Abstract
Membrane contacts between endoplasmic reticulum (ER) and plasma membrane (PM), or ER-PM junctions, are ubiquitous in eukaryotic cells and are platforms for lipid and calcium signaling and homeostasis. Recent studies have revealed proteins crucial to the formation and function of ER-PM junctions in non-neuronal cells, but little is known of the ER-PM junctions prominent in aspiny regions of mammalian brain neurons. The Kv2.1 voltage-gated potassium channel is abundantly clustered at ER-PM junctions in brain neurons and is the first PM protein that functions to organize ER-PM junctions. However, the molecular mechanism whereby Kv2.1 localizes to and remodels these junctions is unknown. We used affinity immunopurification and mass spectrometry-based proteomics on brain samples from male and female WT and Kv2.1 KO mice and identified the resident ER vesicle-associated membrane protein-associated proteins isoforms A and B (VAPA and VAPB) as prominent Kv2.1-associated proteins. Coexpression with Kv2.1 or its paralog Kv2.2 was sufficient to recruit VAPs to ER-PM junctions. Multiplex immunolabeling revealed colocalization of Kv2.1 and Kv2.2 with endogenous VAPs at ER-PM junctions in brain neurons from male and female mice in situ and in cultured rat hippocampal neurons, and KO of VAPA in mammalian cells reduces Kv2.1 clustering. The association of VAPA with Kv2.1 relies on a “two phenylalanines in an acidic tract” (FFAT) binding domain on VAPA and a noncanonical phosphorylation-dependent FFAT motif comprising the Kv2-specific clustering or PRC motif. These results suggest that Kv2.1 localizes to and organizes neuronal ER-PM junctions through an interaction with VAPs.
SIGNIFICANCE STATEMENT Our study identified the endoplasmic reticulum (ER) proteins vesicle-associated membrane protein-associated proteins isoforms A and B (VAPA and VAPB) as proteins copurifying with the plasma membrane (PM) Kv2.1 ion channel. We found that expression of Kv2.1 recruits VAPs to ER-PM junctions, specialized membrane contact sites crucial to distinct aspects of cell function. We found endogenous VAPs at Kv2.1-mediated ER-PM junctions in brain neurons and other mammalian cells and that knocking out VAPA expression disrupts Kv2.1 clustering. We identified domains of VAPs and Kv2.1 necessary and sufficient for their association at ER-PM junctions. Our study suggests that Kv2.1 expression in the PM can affect ER-PM junctions via its phosphorylation-dependent association to ER-localized VAPA and VAPB.
Keywords: ion channel, membrane contact sites, neuron, subcellular
Introduction
Voltage-gated potassium (Kv) channels are key determinants of intrinsic electrical excitability (Trimmer, 2015). The Kv2.1 channel is the major delayed rectifier Kv channel in mammalian brain neurons (Murakoshi and Trimmer, 1999; Guan et al., 2007; Liu and Bean, 2014; Hönigsperger et al., 2017; Palacio et al., 2017) and de novo mutations in Kv2.1 are associated with devastating neurological disorders (Torkamani et al., 2014; Thiffault et al., 2015; de Kovel et al., 2016). Kv2.1 is phosphorylated at more than three dozen in vivo sites (Park et al., 2006; Trimmer and Misonou, 2015) that affect voltage activation (Murakoshi et al., 1997; Ikematsu et al., 2011), plasma membrane (PM) expression (Redman et al., 2007), and PM clustering (Misonou et al., 2004; Bishop et al., 2015). Kv2.1 and its paralog Kv2.2 are present in large clusters on the soma, proximal dendrites, and axon initial segment (AIS) (Trimmer, 1991; Du et al., 1998; Sarmiere et al., 2008; Kihira et al., 2010; Bishop et al., 2015), which represent the aspiny regions of brain neurons (Spruston and McBain, 2007). A short proximal restriction and clustering (PRC) domain within the extensive cytoplasmic C terminus is both necessary and sufficient for Kv2-channel-like clustering (Lim et al., 2000; Bishop et al., 2015; Baker et al., 2016) and includes four amino acids (three serines and a phenylalanine) whose individual mutation eliminates clustering; reversible phosphorylation at some/all of these serine residues contributes to dynamic modulation of Kv2.1 clustering (Lim et al., 2000; Bishop et al., 2015; Cobb et al., 2015). Although molecular mechanisms underlying the highly restricted spatial organization of numerous ion channels at specific sites in brain neurons have been elucidated (Lai and Jan, 2006; Vacher et al., 2008; Nusser, 2012; Trimmer, 2015), those underlying the PRC-mediated clustering of Kv2 channels remain unknown. This information is crucial to understanding the basis of the exceptional localization of these abundant neuronal ion channels and to better inform using the Kv2.1 PRC domain to direct the restricted subcellular localization of optogenetic tools (Wu et al., 2013; Baker et al., 2016).
Neuronal Kv2 channels are clustered at sites where endoplasmic reticulum (ER) forms contact sites with PM (Du et al., 1998; Mandikian et al., 2014; Bishop et al., 2015, 2018), termed ER-PM junctions (Henne et al., 2015; Gallo et al., 2016; Chang et al., 2017; Saheki and De Camilli, 2017), which were originally discovered in electron micrographs of brain neurons (Gray, 1959; Rosenbluth, 1962; Peters et al., 1968) and engaging >10% of somatic PM area in certain neurons (Wu et al., 2017). ER-PM junctions are ubiquitous in eukaryotic cells and act as hubs for lipid exchange and protein trafficking and as specialized Ca2+ signaling microdomains, and are organized by a set of otherwise unrelated ER membrane protein tethers that bind PM phospholipids (Henne et al., 2015; Prakriya and Lewis, 2015; Gallo et al., 2016; Chung et al., 2017; Dickson, 2017). Although many of these ER tethers have high levels of mRNA expression in brain (Nishi et al., 2003; Min et al., 2007; Moccia et al., 2015; Takeshima et al., 2015), their subcellular localization in brain neurons has not been defined.
Kv2.1 is unique among PM proteins in its ability to organize ER-PM junctions (Fox et al., 2015). Proteomic analyses have been crucial in defining protein complexes that mediate synaptic signaling (Grant, 2012, 2013) and that comprise ER-PM junctions in non-neuronal cells (Prakriya and Lewis, 2015; Dickson, 2017). Here, we undertook a novel and unbiased proteomic approach to define vesicle associated membrane protein-associated protein isoforms A and B (VAPA and VAPB) as prominent Kv2-associated proteins. We characterize Kv2:VAP association at ER-PM junctions in heterologous cells, cultured hippocampal neurons (CHNs), and brain. We also define the impact of CRISPR-mediated KO of VAPA expression on Kv2.1 clustering and identify a plausible molecular mechanism for PRC domain-mediated Kv2-VAP interaction that underlies the generation of a prominent form of neuronal ER-PM junctions.
Materials and Methods
Animals.
All procedures involving mice and rats were approved by the University of California Davis Institutional Animal Care and Use Committee and were performed in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All mice and rats were maintained under standard light/dark cycles and allowed to feed and drink ad libitum. Adult male and female mice were used in proteomic and immunohistochemistry experiments. Kv2.1 KO mice (RRID:MGI:3806050) have been described previously (Jacobson et al., 2007; Speca et al., 2014) and were generated from breeding of Kv2.1+/− mice that had been backcrossed on the C57BL/6J background (RRID:IMSR_JAX:000664). Kv2.2-KO mice (Hermanstyne et al., 2010, 2013) were obtained from Drs. Tracey Hermanstyne and Jeanne Nerbonne. All Kv2.2-KO mice used here were obtained from heterozygotic crosses in the C57BL/6J background (RRID:IMSR_JAX:000664). Both male and female mice >12 weeks old were used. Littermates were used when available. Sprague Dawley rats were used as a source of hippocampal neurons for primary culture.
Antibodies.
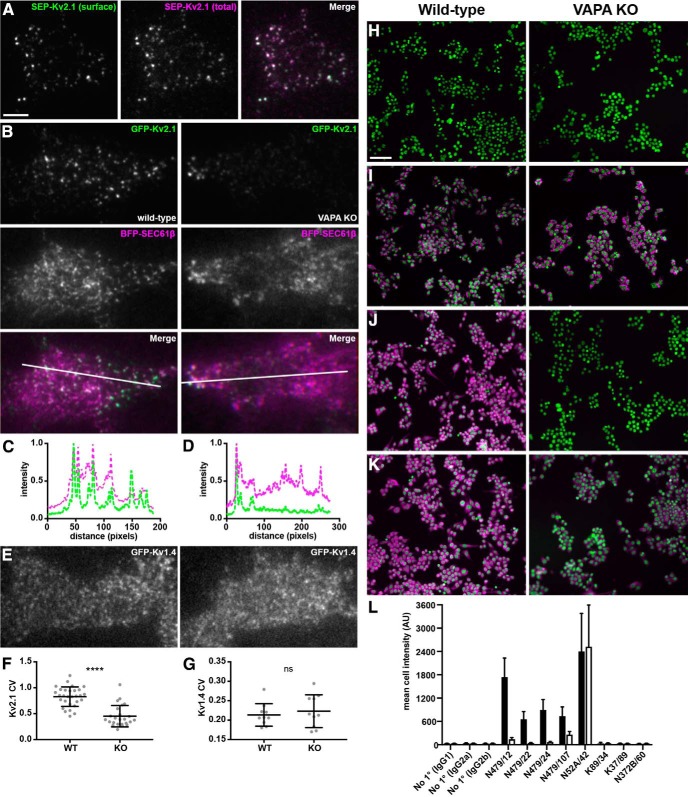
All primary antibodies used in this study are described in Table 1. Validation in KO mice has been demonstrated previously for the anti-Kv2.1 mouse monoclonal antibody (mAb) K89/34 and rabbit polyclonal KC antibodies (Misonou et al., 2006; Mandikian et al., 2014; Speca et al., 2014; Bishop et al., 2015, 2018) and the anti-Kv2.2 mouse mAbs N372B/60 and N372B/1 (Bishop et al., 2015, 2018). The anti-VAPA mouse mAbs were generated for this study. In brief, hybridomas producing these mAbs were generated using standard methods (Trimmer et al., 1985; Bekele-Arcuri et al., 1996) from BALB/C mice immunized with a bacterially expressed GST fusion protein containing an N-terminal fragment (aa 1-219) of rat VAPA encoded by pGEX-T-rnVAPA-1-219 (Addgene plasmid #13395, a gift from Axel Brunger and corresponding to accession number NP_113819.3) that was originally developed for structural studies (Kaiser et al., 2005). The anti-VAPA mouse mAbs N479/12 (VAPA-specific, IgG1, RRID:AB_2722707), N479/22 (VAPA-specific, IgG2a, RRID:AB_2722708), N479/24 (VAPA-specific, IgG2a, RRID:AB_2722709), and N479/107 (VAPA/B-specific, IgG2b, RRID:AB_2722711) were selected by a multistep screening and validation procedure (Bekele-Arcuri et al., 1996; Gong et al., 2016). Briefly, ≈3000 candidates were initially screened by ELISA assays against both the immunogen and fixed heterologous cells expressing full-length VAPA protein. A set of 144 ELISA-positive candidates was then evaluated for specificity on immunoblots against rat brain membrane proteins and extracts of transiently transfected COS-1 cells (which express endogenous VAPs) expressing GFP-VAPA or GFP-VAPB and immunocytochemistry against the same set of transfected heterologous cells. A subset of the subcloned mAbs (the anti-VAPA-specific N479/12, N479/22, N479/24, and VAPA/B-specific N479/107) were further validated for specificity in fluorescence immunocytochemistry by parallel immunolabeling of fixed WT RAW264.7 mouse macrophage cells (which endogenously express both VAPA and VAPB) and VAPA KO RAW 264.7 cells (McCune et al., 2017). Controls included irrelevant mAbs of each mouse IgG subclass (K89/34 IgG1, K37/89 IgG2a, N372B/60 IgG2b), a positive control anti-mortalin mAb N52A/42 IgG1, and no primary antibody controls for each mouse IgG subclass-specific secondary antibody. The extent of immunolabeling was evaluated by fluorescence microscopy using a Nikon Ti-E inverted microscope, equipped with an automated stage and a 20×/0.75 numerical aperture (NA) objective and an Andor Zyla 5.5 scMOS camera controlled by NIS Elements. Quantification of the immunolabeling signaling intensity in each of 1300–2800 cells in each sample was determined by high content analysis. The results of these experiments are shown in Figure 5, H–L. In summary, whereas we observed no difference in the level of immunolabeling for the positive control mAb N52A/42 in the WT versus VAPA KO cells, we observed a sharp reduction in immunolabeling for each of the N479 mAbs tested. For the three VAPA-specific mAbs, N479/12, N479/22, and N479/24, we saw a reduction of the immunolabeling signal in the VAPA KO cells to levels approaching those of subclass-matched irrelevant primary and no primary control levels and, for the VAPA/B mAb N479/107, a smaller but still substantial reduction of signal in the VAPA KO cells.
Table 1.
Antibody information
| Antibody | Immunogen | Manufacturer information | Concentration used | Figure(s) |
|---|---|---|---|---|
| N479/12 | Fusion protein aa 1-219 of rat VAPA | Mouse IgG1 mAb, NeuroMab, RRID: AB_2722707 | Tissue culture supernatant, 1:5 | 6A–F, 9 |
| N479/22 | Fusion protein aa 1-219 of rat VAPA | Mouse IgG2a mAb, NeuroMab, RRID:AB_2722708 | Tissue culture supernatant, neat (Fig. 8), 1:5 (Fig. 6), 1:10 (Fig. 7) | 6G–J, 7 |
| N479/24 | Fusion protein aa 1-219 of rat VAPA | Mouse IgG2a mAb, RRID:AB_2722709 | Tissue culture supernatant, 1:5 | 10 |
| N479/107 | Fusion protein aa 1-219 of rat VAPA | Mouse IgG2b mAb, NeuroMab, RRID:AB_2722711 | Tissue culture supernatant, 1:5 | 7, 8 |
| KC | Synthetic peptide aa 837-853 of rat Kv2.1 | Rabbit pAb, in-house (Trimmer Laboratory), RRID:AB_2315767 | Affinity purified, 1:150 | 8, 9, 10 |
| K89/34 | Synthetic peptide aa 837-853 of rat Kv2.1 | Mouse IgG1 mAb, NeuroMab catalog #73-014, RRID:AB_10672253 | Tissue culture supernatant, 1:10 | 7 |
| 34C | Partially purified chicken pectoral muscle ryanodine receptor | Mouse IgG1 mAb, Thermo Fisher catalog #MA3-925 RRID:AB_2254138 | Purified, 1 μg/ml | 8 |
| 34C | Partially purified chicken pectoral muscle ryanodine receptor | Mouse IgG1 mAb, Developmental Studies Hybridoma Bank, RRID:AB_528457 | Concentrated tissue culture supernatant, 1 μg/ml | 9 |
| N106/36 | Full-length recombinant human ankG | Mouse IgG2b mAb, NeuroMab catalog #75-147, RRID:AB_10675130 | Tissue culture supernatant, 1:5 | 9 |
| N372B/1 | Fusion protein aa 717-907 of rat Kv2.2 long isoform | Mouse IgG1 mAb, NeuroMab catalog #73-369, RRID:AB_2315869 | Tissue culture supernatant, 1:2 | 7, 8 |
| N372B/60 | Fusion protein aa 717-907 of rat Kv2.2 long isoform | Mouse IgG2a mAb, NeuroMab catalog #73-360, RRID:AB_2315867 | Purified, 10 μg/ml | 9, 10, 11 |
| Kv1.5E | Synthetic peptide aa 271-284 of rat Kv1.5 | Rabbit pAb, in-house (Trimmer Laboratory) RRID:AB_2722698 | Affinity purified, 10 μg/ml | 10 |
| 2–2.20.14, DyLight 488 | HA peptide YPYDVPDYA | Mouse IgG1 mAb, Thermo Fisher Scientific catalog #26183-D488; RRID: AB_2533051 | Purified, 0.3 μg/ml | 11 |
Figure 5.
