). Normal NMJs have a normal postsynaptic apparatus and a terminal arborization of the axon on the presynaptic side. Fragmented NMJs are marked by their structure broken up into three or more fragments. Hypertrophic NMJs were characterized by enlarged postsynaptic elements. Flattened NMJs were faintly visible and the area of NMJ was flattened and considerably decreased. The unclassified group of NMJs contains those motor end plates that could not be classified into any of the above groups.
In intact muscles, subterminal axons running to a NMJ do not divide. Sprouts develop when the axon supplying a motor end plate splits before innervating the NMJ (preterminal sprout) or beyond the NMJ (ultraterminal sprout) in any direction. Preterminal and ultraterminal sproutings were determined in 6- and 12-month-old PRMT8-null mice by counting the sprouts in every third section containing the innervations zone of TA and EDL muscles.
Electron microscopy.
Experiments were performed on 3- and 12-month-old PRMT8 knock-out and wild-type mice. The animals were deeply anesthetized with sodium pentobarbital (50 mg/kg, i.p.) and transcardially perfused first with Tyrode's solution (oxygenated with a mixture of 95% O2, 5% CO2), followed by a fixative containing 2.5% glutaraldehyde, 0.5% paraformaldehyde, and 0.2% picric acid dissolved in 0.1 m phosphate buffer (PB, pH 7.4). The L3–L5 segments of the spinal cord, the L4 ventral root, the sciatic and phrenic nerves, and the TA and EDL muscles were removed and postfixed in the same fixative for 1–2 h. Following extensive washes in 0.1 m phosphate buffer, the tissue blocks were treated with 1% OsO4 for 1 h and then dehydrated and embedded into Durcupan ACM resin (catalog #44610-1EA; Sigma-Aldrich). Ultrathin sections were cut, collected on Formwar-coated single-slot grids, and counterstained with uranyl acetate and lead citrate. Sections were investigated in JEOL JEM 1010 transmission electron microscope and photographed with an Olympus Veleta slow scan cooled CCD camera. Images were stored in an IBM PC and processed with Adobe Photoshop CS5 software (SciRes_000161).
PRMT8 antibodies.
Cellular localization of endogenous PRMT8 has been debated due to the lack of a specific antibody (Lee et al., 2005; Kousaka et al., 2009). In this work, we used three antibodies. P90742 PP1/PP13 rabbit polyclonal antibodies were developed by Cell Signaling Technology and used for immunoblotting only.
We also developed a new antibody for immunohistochemical detection of PRMT8. Briefly, the synthetic gene encoding for the N-terminal amino acids (Ala2-Glu55, AENAVESTEVSSAPPQPPQPVIPAKPVQCVHHVSTQPSCPGRGKMSKLLNPEE, mPRMT8-54) was subcloned into pUC57-Simple vector and then further subcloned into pGEX-6P-1 vector (Sigma-Aldrich). pGEX-6P-1-mPRMT8-54 was transformed into Rosetta (DE3) pLysS (Novagen) and transformants were tested for GST-mPRMT8-54 overexpression. The fusion protein was purified via GST affinity chromatography and a subsequent desalting step on Bio-Rad PROFINIA Automated Affinity Purification System. PreScission Protease (GE Healthcare), provided as 2 units/μl in storage buffer (50 mm Tris-HCl, 150 mm NaCl, 10 mm EDTA, 1 mm DTT, pH 8.0, 20% glycerol), was used to digest GST-mPRMT8-54 (Mr 32 kDa). Purity and homogeneity of the mPRMT8-54 peptide preparations was investigated using SDS-PAGE analysis with Coomassie staining. Immunization of two rabbits (rabbit 1341032 and 134033) for mPRMT8 polyclonal antibody development against the N-terminal Ala2-Glu55 peptide was performed by Covalab. Further details are available upon request. To verify the specificity of the antibody, we tested the antibody on mouse brain and spinal cord sections obtained from wild-type and PRMT8 knock-out animals. We could detect strong nuclear localization of PRMT8 in the cortex and in Purkinje cells, confirming essentially a previous report documenting PRMT8 mRNA distribution in these tissues (Taneda et al., 2007).
Immunostaining and confocal microscopy.
The mice were killed under deep ketamine–xylazine anesthesia (ketamine hydrochloride, 110 mg/kg body weight; xylazine [Rompun] 12 mg/kg body weight) and perfused transcardially with 4% paraformaldehyde in PBS. The spinal cords were dissected and the lumbar segments (L4) were identified using the ribs and vertebrae as a guide. Spinal cords, L4 spinal nerves, and phrenic nerves were removed, along with the TA and EDL muscles. Serial transverse sections (25 μm thick) were cut from the spinal cords on a cryostat (Leica CM 1850) and mounted onto gelatin-coated glass slides, which were placed into a −20°C fridge until further use. Nonspecific binding sites were subsequently blocked with 1% solution (v/w) made of skimmed milk powder. Spinal cord cross-sections were incubated overnight at 4°C with anti-PRMT8 (noncommercial, see “PRMT8 antibody” section, VAChT; Synaptic Systems), anti-ChAT (Merck-Millipore, ab-144p) anti-MBP (sc-13914; Santa Cruz Biotechnology), anti-asymmetric di-methyl arginine motif (13522S; Cell Signaling Technology), anti-GFAP (ab4674; Abcam), and anti-γH2AX (AF2288; R&D Systems) antibodies. The antigen–antibody reactions were visualized through the use of various fluorescent-conjugated Alexa Fluor secondary antibodies for 1 h at room temperature. Mounting medium containing DAPI (H1500, Vectashield; Vector Laboratories) was used before covering slides. Fluorescence signals were detected in an Olympus BX50 epifluorescence microscope equipped with a DP70 digital camera (Olympus) or in an Olympus Fluoview FV10c-W3 compact confocal microscope. Quantification of immunohistochemistry results was performed by ImageJ.
Immunoblotting.
Total extracts of the spinal cords or NSC34 cells were prepared by homogenization in RIPA buffer containing protease inhibitors. Then, 20 μg of protein extract was separated by SDS-PAGE in 4–20% gel (Bio-Rad) and then transferred to Immobilon-P Transfer Membrane (Millipore). Membranes were probed with anti-BIP (C50B12; Cell Signaling Technology), anti-γH2AX (AF2288; R&D Systems), anti-asymmetric di-methyl arginine motif (13522S; Cell Signaling Technology), anti-GFAP (ab4674; Abcam), anti-ChAT (NBP1-30052; Novus), anti-FUS (IHC-00074; Bethyl), anti-GAPDH (sc-32233; Santa Cruz Biotechnology), anti-MBP (sc-13914; Santa Cruz Biotechnology), anti-αTubulin (sc-8035; Santa Cruz Biotechnology), anti-CREB1 (06-863; Millipore), anti-FLAG (F1804; Sigma-Aldrich), and anti-GFP (ab290; Abcam) antibodies according to the manufacturer's recommendations. Methylated anti-FUS antibodies were described previously (Suárez-Calvet et al., 2016). Quantification of immunoblot results were performed by ImageJ.
qRT-PCR.
qRT-PCR was performed using real-time PCR equipment (Bio-Rad CFX384 Touch). Gene expression was quantified by the comparative CT method and normalized to 36b4 or Ppia expression. Values are expressed as mean ± SD of the mean. GraphPad Prism version 5.02 was used for data interpretation. The sequences of the primers are available upon request.
RNA sequencing (RNA-Seq).
The RNA-Seq library was prepared from five animals per genetic group by using a TruSeq RNA Sample Preparation Kit (Illumina) according to the manufacturer's protocol. Illumina RNA-Seq was performed using standard procedures at the University of Debrecen.
RNA-Seq samples were analyzed using an in-house pipeline. Briefly, the 50 bp raw single-end reads were aligned using TopHat (Trapnell et al., 2013) to the mm10 genome assembly (GRCm38) and only the uniquely mapped reads were kept using the “−max multihits 1” option; otherwise, the default parameters were used. Samtools version 1.0 was used for indexing the alignment files. Coverage density tracks (wig files) for RNA-Seq data were generated by igvtools with the “count” command and then converted into tdf files using the “toTDF” option. The aligned reads were assembled by Cufflinks using default parameters. Genes with RPKM ≥ 1 (at least in one sample) were considered to be expressed. Heat maps were drawn with the R package pheatmap. Statistically significant difference was considered as FDR <0.1 and FC > = 1.5 using Cufflinks. The raw sequence data have been submitted to the NCBI SRA database (BioProject: PRJNA359090).
ChIP.
ChIP-qPCR experiments were performed as described previously (Barish et al., 2012) with minor modifications. Briefly, cells were cross-linked in two steps: (1) disuccinimidylglutarate (DSG) for 30 min and then (2) 1% methanol-free ultrapure formaldehyde for 10 min at room temperature. Glycine was added for 5 min at a 125 mm final concentration. After fixation, chromatin was sonicated with Diagenode Bioraptor to generate 200–1000 bp fragments. Chromatin was immunoprecipitated with antibodies against preimmune IgG (12-370; Millipore) and CREB1 (06–863; Millipore). The eluted DNA was purified (MinElute; Qiagen) and quantified with Qubit fluorometer (Invitrogen). A more detailed description is available upon request.
ChIP-Seq analysis.
ChIP-Seq analysis was performed as described previously (Simandi et al., 2016). Briefly, primary analysis of the ChIP-Seq raw reads was performed using a ChIP-Seq analyze command line pipeline (Barta, 2011). The Burrows-Wheeler Alignment Tool (Li and Durbin, 2009) was used to align the reads to mm10 genome assembly (GRCm38) with default parameters. MACS2 (Zhang et al., 2008) was used for predicting transcription factor peaks (q-value ≤0.01). Artifacts were removed using the ENCODE blacklist (ENCODE Project Consortium, 2012). Genome coverage files (bedgraph files) for visualization were generated by makeUCSCfile.pl and then converted into tdf files using igvtools with “toTDF” option. De novo motif discovery was performed on the 100 bp vicinity of the peak summits using findMotifsGenome.pl with options “−length −len 10,12,14,16” and “−size 200” on the repeat-masked mouse genome (mm10r) from HOMER. Integrative Genomics Viewer (IGV2.3, Broad Institute) was used for data browsing (Thorvaldsdóttir et al., 2013) and creating representative snapshots.
Cellular viability assay.
Cells were plated at a density of 5000 cells per well on 96-well plates, differentiated in the presence of retinoic acid for 2 d, and then treated with thapsigargin for 24 h. Cell numbers were determined by MTT-based colorimetric assay (Roche). Results are represented relative to the control conditions.
DNA damage repair assay.
HeLa-GFP-FUS cells were a kind gift from Ina Poser. Cells were maintained in DMEM supplemented with 10% FBS and 400 μg/ml G418. For micro-irradiation experiments, cells were plated onto 8-well Ibidi chambered coverslips 2 d before measurements at a density of 15,000 cells in 300 μl of phenol red free DMEM per well. Medium was changed for DMEM containing 40 μm ADOX. Micro-irradiation studies were performed on a Zeiss LSM 880 confocal laser scanning microscope. Briefly, 20 min before the irradiation, cells were presensitized with 2 μg/ml HOECHST 33258 dye. A 150 × 1 pixel (step size: 70 nm) ROI inside the cell nucleus was selected and micro-irradiated by a 405 nm diode laser at 100% intensity (32 μW at the objective) using the bleach function of ZEN software with 8 iterations and 33 μs pixel dwell time, resulting in a total delivered energy of 8.44 nJ/pixel. Cells were scanned before and immediately after micro-irradiation and then a time series of images was recorded with 30 s intervals for 5 min. We used FiJi Software for subsequent data analysis. The fluorescence intensity ratio characterizing the enrichment of GFP-FUS in the micro-irradiated ROI was calculated by determining the average fluorescence intensity per pixel for GFP-FUS in the ROI and dividing this value with the average pixel intensity detected in the whole cell.
Statistical analysis.
Results are presented as the mean ± SEM. Comparisons between two groups were performed using Student's t test for unpaired data. A p-value of <0.05 was considered significant. All data were analyzed using GraphPad Prism software.
Study approval.
All animal study protocols were approved by the Animal Care and Protection Committee at the Universities of Debrecen and Szeged and were performed in accordance with the European Community Council Directives.
Results
ADMA level is restricted to postmitotic spinal cord MNs
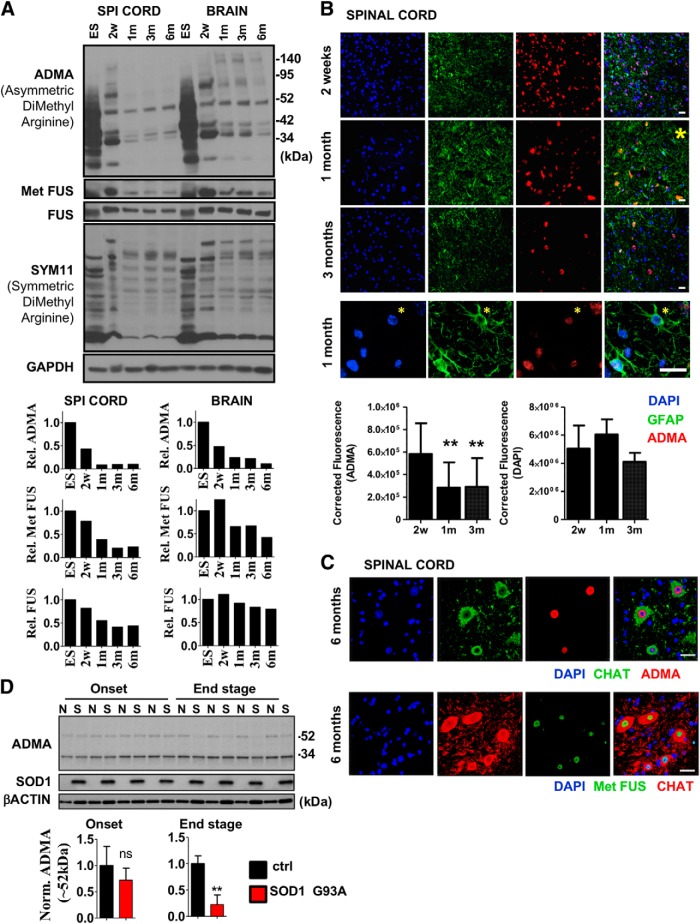
Biological functions of arginine methylation have been previously studied mostly in the context of aberrant cellular proliferation, including many types of cancers (for review, see Wei et al., 2014; Poulard et al., 2016). Therefore, the function of arginine methylation and the enzymes producing it remain largely uncharted in terminally differentiated, postmitotic cells. Comparing total levels of asymmetric and symmetric arginine dimethylation of highly proliferating embryonic stem cells (ESCs) versus brain or spinal cord tissues derived from 2-week-old (juvenile), 1-month-old (early adult), and 3- or 6-month-old (adult) mice revealed that the level of ADMA, catalyzed by type I methyltransferases, was greatly reduced during early embryonic development and further decreased in the transition from juvenile to adult stage (Fig. 1A). FUS, a well established substrate of asymmetric arginine methyltransferases (Dormann et al., 2012; Scaramuzzino et al., 2013; Scekic-Zahirovic et al., 2017), also showed reduction in its arginine methylation level as detected by a methylated FUS-specific antibody (Dormann et al., 2012) (Fig. 1A). These changes in the ADMA and Met FUS levels were more pronounced in the spinal cord (Fig. 1A). In contrast, symmetric arginine dimethylation, detected by SYM11 antibody, was relatively constant between the compared stages (Fig. 1A).
Figure 1.
Asymmetric arginine dimethylation is highly abundant in postmitotic spinal cord MNs. A, Immunoblot analysis of FUS, methylated FUS, symmetric (SYM11) and asymmetric (ADMA) arginine dimethylated protein levels in ESCs versus spinal cord samples derived from 2-week-old (2w) to 6-month-old (6m) mice. Representative result of at least three independent immunoblot analyses and its quantification are shown. Note that FUS in ESCs show slightly different molecular weight. B, Coimmunostaining of mouse spinal cord cross sections derived from 2-week-old, 1-month-old, and 3-month-old animal for ADMA-containing proteins and GFAP (astrocyte marker). Magnification of a region marked by yellow star on the section of the 1-month-old sample is shown below the panel. Representative results of at least three independent analyses and their quantification are shown. Scale bar, 25 μm. C, Coimmunostaining of spinal cord cross sections derived from 6-month-old mouse for ADMA-containing proteins, methylated FUS (Met FUS), and ChAT (MN marker). Representative results of at least three independent analyses are shown. Scale bar, 25 μm. D, Immunoblot analysis of ADMA levels in G93A SOD1 transgenic (S) and control (N) mice (n = 4 in each group). Onset ages of the disease were determined as the time when the animals reached peak body weight and the end stage of the disease were determined when the animals could not right themselves within 10 s when placed on their side (typically 12–15 weeks). Data are presented as mean ± SEM. **p ≤ 0.01 (Student's t-test).
Further immunohistochemistry analysis showed that, whereas many cells, including GFAP+ astrocytes, show strong staining for ADMA in the spinal cord of 2-week-old and 1-month-old mice; ADMA signal become restricted to very few cells by 3 months of age (Fig. 1B). These results suggest that the low abundance of total ADMA in adult mice is not due to uniformly reduced levels of asymmetrically dimethylated proteins in the various cell types. In fact, ADMA signals detected in total spinal cord lysates appears to be derived mostly from ChAT+ MNs (Fig. 1C). These postmitotic cells, representing only a small percentage of all the spinal cord cells, showed strong ADMA staining in their nuclei (Fig. 1C). Strikingly, methylated FUS also showed similar neuron-restricted expression in the spinal cord of 6-month-old mice (Fig. 1C).
We hypothesized that spinal cord ADMA level would likely decrease in certain neurodegenerative diseases such as ALS simply due to the gradual decrease in the number of ADMA+ MNs with the disease progression. To test this, we compared ADMA levels in spinal cord samples of control and mutant G93A-SOD1 transgenic mice and found that ADMA level was indeed decreased in the end stage (Fig. 1D), further confirming that MNs could be the primary source of ADMA detected in total tissue lysates.
Inhibition of methyltransferase activity induces accumulation of unrepaired DNA DSBs and decreases cell viability
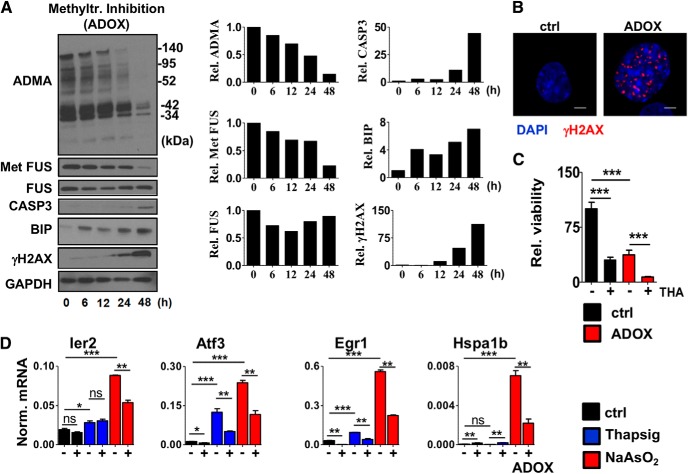
The striking cell-type-restricted distribution of ADMA in adult spinal cord raised the question of whether the residual ADMA is required for viability of MNs and their response to external stimuli. To address this question, we used retinoic acid-differentiated NSC34 MN cells and treated them with previously described methyltransferase inhibitor ADOX. Untreated NSC34 cells showed high ADMA level, which was completely diminished by ADOX treatment (Fig. 2A). In parallel, ADOX-dependent inhibition of methyltransferase activity resulted in gradual accumulation of BIP (marker of ER stress), γH2AX (marker of unrepaired DNA DSBs), and cleaved form of Caspase-3 (marker of apoptosis), implicating methylation in the control of ER stress and in the maintenance of genome and cellular integrity (Fig. 2A,B). Consistent with this, cellular viability and stress resistance was also decreased in ADOX–treated cells (Fig. 2C).
Figure 2.