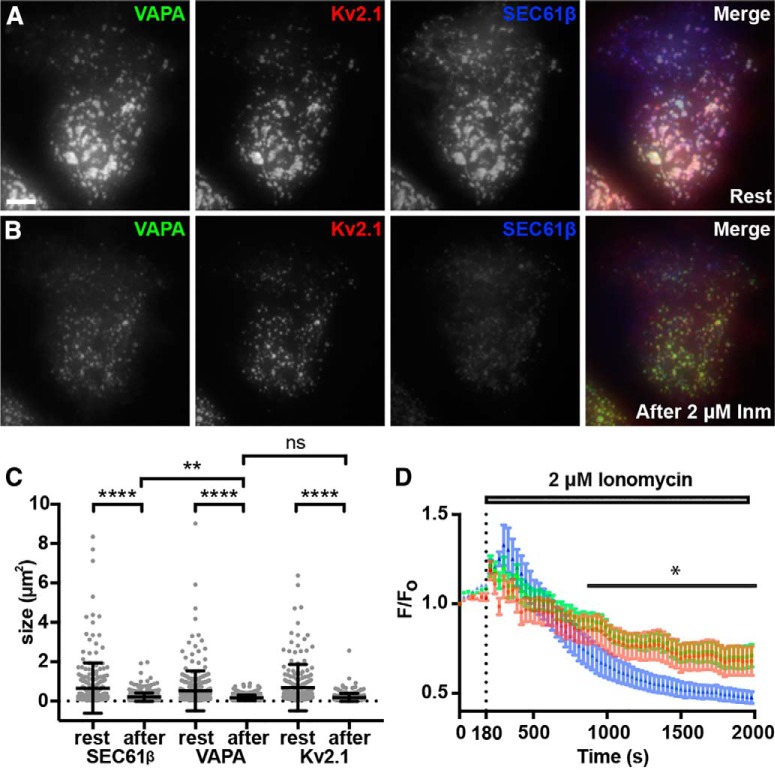
CRISPR-mediated KO of VAPA expression affects Kv2.1 localization in RAW264.7 cells A. Representative TIRF images of SEP-Kv2.1 expression in a live WT RAW264.7 cell expressing SEP-Kv2.1 (green is cell surface pHluorin fluorescence, magenta is total cellular mCherry fluorescence). Scale bar is 5 μm and holds for all panels in figure. B, Representative TIRF images of GFP-Kv2.1 (green) and BFP-SEC61β (magenta) in live WT (left) or VAPA KO (right) RAW264.7 cells coexpressing GFP-Kv2.1 and BFP-SEC61β. Note the reduction in clustered Kv2.1 expression in the VAPA KO cell. C, Line scan analysis of selection indicated in merged image of WT RAW264.7 cell coexpressing GFP-Kv2.1 and BFP-SEC61β. D, Line scan analysis of selection indicated in merged image of VAPA KO RAW264.7 cell coexpressing GFP-Kv2.1 and BFP-SEC61β. E, Representative TIRF image of live WT (left) or VAPA KO (right) RAW264.7 cells expressing GFP-Kv1.4. F, Summary graph of CV measurements of GFP-Kv2.1 in WT and VAPA KO RAW264.7 cells (****p = 6.648 × 10−9, n = 29 cells, two-tailed unpaired t test). G, Summary graph of CV measurement of GFP-Kv1.4 in WT and VAPA KO RAW264.7 cells (ns, 0.5603, n = 10 cells, two-tailed unpaired t test). H-K. Images of WT (left) or VAPA KO (right) RAW264.7 cells used for high-content analysis of immunolabeling (magenta) and with Hoechst labeling of nuclei (green). H, No primary (anti-IgG2b secondary antibody). I, Positive control anti-mortalin mAb N52A/42 (IgG1). J, Anti-VAPA mAb N479/22 (IgG2a). K, Anti-VAPA/B mAb N479/107 (IgG2b). Scale bar in H is 65 μm and holds for H–K. L, Mean immunolabeling intensity of all samples across 1300–2800 individual cells in each sample. See Materials and Methods for further details of these antibody validation results.
Immunopurification and proteomic analyses.
Mice were acutely decapitated in the absence of anesthesia and brains were rapidly excised and homogenized in 5 ml of ice-cold homogenization buffer containing 1 mm DSP [Lomant's reagent, dithiobis(succinimidyl propionate); Thermo Fisher Scientific catalog #22585], 320 mm sucrose (Sigma-Aldrich catalog #S0389), 5 mm Na phosphate, pH 7.4, 0.1 m sodium fluoride, 1 mm PMSF, and protease inhibitors. Following a 1 h incubation on ice, DSP was quenched using 1 m Tris pH 7.4 (JT Baker catalog #4109-01: Tris base and 4103-01: Tris-HCl), added to a final concentration of 0.02 m. Then, 2 ml of this brain homogenate was added to an equal volume of ice-cold 2× radioimmunoprecipitation assay (RIPA) buffer (final detergent concentrations: 1% TX-100, 0.5% deoxycholate, 0.1% SDS) and incubated for 30 min on a tube rotator at 4°C. Following this incubation, insoluble material was pelleted by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was incubated with primary antibodies overnight at 4°C using either the anti-Kv2.1 rabbit polyclonal antibody KC (Trimmer, 1991) directed against the Kv2.1 C terminus (for KO validation, see Misonou et al., 2006 and Speca et al., 2014) and the anti-Kv2.2 mouse monoclonal antibody N372B/60 (Bishop et al., 2015, 2018). This was followed by the addition 200 μl of protein G beads (GE Life Sciences catalog #17061802) for 1 h on a tube rotator at 4°C. The beads were then washed 6 times in 1× RIPA buffer and eluted by boiling in reducing sample buffer for 5 min. For analysis, immunopurified (IP) eluates were run on a 9% SDS polyacrylamide gel which was then stained with SYPRO Ruby (Thermo Fisher catalog #S12000) according to the manufacturer's protocol. For preparation of samples for proteomic profiling by tandem mass spectrometry, IP eluates were run shortly into the resolving gel of a 7.5% SDS polyacrylamide gel and stained with a colloidal Coomassie stain containing 0.08% Coomassie brilliant blue G250 (JT Baker catalog #F78903), 1.6% phosphoric acid, 8% ammonium sulfate, and 20% methanol overnight at room temperature. Gels were destained in Milli-Q (Millipore) water and a single gel band containing all eluted proteins was excised from each gel. Gel bands were prepared essentially as described previously (Saveliev et al., 2013). Briefly, gel bands were cut into small ≈1 mm3 pieces and washed in 50 mm ammonium bicarbonate. Gel bands were desiccated using repetitive acetonitrile washes and reduced with 10 mm dithiothreitol in 50 mm ammonium bicarbonate. Gel bands were further desiccated and alkylated with 55 mm iodoacetamide in 50 mm ammonium bicarbonate. Proteins were trypsinized in gel overnight at 37°C and extracted the next day with 0.1% trifluoroacetic acid in 60% acetonitrile. Extractions were lyophilized and resuspended in 0.1% trifluoroacetic acid in 60% acetonitrile.
Proteomic profiling of gel band extracts was performed at the University of California–Davis proteomic facility. Digested peptides were analyzed by LC-MS/MS on a Thermo Fisher Scientific Q Exactive Plus Orbitrap Mass spectrometer in conjunction Proxeon Easy-nLC II HPLC and Proxeon nanospray source. The digested peptides were loaded on a 100 μm × 25 mm Magic C18 100Å 5U reverse phase trap where they were desalted online before being separated using a 75 μm × 150 mm Magic C18 200Å 3U reverse phase column. Peptides were eluted using a 60 min gradient with a flow rate of 300 nl/min. An MS survey scan was obtained for the m/z range 350–1600 and MS/MS spectra were acquired using a top 15 method in which the top 15 ions in the MS spectra were subjected to high energy collisional dissociation. An isolation mass window of 1.6 m/z was for the precursor ion selection and normalized collision energy of 27% was used for fragmentation. A 15 s duration was used for the dynamic exclusion.
Tandem mass spectra were extracted and charge state deconvoluted by Proteome Discoverer (Thermo Fisher Scientific) All MS/MS samples were analyzed using X! Tandem (the GPM, thegpm.org; version Alanine 2017.2.1.4). X! Tandem was set up to search the UniProt Mouse database (May 2017, 103089 entries), the cRAP database of common proteomic contaminants (www.thegpm.org/crap; 114 entries), the ADAR2 catalytic domain sequence plus an equal number of reverse protein sequences assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 ppm and a parent ion tolerance of 20 ppm. The iodoacetamide derivative of cysteine was specified in X! Tandem as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulfone of methionine, tryptophan oxidation to formylkynurenin of tryptophan and acetylation of the N terminus were specified in X! Tandem as variable modifications.
Scaffold (version Scaffold_4.8.4; Proteome Software) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they exceeded specific database search engine thresholds. X! Tandem identifications required at least −log (expect scores) scores of >2.0 with a mass accuracy of 5 ppm. Protein identifications were accepted if they contained at least two identified peptides. Our threshold for peptide acceptance was >95% probability.
Preparation of mouse brain sections for immunohistochemistry.
We used adult male and female WT mice generated from the breeding of Kv2.1+/− mice. Mice were deeply anesthetized with 90 mg/kg Na-pentobarbital salt (Sigma-Aldrich catalog #P3761) in 0.9% NaCl solution through intraperitoneal injections, followed by boosts as needed. Once mice were completely anesthetized, they were transcardially perfused with 4.5 ml of ice-cold PBS containing 150 mm NaCl, 10 mm sodium phosphate buffer (PB), pH 7.4, containing 10 U/ml heparin, followed by an ice-cold fixative solution of 4% formaldehyde (freshly prepared from paraformaldehyde, Sigma-Aldrich catalog #158127) in 0.1 m sodium PB, pH 7.4, using a volume of 1 ml of fixative solution per gram of mouse weight. Following perfusions, brains were removed from the skull and cryoprotected in 10% sucrose, 0.1 m PB overnight at 4°C and then transferred to a solution of 30% sucrose and 0.1 m PB until they sank to the bottom of the tube (24–48 h). Following cryoprotection, all brains were frozen, and cut on a freezing stage sliding microtome (Richard Allen Scientific) to obtain 30-μm-thick sagittal sections. Sections were collected in 0.1 m PB and processed for immunohistochemistry as free-floating sections.
Multiplexed fluorescence immunohistochemistry.
For a list of antibodies used in this study, see Table 1. Multiplex immunofluorescence labeling of mouse brain sections was performed essentially as described previously (Manning et al., 2012). Briefly, free-floating sections were washed 3× in 0.1 m PB and 10 mm sodium azide at room temperature with slow agitation. All subsequent incubations and washes were at room temperature with slow agitation unless stated otherwise. Sections were incubated in blocking buffer (10% goat serum in 0.1 m PB, 0.3% Triton X-100, and 10 mm sodium azide) for 1 h. Immediately after blocking, sections were incubated with primary antibody combinations (diluted in blocking buffer) overnight at 4°C. Following incubation, sections were washed 3 × 10 min each in 0.1 m PB and incubated for 1 h in blocking buffer containing affinity-purified goat anti-rabbit and/or goat anti-mouse IgG-subclass-specific Alexa Fluor-conjugated secondary antibodies (all secondary antibodies from Thermo Fisher Scientific) at a dilution of 1:1500 (for Alexa Fluor 647 conjugates) and 1:2000 (for Alexa Fluor 488 and 555 conjugates) and the DNA-specific dye Hoechst 33258 at 200 ng/ml. After 3 × 10 min washes in 0.1 m PB, sections were mounted and dried onto gelatin-coated slides, treated with 0.05% Sudan black (EM Sciences catalog #21610) in 70% ethanol for 1.5 min, extensively washed in water, and mounted with ProLong Gold (Thermo Fisher Scientific catalog #P36930). All immunolabeling reported is representative of three animals (biological replicates). Brain sections from all biological replicates were labeled, treated, and mounted in parallel.
Low-magnification wide-field images were acquired on a Zeiss AxioObserver Z1 microscope with an X-Cite 120 lamp as the fluorescent light source and equipped with an AxioCam MRm digital camera. A 10×/0.5 NA Fluar objective was used and images were reconstructed as tiled mosaics using Axiovision version 4.8.2 acquisition software (Carl Zeiss, RRID:SciRes_000111). Image processing was performed in Axiovision (Carl Zeiss) and NIH Fiji version 2.0.0-rc-43/1.51 (Schindelin et al., 2012). For superresolution light microscopy (N-SIM, Nikon) imaging of brain immunohistochemistry samples, images were acquired using a Hamamatsu ORCA-ER CCD camera installed on a SIM/wide-field equipped Nikon Eclipse Ti microscope using an EXFO X-Cite metal halide light source and a 100× PlanApo total internal reflection fluorescence (TIRF)/1.49 objective run with NIS Elements software (Nikon). SIM images postprocessing was also performed in NIS Elements software (Nikon). Exposure time was optimized for every image independently.
Culture and transfection of rat hippocampal neurons.
Hippocampi were dissected from embryonic day 18 rat embryos and dissociated enzymatically for 20 min at 37°C in 0.25% (w/v) trypsin (Thermo Fisher Scientific catalog #15050065) in HBSS and dissociated mechanically by triturating with glass-polished Pasteur pipettes. Dissociated cells were suspended in plating medium containing Neurobasal medium (Thermo Fisher Scientific catalog #21103049) supplemented with 10% FBS (Invitrogen catalog #16140071), 2% B27 (Invitrogen catalog #17504044), 2% GlutaMAX (Thermo Fisher Scientific catalog #35050061), and 0.001% gentamycin (Invitrogen catalog #15710064) and plated at 60,000 cells per dish in glass bottom dishes (MatTek catalog #P35G-1.5–14-C) coated with poly-l-lysine (Sigma-Aldrich catalog #P2636). At 7 d in vitro (DIV), cytosine-d-arabinofuranoside (Millipore catalog #251010) was added to inhibit non-neuronal cell growth. Neurons were transiently transfected at DIV 7–10 using Lipofectamine 2000 (Invitrogen catalog #11668019) for 1.5 h as described previously (Lim et al., 2000). Neurons were imaged 40–48 h after transfection.
Cell culture, reagents, and transfection.
HEK293T cells were maintained in DMEM supplemented with 10% Fetal Clone III (HyClone catalog #SH30109.03), 1% penicillin/streptomycin, and 1× GlutaMAX (Thermo Fisher Scientific catalog #35050061) in a humidified incubator at 37°C and 5% CO2. RAW 264.7 WT and VAPA KO cells were identically maintained in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, 10 mm HEPES, and 1× GlutaMAX in a humidified incubator at 37°C and 5% CO2. HEK293T and RAW264.7 cells were transfected as described previously (Bishop et al., 2018). Briefly, cells were split to 15% confluence on glass bottom dishes (MatTek catalog #P35G-1.5-14-C) or number 1.5 glass coverslips coated with poly-L-lysine and then transiently transfected using Lipofectamine 2000 following the manufacturer's protocol. Cells were transiently transfected in DMEM without supplements and then returned to regular growth medium 4 h after transfection. Cells were used 40–48 h after transfection.
Plasmid constructs.
For clarity and consistency with most previous studies, we used the original (Frech et al., 1989) amino acid numbering of rat Kv2.1 (accession number NP_037318.1) throughout. DsRed-Kv2.2 and GFP-Kv2.2 were generated using Gibson assembly to insert full-length rat Kv2.2 (accession number NP_446452.2), also termed Kv2.2long (Kihira et al., 2010), into the DsRed-C1 or EGFP-C1 vectors (ClonTech), respectively, resulting in fusion of DsRed or EGFP to the N terminus of full-length rat Kv2.2. GFP-Kv2.2 S605A was generated using QuikChange PCR-based mutagenesis of GFP-Kv2.2, as described previously for generation of Kv2.2 S605A in the RBG4 vector (Bishop et al., 2015). DsRed-Kv2.1 S586A and DsRed-Kv2.1 P404W were generated using QuikChange PCR-based mutagenesis using DsRed-Kv2.1 (Bishop et al., 2018) as a template. GFP-Kv2.1 S586A and GFP-Kv2.1 P404W were similarly generated using GFP-Kv2.1 (Antonucci et al., 2001) as a template. The generation of rat Kv2.1 truncation, internal deletion, and point mutants, as well as Kv1.5N-Kv2.1C chimeras, has been described previously (Lim et al., 2000; Mohapatra and Trimmer, 2006). Mutations were confirmed with sequencing. Plasmids encoding GFP-VAPA (Addgene plasmid #18874) and GFP-VAPA K87D/M89D (Addgene plasmid #18875) were a generous gift from Dr. Alex Brunger. The plasmid encoding DsRed2-ER5 was a generous gift from Dr. Michael Davidson (Addgene plasmid #55836). The plasmid encoding GFP-VAPB was generated by amplifying mouse VAPB (Origene catalog #MR203021) by PCR using primers to introduce a KpnI site upstream of the start codon and an AgeI site on the 3′ end of VAPB. The isolated PCR product was then digested with KpnI and AgeI and inserted into the corresponding sites in pEGFP-N1 (Clontech), producing pEGFP-N1-VAPB, the sequence integrity of which was confirmed by sequencing. The plasmid encoding BFP-SEC61β was a generous gift from Dr. Jodi Nunnari (University of California–Davis). The plasmid encoding mCherry-Esyt3 was a generous gift from Dr. Pietro De Camilli (Yale University School of Medicine). The plasmid encoding mCherry-Junctophilin 4 (JP4) was a generous gift from Dr. Yousang Gwack (University of California–Los Angeles).
Cell fixation, immunocytochemistry, and fixed-cell imaging.
For a list of antibodies used in this study, see Table 1. Briefly, HEK293T cells were fixed in 3.2% formaldehyde (freshly prepared from paraformaldehyde; Sigma-Aldrich catalog #158127) and 0.1% glutaraldehyde (Ted Pella, catalog #18426) for 30 min at room temperature, washed 3 × 5 min in PBS, and quenched with 1% sodium borohydride in PBS for 15 min at room temperature. RAW 264.7 cells were fixed in ice cold 4% formaldehyde in PBS for 15 minutes at 4°C. Cells were blocked and permeabilized in 4% nonfat milk powder in TBS containing 0.1% Triton X-100. Neurons were fixed in ice-cold 4% formaldehyde/4% sucrose in PBS for 15 min at 4°C, washed 3 × 5 min in PBS, and blocked and permeabilized in 4% nonfat milk powder in TBS containing 0.1% Triton X-100. Primary antibody incubation was performed in blocking solution for 1 h at room temperature. Following primary antibody incubation and 3 × 5 min washes in blocking solution at room temperature, coverslips were immunolabeled in blocking solution with species- and or mouse IgG subclass-specific Alexa Fluor-conjugated goat anti-mouse IgG subclass-specific (Manning et al., 2012) or goat anti-rabbit IgG secondary antibodies (all secondary antibodies from Thermo Fisher Scientific) at a 1:1500 dilution and Hoechst 33258 (Thermo Fisher Scientific catalog #H1399) at 200 ng/ml for 1 h, washed 3 × 5 min in PBS, and mounted onto microscope slides using Prolong gold (Thermo Fisher Scientific, catalog #P36930) or, for samples prepared for TIRF, imaged in imaging saline.
For conventional fluorescence imaging (used in Fig. 10A,B) images were acquired with an AxioCam MRm digital camera installed on a Zeiss AxioImager M2 microscope or with an AxioCam HRm digital camera installed on a Zeiss AxioObserver Z1 microscope with a 63×/1.40 NA plan-Apochromat oil-immersion objective or a 20×/0.8 NA plan-Apochromat objective and an ApoTome coupled to Axiovision software (Zeiss). For TIRF imaging of fixed cells, imaging was identical to that used in live-cell TIRF experiments but in the absence of a heated stage/objective heater. Images were obtained with an Andor iXon EMCCD camera installed on a TIRF/wide-field equipped Nikon Eclipse Ti microscope using a Nikon LUA4 laser launch with 405, 488, 561, and 647 nm lasers and a 100× PlanApo TIRF/1.49 NA objective run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images. For superresolution light microscopy (N-SIM; Nikon) imaging of fixed cells, images were acquired using a Hamamatsu ORCA-ER CCD camera installed on a SIM/wide-field equipped Nikon Eclipse Ti microscope using an EXFO X-Cite metal halide light source and a 100× PlanApo TIRF/1.49 objective run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images. SIM analysis was performed in NIS Elements. For superresolution light microscopy (Airyscan; Zeiss) imaging of fixed cells, images were acquired with a Zeiss LSM 880 confocal laser scanning microscope (Carl Zeiss) equipped with an Airyscan detection unit and a Plan-Apochromat 63×/1.40 Oil DIC M27 objective.
Figure 10.
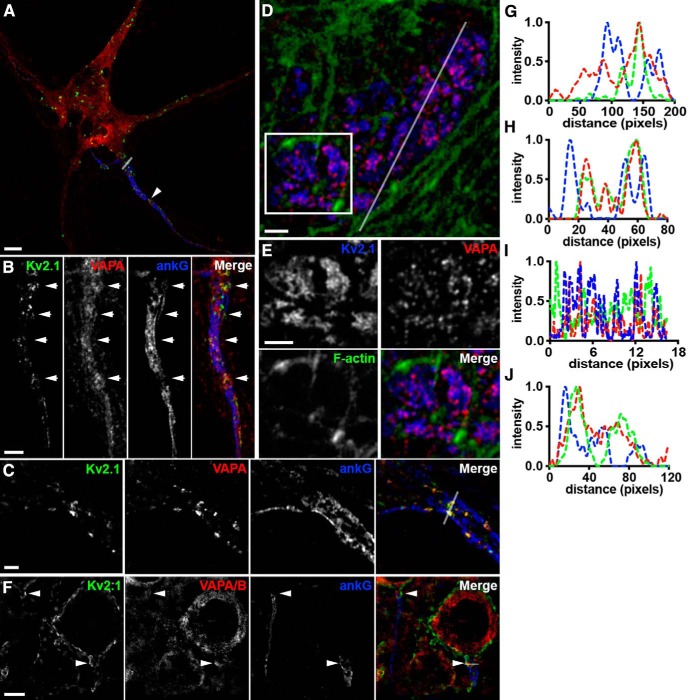
Endogenous complexes of Kv2.1 and VAPs represent specialized domains within the cortical actin cytoskeleton. A, Representative image of a single fixed CHN immunolabeled for Kv2.1 (green), VAPA (red), and ankG (blue) and imaged with superresolution (Airyscan; Zeiss) microscopy. Arrowhead denotes region of AIS enlarged in B. Scale bar, 5 μm. B, Magnified view of axon initial segment indicated in A. Arrows point to sites of colocalized Kv2.1/VAPA in “voids” in ankG. Scale bar, 3 μm. C, AIS of a distinct fixed CHN immunolabeled for Kv2.1 (green), VAPA (red), and ankG (blue) and imaged with superresolution (N-SIM; Nikon) microscopy. Scale bar, 1.5 μm. D, Representative image of a single fixed HEK293T cell expressing GFP-Kv2.1 (blue) immunolabeled for endogenous VAPA (red) and phalloidin stained for filamentous actin (green). Scale bar, 1.25 μm. E, Magnified view of selection indicated in D. Scale bar, 1.25 μm. F, Representative images of cortical neurons (layer 5) from intact mouse brain sections immunolabeled for Kv2.1 (green), VAPA/B (red), and ankG (blue). Arrows point to sites of associated Kv2.1 and VAPA/B puncta in “voids” in ankG. Scale bar, 5 μm. G, Line scan analysis of intensity of selection indicated in A. H, Line scan analysis of intensity of selection indicated in merged image of C. I, Line scan analysis of intensity of selection indicated in D. J, Line scan analysis of intensity of merged image of F.
Live-cell TIRF imaging.
TIRF microscopy was performed at the University of California–Davis MCB Imaging Facility as described previously (Bishop et al., 2018). Live transfected HEK293T cells cultured on glass-bottom dishes were imaged in a physiological saline solution containing the following (in mm): 4.7 KCl, 146 NaCl, 2.5 CaCl2, 0.6 MgSO4, 1.6 NaHCO3, 0.15 NaH2PO4, and 20 HEPES, pH 7.4, containing 8 mm glucose and 0.1 mm ascorbic acid or, in the case of experiments involving ionomycin (Inm) treatment, HBSS (Invitrogen catalog #14025076). Cells were maintained at 37°C during the course of imaging with a heated stage and objective heater. For experiments involving Inm (Millipore catalog #56092–82-1), Inm was diluted to 4 μm in HBSS and added by pipette to glass-bottom dishes already containing HBSS to a final concentration of 2 μm. Images were obtained with an Andor iXon EMCCD camera installed on a TIRF/wide-field equipped Nikon Eclipse Ti microscope using a Nikon LUA4 laser launch with 405, 488, 561, and 647 nm lasers and a 100× PlanApo TIRF, 1.49 NA objective run with NIS Elements software (Nikon). Images were collected within NIS Elements as ND2 images. Fixed cells were imaged identically in the absence of a stage heater.
Experimental design and statistical analysis.
For all datasets presented in this study for which statistical analyses were performed, measurements were imported into GraphPad Prism for presentation and statistical analysis as noted in each figure legend. Data were summarized as mean ± SD and these values are noted in each figure legend. Exact p-values are also noted in each figure legend. Results reported from experiments involving proteomics on brain samples from WT and Kv2.1 KO mice were observed in three independent sets of sex- and age-matched adult littermates (mice used in trial 1 were female and mice used in trials 2 and 3 were male). Results reported from experiments involving proteomics on brain samples from WT and Kv2.2 KO mice were observed in one set of age-matched adult male littermates. Results reported from experiments involving immunohistochemistry on brain sections were observed in samples from at least three independent adult animals (both male and female mice were used in this study). For experiments involving HEK293T cells and mixed-sex cultured rat hippocampal neurons, the number of samples (n) indicates the number of cells analyzed and is noted in each figure legend.
All colocalization analyses were performed within Nikon NIS Elements using ND2 files. A region of interest (ROI) was drawn within a cell and Pearson's correlation coefficient (PCC) and Mander's overlap coefficient values were collected. Measurements of structure sizes were quantified automatically within Fiji essentially as described previously (Dickson et al., 2016). ND2 files of BFP-SEC61β, DsRed-Kv2.1, GFP-VAPA, or immunolabeling of endogenous VAPs collected in TIRF were imported directly into Fiji, background subtracted, converted into an 8-bit image, and automatically converted into a binary mask using auto local thresholding. An ROI with identical dimensions was drawn within each cell analyzed. The number of individual ER-PM junctions, average ER-PM junction size, and percentage PM occupancy were quantified automatically using the “analyze particles” function in Fiji. Signals smaller than 0.04 μm2 were excluded from this analysis. An identical approach was taken in whole-cell analysis.
Statistical analyses of the normalized Kv2.1 isoform-dependent fold increases in VAPA fluorescence intensity were performed on log-transformed data. Quantification of intensity was also performed within ImageJ. ND2 or ZVI (Zeiss) files were imported directly into Fiji and an ROI of identical dimensions was used to collect mean intensity measurements of VAP labeling intensity from images of cells expressing various Kv2 isoforms or control cells. Coefficient of variation is defined as SD of intensity divided by mean intensity as described previously (Bishop et al., 2015; Jensen et al., 2017). Quantification of coefficient of variation and intensity measurements were collected in Fiji. An ROI was drawn around a cell and SD of intensity and mean intensity values were collected. For line scan analysis of intensity, raw intensity values were collected within Fiji and normalized to the maximum value collected.
Results
Kv2 channels and VAPs associate in mouse brain
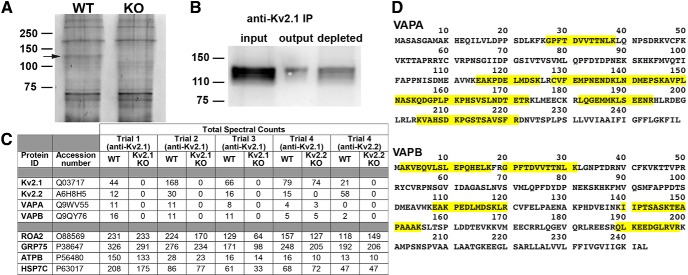
Neuroproteomic characterization of the supramolecular complex at the postsynaptic density (Husi et al., 2000) provided a foundation for important insights into the dynamic signaling events mediated by the constituent proteins in normal synaptic function and plasticity (Frank and Grant, 2017) and how this is disrupted in disease (Grant, 2012). Previous attempts (Chung and Li, 2005 and our unpublished data) to use this approach to identify Kv2.1-interacting proteins using the nonionic detergent Triton X-100 (TX-100) did not yield candidate proteins that defined the clustered localization of Kv2.1. We therefore took a novel proteomics-based approach to identify candidate interacting proteins at Kv2-containing ER-PM junctions in mammalian brain neurons. Although similar to previous attempts in using IP using KO-validated anti-Kv2.1 and Kv2.2 antibodies, in this case, we instituted three modifications to these earlier approaches. First, we used as starting material brain homogenates that had been subjected to DSP-mediated chemical cross-linking (Lomant and Fairbanks, 1976) during homogenization. Second, we extracted these cross-linked brain homogenates under harsher detergent conditions (1% TX-100, 0.5% deoxycholate, and 0.1% SDS) than used in these previous studies. Importantly, we also performed side-by-side IPs on paired samples from WT mice and from KO littermates lacking either expression of Kv2.1 (Speca et al., 2014) or Kv2.2 (Hermanstyne et al., 2013) to rigorously define the specificity of proteins co-IPing with Kv2.1 and Kv2.2. Figure 1A shows an SDS gel analysis of the outcome of one such IP reaction, in this case for Kv2.1, for which staining for total protein reveals the presence of a band with the characteristic electrophoretic mobility of Kv2.1 (≈120 kDa) in the sample from the brain of a WT mouse. Importantly, this band was not present in an identically prepared sample from a Kv2.1 KO mouse (Fig. 1A). Immunoblot analyses of IP product supported that Kv2.1 was present in the IP from the WT mouse brain starting material (Fig. 1B).