Inhibition of methyltransferase activity induces accumulation of unrepaired DNA DSBs. A, Immunoblot analysis of BIP (ER stress), cleaved CASP3 (apoptosis), FUS/Met FUS, ADMA, and γH2AX (DNA DSB) in differentiated NSC34 cells treated with the methyltransferase inhibitor ADOX (40 μm) for the indicated times. Representative results of at least three independent immunoblot analyses and their quantification are shown. B, Immunostaining of γH2AX in cells treated with ADOX for 24 h (40 μm). Representative cells were chosen to show at high magnification. Scale bar, 5 μm. C, Cellular viability of NSC34 cells measured by MTT assay. Differentiating cells were pretreated with ADOX for 24 h (40 μm) at day 3 and then subjected to thapsigargin (ER stress, 0.2 μm) for 24 h. Averages of eight biological replicates are shown. D, qRT-PCR analysis of gene expression in differentiated NSC34 cells pretreated with ADOX for 24 h (40 μm) from day 3 and then subjected to thapsigargin (ER stress) or sodium arsenite (oxidative stress) for 3 h at day 4. Averages of three biological replicates are shown. Data are presented as mean ± SEM. ns, Nonsignificant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student's t test).
Previous studies extensively documented the role of arginine methyltransferases in gene expression regulation in various cellular contexts (Lee and Stallcup, 2009). As part of the normal stress response upon brain or spinal cord injury, the activation of immediate early genes (IEGs) (e.g., Ier2, Fos, Myc, Egr1, Atf3) has a pivotal role (Minatohara et al., 2015). Dysregulated control of these genes has been linked to decreased synaptic plasticity of neurons in Alzheimer's disease, ALS, and frontotemporal dementia (FTD) (Lederer et al., 2007; Seijffers et al., 2014; Minatohara et al., 2015), suggesting a general involvement of these genes in neurodegenerative diseases. To test the role of arginine methylation in transcriptional response of MNs, we pretreated differentiated NSC34 cells with ADOX and then treated the cells with thapsigargin or sodium arsenite for 3 h to elicit ER stress or oxidative stress, respectively. Pretreatment with methyltransferase inhibitor reduced the stress-mediated induction of many IEGs (e.g., Ier2, Egr1, Atf3), as well as other stress-activated gene transcription (e.g., Hspa1b), indicating a broad dysregulation of the stress-induced neuroprotective gene networks (Fig. 2D).
Overall, these results demonstrate the spatiotemporal presence of ADMA in the developing spinal cord and cell-type-restricted occurrence in the adult MNs. Considering the general effect of ADOX on methylation processes, these results also provide hints regarding the potential role of arginine methylation in the regulation of cellular stress tolerance.
PRMT8 is a tissue-specific arginine methyltransferase in spinal cord MNs
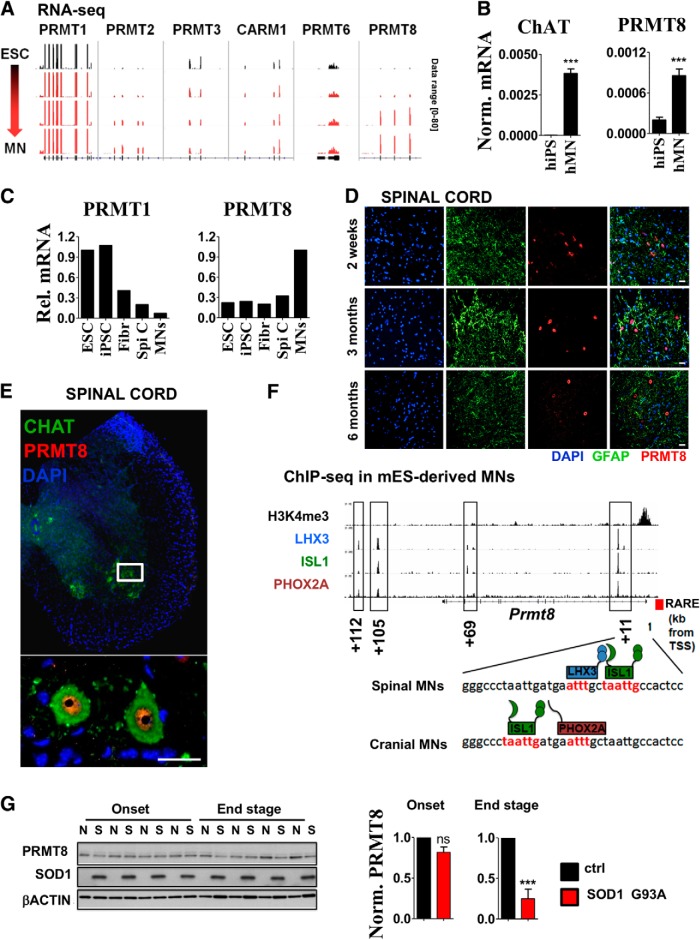
To mechanistically characterize the role of asymmetric arginine dimethylation in MNs, we decided to identify the arginine methyltransferases expressed in MNs. First, we compared the expression of known PRMTs during MN differentiation. Reanalysis of transcriptomic data obtained during direct reprogramming of mouse embryonic stem cells to MNs (Mazzoni et al., 2013) revealed the high expression of Prmt1 at each stage (Fig. 3A). Interestingly, its closest homolog, Prmt8 (Lee et al., 2005), showed robust induction in the differentiating MNs (Fig. 3A). Human PRMT8 expression showed similar induction along the differentiation of human iPSCs to MNs (Fig. 3B). In addition, reanalysis of gene expression data from Ho et al. (2016) and comparison of PRMT8 expression in total human spinal cord versus LCM-isolated MNs confirmed the relative enrichment of PRMT8 in MNs over other cell types of the human spinal cord (Fig. 3C), whereas PRMT1 expression was more robust in undifferentiated human ESCs. Overall, these findings, partially based on previously published human and mouse transcriptomic data, led us to focus on PRMT8 and further investigate its role in the CNS for two reasons: (1) cell-restricted expression of Prmt8 indicated a unique role of this arginine methyltransferase in adult spinal cord MNs and (2) cell-type-selective expression of Prmt8 makes it a potentially exploitable target for therapeutic interventions.
Figure 3.
PRMT8 is a tissue-specific arginine methyltransferase in spinal cord MNs. A, IGV genome browser view of RNA-Seq data comparing expression levels of type I arginine methyltransferases along ESC to MN differentiation. RNA-Seq experiment was performed by Mazzoni et al. (2013) and data have been reanalyzed to compare expression levels of type I PRMTs. Data range used for visualization was set to [0–80] for each track. B, CHAT and PRMT8 expression in human iPSCs vs iPSC-derived MNs as measured by qRT-PCR. Averages of three biological replicates are shown. C, Relative expression levels of PRMT1 and PRMT8 in human ESCs and iPSCs versus spinal cord and LCM-captured postmortem human MNs. Raw data were obtained from Ho et al. (2016). Fibr, Fibroblast; Spi C, spinal cord. D, Coimmunostaining of spinal cord cross-sections derived from 2-week-old, 3-month-old, and 6-month-old mice for PRMT8 and GFAP (astrocyte marker). Representative results of at least three independent analyses are shown. Scale bar, 25 μm. E, Immunostaining of spinal cord cross sections derived from 6-month-old mouse for PRMT8 and MN marker ChAT. Scale bar, 25 μm. F, Occupancy of LHX3, ISL1, and PHOX2A transcription factors in the proximity of Prmt8 encoding gene as detected by ChIP-Seq. Primary ChIP-Seq data were obtained from Mazzoni et al. (2013). Previously described (Simandi et al., 2015) retinoic acid response element (RARE) in the proximity of the transcription start site (TSS) is also marked with a red box. G, Immunoblot analysis of PRMT8 in G93A SOD1 transgenic (S) and control (N) mice (n = 4 in each group). Samples are identical to the ones shown in Figure 1D. Quantification of relative PRMT8 levels in the onset and end stages are shown. Data are presented as mean ± SEM. ns, Nonsignificant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student's t test).
To characterize the cellular localization and distribution of PRMT8 protein, we developed an antibody against mouse PRMT8 (see Materials and Methods for more details). PRMT8 expression was restricted to a subset of cells in the spinal cord at all studied ages (2 weeks to 12 months) (Fig. 3D,E). PRMT8 costaining with ChAT showed the presence of PRMT8 in mouse spinal cord MNs and revealed its predominantly nuclear localization (Fig. 3E).
Next, we sought to understand the regulation of cell-type-restricted PRMT8. MN cell identity and cell-type-specific gene expression appears to be strictly coordinated by lineage-specific transcription factors [e.g., by LHX3, ISL1 (spinal MNs) or LHX3 and PHOX2A (cranial MNs) Sockanathan and Jessell, 1998; Mazzoni et al., 2013]. Analysis of ChIP-Seq data obtained from ESC-derived MNs (Mazzoni et al., 2013) and from our previous report (Simandi et al., 2015) revealed that PRMT8 is likely under the control of RAR:RXR, LHX3, PHOX2A, and ISL1, providing the basis of its cell-type-specific expression (Fig. 3F).
Cell type-restricted expression of PRMT8 led us to investigate the consequences of disease-related loss of MNs to the expression of PRMT8. Using the same sample set as shown in Figure 1D, we found that the PRMT8 level significantly decreased in the end stage, further supporting the notion that predominantly MNs express PRMT8 (Fig. 3G).