Figure 1.
VAPA and VAPB associate with Kv2 channels in mouse brain. A, SYPRO Ruby-stained SDS-polyacrylamide gel of protein recovered from IPs using WT and Kv2.1 KO mouse brain. Arrow points to a band that is likely Kv2.1 based on electrophoretic mobility and absence from Kv2.1 KO IP. B, Immunoblot analysis of input, output, and depleted fractions from a single trial of Kv2.1 IPs performed using WT mouse brain. Note the presence of Kv2.1 in the output fraction. C, Total spectral counts from proteins recovered from three separate trials of Kv2.1 IPs from WT and Kv2.1 KO mouse brain, from a Kv2.1 IP from a Kv2.2 KO brain sample, and from a Kv2.2 IP. Note the specific presence of VAPA and VAPB in the Kv2.1 IP samples from WT but not in Kv2.1 KO brain. D, Sequence coverage of VAPA and VAPB from the Kv2.1 IPs.
We subjected the products of three successive rounds of parallel Kv2.1 IP reactions performed on samples prepared from three pairs of WT and Kv2.1 KO mice (i.e., six mice total) to proteomic analysis of constituent proteins by liquid chromatography tandem mass spectrometry (LC-MS/MS). The specificity of these analyses was revealed by the robust presence of Kv2.1-derived tryptic peptide spectra in the independent Kv2.1 IP samples from the brains of three WT mice and their absence in the parallel samples from the Kv2.1 KO mice (Fig. 1C). The Kv2.1 paralog Kv2.2, which colocalizes and associates with Kv2.1 in a subset of brain neurons (Kihira et al., 2010; Bishop et al., 2015, 2018), was also specifically present in Kv2.1 IPs performed on the WT but not Kv2.1 KO samples (Fig. 1C). Most prominent among all other proteins that were found specifically in the Kv2.1 IPs (Fig. 1C) were the VAP isoforms A (VAPA; UniProt Q9WV55) and B (VAPB; UniProt Q9QY76). Overall sequence coverage of VAPA and VAPB (Fig. 1D) from these Kv2.1 IPs was extensive (39.3% and 28.0%, respectively). No other proteins that represent apparent intermediaries for association of Kv2.1 with the VAP proteins were found, although we note that the amphoterin-induced protein 1 (AMIGO-1) auxiliary subunit of Kv2 channels (Peltola et al., 2011; Bishop et al., 2018) was also not detectably present in the purified material. The importance of the parallel analysis of the Kv2.1 KO samples is demonstrated by the robust and reproducible signals from a large number of nonspecific proteins present in the Kv2.1 IP products from both WT and Kv2.1 KO mouse brain samples, the values for the most abundant of which (ROA2, GRP75, ATPB, and HSP7C) are shown in Figure 1C. Signals from these abundant cytoplasmic and nuclear proteins is well in excess of the signals from the target Kv2.1 P.M. ion channel and the ER-localized VAP membrane proteins.
We undertook a similar approach to IP Kv2.2 from cross-linked samples from WT and Kv2.2 KO mice. In addition to Kv2.2 and Kv2.1, a small number of peptides from VAPB were specifically returned. Importantly, both VAPA and VAPB were also present in Kv2.1 IPs performed on samples from Kv2.2 KO brains demonstrating that Kv2.1 is associated with the VAP proteins in the absence of Kv2.2. These data demonstrate that both VAPA and VAPB biochemically associate with Kv2 channels in chemically cross-linked mouse brain homogenate.
Kv2.1 recruits coexpressed VAP proteins to Kv2.1-containing ER-PM junctions
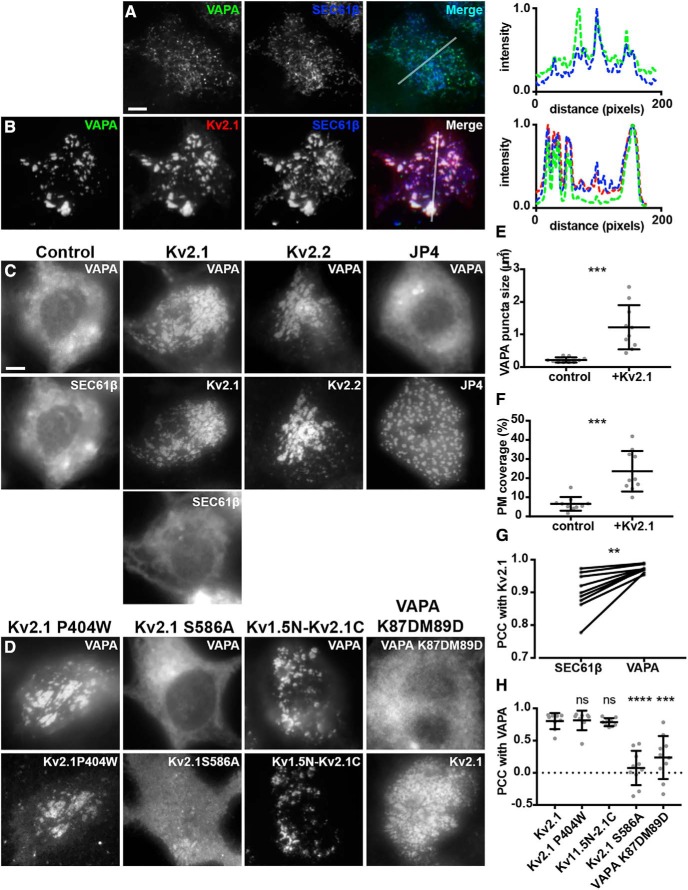
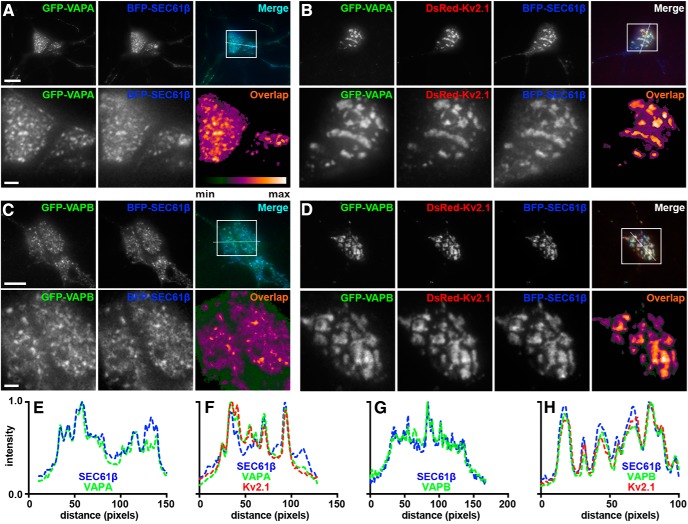
To begin to define the impact of the association of Kv2.1 with VAP proteins identified in the proteomic analyses described above, we coexpressed these proteins in HEK293T cells, which although of the neuronal lineage (Shaw et al., 2002), lack endogenous Kv2 channel expression (Yu and Kerchner, 1998), and in which Kv2.1 is clustered at ER:PM junctions (Fox et al., 2015; Bishop et al., 2018). To selectively visualize exogenous VAPs present at ER-PM junctions in live HEK293T cells, we made use of TIRF microscopy, in conjunction with expression of the general ER marker BFP-SEC61β as described previously (Fox et al., 2015; Besprozvannaya et al., 2018; Bishop et al., 2018). In HEK293T cells expressing VAPA in the absence of Kv2.1, VAPA was apparent in TIRF imaging as a ramified system of near-PM puncta and tubules that colocalized with the SEC61β ER marker (Fig. 2A) and that represent ER-PM junctions (Fox et al., 2015; Besprozvannaya et al., 2018; Bishop et al., 2018). The subcellular localization of VAPA was fundamentally altered by coexpression with Kv2.1. In HEK293T cells in which Kv2.1 was coexpressed, VAPA colocalized with Kv2.1 at the enlarged ER-PM junctions mediated by Kv2.1 overexpression (Fig. 2B). To quantify these observations, we analyzed the “TIRF footprint” of VAPA and found that Kv2.1 expression yielded a significant increase in both VAPA puncta size (Fig. 2E) and the surface area of the PM that VAPA occupied (Fig. 2F) compared with cells lacking Kv2.1 expression. As we showed previously (Bishop et al., 2018), the localization of SEC61β was also altered in cells expressing Kv2.1, consistent with an increase in ER-PM junction size mediated by Kv2.1 expression (Fig. 2). Similar results were obtained from cells coexpressing GFP-tagged VAPB instead of VAPA (data not shown). Despite the overall extensive overlap between Kv2.1, VAPA, and SEC61β in TIRF, the distinct relationship between Kv2.1 and VAPA was apparent in that they exhibited significantly higher colocalization with one another, as measured with PCCs, than did Kv2.1 and SEC61β in the same cells (Fig. 2G).
Figure 2.
Kv2 channels colocalize with and redistribute VAPA in coexpressing HEK293T cells. A, Representative images of a single live HEK293T cell coexpressing GFP-VAPA (green) and BFP-SEC61β (blue) and imaged with TIRF. Line scan analysis of selection indicated in merged image shown to the right. Scale bar, 5 μm for A and B. Line scan analysis of selection indicated in merged image shown to the right. B, Representative images of a single live HEK293T cell coexpressing GFP-VAPA (green), DsRed-Kv2.1 (red), and BFP-SEC61β (blue) and imaged with TIRF. Line scan analysis of selection indicated in merged image shown to the right. C, Representative wide-field images of VAPA and SEC61β expression in live HEK293T cells coexpressing BFP-SEC61β and GFP-VAPA alone (control) or coexpressing Kv2.1, Kv2.2, or JP4 as indicated. Note the redistribution of GFP-VAPA, but not BFP-SEC61β, in cell coexpressing Kv2.1 relative to control cell. Note the redistribution of GFP-VAPA in cells coexpressing Kv2.1 or Kv2.2, but not JP4, relative to control cell. Scale bar is 5 μm and holds for panels C and D. D, Representative wide-field images of VAPA expression in live HEK293T cells coexpressing GFP-VAPA and mutant Kv2.1 isoforms as indicated or GFP-VAPA K87D/M89D and WT Kv2.1. Note the redistribution of GFP-VAPA in cells coexpressing Kv2.1 P404W and Kv1.5N-Kv2.1C but not Kv2.1 S586A. Note that coexpression of Kv2.1 with VAPA K87D/M89D does not lead to redistribution of VAPA K87D/M89D to the same extent as WT VAPA. E, Summary graph of VAPA puncta size measured from cells coexpressing GFP-VAPA and DsRed-Kv2.1 (+Kv2.1) or expressing GFP-VAPA alone (***p = 0.0001996, n = 10 cells, two-tailed unpaired t test). F, Summary graph of PM coverage measured from same cells as in E (***p = 0.0001392, n = 10 cells, two-tailed unpaired t test;). G, PCC measurements between Kv2.1 and VAPA or Kv2.1 and BFP-SEC61β from the same cells (**p = 0.003276, n = 9 cells, two-tailed paired t test). H, Summary graph of PCC measurements between VAPA and various Kv2 isoforms as indicated (ns, p = 0.8827, n = 8 cells; ns, p = 0.7205, n = 9 cells; ****p = 0.0000009074, n = 10 cells; ***p = 0.0001768, n = 10 cells; two-tailed unpaired t test vs Kv2.1:VAPA) Note the significant reduction in colocalization in Kv2.1 S586A and VAPA K87D/M89D.
We also imaged live HEK293T cells in conventional wide-field mode to allow for visualization of the populations of VAPA and SEC61β throughout the cell. In these wide-field images, we observed that expression of Kv2.1 was sufficient to induce a redistribution of the whole-cell VAPA signal from bulk ER to prominent clusters that colocalized with Kv2.1 (Fig. 2C). This fundamental change in the subcellular localization of VAPA induced by Kv2.1 coexpression was not seen for the ER protein SEC61β in the same cells (Fig. 2C) or for DsRed2-ER5, another marker of bulk ER (data not shown), showing that this is not a general change in ER morphology but a specific recruitment of VAPA to Kv2.1 clusters. Coexpression with Kv2.2 also led to this prominent change in VAPA localization (Fig. 2C). Conversely, the generation of enhanced ER-PM junctions by expression of JP4 (Fig. 2C) or Extended-Synaptotagmin 3 (data not shown) did not result in recruitment of VAPA to these sites, showing that the formation of ER-PM junctions in itself is not sufficient to recruit VAPA, but rather is specific to Kv2 channels, and demonstrating that this phenomenon is a unique and conserved feature of the Kv2 family.
To begin to define the molecular requirements of Kv2-mediated redistribution of VAPs, we expressed mutant Kv2 isoforms with VAPA in the same assay. Interestingly, a distinct point mutant (P404W), which leads to expression of Kv2.1 channels that no longer conduct K+ (Lee et al., 2003) but have an intact PRC motif, remains capable of recruiting VAPA into large puncta that colocalized with this mutant Kv2.1 isoform (Fig. 2D), indicating that ion channel conduction is not necessary for the ability of Kv2.1 to redistribute VAPA. Importantly, a point mutation within the PRC motif (S586A) that eliminates clustering of Kv2.1 (Lim et al., 2000) also eliminates the ability of Kv2.1 to mediate this profound redistribution of VAPA (Fig. 2D), indicating that an intact PRC domain within the Kv2.1 C terminus is necessary for the ability of Kv2.1 to redistribute VAPA. To determine whether the Kv2.1-C terminus is also sufficient to recruit VAPA to these near-PM clusters, we examined a chimeric protein in which the entire Kv2.1 C terminus (including the PRC motif) is appended onto the unrelated Kv1.5 channel. Unlike Kv1.5, this Kv1.5N-2.1C chimera exhibits Kv2.1-like clustering in CHNs (Lim et al., 2000) and HEK293T cells (Mohapatra and Trimmer, 2006). We found that this chimera is able to recruit exogenous VAPA from bulk ER to the sites of Kv1.5N-2.1C clustering (Fig. 2D), demonstrating that the Kv2.1 C terminus is sufficient to induce Kv2.1-like remodeling of VAPA. Finally, because VAPs associate with numerous proteins that form the so-called “VAPome,” many but not all of which contain a “FFAT” motif that binds to an FFAT-binding domain present on the VAP cytoplasmic N terminus (Murphy and Levine, 2016), we examined a double point mutant of VAPA (K87D/M89D) that is deficient in binding FFAT motifs (Kaiser et al., 2005). We found that this mutant was significantly reduced in its recruitment to Kv2.1-containing ER-PM junctions (Fig. 2D). To quantify these observations, we measured PCC values between VAPA and Kv1.5N-2.1C or Kv2.1 P404W and found no significant difference between colocalization relative to WT Kv2.1 and VAPA (Fig. 2H). We did find significantly lower colocalization between Kv2.1 S586A and WT VAPA and WT Kv2.1 and VAPA K87D/M89D relative to that seen for the WT isoforms of Kv2.1 and VAPA (Fig. 2H).