PRMT8-null animals show progressive muscle atrophy and NMJ fragmentation
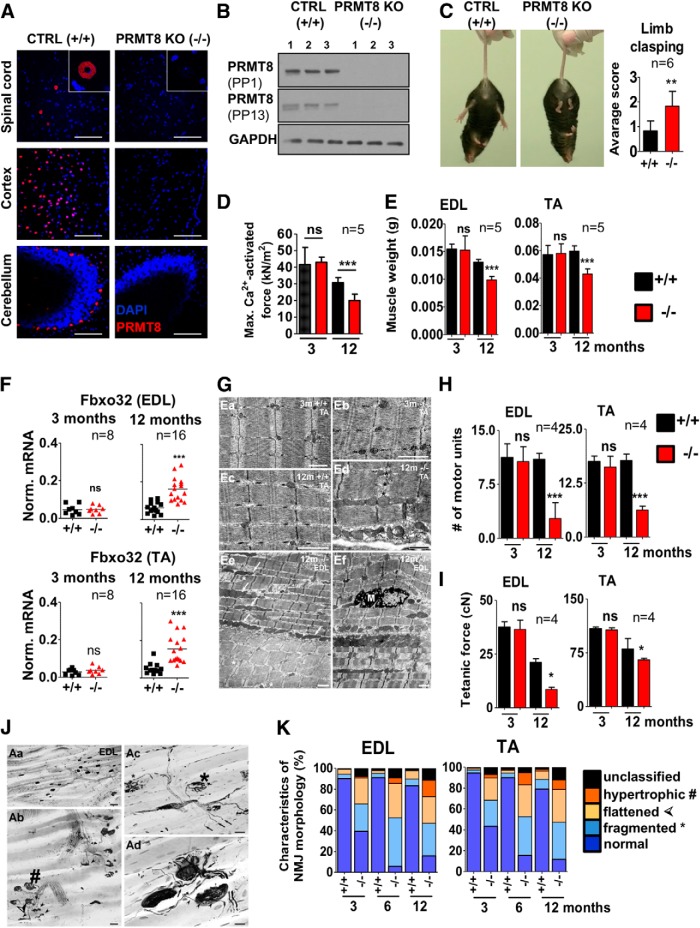
To elucidate the role of PRMT8 in vivo in spinal cord MNs, we used a knock-out line generated by the European Conditional Mouse Mutagenesis Program and Knock-out Mouse Project (EUCOMM/KOMP). As described recently, these animals are viable, suggesting that PRMT8 per se is not required for the development of the CNS (Kim et al., 2015; Simandi et al., 2015). We validated the knock-out by PRMT8 immunostainings of the spinal cord and various brain regions (Fig. 4A) and also by Western blot (Fig. 4B). To assess the motor function of mice, we used 3-, 6-, and 12- to 15-month-old littermate control and PRMT8-null animals. A limb-clasping test revealed that PRMT8-null animals show decreased motor performance, which was more pronounced in 12-month-old mice (Fig. 4C, Movies 1 and 2). Muscle force measurement on isolated muscle fibers revealed that 12-month-old PRMT8-null animals exhibit decreased muscle strength [30.7 ± 3.1 kN/m2 (control) vs 20.6 ± 1.9 kN/m2 (PRMT8-null); Fig. 4D].
Figure 4.
Aged PRMT8-null mice develop muscle weakness and progressive fragmentation of NMJs. A, Immunostaining of spinal cord (anterior horn), cortex, and cerebellum isolated from control and PRMT8-null mice for PRMT8. Scale bars, 100 μm. B, Immunoblot analysis of PRMT8 in brain samples of control and PRMT8 knock-out mice. Further details on antibodies PP1 and PP13 are presented in the Materials and Methods. C, Control animals at 15 months of age have normal limb reflexes when suspended by the tail, whereas age-matched PRMT8-null animals show an abnormal and weak limb clasping (n = 6 in each group). Scoring is based on Guyenet et al. (2010). See also Movies 1 (ctrl) and 2 (PRMT8 knock-out). D, Comparison of maximal Ca2+-activated force levels of single isolated EDL myocytes. Force development during isometric contractions was measured in fast skeletal muscle fibers from EDL muscles of 3- and 12-month-old control and PRMT8-null mice (n = 5 in each group). E, Muscle weight loss in TA and EDL muscles of aging animals (n = 5 in each group). Note the significant muscle weight loss in 12-month-old PRMT8-null animals compared with their age-matched controls. F, qRT-PCR analysis of muscle atrophy marker Fbxo32. Total RNA was extracted from TA and EDL muscles of 3- and 12-month-old control and PRMT8-null mice (n = 8–16 in each group). G, Electron microscopic ultrastructural analysis of 3- and 12-month-old control and PRMT8-null TA and EDL muscles. Shown are normal structure of a control (Ea) and PRMT8-null (Eb) TA muscles; also shown is a TA muscle sample taken from the 12-month-old control animals (Ec). Note that there is no obvious difference compared with 3-months samples (Ea). Ed is a TA muscle sample taken from 12-month-old PRMT8-null animals. Note the obvious morphological changes including uneven sizes and a fuzzy appearance of the cross-striation pattern of the sarcomers, enlarged mitochondria, and occasional loss of Z discs. The presumed location of a missing Z disc is marked by asterisks. Ee, Ef, Disintegrated structure of EDL muscles taken from 12-month-old PRMT8-null animals. Note the enlarged mitochondria (arrows), uneven staining density of fibers, and splitting myofibrils (asterisks) in Ee. Ef shows a macrophage (M) among the myofibrils of a disorganized muscle fiber. Scale bars, 1 μm. H, Number of motor units as determined by counting the recruited individual twitch contractions elicited by increasing stimulation of the supplying nerve (n = 4 in each group). Note the dramatic loss of functional motor units in both TA and EDL muscles of PRMT8-null animals. I, Smaller numbers of motor units produced smaller maximum tetanic forces (cN) in muscles of aging PRMT8-null animals, especially in the EDL muscle (n = 4 in each group). J, Histological analysis of NMJs in EDL muscles of 12-month-old animals. Aa, Distribution of motor end plates and axons in the innervation zone of an intact EDL muscle. Ab, Various pathological forms of degenerating NMJs in an EDL muscle. Double arrowhead, right upper corner: intact-looking NMJ. Hatchmark indicates hypertrophic NMJs; asterisks, fragmented NMJs; and arrowheads flattened NMJs. Ac, Higher-magnification view of two fragmented NMJs (asterisks) in an EDL muscle. Small arrows point to axons with bulb-like endings, which do not belong to motor end plates. Ad, Various forms of sprouting in EDL muscles. Arrows indicate preterminal sprouting and arrowheads ultraterminal sprouting. Scale bars: Aa, 50 μm; Ab, 25 μm; Ac, 20 μm; Ad, 10 μm. K, Quantification of NMJ morphology data. y-axis represents the percentage of NMJs appearing either pretzel-like (i.e., normal) and pathological NMJs. Note the high proportion of pathological NMJs in PRMT8-null animals, grouped as fragmented, hypertrophic, or flattened. Morphological characterization of NMJ changes was scored based according to recent studies (Valdez et al., 2010; Bruneteau et al., 2015). Data are presented as mean ± SEM. ns, Nonsignificant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student's t test).
Control animals at 15 months of age have normal limb reflexes when suspended by the tail.

PRMT8-null animals show abnormal and weak limb clasping.

Wet muscle weights of control versus PRMT8-null animals indicated an accelerated muscle weight loss in the knock-out mice (Fig. 4E). Accordingly, transcriptional levels of Fbxo32 (Atrogin-1) (a muscle atrophy-induced gene) (Bonaldo and Sandri, 2013) showed a remarkably elevated level in 12-month-old PRMT8-null animals compared with controls (Fig. 4F), suggesting an extensive atrophy in the muscle of PRMT8-null mice during aging.
Next, we used electron microscopy to evaluate morphological changes of the muscle structure. No differences were identified in young animals; however, lack of PRMT8 resulted in ultrastructural changes, including disorganized sarcomer structure, uneven size and staining density of individual fibers and myofibrils, enlarged mitochondria, and occasional loss of Z discs and appearance of splitting myofibrils in 12-month-old animals (Fig. 4G). Numbers of motor units were significantly lower in 12-month-old PRMT8-null mice (Fig. 4H). Determination of motor units belonging to control and PRMT8-null TA and EDL muscles revealed a significant age-dependent decrease in PRMT8-null animals (EDL: 10.6 ± 1 vs 2.75 ± 1.1, TA: 16.25 ± 1.2 vs 6.25 ± 0.5 in 3 in and 12-month-old animals, respectively, see Fig. 4H), whereas the number of motor units remained constant in the 12-month-old control animals. Smaller numbers of motor units produced smaller tetanic forces in 12-month-old PRMT8-null animals compared with their age-matched controls (PRMT8-null EDL: 36.5 ± 4.3 cN vs 8.6 ± 1.0cN, wild-type EDL: 37.6 ± 2.4cN vs 21.2 ± 1.7 cN in 3- and 12-month-old animals, respectively, Fig. 4I), especially in EDL muscles. A marked reduction of tetanic forces was also observed in aging control animals and this change is likely to be attributable to the aging processes of the muscle fibers as reported previously (Nair, 2005).
Because Prmt8 is not expressed in the muscle, the observed age-dependent muscle phenotype is most likely neurogenic due to loss of PRMT8 in the MNs and develops on the basis of reduced neuromuscular activity (Bütikofer et al., 2011). To test this hypothesis, we next analyzed the number and morphology of motor end plates in TA and EDL muscles. Several morphological characteristics of NMJs from control and PRMT8-null mice were assessed. In wild-type animals clusters of NMJs exhibited typical pretzel-like appearance and they were supplied by regularly arranged bundles of motor axons (Fig. 4J, Aa). In contrast, NMJs of PRMT8-null mice showed various pathological alterations, including hypertrophy, fragmentation, and flattened morphology (Fig. 4J, Ab,Ac). More than 80% of the NMJs in 12-month-old knock-out animals were found to be pathological, whereas only up to 20% of NMJs were affected in age-matched control animals (Fig. 4K).
Impaired axonal transport and premature accumulation of aging pigments in MNs of PRMT8 knock-out animals
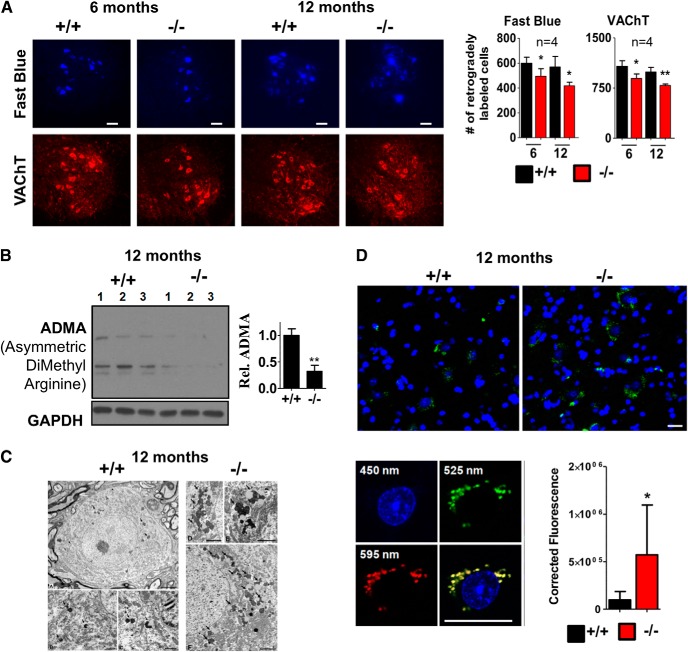
We next performed histological analysis of MNs in the lumbar spinal cord using VAChT-specific antibody. The numbers of motor neurons (VAChT+) in the lumbar ventral horn (L4–L5) supplying the sciatic nerve decreased in PRMT8-null animals by 16.8% and 20.7% in 6- and 12-month-old animals compared with their age-matched controls, respectively (Fig. 5A). Retrograde labeling of the same motor pool with Fast Blue also revealed a mild decrease (17.4% and 26.8% in 6- and 12-month-old animals, respectively), suggesting an impaired axonal transportin PRMT8-null animals (Fig. 5A, right). Overall, these data suggested that more than one-fourt of aging MNs were defunct in 12-month-old PRMT8-null mice. The relatively mild impacts of PRMT8 depletion on the number of MNs led us to investigate whether loss of PRMT8 is associated with decreased ADMA level and if these cells are intact. Immunoblot comparison revealed decreased arginine methylation (Fig. 5B), suggesting that, even if other type I arginine methyltransferases (e.g., Prmt1) are expressed in adult spinal cord MNs, loss of Prmt8 is not fully compensated by them.
Figure 5.
Age-dependent dysfunction of axonal transport and decreased MN numbers in PRMT8-null mice. A, Left, Representative images taken from cross-sections ofL4 spinal cord segments showing Fast Blue-labeled and VAChT+ MNs of a control and PRMT8-null animals at 6 and 12 months of age. Right, Quantification of Fast Blue-labeled and VAChT+ MNs (n = 4 in each group). Note the moderate loss of VAChT+ (surviving) MNs and the more severe loss of capacity to retrogradely transport Fast Blue (neurons with lost or degenerating axons) in these neurons. Scale bar, 100 μm. B, Immunoblot analysis of spinal cord samples taken from 12-month-old mice for ADMA (n = 3 in each group). C, Electron micrographs of MNs from the L4 spinal segment of 12-month-old control and PRMT8-null mice. MNs show normal ultrastructural morphology but lipofuscin granules (some of the labeled with arrowheads) are much more numerous in the PRMT8-null MNs. Scale bars: (A): 5 μm; (B, C): 1 μm; (D–F): 2 μm. D, Quantification of autofluorescent signals detected in spinal cord samples of 12-month-old control and PRMT8 knock-out mice. Representative results of at least three independent analyses are shown. Data are presented as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01 (Student's t-test).
Electron microscopy revealed premature accumulation of abnormal amount of lipofuscin in the MNs (Fig. 5C). Further analysis quantitatively confirmed the increased level of autofluorescent aging pigments in the PRMT8 knock-out spinal cord MNs (Fig. 5D), a pathological change that also appears prematurely in ALS patients (McHolm et al., 1984).
Accumulation of DNA DSBs in MNs of aged PRMT8-null animals
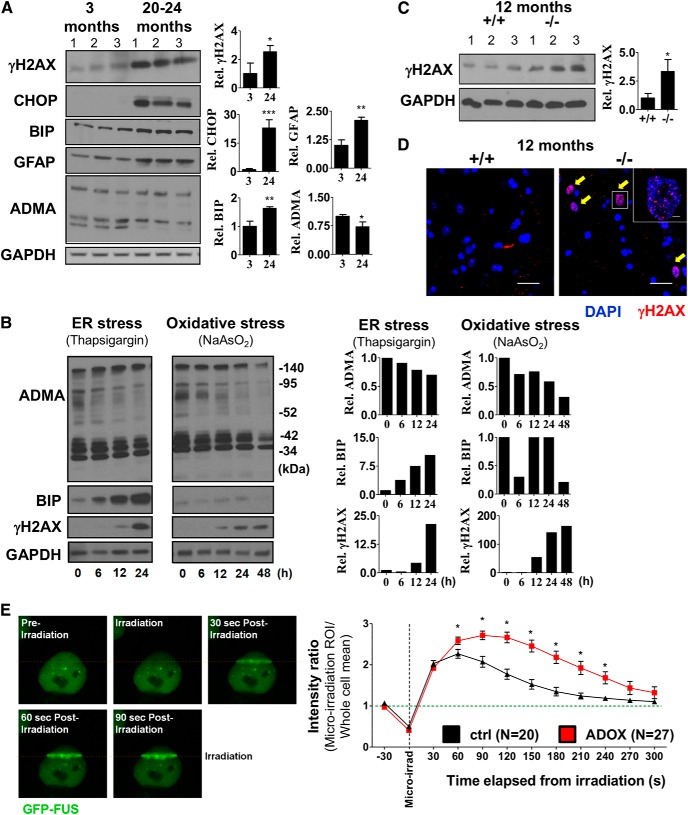
Arginine methylation has been previously linked to DNA DSB repair in proliferating cells (Yu et al., 2009, 2012) and, according to our data, the ADOX-treated NSC34 cells showed accumulation of γH2AX (Fig. 2B). Recent findings in neurodegenerative diseases revealed that even small changes in the machinery of DNA repair can affect the age of onset of neurodegeneration, raising the possibility that compromised DNA repair is a common risk in the progression of such diseases (Madabhushi et al., 2014). Comparison of spinal cord tissues isolated from 3-month-old and 24-month-old mice revealed the increased level of γH2AX, as well as presence of CHOP (ER stress marker) and increased GFAP level (astrocyte activation) in aging mice, likely as consequences of aging-associated stress (Fig. 6A). ADMA level showed a mild reduction with aging (Fig. 6A). To test how ADMA level changes upon stress in in vitro conditions, we treated MN cells with thapsigargin (ER stress) and sodium arsenite (oxidative stress). Strikingly, persistent ER stress or oxidative stress reduced total ADMA levels and in parallel induced DNA DSB response (γH2AX) (Fig. 6B). To determine whether loss of PRMT8 may lead to premature accumulation of DSBs in spinal cord MNs, we compared γH2AX levels. Immunoblot analysis revealed an increased γH2AX level in 12-month-old PRMT8 knock-out mice, indicating insufficient DNA repair (Fig. 6C). Immunohistochemistry confirmed the accumulation of γH2AX+ cells in the PRMT8 knock-out spinal cords, but not in age-matched controls (Fig. 6D).
Figure 6.
Age-dependent accumulation of unrepaired DNA DSBs in MNs of PRMT8-null mice. A, Immunoblot analysis of ADMA, CHOP (ER stress), BIP (ER stress marker), GFAP (astrocyte activation), and γH2AX (DNA repair) in spinal cord samples of 3-month-old and 20- to 24-month-old mice. B, Immunoblot analysis of ADMA, BIP, and γH2AX. Differentiated NSC34 MN cells were treated with thapsigargin or sodium arsenite for the indicated times. GAPDH served as a loading control. Note the unexpected accumulation of γH2AX upon ER stress. C, Immunoblot analysis of spinal cord samples taken from 12-month-old mice for γH2AX (n = 3 in each group). D, Immunostaining of spinal cord samples from 12-month-old mice for γH2AX. Representative images are shown. Immunoblot presented in C serves as a quantitative comparison. E, Left, Micro-irradiation-induced FUS recruitment to DNA DSBs. HeLa cells expressing GFP-FUS constitutively were micro-irradiated at 405 nm in a 150 × 1 pixel ROI (step size: 70 nm). Right, Comparative analysis of FUS kinetics in control versus ADOX-pretreated cells. Data are presented as mean ± SEM. ns, Nonsignificant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student's t test).
Previous studies already proposed the role of arginine methylation in DNA repair in proliferating cells. Importantly, the mutant form of FUS, in which the mutation affects the arginine methylated site, results in impaired DSB repair (Wang et al., 2013). Using a GFP-FUS-expressing stable cell line, we confirmed the dynamic recruitment of FUS to micro-irradiation-induced DSBs (Fig. 6E). Moreover, we found that ADOX pretreatment of the cells strongly increased the residence time of FUS at the sites of DSBs, indicating impaired function and altered kinetics of hypomethylated FUS (Fig. 6E). Collectively, these data suggest impaired DNA DSB repair in aging PRMT8-null animals.
Loss of PRMT8 disrupts the CREB1-dependent neuroprotective transcriptional network
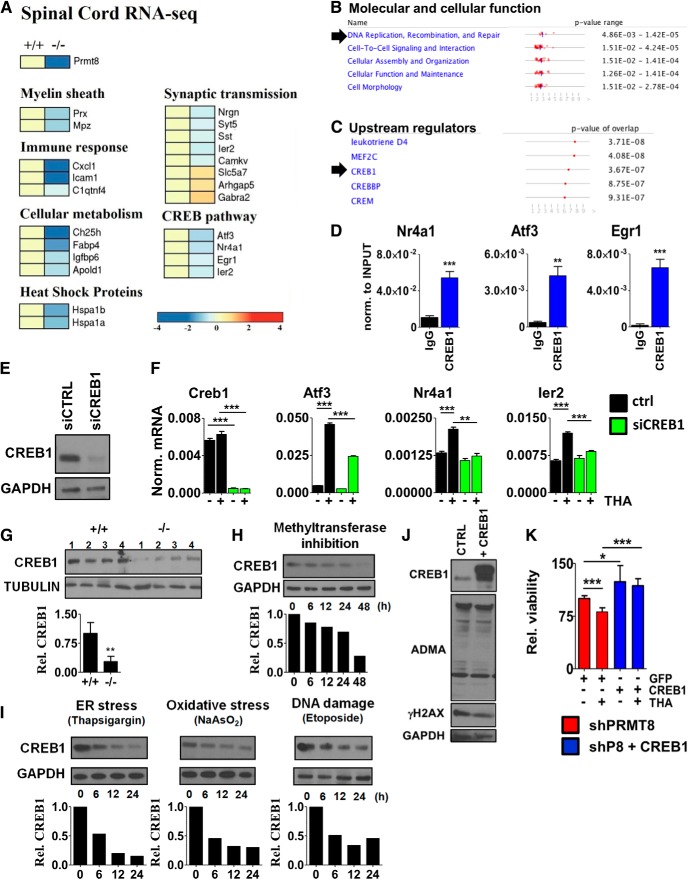
In previous studies, profiling of gene expression has been widely used in MN disease models at several stages during the course of disease and revealed early gene expression changes related to inflammation, apoptosis, ATP biosynthesis, myelination, axonal transport, and synaptic transmission (de Oliveira et al., 2013; Scekic-Zahirovic et al., 2016). To identify components affected in spinal cords of aged PRMT8-null animals in an unbiased manner, we applied RNA-Seq on total RNA extracted from the lumbar spinal cord regions of five mice per group. We considered those genes differentially expressed that showed at least 1.5-fold change in their expression at least in 4 mice per group. As expected, Prmt8 expression was completely diminished in the knock-out mice (Fig. 7A). Several genes with well established roles in myelinization (e.g., Mpz, Prx) were dysregulated (Fig. 7A). Genes involved in synaptic plasticity (Nrgn, Syt5, Sst, Camkv, Gabra2) and cellular metabolism (Ch25h, Igfbp6) were also decreased in PRMT8-null mice (Fig. 7A). The list of dysregulated genes included many genes previously shown to be differentially expressed in human ALS or FTD (e.g., Nrgn, Gabra2, Hspa1b, Gucy1a2) (Lederer et al., 2007; Comley et al., 2015) and several IEGs (e.g., Atf3, Egr1, Ier2). Pathway analysis of differentially expressed genes by ingenuity pathway analysis (IPA) indicated DNA replication, recombination, and repair as one of the top cellular function affected in PRMT8 knock-out tissues (Fig. 7B), consistent with the accumulation of γH2AX detected in spinal cord lysates of PRMT8 knock-out mice (Fig. 6). To better understand the molecular mechanisms impaired in PRMT8-depleted MNs, we used IPA to predict potential upstream transcriptional regulators causing dysregulated gene expression. This analysis revealed that the function of CREB is likely affected in the absence of PRMT8 (Fig. 7C).