These data together demonstrate that expression of PM Kv2 ion channels results in a specific and profound redistribution of VAPA to Kv2-mediated EPJs in HEK293T cells and that the PRC domain contained within the Kv2.1 C terminus is both necessary and sufficient for this activity. We also found that an intact FFAT-binding domain contained on the VAPA N terminus is also necessary for this activity, but K+ conduction by Kv2.1 is not.
Kv2.1 recruits coexpressed VAP proteins to Kv2.1-containing ER-PM junctions in cultured hippocampal neurons
Similar to our imaging experiments in HEK293T cells, we performed parallel experiments in CHNs. In CHNs in which GFP-tagged VAPA or VAPB was exogenously expressed with BFP-SEC61β, TIRF imaging revealed that the VAPs were present as a ramified system of near-PM reticules and tubules on the soma and proximal processes that colocalized with SEC61β (VAPA: Fig. 3A,E; VAPB: Fig. 3C,G). Upon expression of exogenous DsRed2-Kv2.1, the exogenous VAPs underwent a dramatic redistribution such that they were now present in large, near-PM clusters at which they were colocalized with Kv2.1 at the enhanced ER-PM junctions mediated by Kv2.1 overexpression (VAPA: Fig. 3B,F; VAPB: Fig. 3D,H), demonstrating that, similar to our results in HEK293T cells, exogenously expressed VAPA and VAPB are recruited to and colocalize with Kv2.1 at ER-PM junctions in CHNs.
Figure 3.
VAPA and VAPB colocalize with Kv2.1 at ER-PM junctions in overexpressing cultured hippocampal neurons. A, Representative images of a live CHN coexpressing GFP-VAPA (green) and BFP-SEC61β (blue) and imaged with TIRF. Scale bar indicates 10 μm and also holds for low-magnification panels in B. Magnified view of selection indicated in merged image shown in lower panels. Scale bar is 2.5 μm and also holds for magnified panels of B. Pixel overlap analysis shown in bottom right panel. B, Representative images of a live CHN coexpressing GFP-VAPA (green), DsRed-Kv2.1 (red), and BFP-SEC61β (blue) and imaged with TIRF. Magnified view of selection indicated in merged image shown in lower panels. Pixel overlap analysis shown in bottom right panel. C, Representative images of a live CHN coexpressing GFP-VAPB (green) and BFP-SEC61β (blue) and imaged with TIRF. Scale bar is 10 μm and also holds for low-magnification panels of D. Magnified view of selection indicated in merged image shown in lower panels. Scale bar is 2.5 μm and also holds for magnified panels of D. Pixel overlap analysis shown in bottom right panel. D, Representative images of a live CHN coexpressing GFP-VAPB (green), DsRed-Kv2.1 (red), and BFP-SEC61β (blue) and imaged with TIRF. Magnified view of selection indicated in merged image shown in lower panels. Pixel overlap analysis is shown in bottom right panel. E, Line scan analysis of selection indicated in merged image of A. F, Line scan analysis of selection indicated in merged image of B. G, Line scan analysis of selection indicated in merged image of C. H, Line scan analysis of selection indicated in merged image of D.
Dispersal of Kv2.1 clusters results in a coordinated dispersal of associated VAPA from ER-PM junctions
Kv2.1 clustering (Misonou et al., 2004, 2005b, 2006; Mohapatra and Trimmer, 2006; Cerda and Trimmer, 2011; Bishop et al., 2015) and localization to ER-PM junctions (Fox et al., 2015) in both HEK293T cells and CHNs is disrupted in response to stimuli that elevate intracellular calcium leading to calcineurin-dependent dephosphorylation of Kv2.1. To determine whether such treatments would coordinately affect the Kv2.1-mediated redistribution of VAPA, we treated HEK293T cells coexpressing DsRed-Kv2.1, GFP-VAPA, and BFP-SEC61β with the calcium ionophore Inm. Before Inm treatment, TIRF imaging revealed that near-PM VAPA was present in large clusters in which it colocalized with Kv2.1 (Fig. 4A). Upon Inm addition, we observed a simultaneous and coordinate dispersal of VAPA and Kv2.1 from ER-PM junctions (Fig. 4B) and a significant reduction in ER-PM junction size as revealed by the near-PM SEC61β signal (Fig. 4C). This was accompanied by a significant and coordinate reduction in the sizes of the near-PM VAPA and PM Kv2.1 (Fig. 4C) puncta. The responses of the near-PM SEC61β and VAPA puncta to Inm treatment were distinct such that there was a significant difference in the sizes of the near-PM SEC61β and VAPA puncta after (p = 0.0044), but not before (p = 0.1281), Inm treatment (Fig. 4C).
Figure 4.
Dispersal of Kv2.1 from ER-PM junctions via elevation of intracellular Ca2+ results in a coordinated reduction in ER-PM junction and VAPA puncta size. A, Representative images of a single live HEK293T cell coexpressing GFP-VAPA (green), DsRed-Kv2.1 (red), and BFP-SEC61β (blue), imaged with TIRF, before 2 μm Inm treatment (Rest). Scale bar is 5 μm and holds for all panels. B, Same cell as in A after 30 min incubation in 2 μm Inm. C, Summary graph of the impact of Inm treatment on the sizes of ER-PM junctions (****p = 4 × 10−15), VAPA puncta size (****p = 5.254 × 10−12), and Kv2.1 cluster size (****p = 2 × 10−14). All comparisons were with two-tailed unpaired t tests of values before versus after Inm treatment from n = 3 cells each. The changes in the sizes of ER-PM junctions and VAP clusters are significantly different (**p = 0.0044). D, Normalized peak fluorescence intensity measurements of GFP-VAPA (green), DsRed-Kv2.1 (red), and BFP-SEC61β (blue) over the course of Inm treatment. Note the significant difference between VAPA and SEC61β intensity following Inm treatment for all time points following 900 s (0.005969 ≤ *p ≤ 0.0333, n = 3 cells, two-tailed paired t test).
We next analyzed the time course of these responses by measuring fluorescence intensity normalized to initial fluorescence intensity (F/F0) in the near-PM TIRF imaging field, which for Kv2.1 decreases upon lateral dispersion of the high-density PM clusters into a more uniform distribution (Misonou et al., 2004, 2006), and for the ER proteins upon retraction from their near-PM localization in the TIRF imaging field resulting from their localization at ER-PM junctions (Fox et al., 2015). After a transient increase in the F/F0 values of all three proteins just after Inm addition, the basis of which remains unknown, we found that the responses of VAPA and Kv2.1 to Inm in terms of a gradual decrease in F/F0 were overlapping and distinct from the more prominent decrease in in F/F0 for SEC61β (Fig. 4D). This coordinate regulation of VAPA and Kv2.1 provides further support for a specific association of these ER and PM proteins which colocalize at ER-PM junctions.
CRISPR-mediated KO of VAPA reduces Kv2.1 clustering
We next investigated the impact of reducing VAP expression on Kv2.1 clustering. VAPA KO mice exhibit embryonic lethality (McCune et al., 2017), preventing studies in brain. We instead took advantage of a recently generated mammalian cell line (mouse macrophage RAW 264.7 cells) in which VAPA expression was eliminated via CRISPR-mediated gene editing (McCune et al., 2017). We first expressed a recombinant isoform of Kv2.1 tagged with cytoplasmic mCherry and an extracellular pHluorin as a reporter of cell surface Kv2.1 termed SEP-Kv2.1 (Jensen et al., 2017) in WT RAW 264.7 cells. We found that SEP-Kv2.1 was efficiently expressed on the cell surface, where it was present in prominent clusters (Fig. 5A). WT RAW 264.7 cells expressing GFP-Kv2.1 exhibited robust GFP-Kv2.1 clusters including at the basal membrane visible in TIRF imaging (Fig. 5B). These clusters were present at ER-PM junctions as visualized by their colocalization with the near-PM SEC61β signals visible in TIRF (Fig. 5B,C). In the CRISPR-edited VAPA KO RAW 264.7 cells, clustering of GFP-Kv2.1 was reduced (Fig. 5B), resulting in reduced colocalization with SEC61β (Fig. 5D) and in a significant reduction in the CV values of GFP-Kv2.1 signal intensity (Fig. 5F). In contrast, there was no apparent change in the subcellular distribution of GFP-Kv1.4, a related Kv channel that is not localized to ER-PM junctions in neurons (Trimmer, 2015), in WT versus VAPA KO RAW 264.7 cells (Fig. 5E) and no significant change in the extent of GFP-Kv1.4 clustering, as reported by CV (Fig. 5G). These data demonstrate that VAPA KO in mammalian cells results in a specific and significant reduction in Kv2.1 clustering and consistent with the Kv2:VAP interaction underlying the co-clustering of these proteins at ER-PM junctions.
Kv2.1 expression recruits endogenous VAPs to Kv2.1-containing ER-PM junctions
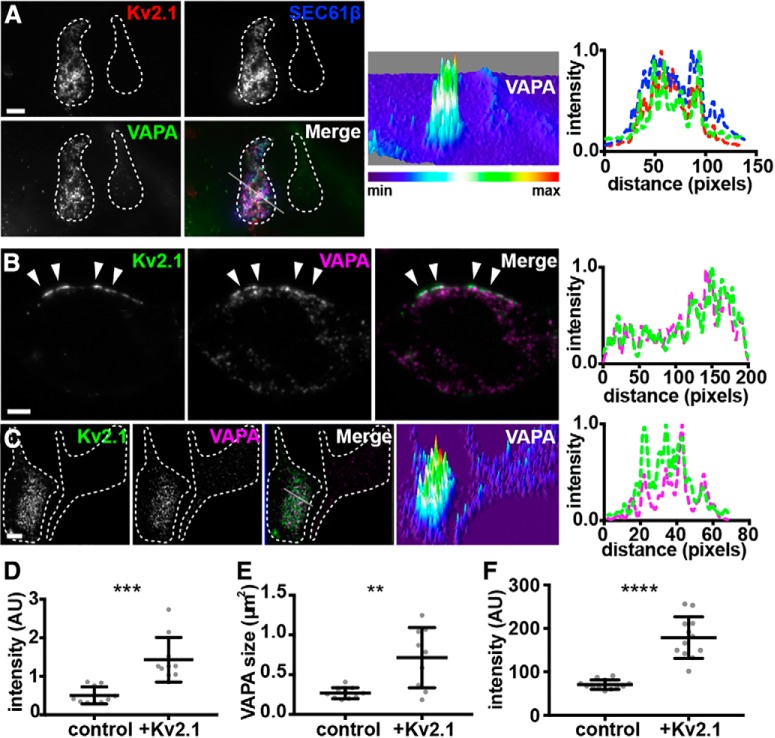
Given the effects of Kv2.1 expression on recruiting coexpressed VAP proteins to Kv2.1-containing ER-PM junctions, we next addressed the impact of Kv2.1 on endogenous VAPs that are expressed (Geiger et al., 2012; Huttlin et al., 2015) in HEK293T cells. We expressed GFP-Kv2.1 and immunolabeled endogenous VAPA using a recently developed anti-VAPA mouse monoclonal antibody (N479/12). We found that Kv2.1 expression led to a significant increase in signal intensity of the near-PM population of endogenous VAPA that was visible in TIRF imaging (Fig. 6A,D). The size of near-PM puncta of endogenous VAPA was also significantly increased upon Kv2.1 expression (Fig. 6E). In optical sections (Apotome; Zeiss) taken through the center of these cells, Kv2.1 clusters could be found juxtaposed to projections of endogenous VAPA that terminated at the cell cortex underneath Kv2.1 clusters (Fig. 6B). To complement the TIRF imaging results in Figure 6, A, D–E, we also used optical sectioning to image the basal PM, the site of robust Kv2.1 clustering in cultured cells (Antonucci et al., 2001; Mohapatra and Trimmer, 2006). We found that Kv2.1 expression was sufficient to recruit endogenous VAPA (labeled with mAb N479/22) to this subcellular location (Fig. 6C). This was seen in the larger structures that matched the morphology of Kv2.1 clusters (Fig. 6C). We additionally found a significant increase in endogenous VAPA signal intensity in the basal PM of cells expressing GFP-Kv2.1 relative to control (nontransfected) cells (Fig. 6F). These data together demonstrate that expression of Kv2.1 is sufficient to recruit endogenous VAPs to Kv2.1-containing ER-PM junctions.
Figure 6.
Kv2.1 colocalizes with and recruits endogenous VAPs to Kv2.1-mediated ER-PM junctions in HEK293T cells. A, TIRF image of a pair of fixed HEK293T cells, the left of which is coexpressing DsRed-Kv2.1 (red) and BFP-SEC61β (blue), fixed, and immunolabeled for endogenous VAPA (green). Scale bar, 5 μm. Surface plot of VAPA intensity shown in right panel. Note the difference in VAPA intensity in left versus right cell. Line scan analysis of selection indicated in merged image of F is shown to the far right. B, Single optical section (Apotome; Zeiss) taken through the center of a fixed HEK293T cell expressing GFP-Kv2.1 (green) and immunolabeled for endogenous VAPA (magenta). Note the presence of PM Kv2.1 clusters overlaying projections of VAPA toward the cell cortex. Scale bar, 5 μm. Line scan analysis of selection indicated by arrows in merged image of B is shown to the far right. C, Single optical section (Apotome) of a pair of fixed HEK293T cells, the left of which is expressing GFP-Kv2.1 (green) and immunolabeled for endogenous VAPA (magenta). Note the difference in VAPA intensity in left versus right cell. Scale bar, 5 μm. Surface plot of VAPA intensity shown in right panel. Line scan analysis of the merged image in C is shown to the far right. D, Summary graph of VAPA intensity (TIRF) measurements from Kv2.1-expressing versus nontransfected cells (**p = 0.001659, n = 10 cells, two-tailed unpaired t test). E, Summary graph of VAPA puncta size (TIRF) measurements from Kv2.1-expressing versus nontransfected cells (**p = 0.001894, n = 10 cells, two-tailed unpaired t test). F, Summary graph of VAPA intensity (Apotome) measurements from the basal PM of Kv2.1-expressing versus nontransfected cells (**p = 0.001659, n = 10 cells, two-tailed unpaired t test). ***p = 0.000166, ****p = 1.312 × 10−8.
Endogenous Kv2 channels and VAPs colocalize in mammalian brain neurons
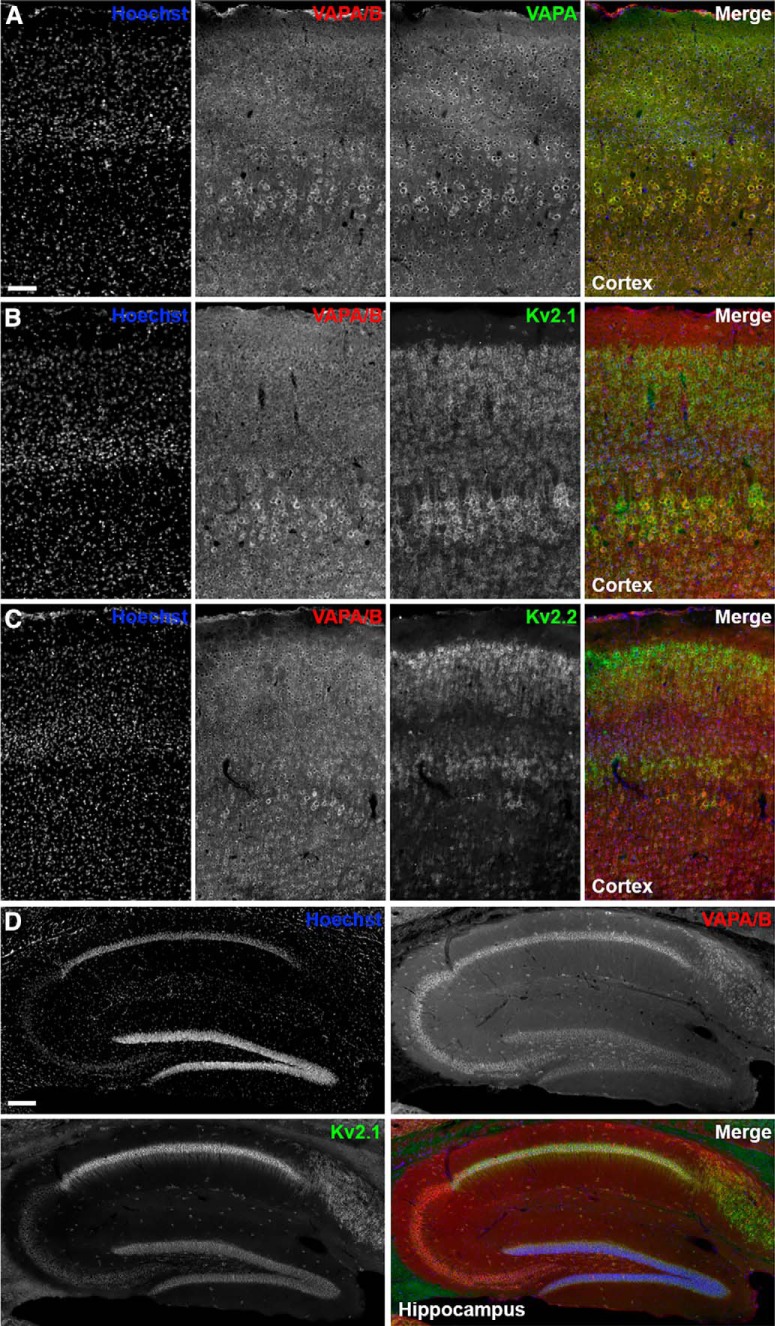
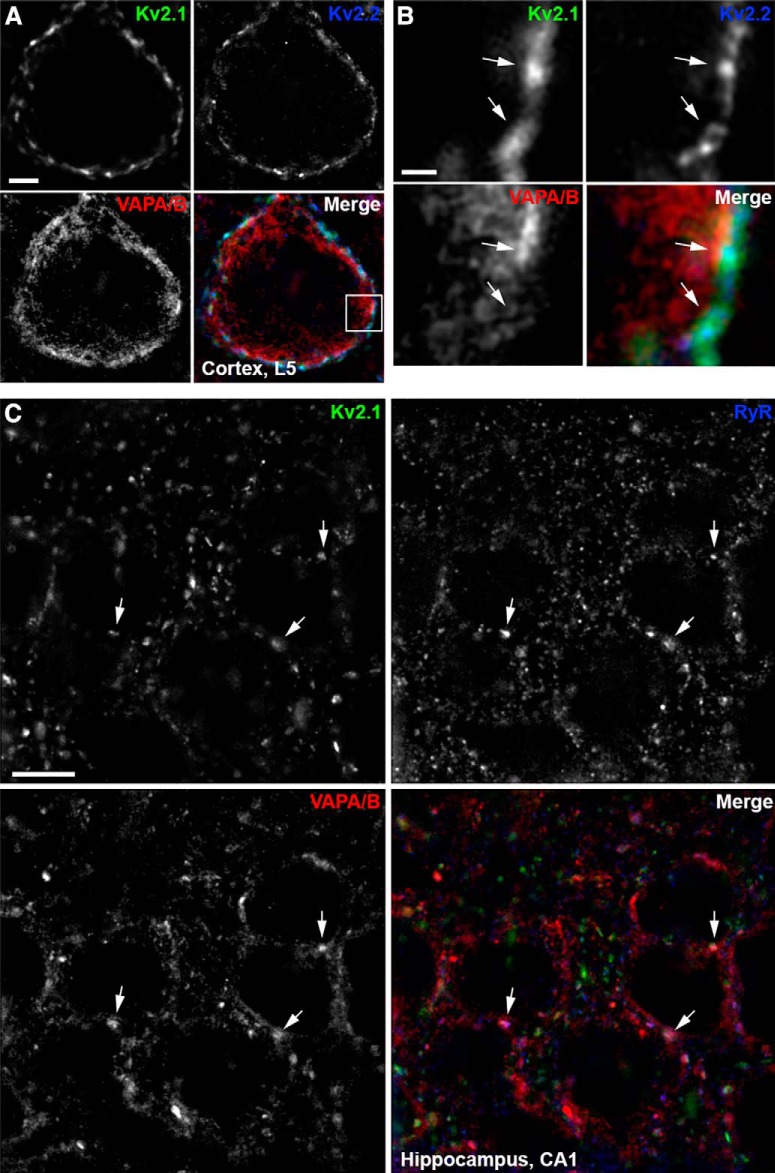
To extend our results that VAPA and VAPB copurify with Kv2.1 and Kv2.2 from mouse brain, and that Kv2.1 or Kv2.2 expression in CHNs and HEK293T cells affects VAP localization, we next examined the localization of endogenous Kv2 channels and VAPs in intact/native neurons. In low-magnification imaging of brain sections from adult mice, we found that VAPA and VAPA/B immunolabeling was similar in neocortex (Fig. 7A), suggesting that VAPA immunoreactivity dominates in this brain region. As expected for abundant ER proteins with numerous and diverse functions (Murphy and Levine, 2016), the anti-VAPA and anti-VAPA/B immunoreactivity in neocortex and hippocampus was present as a dense somatic signal in numerous cell types (Fig. 7). Immunolabeling for VAPs exhibited extensive overlap with that of Kv2.1 and Kv2.2 in both neocortex (Fig. 7B,C) and hippocampus (Fig. 7D). In high-magnification/superresolution optical sections taken through the center of layer 5 cortical neurons, Kv2.1 and Kv2.2 clusters could be found juxtaposed to projections of VAPA/B that terminated at the cell cortex underneath Kv2.1 clusters (Fig. 8A,B).
Figure 7.
Cellular expression of Kv2 channels and VAPs in mouse brain. A, Representative images of mouse cortex stained with Hoechst (blue) and immunolabeled for VAPA/B (red) and VAPA (green). Scale bar, 75 μm. B, Representative images of mouse cortex stained with Hoechst (blue) and immunolabeled for VAPA/B (red) and Kv2.1 (green). C, Representative images of mouse cortex stained with Hoechst (blue) and immunolabeled for VAPA/B (red) and Kv2.2 (green). D, Representative images of mouse hippocampus stained with Hoechst (blue) and immunolabeled for VAPA/B (red) and Kv2.1 (green). Scale bar, 150 μm.
Figure 8.
Endogenous Kv2 channels, VAPs, and RyRs colocalize in mouse brain neurons. A, Single optical section taken through the center of the soma of a single mouse brain cortical (layer 5) neuron immunolabeled for Kv2.1 (green), Kv2.2 (blue), and VAPA/B (red), and imaged with superresolution (N-SIM; Nikon) microscopy. Scale bar, 5 μm. B, Magnification of selection indicated in merged image of A. Arrows point to associated Kv2.1, Kv2.2, and VAPA/B immunolabeling. Scale bar, 0.625 μm. C, Single optical section taken through the center of multiple neurons within the CA1 region of mouse brain hippocampus immunolabeled for Kv2.1 (green), RyR (blue), and VAPA/B (red) and imaged with superresolution (N-SIM; Nikon) microscopy. Arrows point to associated Kv2.1, RyR, and VAPA/B immunolabeling. Scale bar, 5 μm.
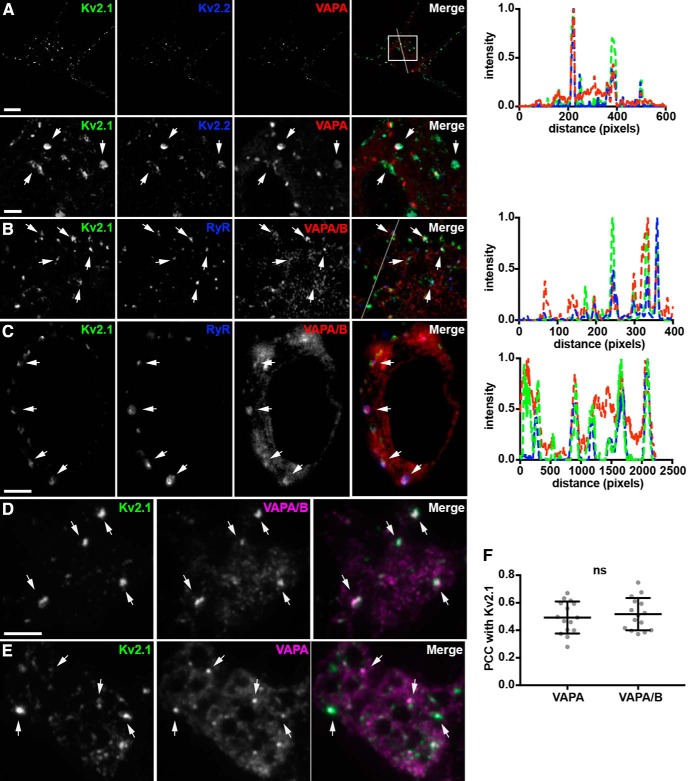
To gain further insights into the spatial relationship between VAPs and Kv2 channels endogenously expressed in neurons, we used three different imaging modalities (Airyscan: Zeiss; N-SIM, TIRF: Nikon) to image immunolabeled CHNs. Airyscan superresolution imaging (Kolossov et al., 2018) of immunolabeled CHNs revealed found prominent co-clusters of VAPs with both Kv2.1 and Kv2.2 on the soma and proximal processes in CHNs (Fig. 9A). We also observed VAP puncta (red signals in the merged panels of Figs. 9A) that did not specifically colocalize with clusters of Kv2 channels.
Figure 9.
Endogenous VAPs colocalize with Kv2 channels and RyRs at ER-PM junctions in cultured hippocampal neurons. A, Representative image of a single fixed CHN immunolabeled for endogenous Kv2.1 (green), Kv2.2 (blue), and VAPA (red), and imaged with superresolution (Airyscan; Zeiss) microscopy. Scale bar, 5 μm. Magnified view of selection indicated in merged image shown in lower panels with arrows pointing at associated puncta of Kv2 and VAPA immunolabeling. Scale bar, 2.5 μm (also applies to B). Line scan analysis of selection indicated in merged image of A shown to right. B, Representative image of the soma of a single fixed CHN immunolabeled for endogenous Kv2.1 (green), RyR (blue), and VAPA/B (red) and imaged with superresolution (Airyscan) microscopy. Arrows point to associated Kv2.1, RyR, and VAPA/B immunolabeling. Line scan analysis of selection indicated in merged image of B shown to right. C, Single optical section taken through the center of the soma of a fixed CHN immunolabeled for endogenous Kv2.1 (green), RyR (blue), and VAPA/B (red) and imaged with superresolution (N-SIM; Nikon) microscopy. Arrows point to associated puncta of Kv2.1, RyR, and VAPA/B immunolabeling. Scale bar, 5 μm. Line scan analysis of selection indicated with arrows in merged image of C shown to right. D, Representative images of the soma of a single fixed CHN immunolabeled for endogenous Kv2.1 (green) and VAPA/B (magenta) and imaged with TIRF. Arrows point to puncta of associated Kv2.1 and VAPA/B immunolabeling. Scale bar, 10 μm (also holds for E). E, Representative images of the soma of a single fixed CHN immunolabeled for endogenous Kv2.1 (green) and VAPA (magenta) and imaged with TIRF. Arrows point to puncta of associated Kv2.1 and VAPA/B immunolabeling. F, Summary graph of PCC measurements between Kv2.1 and VAPA and Kv2.1 and VAPA/B (ns, p = 0.5719, n = 14 neurons, two-tailed unpaired t test).
We also immunolabeled endogenous VAPs in CHNs for ryanodine receptor (RyR) intracellular calcium-release channels, which are prominent components of ER-PM junctions in many neuronal and non-neuronal cell types (Carrasco and Meyer, 2011), including Kv2.1-containing ER-PM junctions in certain classes of brain neurons (Antonucci et al., 2001; King et al., 2014; Mandikian et al., 2014). Airyscan superresolution imaging revealed overlap of immunolabeling for VAPs and RyRs at a subset of Kv2.1 clusters (Fig. 9B). We again observed VAP puncta (red signals in the merged panel of Fig. 9B, also apparent in the corresponding line scan) that did not colocalize with clusters of Kv2 channels. Optical sections taken through the center of a CHN using N-SIM superresolution imaging showed co-clusters of PM Kv2.1 overlaying RyR-containing VAP projections (Fig. 9C). Similar spatial distributions of Kv2.1 and RyR clusters and VAPA/B puncta were observed in superresolution imaging of hippocampal CA1 neurons in mouse brain sections (Fig. 8C). These data demonstrate that endogenous Kv2 clusters, including those that contain RyRs, colocalize with endogenous VAPs, in CHNs.
Finally, we used TIRF imaging to restrict our analysis to near-PM immunolabeling of endogenous neuronal VAPs. We found large puncta or reticular projections of VAPs that colocalized with the large clusters of endogenous Kv2.1 present in these CHNs (Fig. 9D,E), although VAP puncta that did not colocalize with clusters of Kv2 channels were also seen (magenta signals in the merged panel of Fig. 9D,E). There were no significant differences in the colocalization of the immunolabeling obtained with VAPA-specific versus VAPA/B-specific antibodies with Kv2.1 (Fig. 9F). These experiments together demonstrate that, in mammalian brain neurons, endogenous VAPs are localized to Kv2 containing ER-PM junctions that can also contain RyRs.
Sites of VAP-Kv2 colocalization represent specialized domains within the cortical actin cytoskeleton
In addition to its prominent localization on the soma and proximal dendrites, Kv2.1 is also present in robust clusters on the AIS (Sarmiere et al., 2008; King et al., 2014). These Kv2.1 clusters on the AIS are present at sites deficient in the actin-associated scaffolding protein ankyrinG or ankG (King et al., 2014) and that represent locations at which the ER present in the AIS, termed the cisternal organelle, comes into close apposition to the PM (Sánchez-Ponce et al., 2011; King et al., 2014; Schlüter et al., 2017). Kv2.1 clusters are also expressed in “voids” in the cortical actin cytoskeleton when exogenously expressed in Madin–Darby Canine Kidney (MDCK) cells (Scannevin et al., 1996) and in HEK293 cells (Tamkun et al., 2007). Both Airyscan (Fig. 10A,B) and N-SIM (Fig. 10C) superresolution imaging revealed that endogenous VAP puncta on the AIS were often present at sites of Kv2.1 clustering and that these sites were deficient in ankG immunolabeling, as evident in the corresponding line scans (Fig. 10G,H). Similar results were obtained from cortical neurons in brain sections (Fig. 10F,J).
We also investigated the relationship between the actin cytoskeleton and the clusters of endogenous VAPs that were generated upon expression of Kv2.1 in HEK293T cells. We found by SIM imaging that the sites to which VAP was recruited by Kv2.1 expression were, like Kv2.1, located in zones depleted for F-actin (Fig. 10D,E), as is also evident in the corresponding line scan analysis of these cells (Fig. 10I,J). These data further support the concordance of the highly restricted spatial distributions of Kv2 channels and endogenous VAPs in neurons and heterologous cells.