Figure 7.
Compromised CREB1-dependent activation of stress signaling pathways. A, RNA-Seq analysis of spinal cord samples isolated from 6-month-old control and PRMT8-null mice (n = 5 in each group). B, C, Prediction of PRMT8-dependent molecular and cellular functions and upstream regulators using IPA based on RNA-Seq data obtained in control and PRMT8-null spinal cord samples. D, ChIP-qPCR validation of CREB1 binding in differentiated NSC34 cells. E, Analysis of siCREB1 efficiency by immunoblot. Differentiating NSC34 cells were transfected and incubated for 2 d. F, qRT-PCR analysis of Creb1and target gene expression in CREB1-depleted cells upon thapsigargin-induced stress. G, Immunoblot analysis of spinal cord samples taken from 12-month-old control and PRMT8-null animals for CREB1 (n = 4 in each group). H, Immunoblot analysis of CREB1 in ADOX-treated differentiated NSC34 cells. I, Changes of CREB1 protein level upon ER stress (thapsigargin, 1 μm), oxidative stress (sodium arsenite, 0.2 μm), and DNA DSB induction (etoposide, 5 μm). Representative results of at least three independent experiments are shown. J, Immunoblot analysis of ADMA level in control versus CREB1-overexpressing NSC34 cells. Representative results of at least three independent immunoblot analyses and their quantification are shown. K, Cellular viability of PRMT8-depleted cells transfected with GFP (control) or CREB1 as measured by MTT assay. Data are presented as mean ± SEM. ns, Nonsignificant, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student's t test).
CREB1 is a constitutively expressed transcription factor that regulates the expression of genes involved in neuronal survival and function (Sakamoto et al., 2011). To assess whether CREB1 directly regulates the genes differentially expressed in the spinal cord of PRMT8-null mice (e.g., Atf3, Nr4a1, Egr1, Ier2; Fig. 7A), we analyzed binding of CREB1 in the proximity of their transcription start site by ChIP-qPCR. These results confirmed that CREB1 is enriched at these sites (Fig. 7D). The requirement of CREB1 for the induction of these IEGs upon cellular stress was validated by loss-of-function experiments in differentiated NSC34 cells (Fig. 7E,F).
To understand why the CREB1-dependent gene expression network is dysregulated in PRMT8-null mice, we next analyzed CREB1 at the protein level. This comparison revealed a reduction of CREB1 protein levels in in vivo in 12-month-old PRMT8 knock-out spinal cord samples (Fig. 7G). Comparison of CREB1 levels following inhibition of methyltransferase activity by ADOX confirmed a gradual decrease in CREB1 protein levels (Fig. 7H), formally suggesting that arginine methylation is required to maintain CREB1 level. Exposing NSC34 cells to persistent stress also reduced the level of CREB1 (Fig. 7I) in addition to reducing the ADMA level (Fig. 6B), suggesting a positive correlation between the level of ADMA and CREB1.
We next wanted to determine whether ectopic expression of PRMT8 or CREB1 is sufficient to increase cell survival and stress tolerance. Overexpression of CREB1 failed to increase the level of ADMA, suggesting that mechanistically PRMT8 is upstream of CREB1 (Fig. 7J). However, restoration of CREB1 levels could increase the stress tolerance and survival rate of PRMT8-depleted MNs (Fig. 7K). Together, these results indicate that overexpression of CREB1 or wild-type PRMT8 results in a cellular phenotype with increased stress tolerance.
Discussion
Tissue integrity in multicellular organisms is highly dependent on a coordinated response to external and internal insults during aging and disease processes. Recent findings suggest that stress response signaling can be manipulated and altered to extend life span and delay the onset of age-related pathologies such as neurodegenerative diseases, suggesting an entirely new therapeutic approach for these degenerative disorders (Epel and Lithgow, 2014). Therefore, identification of tissue- and cell-type-specific regulators of stress response is of great interest and holds therapeutic potential. It would be of particular relevance to determine whether if tissue-specific mechanisms and ones selective to postmitotic states could be uncovered and targeted safely in aging and disease. As part of our systematic analysis of neural differentiation, we have identified such a mechanism and a protein responsible for that. Here, we show that PRMT8 is highly and selectively expressed in postmitotic spinal cord MNs and, although it is not essential for their differentiation per se, it acts as a key modulator of the stress tolerance and plasticity of these cells by enabling CREB1-dependent transcriptional activation of IEGs and DNA damage response induced as protective mechanisms in the aging tissues. These findings provide several important insights that will help us to better understand common mechanisms of aging and neurodegeneration-related pathologies.
First, despite extensive research, initial mechanisms leading to aging or disease-related progressive loss of MNs remain largely unknown. An emerging concept on the field of MN diseases is the existence of a “dying-back” process (Cavanagh, 1964; Fischer and Glass, 2007; Dadon-Nachum et al., 2011). The extensive neuropathological analyses presented in this study regarding the aging PRMT8-null mice support that the pathological process commences at the most distal part of the MN, the NMJ. Accordingly, the ratio of affected and pathologically altered motor end plates is very high (80% at 12 onths). As the degeneration extends proximally, the number of surviving (VAChT+) MNs decreases by 20.7% and that of those which are able to transport Fast Blue retrogradely, by 26.8%. These data suggest that 12-month-old PRMT8-null animals may suffer a loss of only one-fifth of their sciatic motor pool accompanied by a higher number of degenerated or lost axons, but the complete disintegration at the level of motor end plates renders the muscle atrophic and thus defunct. This dying-back mechanism of neurodegeneration has been linked to neuronal loss in many human degenerative and traumatic nervous system lesions, including Charcot-Marie Tooth disease, Alzheimer's disease, autoimmune diseases (e.g., myasthenia gravis), intoxication, or death of MNs (e.g., spinal muscular atrophy, ALS; Dadon-Nachum et al., 2011). Age-associated degeneration can also affect the integrity of NMJs, resulting in progressive muscle weakness (Coleman, 2005). An important issue is the relevance of PRMT8 function to these human degenerative diseases and aging-associated muscle weakness. So far, no studies have linked PRMT8 mutations to any of these neurodegenerative disorders, suggesting formally that either PRMT8 mutations have no significant contribution to human disease progression or PRMT8 is essential for human development and its total loss is incompatible with life. Indeed, based on the Broad's Human KO Project, the predictions suggest that loss-of-function variants for PRMT8 are not tolerated in humans (Narasimhan et al., 2016). This also raises important evolutionary questions, suggesting that PRMT8 obtained additional functions during evolution and it became required for life in humans. Further experiments in human stem-cell-derived neurons, including those taken from ALS patients, will be required to address this possibility adequately.
Second, we discovered a regulatory relationship by which sufficient CREB1 levels are maintained in normal, healthy cells and regulate cellular plasticity and regenerative transcriptional program through the activation of IEGs. CREB is a bona fide neuron survival factor known to integrate several signaling cascades, including a large number of receptors and kinases (Lonze and Ginty, 2002). Overexpression of dominant-negative CREB mutants causes apoptosis of postmitotic neurons and genetic disruption of CREB in mice results in axonal growth defects and perinatal lethality (Lonze and Ginty, 2002). The anti-apoptotic functions of CREB are only partially understood. CREB-dependent transcription of prosurvival genes such as Bcl2 and neuron growth factors has been identified (Lonze and Ginty, 2002). ChIP analysis of CREB1 binding presented in this study and loss-of-function approaches have extended our knowledge on CREB1-dependent protective pathways, revealing that CREB1 regulates the activation of IEGs in MNs. Moreover, CREB1 overexpression experiments revealed that CREB1 exhibits a neuroprotective function against oxidative and ER stress in spinal cord MNs, similarly, as shown previously in the hippocampus (Mantamadiotis et al., 2002). PRMT8 is required for the maintenance of CREB1 levels and this mechanism is likely indispensable for the maintenance and reinnervation of NMJs during the course of life. Loss of PRMT8 results in dysregulated activation pattern of IEGs (e.g., Atf3, Nr4a1, Egr1) due to the decreased total level of CREB1. We believe that there is no direct one-to-one molecular link by which PRMT8 controls protective CREB1 protein levels. Our results suggest that CREB1 level is generally negatively controlled by persistent stress signals. These findings also explain previous results showing a downregulation of CREB1 protein in Alzheimer's brain (Pugazhenthi et al., 2011). A very likely scenario is that DNA repair mechanisms in PRMT8-depleted cells are disturbed and the cells accumulate DNA DSBs (reflected by the increased γH2AX level), which in turn reduces CREB1 levels (similarly to etoposide), leading to cell death. Indeed, previous studies already linked other PRMTs to the control of DNA damage response (Boisvert et al., 2005a). Further studies will be required to clarify whether PRMT8 can also modulate DNA DSB repair mechanisms in postmitotic neurons and, through these mechanisms, protect them against oxidative-stress-induced DNA damage and cell death. It is tempting to propose the role of methylated FUS in this process. Our results confirmed those of previous studies documenting the recruitment of FUS to DSBs (Wang et al., 2013) and we found that hypomethylated FUS showed altered kinetics. Recent studies showed that FUS and other RBPs create liquid-like compartments at the sites of DNA damage by liquid demixing (Patel et al., 2015; Kai, 2016). According to our Western blot analysis, treatment with ADOX reduced the number of methylated FUS molecules. We could hypothesize that the formation of these compartments might disturbed by methylation inhibition, which could reduce the dynamics of FUS. Alternatively, nonmethylated FUS may bind to sites of DNA damage with lower affinity; it may also recruit DNA repair enzymes with a lower efficiency, whereby the kinetics of the repair process may become slower. Further studies are needed to clarify the mechanism of methylation inhibition on FUS function.