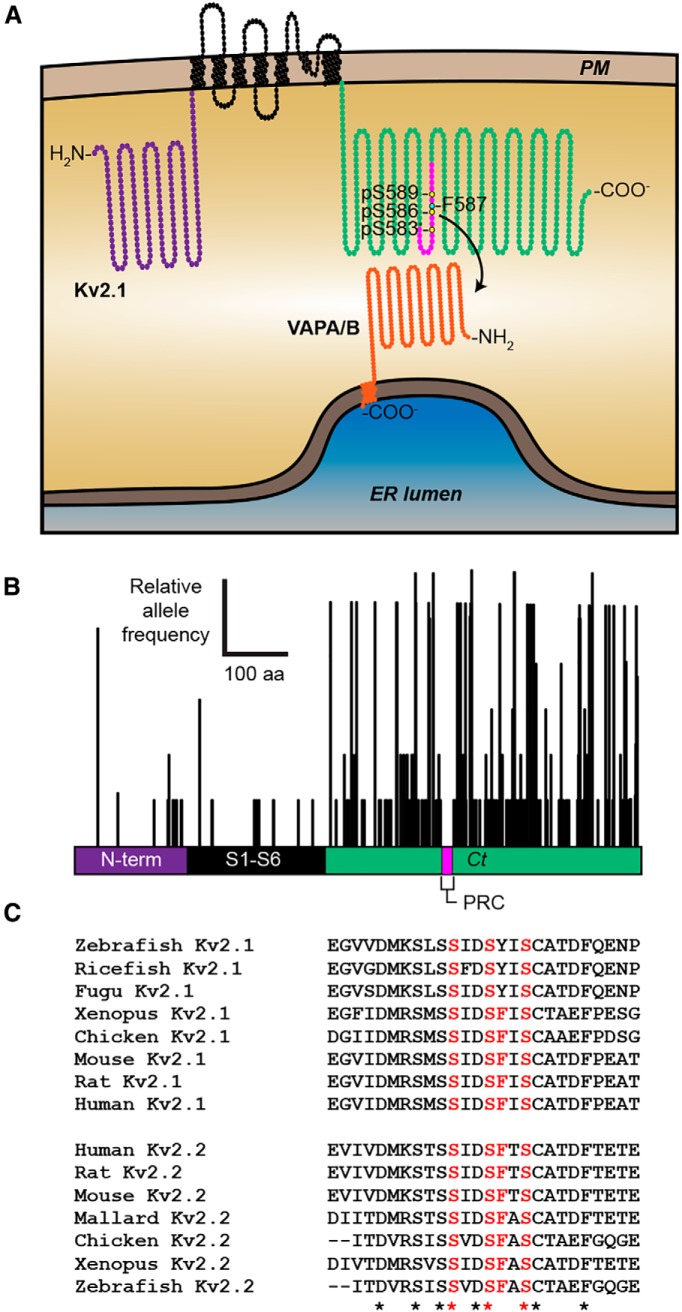
Kv2.1-mediated recruitment of VAPs is dependent on the phosphoserine- and phenylalanine-containing PRC motif of the Kv2.1 C terminus
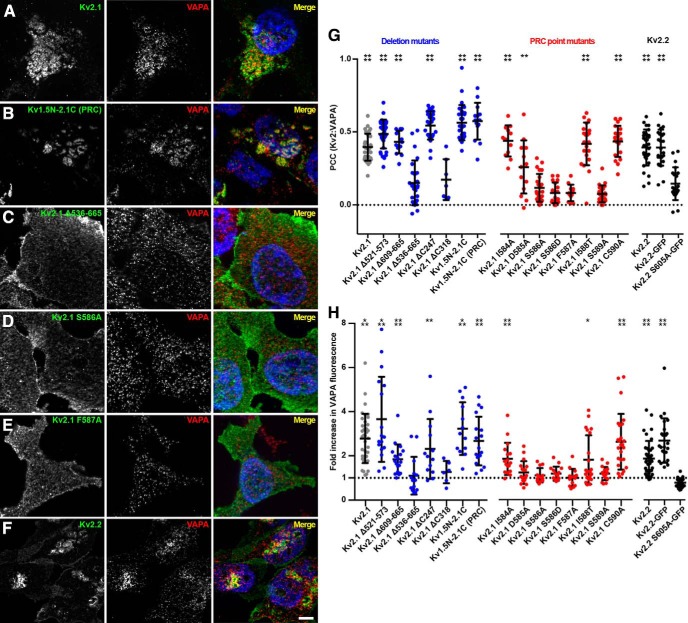
VAPs associate with numerous proteins that form the so-called “VAPome,” many but not all of which contain a FFAT motif that binds to an FFAT-binding domain present on the VAP cytoplasmic N terminus (Murphy and Levine, 2016). Our observation that the VAPA FFAT-binding domain mutant VAPA K87D/M89D (which lacks the FFAT motif-binding capacity of WT VAPA) had significantly reduced recruitment to Kv2.1 clusters relative to WT VAPA (Fig. 2) suggested that the recruitment of VAPs by Kv2.1 is FFAT motif dependent. Although Kv2.1 has no canonical FFAT motifs, the PRC motif that is required for clustering of Kv2 channels and that is sufficient to confer Kv2.1 clustering to other proteins (Lim et al., 2000) may represent a noncanonical FFAT motif of the type present on other VAP binding proteins (Murphy and Levine, 2016). Within the PRC motif are four amino acids whose single mutation eliminates Kv2.1 clustering (Lim et al., 2000), including phenylalanine F587, and serine residues S583, S586, and S589 (highlighted in Fig. 9A). Both S586 (Park et al., 2006; Cobb et al., 2015) and S589 (Ikematsu et al., 2011) have been shown to be phosphorylated in vivo, which would provide an acidic context to these otherwise uncharged amino acids. Because of these considerations, we tested whether the colocalization of VAPs with Kv2 channels required an intact PRC motif.
On the basis of our observation that Kv2.1 expression was sufficient to significantly redistribute endogenous VAPs, which are otherwise distributed throughout bulk ER, to Kv2.1 clusters located in the basal PM in HEK293T cells (Fig. 6C,F), we assayed a battery of internal deletion, truncation, and point mutations in the Kv2.1 C terminus for their ability to colocalize with or redistribute endogenous VAPA in HEK293T cells (representative images are shown in Fig. 11A–E). From these cells, we measured PCC values between Kv2.1 and endogenous VAPA as well as VAPA fluorescence intensity in the basal PM (values summarized in Fig. 11G,H). We found that an intact PRC domain was necessary for VAPA recruitment such that elimination of the PRC (in ΔC318 or Δ536–665) or mutation of residues within the PRC that are critical for Kv2.1 clustering (including phenylalanine F587 and serine residues S586 and S589) resulted in a significant reduction in both VAPA:Kv2.1 colocalization (Fig. 11G) and the VAPA fluorescence intensity (Fig. 11H). Similarly, Kv2.2 expression was also sufficient to recruit VAPA to Kv2.2 clusters and a mutant isoform of Kv2.2 harboring the analogous PRC mutation as Kv2.1 S586A (Kv2.2 S605A) had no impact on VAPA morphology relative to nontransfected cells (Fig. 11F–H). Similar to our findings with exogenous VAPA (Fig. 2D), expression of the Kv1.5N-2.1C chimera was sufficient to redistribute endogenous VAPA (Fig. 11G,H). Interestingly, expression of another Kv1.5–2.1 chimeric channel in which a shorter fragment of the Kv2.1 C terminus (aa 536–600) including the PRC domain is appended onto Kv1.5 [termed Kv1.5N-2.1 (PRC) was also sufficient to redistribute endogenous VAPA (Fig. 11B,G,H). These data together demonstrate that the Kv2 PRC domain is both necessary and sufficient for the unique ability of Kv2.1 (and Kv2.2) to recruit endogenous VAPA to ER-PM junctions in HEK293T cells and that mutation of a single phenylalanine present within an acidic tract constituted by phosphorylated serine residues or of the phosphorylated serine residues themselves is sufficient to abolish this activity.
Figure 11.
The Kv2 channel PRC domain is both necessary and sufficient for VAPA recruitment to Kv2-mediated ER-PM junctions. A–F, Representative optical sections (ApoTome; Zeiss) taken through the basal PM of fixed HEK293T cells expressing the indicated WT, mutant and chimeric Kv2.1 and Kv2.2 isoforms and immunolabeled for the Kv2.1 and Kv2.2 isoforms (green) and endogenous VAPA (red). The merged panels also show Hoechst nuclear staining (blue). Scale bar, 5 μm. G, Summary graph of PCC measurements taken between Kv2.1 and Kv2.2 isoforms and VAPA (****p = 0.0001; **p = 0.0021; n = 7–34 cells; one-way ANOVA followed by Dunnett's test vs Kv2.1 S586A for Kv2.1 isoforms, or versus Kv2.2 S605A-GFP for Kv2.2 isoforms). H, Summary graph of the fold increase in normalized fluorescence intensity of VAPA in the basal PM of Kv2.1 and Kv2.2 isoform-expressing cells relative to nontransfected cells. (Kv2.1 WT: ***p = 0.00017; Kv2.1 Δ521–573: ***p = 0.00046; Kv2.1 Δ609–665: ***p = 4.49 × 10−6; Kv2.1 ΔC247: **p = 0.0053; Kv1.5N-2.1C: ***p = 0.00013; Kv1.5N-2.1C (PRC): ****p = 1.12 × 10−7; Kv2.1 I584A: ****p = 1.74 × 10−6; Kv2.1 I588: *p = 0.024; Kv2.1 C590A: ****p = 1.09 × 10−5; Kv2.2: ****p = 2.88 × 10−5; Kv2.2-GFP, ****p = 1.59 × 10−8; n = 6–38 cells; two-tailed unpaired t test versus nontransfected cells).
Discussion
Previous attempts to use an IP-based proteomics approach to identify Kv2.1-interacting proteins yielded quantities of Kv2.1 sufficient for phosphorylation site identification (Park et al., 2006; Ikematsu et al., 2011; Song et al., 2012), but did not yield candidate associated proteins. Here, we used chemical cross-linking from WT and KO brain samples and harsher detergent extraction conditions to identify VAPA and VAPB as proteins associated with brain Kv2 channels. Chemical cross-linking, although not free of artifacts (Middaugh et al., 1983), has allowed for identification of numerous bona fide interacting partners. That VAPA and VAPB were selectively copurified with Kv2.1 from brain samples prepared from WT but not KO mice, and that our subsequent analyses revealed their extensive association and colocalization supports that they are likely associated with Kv2-containing ER-PM junctions. Our results provide valuable insights into the composition of native ER-PM junctions, especially prominent in brain neurons (Rosenbluth, 1962; Wu et al., 2017) in aspiny regions such as the soma, proximal dendrites and AIS (Spruston and McBain, 2007). That VAPA and VAPB were the most abundant Kv2.1-associated proteins copurified from whole brain suggests that the Kv2:VAP association is abundant and widespread. VAPA and VAPB mRNA are expressed throughout the brain (e.g., Allen Mouse Brain Atlas, http://mouse.brain-map.org/), which was expected because VAP proteins are ubiquitously present in mammalian cells (Murphy and Levine, 2016) and exhibit prominent overlap with Kv2.1 in forebrain neurons that express high levels of Kv2.1 (Trimmer, 1991; Maletic-Savatic et al., 1995; Bishop et al., 2015, 2018; Palacio et al., 2017). There likely exists heterogeneity in ER-PM junctions in different types of brain neurons in the extent to which Kv2.1 and Kv2.2 are associated with VAPA and VAPB. Moreover, the relationship between the overall molecular composition and function of these ER-PM junctions in brain neurons relative to those extensively characterized in non-neuronal cells (Henne et al., 2015; Prakriya and Lewis, 2015; Gallo et al., 2016; Chung et al., 2017; Dickson, 2017) remains to be elucidated.
That VAPs are critical ER resident proteins for clustering of PM Kv2 channels at ER-PM junctions is supported by our findings that Kv2.1 clustering and expression is diminished in CRISPR-edited VAPA KO cells. The parallel CRISPR-edited VAPA KO mice exhibited embryonic lethality (McCune et al., 2017) such that there is no currently available model for studying the impact of VAPA KO on Kv2 clustering in brain neurons in situ. The fact that Kv2.1 clustering is significantly reduced in the VAPA KO cells suggests that the VAPB expressed in these cells (McCune et al., 2017) does not compensate for the loss of VAPA. That mutations in the FFAT-binding domain of VAPA that eliminate binding to other VAP interactors significantly reduced the Kv2-VAP association at ER-PM junctions further supports the necessity of this interaction for Kv2 clustering at these sites.
Our findings support a model whereby the PRC domain that is necessary and sufficient for clustering of Kv2 channels binds to the FFAT-binding domain of VAPs. This model posits that their respective extended cytoplasmic domains (the VAP N terminus and the Kv2 C terminus) would need to span the 7–30 nm cleft between ER and PM to mediate this binding (Fig. 12A). This is plausible given that the low-resolution cryo-EM structure of Kv2.1 suggests a 7- to 9-nm-long cytoplasmic domain (Adair et al., 2008) and the VAPA structure a cytosolic N terminus length of 8–30 nm, depending on the exact nature of unstructured linker domains (Kaiser et al., 2005; Murphy and Levine, 2016). That VAPs form dimers that bind two FFAT motifs in vitro (Kaiser et al., 2005) suggests a plausible mechanism for why certain protein backbones engineered to contain a PRC domain do not exhibit clustered localization at neuronal ER-PM junctions when expressed in CHNs (Lim et al., 2000), whereas oligomeric backbones such as Kv1.5 support both association with VAPs and PRC-mediated clustering at ER-PM junctions (Lim et al., 2000; Mohapatra and Trimmer, 2006). The model that emerges from these studies is that phosphorylation of the otherwise uncharged serine residues (S583, S586, and S589) within the PRC confers the negative charges that define the “acidic tract” adjacent to F587 in this noncanonical FFAT motif to constitute the minimal interaction motif with VAPs, thereby providing the molecular basis for Kv2.1 clustering and organization of ER-PM junctions via a conditional interaction with VAPs (Fig. 12A). Association of other proteins with VAPs is modulated by their phosphorylation state, although in regions outside of the FFAT domain itself (Goto et al., 2012; Kumagai et al., 2014). Interestingly, cytoplasmic domains outside of the PRC are involved in phosphorylation-dependent regulation of the PRC-mediated clustering of Kv2.1 (Mohapatra and Trimmer, 2006; Jensen et al., 2017) such that phosphorylation at distinct sites within and outside of the PRC domain may underlie the dynamic regulation of Kv2-VAP association and, subsequently, ER-PM junctions. Future studies of the structure of the VAP-Kv2 complex, as accomplished for VAPA with other interactors (Kaiser et al., 2005; Furuita et al., 2010), will provide critical details of how changes in Kv2.1 phosphorylation state affect its localization at ER-PM junctions.
Figure 12.