An important conclusion of our study is that the predominant arginine methyltransferase PRMT1, which is ubiquitously expressed in every tissue, is insufficient alone to maintain genome integrity and provide stress resistance in aging neurons. PRMT1 displays a high degree of sequence identity to PRMT8, formally suggesting that these two enzymes have common substrates (e.g., FUS; Scaramuzzino et al., 2013) and may function redundantly in neurons. Therefore, in postmitotic neurons, one of the most vulnerable cell types in our body, the tissue-specific appearance of PRMT8 in addition to PRMT1 most likely had major evolutionary benefits for survival and function over an entire life span. However, the decreased expression of Prmt1 in postmitotic MNs versus the rapidly dividing ESCs and the exactly opposite pattern of Prmt8 expression (Fig. 3C) suggest that these two enzymes may have context-dependent, nonoverlapping functions as well. In fact, ADMA level significantly decreased in PRMT8 knock-out mice, suggesting that PRMT1 or other arginine methyltransferases cannot fully compensate for the loss of PRMT8. Further studies will be needed to clarify the interrelationship between PRMT1 and PRMT8 and whether PRMT1 also regulates cellular stress tolerance of MNs similarly to PRMT8.
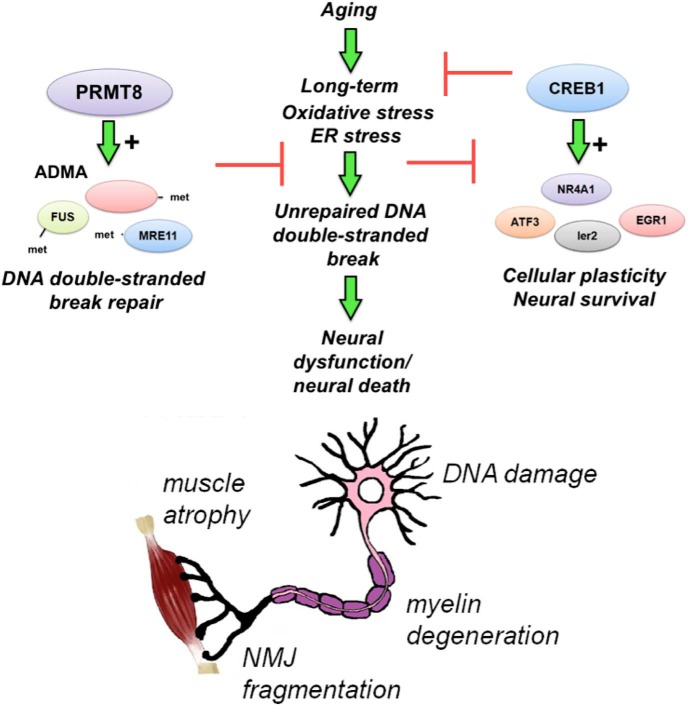
In summary, we provide evidence for the existence of a novel mechanism by which asymmetrical arginine dimethylation produced by PRMT8 can provide protection against age-related cellular stress through maintaining genome integrity and CREB1-dependent activation of IEGs (Fig. 8). Importantly, this supports the idea that tissue-specific regulators of stress response exist and function in mature cells, so cell-type-specific selective drug targeting may be possible in various diseases, including enhancement of MN stress tolerance and consequently life span in neurodegenerative disorders such as ALS and/or MN disease.
Figure 8.
PRMT8 provides cellular stress tolerance in aging MNs. ADMA provides protection against age-related accumulation of DNA damage and cell death in postmitotic neurons. Loss of PRMT8 results in accumulation of DNA DSBs, decreased level of CREB1, and dysregulated activation of IEGs in aging cells. As a consequence, loss of synaptic plasticity, NMJ fragmentation, and eventual loss of MNs occur at a premature age.
Footnotes
This work was supported by the research program GINOP-2.3.2-15/2016-00043 (T.H.), KTIA_13_NAP-A_II/7 (T.H.), GINOP 2.3.2-15-2016-00034 (A.N.), KTIA_NAP_13-1-2013-0001 (M.A.), and MTA- TKI 242 (M.A.). L.N. is supported by grants from the Hungarian Scientific Research Fund (OTKA 116855, 124298, 126885) and TAMOP 422_2012_0023 VÉD-ELEM implemented through the New Hungary Development Plan cofinanced by the European Social Fund and the European Regional Development Fund and the Hungarian Brain Research Program (Grant KTIA_13_NAP-A-I/9). D.D. is supported by the Deutsche Forschungsgemeinschaft, within the framework of Munich Cluster Systems Neurology (EXC 1010 SyNergy) and the Emmy Noether program (DO 1804/1-1). H.X. is supported by the National Institutes of Health (Grants R21 AG048519, R01 AG021173, R01 AG038710, R01 AG044420, R01 NS046673, RF1 AG056130, and RF1 AG056114), the Tanz Family Fund, and Cure Alzheimer's Fund. We thank Drs. Randy Kaufman, Barbara Ranscht (Sanford Burnham Prebys Medical Discovery Institute), Peter Somogyi (University of Oxford), and members of the Nagy laboratory for discussions and comments on the manuscript and Mrs. Heja Aga and Dr. Andras Szabo for technical assistance with imunohistochemistry.
The authors declare no competing financial interests.
References
- Auburger G, Gispert S, Brehm N (2016) Methyl-arginine profile of brain from aged PINK1-KO+A53T-SNCA mice suggests altered mitochondrial biogenesis. Parkinsons Dis 2016:4686185. 10.1155/2016/4686185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, Tai LJ, Leblanc M, Diehl C, Cerchietti L, Miller YI, Witztum JL, Melnick AM, Dent AL, Tangirala RK, Evans RM (2012) The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab 15:554–562. 10.1016/j.cmet.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta E. (2011) Command line analysis of ChIP-Seq results. J EMBnet 17:13–17. 10.14806/ej.17.1.209 [DOI] [Google Scholar]
- Bedford MT, Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Mol Cell 33:1–13. 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc RS, Richard S (2017) Arginine methylation: the coming of age. Mol Cell 65:8–24. 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Déry U, Masson JY, Richard S (2005a) Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev 19:671–676. 10.1101/gad.1279805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Rhie A, Richard S, Doherty AJ (2005b) The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4:1834–1841. 10.4161/cc.4.12.2250 [DOI] [PubMed] [Google Scholar]
- Bonaldo P, Sandri M (2013) Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6:25–39. 10.1242/dmm.010389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T (2013) Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol Biosyst 9:2231–2247. 10.1039/c3mb00009e [DOI] [PubMed] [Google Scholar]
- Bruneteau G, et al. (2015) Endplate denervation correlates with nogo-A muscle expression in amyotrophic lateral sclerosis patients. Ann Clin Transl Neurol 2:362–372. 10.1002/acn3.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bütikofer L, Zurlinden A, Bolliger MF, Kunz B, Sonderegger P (2011) Destabilization of the neuromuscular junction by proteolytic cleavage of agrin results in precocious sarcopenia. FASEB J 25:4378–4393. 10.1096/fj.11-191262 [DOI] [PubMed] [Google Scholar]
- Cavanagh JB. (1964) The significance of the dying back process in experimental and human neurological disease. Int Rev Exp Pathol 3:219–267. [PubMed] [Google Scholar]
- Coleman M. (2005) Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci 6:889–898. 10.1038/nrn1788 [DOI] [PubMed] [Google Scholar]
- Comley L, Allodi I, Nichterwitz S, Nizzardo M, Simone C, Corti S, Hedlund E (2015) Motor neurons with differential vulnerability to degeneration show distinct protein signatures in health and ALS. Neuroscience 291:216–229. 10.1016/j.neuroscience.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Dadon-Nachum M, Melamed E, Offen D (2011) The dying-back phenomenon of motor neurons in ALS. J Mol Neurosci 43:470–477. 10.1007/s12031-010-9467-1 [DOI] [PubMed] [Google Scholar]
- Dantuma NP, van Attikum H (2016) Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J 35:6–23. 10.15252/embj.201592595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Rumsey JW, Gregory CA, Bhargava N, Kang JF, Molnar P, Riedel L, Guo X, Hickman JJ (2007) Embryonic motoneuron-skeletal muscle co-culture in a defined system. Neuroscience 146:481–488. 10.1016/j.neuroscience.2007.01.068 [DOI] [PubMed] [Google Scholar]
- de Oliveira GP, Alves CJ, Chadi G (2013) Early gene expression changes in spinal cord from SOD1(G93A) amyotrophic lateral sclerosis animal model. Front Cell Neurosci 7:216. 10.3389/fncel.2013.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, Haass C (2012) Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J 31:4258–4275. 10.1038/emboj.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel ES, Lithgow GJ (2014) Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J Gerontol A Biol Sci Med Sci 69:S10–S16. 10.1093/gerona/glu055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer LR, Glass JD (2007) Axonal degeneration in motor neuron disease. Neurodegener Dis 4:431–442. 10.1159/000107704 [DOI] [PubMed] [Google Scholar]
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 13:372–387. 10.1074/mcp.O113.027870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet S. J., Furrer S. A., Damian VM, Baughan TD, La Spada AR, Garden GA (2010) A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp 39:1787. 10.3791/1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA (2010) The aging stress response. Mol Cell 40:333–344. 10.1016/j.molcel.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart-Smith G, Low JK, Erce MA, Wilkins MR (2012) Enhanced methylarginine characterization by post-translational modification-specific targeted data acquisition and electron-transfer dissociation mass spectrometry. J Am Soc Mass Spectrom 23:1376–1389. 10.1007/s13361-012-0417-8 [DOI] [PubMed] [Google Scholar]
- Herrup K, Yang Y (2007) Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci 8:368–378. 10.1038/nrn2124 [DOI] [PubMed] [Google Scholar]
- Ho R, Sances S, Gowing G, Amoroso MW, O'Rourke JG, Sahabian A, Wichterle H, Baloh RH, Sareen D, Svendsen CN (2016) ALS disrupts spinal motor neuron maturation and aging pathways within gene co-expression networks. Nat Neurosci 19:1256–1267. 10.1038/nn.4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M. (2016) Roles of RNA-binding proteins in DNA damage response. Int J Mol Sci 17:310. 10.3390/ijms17030310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JD, Kako K, Kakiuchi M, Park GG, Fukamizu A (2008) EWS is a substrate of type I protein arginine methyltransferase, PRMT8. Int J Mol Med 22:309–315. [PubMed] [Google Scholar]
- Kim JD, Park KE, Ishida J, Kako K, Hamada J, Kani S, Takeuchi M, Namiki K, Fukui H, Fukuhara S, Hibi M, Kobayashi M, Kanaho Y, Kasuya Y, Mochizuki N, Fukamizu A (2015) PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci Adv 1:e1500615. 10.1126/sciadv.1500615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole AJ, Annis RP, Deshmukh M (2013) Mature neurons: equipped for survival. Cell Death Dis 4:e689. 10.1038/cddis.2013.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N (2011) Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J 30:2520–2531. 10.1038/emboj.2011.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousaka A, Mori Y, Koyama Y, Taneda T, Miyata S, Tohyama M (2009) The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience 163:1146–1157. 10.1016/j.neuroscience.2009.06.061 [DOI] [PubMed] [Google Scholar]
- Lake AN, Bedford MT (2007) Protein methylation and DNA repair. Mutat Res 618:91–101. 10.1016/j.mrfmmm.2006.09.010 [DOI] [PubMed] [Google Scholar]
- Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S (2007) Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics 8:26. 10.1186/1471-2164-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem 280:32890–32896. 10.1074/jbc.M506944200 [DOI] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR (2009) Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol 23:425–433. 10.1210/me.2008-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G (1995) In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol 15:2800–2808. 10.1128/MCB.15.5.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD (2002) Function and regulation of CREB family transcription factors in the nervous system. Neuron 35:605–623. 10.1016/S0896-6273(02)00828-0 [DOI] [PubMed] [Google Scholar]
- Madabhushi R, Pan L, Tsai LH (2014) DNA damage and its links to neurodegeneration. Neuron 83:266–282. 10.1016/j.neuron.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schütz G (2002) Disruption of CREB function in brain leads to neurodegeneration. Nat Genet 31:47–54. 10.1038/ng882 [DOI] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Closser M, Morrison CA, Nedelec S, Williams DJ, An D, Gifford DK, Wichterle H (2013) Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci 16:1219–1227. 10.1038/nn.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHolm GB, Aguilar MJ, Norris FH (1984) Lipofuscin in amyotrophic lateral sclerosis. Arch Neurol 41:1187–1188. 10.1001/archneur.1984.04050220085023 [DOI] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, Okuno H (2015) Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol Neurosci 8:78. 10.3389/fnmol.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair KS. (2005) Aging muscle. Am J Clin Nutr 81:953–963. 10.1093/ajcn/81.5.953 [DOI] [PubMed] [Google Scholar]
- Namba T, Nakamura T, Grob D (1967) Staining for nerve fiber and cholinesterase activity in fresh frozen sections. Am J Clin Pathol 47:74–77. 10.1093/ajcp/47.1.74 [DOI] [PubMed] [Google Scholar]
- Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, Barnett AH, Bates C, Bellary S, Bockett NA, Giorda K, Griffiths CJ, Hemingway H, Jia Z, Kelly MA, Khawaja HA, Lek M, McCarthy S, McEachan R, O'Donnell-Luria A, et al. (2016) Health and population effects of rare gene knockouts in adult humans with related parents. Science 352:474–477. 10.1126/science.aac8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nógrádi A, Szabó A, Pintér S, Vrbová G (2007) Delayed riluzole treatment is able to rescue injured rat spinal motoneurons. Neuroscience 144:431–438. 10.1016/j.neuroscience.2006.09.046 [DOI] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H (2008) Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins 72:1125–1137. 10.1002/prot.22004 [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Penney J, Seo J, Kritskiy O, Elmsaouri S, Gao F, Pao PC, Su SC, Tsai LH (2017) Loss of protein arginine methyltransferase 8 alters synapse composition and function, resulting in behavioral defects. J Neurosci 37:8655–8666. 10.1523/JNEUROSCI.0591-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintér S, Gloviczki B, Szabó A, Márton G, Nógrádi A (2010) Increased survival and reinnervation of cervical motoneurons by riluzole after avulsion of the C7 ventral root. J Neurotrauma 27:2273–2282. 10.1089/neu.2010.1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulard C, Corbo L, Le Romancer M (2016) Protein arginine methylation/demethylation and cancer. Oncotarget 7:67532–67550. 10.18632/oncotarget.11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugazhenthi S, Wang M, Pham S, Sze CI, Eckman CB (2011) Downregulation of CREB expression in Alzheimer's brain and in abeta-treated rat hippocampal neurons. Mol Neurodegener 6:60. 10.1186/1750-1326-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Karelina K, Obrietan K (2011) CREB: a multifaceted regulator of neuronal plasticity and protection. J Neurochem 116:1–9. 10.1111/j.1471-4159.2010.07080.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaramuzzino C, Monaghan J, Milioto C, Lanson NA Jr, Maltare A, Aggarwal T, Casci I, Fackelmayer FO, Pennuto M, Pandey UB (2013) Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One 8:e61576. 10.1371/journal.pone.0061576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J, et al. (2016) Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J 35:1077–1097. 10.15252/embj.201592559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J, Oussini HE, Mersmann S, Drenner K, Wagner M, Sun Y, Allmeroth K, Dieterlé S, Sinniger J, Dirrig-Grosch S, René F, Dormann D, Haass C, Ludolph AC, Lagier-Tourenne C, Storkebaum E, Dupuis L (2017) Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol 133:887–906. 10.1007/s00401-017-1687-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Zhang J, Matthews JC, Chen A, Tamrazian E, Babaniyi O, Selig M, Hynynen M, Woolf CJ, Brown RH Jr (2014) ATF3 expression improves motor function in the ALS mouse model by promoting motor neuron survival and retaining muscle innervation. Proc Natl Acad Sci U S A 111:1622–1627. 10.1073/pnas.1314826111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Taylor JP (2013) Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis 1:e25200. 10.4161/rdis.25200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simandi Z, Czipa E, Horvath A, Koszeghy A, Bordas C, Póliska S, Juhász I, Imre L, Szabó G, Dezso B, Barta E, Sauer S, Karolyi K, Kovacs I, Hutóczki G, Bognár L, Klekner Á, Szucs P, Bálint BL, Nagy L (2015) PRMT1 and PRMT8 regulate retinoic acid-dependent neuronal differentiation with implications to neuropathology. Stem Cells 33:726–741. 10.1002/stem.1894 [DOI] [PubMed] [Google Scholar]
- Simandi Z, Horvath A, Wright LC, Cuaranta-Monroy I, De Luca I, Karolyi K, Sauer S, Deleuze JF, Gudas LJ, Cowley SM, Nagy L (2016) OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol Cell 63:647–661. 10.1016/j.molcel.2016.06.039 [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Jessell TM (1998) Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell 94:503–514. 10.1016/S0092-8674(00)81591-3 [DOI] [PubMed] [Google Scholar]
- Suárez-Calvet M, Neumann M, Arzberger T, Abou-Ajram C, Funk E, Hartmann H, Edbauer D, Kremmer E, Göbl C, Resch M, Bourgeois B, Madl T, Reber S, Jutzi D, Ruepp MD, Mackenzie IR, Ansorge O, Dormann D, Haass C (2016) Monomethylated and unmethylated FUS exhibit increased binding to transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol 131:587–604. 10.1007/s00401-016-1544-2 [DOI] [PubMed] [Google Scholar]
- Taneda T, Miyata S, Kousaka A, Inoue K, Koyama Y, Mori Y, Tohyama M (2007) Specific regional distribution of protein arginine methyltransferase 8 (PRMT8) in the mouse brain. Brain Res 1155:1–9. 10.1016/j.brainres.2007.03.086 [DOI] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S (2013) Defining the RGG/RG motif. Mol Cell 50:613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tradewell ML, Yu Z, Tibshirani M, Boulanger MC, Durham HD, Richard S (2012) Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet 21:136–149. 10.1093/hmg/ddr448 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-Seq. Nat Biotechnol 31:46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez G, Tapia JC, Kang H, Clemenson GD Jr, Gage FH, Lichtman JW, Sanes JR (2010) Attenuation of age-related changes in mouse neuromuscular synapses by caloric restriction and exercise. Proc Natl Acad Sci U S A 107:14863–14868. 10.1073/pnas.1002220107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16:1383–1391. 10.1038/nn.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Mundade R, Lange KC, Lu T (2014) Protein arginine methylation of non-histone proteins and its role in diseases. Cell Cycle 13:32–41. 10.4161/cc.27353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kitajo K (2012) The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS One 7:e49267. 10.1371/journal.pone.0049267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen T, Hébert J, Li E, Richard S (2009) A mouse PRMT1-null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol 29:2982–2996. 10.1128/MCB.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Vogel G, Coulombe Y, Dubeau D, Spehalski E, Hébert J, Ferguson DO, Masson JY, Richard S (2012) The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res 22:305–320. 10.1038/cr.2011.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Control animals at 15 months of age have normal limb reflexes when suspended by the tail.

PRMT8-null animals show abnormal and weak limb clasping.