The putative noncanonical FFAT motif in the Kv2 channel PRC domain is highly conserved. A, Illustration depicting the proposed phosphorylation-dependent interaction of the Kv2.1 PRC domain with VAPA/B, enabling recruitment of VAPs to Kv2 clusters and ER-PM junction formation (*necessity of S583 for Kv2.1 clustering has previously been described but was not explicitly tested for a role in recruiting VAPA/B in the present study; see text for details). B, Plot of nonsynonymous genic variation in the coding region of the human KCNB1 gene derived from genome and whole exome sequences from >61,000 individuals in the Broad Institute ExAC database. Note the lack of variation in the core S1-S6 and PRC domains of Kv2.1. C, Protein sequence alignments of the putative FFAT motif in vertebrate Kv2.1 and Kv2.2 channel PRC domains. Residues highlighted in red correspond to the critical S583, S586, F587, and S589 residues of rat Kv2.1, and the corresponding S602, S605, F609, and S611 of rat Kv2.2.
To gain further insights into the Kv2-VAP interaction, we analyzed functional variation in the human KCNB1 (Kv2.1) gene in genomic and whole exome sequences from >61,000 unrelated individuals (http://exac.broadinstitute.org/). In general, there is a lack of variation (i.e., genic intolerance) in regions of exomes encoding critical functional domains of proteins, which are sites of most disease-causing mutations (Gussow et al., 2016). We found that the Kv2.1 N-terminal subunit assembly and core regions are highly invariant in humans (Fig. 12B). Mutations within the S1-S6 “core” domain profoundly affect voltage sensing and ion conductance of Kv channels (Swartz, 2004; Jan and Jan, 2012) and the bulk of disease-associated mutations are also in the S1–S6 region (Torkamani et al., 2014; Saitsu et al., 2015; Thiffault et al., 2015). In contrast, the Kv2.1 C terminus has a high degree of variation (Fig. 12B), with the exception of the highly invariant PRC domain. It is intriguing that three disease-associated de novo nonsense mutations have been identified (de Kovel et al., 2016; Marini et al., 2017) that truncate the cytoplasmic C terminus just upstream of the PRC domain in locations similar to truncation mutants that have been well characterized in vitro (VanDongen et al., 1990; Scannevin et al., 1996; Murakoshi et al., 1997; Park et al., 2006) and that disrupt Kv2.1 clustering but have little effect on its expression and function. The PRC domain is also exceptional within the C terminus of Kv2 channels as being highly conserved across diverse species (Fig. 12C), including the four positions (in red) at which single point mutations disrupt clustering. Together, these considerations support the importance of the Kv2-specific PRC domain, which is both necessary and sufficient for Kv2 channel-like clustering at ER-PM junctions and association with VAPs.
The association of Kv2 channels with VAPs expands the already extensive “VAPome” (Murphy and Levine, 2016) to now include PM proteins. VAPs were originally identified as VAMP-associated proteins, but their prominent biological roles comprise interaction with FFAT-containing proteins important in regulation of ER structure, the unfolded protein response, and lipid transport and metabolism (Murphy and Levine, 2016). Most ER-PM junctions are mediated by ER tethers binding to specific PM phospholipids whose generation and cleavage are dynamically regulated. The only known example of protein–protein interactions mediating ER-PM junctions is the conditional association of ER-localized STIMs with PM Orai Ca2+ channels (Prakriya and Lewis, 2015) triggered by depletion of ER Ca2+ stores (Zhang et al., 2005; Wu et al., 2006). There exists a direct interaction of PM L-type Ca2+ channels and ER-localized RyRs at junctional triads that mediate E-C coupling in skeletal muscle (Franzini-Armstrong and Jorgensen, 1994; Sun et al., 1995), although the ER-PM junction itself is maintained by members of the Junctophilin family of phospholipid-binding ER tethers (Henne et al., 2015; Gallo et al., 2016; Chang et al., 2017; Saheki and De Camilli, 2017). We showed here that a subset of native neuronal Kv2-VAP containing ER-PM junctions may also contain RyRs, consistent with our previous studies (Antonucci et al., 2001; Misonou et al., 2005a; Mandikian et al., 2014) and ER-PM junctions formed upon exogenous expression of Kv2.1 can contain STIM1 and Orai1 (Fox et al., 2015). This suggests that the diversity of neuronal ER-PM junctions seen in electron micrographs (Wu et al., 2017) may have functional diversity, not only in Ca2+ signaling proteins such as RyRs, but also in VAP-interacting proteins that mediate lipid signaling and homeostasis (Murphy and Levine, 2016). The dynamic activity-dependent regulation of Kv2.1 phosphorylation state (Misonou et al., 2004, 2006; Cerda and Trimmer, 2011) suggests plasticity in the organization and function of neuronal ER-PM junctions mediated by Kv2:VAP association. Finally, we note that the clustering of PM ion channels is typically accomplished by their interaction with and immobilization by cytoskeletal-associated anchoring proteins (Lai and Jan, 2006; Trimmer, 2015; Zhang and Rasband, 2016). Kv2 channels are distinct from these anchored ion channels by being mobile within the cluster (O'Connell et al., 2006), consistent with a clustering mechanism based on association of two integral membrane proteins within the fluid mosaic of their respective ER and PM lipid bilayers.
Footnotes
This work was supported by the National Institutes of Health (Grants R01 NS042225 and U01 NS090581 to J.S.T. and Grant T32 GM0007377 to M.K.). We thank Ben Johnson, Eamonn Dickson, Karl Murray, Jodi Nunnari, Jon Sack, and Michael Tamkun for helpful advice and critical discussions; Michelle Salemi and the UC Davis Proteomics Core for expert advice on proteomics; Camelia Dumitras, Taryn Gillis, and Belvin Gong for assistance with monoclonal antibodies; Michael Paddy and the UC Davis MCB Imaging Facility and Colleen Manning at Zeiss for expert advice on imaging; Grace Or for help in preparation of cultured hippocampal neurons; and Skip Virgin for RAW 264.7 cells.
The authors declare no competing financial interests.
References
- Adair B, Nunn R, Lewis S, Dukes I, Philipson L, Yeager M (2008) Single particle image reconstruction of the human recombinant Kv2.1 channel. Biophys J 94:2106–2114. 10.1529/biophysj.107.118562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonucci DE, Lim ST, Vassanelli S, Trimmer JS (2001) Dynamic localization and clustering of dendritic Kv2.1 voltage-dependent potassium channels in developing hippocampal neurons. Neuroscience 108:69–81. 10.1016/S0306-4522(01)00476-6 [DOI] [PubMed] [Google Scholar]
- Baker CA, Elyada YM, Parra A, Bolton MM (2016) Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. Elife 5. pii: e14193. 10.7554/eLife.14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekele-Arcuri Z, Matos MF, Manganas L, Strassle BW, Monaghan MM, Rhodes KJ, Trimmer JS (1996) Generation and characterization of subtype-specific monoclonal antibodies to K+ channel alpha- and beta-subunit polypeptides. Neuropharmacology 35:851–865. 10.1016/0028-3908(96)00128-1 [DOI] [PubMed] [Google Scholar]
- Besprozvannaya M, Dickson E, Li H, Ginburg KS, Bers DM, Auwerx J, Nunnari J (2018) GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife 7. pii: e31019. 10.7554/eLife.31019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop HI, Guan D, Bocksteins E, Parajuli LK, Murray KD, Cobb MM, Misonou H, Zito K, Foehring RC, Trimmer JS (2015) Distinct cell- and layer-specific expression patterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J Neurosci 35:14922–14942. 10.1523/JNEUROSCI.1897-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop HI, Cobb MM, Kirmiz M, Parajuli LK, Mandikian D, Philp AM, Melnik M, Kuja-Panula J, Rauvala H, Shigemoto R, Murray KD, Trimmer JS (2018) Kv2 ion channels determine the expression and localization of the associated AMIGO-1 cell adhesion molecule in adult brain neurons. Front Mol Neurosci 11:1. 10.3389/fnmol.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco S, Meyer T (2011) STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem 80:973–1000. 10.1146/annurev-biochem-061609-165311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O, Trimmer JS (2011) Activity-dependent phosphorylation of neuronal Kv2.1 potassium channels by CDK5. J Biol Chem 286:28738–28748. 10.1074/jbc.M111.251942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Chen YJ, Liou J (2017) ER-plasma membrane junctions: why and how do we study them? Biochim Biophys Acta 1864:1494–1506. 10.1016/j.bbamcr.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JJ, Li M (2005) Biochemical characterization of the native Kv2.1 potassium channel. FEBS J 272:3743–3755. 10.1111/j.1742-4658.2005.04802.x [DOI] [PubMed] [Google Scholar]
- Chung WY, Jha A, Ahuja M, Muallem S (2017) Ca2+ influx at the ER/PM junctions. Cell Calcium 63:29–32. 10.1016/j.ceca.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MM, Austin DC, Sack JT, Trimmer JS (2015) Cell cycle-dependent changes in localization and phosphorylation of the plasma membrane Kv2.1 K+ channel impact endoplasmic reticulum membrane contact sites in COS-1 cells. J Biol Chem 290:29189–29201. 10.1074/jbc.M115.690198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Brilstra EH, van Kempen MJ, Van't Slot R, Nijman IJ, Afawi Z, De Jonghe P, Djémié T, Guerrini R, Hardies K, Helbig I, Hendrickx R, Kanaan M, Kramer U, Lehesjoki AE, Lemke JR, Marini C, Mei D, Møller RS, Pendziwiat M, et al. (2016) Targeted sequencing of 351 candidate genes for epileptic encephalopathy in a large cohort of patients. Mol Genet Genomic Med 4:568–580. 10.1002/mgg3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson EJ. (2017) Endoplasmic reticulum-plasma membrane contacts regulate cellular excitability. Adv Exp Med Biol 997:95–109. 10.1007/978-981-10-4567-7_7 [DOI] [PubMed] [Google Scholar]
- Dickson EJ, Jensen JB, Vivas O, Kruse M, Traynor-Kaplan AE, Hille B (2016) Dynamic formation of ER-PM junctions presents a lipid phosphatase to regulate phosphoinositides. J Cell Biol 213:33–48. 10.1083/jcb.201508106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tao-Cheng JH, Zerfas P, McBain CJ (1998) The K+ channel, Kv2.1, is apposed to astrocytic processes and is associated with inhibitory postsynaptic membranes in hippocampal and cortical principal neurons and inhibitory interneurons. Neuroscience 84:37–48. 10.1016/S0306-4522(97)00519-8 [DOI] [PubMed] [Google Scholar]
- Fox PD, Haberkorn CJ, Akin EJ, Seel PJ, Krapf D, Tamkun MM (2015) Induction of stable ER-plasma-membrane junctions by Kv2.1 potassium channels. J Cell Sci 128:2096–2105. 10.1242/jcs.166009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RA, Grant SG (2017) Supramolecular organization of NMDA receptors and the postsynaptic density. Curr Opin Neurobiol 45:139–147. 10.1016/j.conb.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Jorgensen AO (1994) Structure and development of E-C coupling units in skeletal muscle. Annu Rev Physiol 56:509–534. 10.1146/annurev.ph.56.030194.002453 [DOI] [PubMed] [Google Scholar]
- Frech GC, VanDongen AM, Schuster G, Brown AM, Joho RH (1989) A novel potassium channel with delayed rectifier properties isolated from rat brain by expression cloning. Nature 340:642–645. 10.1038/340642a0 [DOI] [PubMed] [Google Scholar]
- Furuita K, Jee J, Fukada H, Mishima M, Kojima C (2010) Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies. J Biol Chem 285:12961–12970. 10.1074/jbc.M109.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Vannier C, Galli T (2016) Endoplasmic reticulum-plasma membrane associations:structures and functions. Annu Rev Cell Dev Biol 32:279–301. 10.1146/annurev-cellbio-111315-125024 [DOI] [PubMed] [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, Mann M (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics 11:M111.014050. 10.1074/mcp.M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Murray KD, Trimmer JS (2016) Developing high-quality mouse monoclonal antibodies for neuroscience research: approaches, perspectives and opportunities. N Biotechnol 33:551–564. 10.1016/j.nbt.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Liu X, Robinson CA, Ridgway ND (2012) Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Mol Biol Cell 23:3624–3635. 10.1091/mbc.e12-04-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SG. (2012) Synaptopathies: diseases of the synaptome. Curr Opin Neurobiol 22:522–529. 10.1016/j.conb.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Grant SG. (2013) SnapShot: Organizational principles of the postsynaptic proteome. Neuron 80:534–e531. 10.1016/j.neuron.2013.10.014 [DOI] [PubMed] [Google Scholar]
- Gray EG. (1959) Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex. Nature 183:1592–1593. 10.1038/1831592a0 [DOI] [PubMed] [Google Scholar]
- Guan D, Tkatch T, Surmeier DJ, Armstrong WE, Foehring RC (2007) Kv2 subunits underlie slowly inactivating potassium current in rat neocortical pyramidal neurons. J Physiol 581:941–960. 10.1113/jphysiol.2007.128454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB (2016) The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol 17:9. 10.1186/s13059-016-0869-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Liou J, Emr SD (2015) Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol 35:123–130. 10.1016/j.ceb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Hermanstyne TO, Kihira Y, Misono K, Deitchler A, Yanagawa Y, Misonou H (2010) Immunolocalization of the voltage-gated potassium channel Kv2.2 in GABAergic neurons in the basal forebrain of rats and mice. J Comp Neurol 518:4298–4310. 10.1002/cne.22457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanstyne TO, Subedi K, Le WW, Hoffman GE, Meredith AL, Mong JA, Misonou H (2013) Kv2.2: a novel molecular target to study the role of basal forebrain GABAergic neurons in the sleep-wake cycle. Sleep 36:1839–1848. 10.5665/sleep.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönigsperger C, Nigro MJ, Storm JF (2017) Physiological roles of Kv2 channels in entorhinal cortex layer II stellate cells revealed by guangxitoxin-1E. J Physiol 595:739–757. 10.1113/JP273024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 3:661–669. 10.1038/76615 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162:425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Dallas ML, Ross FA, Lewis RW, Rafferty JN, David JA, Suman R, Peers C, Hardie DG, Evans AM (2011) Phosphorylation of the voltage-gated potassium channel Kv2.1 by AMP-activated protein kinase regulates membrane excitability. Proc Natl Acad Sci U S A 108:18132–18137. 10.1073/pnas.1106201108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DA, Kuznetsov A, Lopez JP, Kash S, Ammälä CE, Philipson LH (2007) Kv2.1 ablation alters glucose-induced islet electrical activity, enhancing insulin secretion. Cell Metab 6:229–235. 10.1016/j.cmet.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN (2012) Voltage-gated potassium channels and the diversity of electrical signalling. J Physiol 590:2591–2599. 10.1113/jphysiol.2011.224212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CS, Watanabe S, Stas JI, Klaphaak J, Yamane A, Schmitt N, Olesen SP, Trimmer JS, Rasmussen HB, Misonou H (2017) Trafficking of Kv2.1 channels to the axon initial segment by a novel nonconventional secretory pathway. J Neurosci 37:11523–11536. 10.1523/JNEUROSCI.3510-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT (2005) Structural basis of FFAT motif-mediated ER targeting. Structure 13:1035–1045. 10.1016/j.str.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Kihira Y, Hermanstyne TO, Misonou H (2010) Formation of heteromeric Kv2 channels in mammalian brain neurons. J Biol Chem 285:15048–15055. 10.1074/jbc.M109.074260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AN, Manning CF, Trimmer JS (2014) A unique ion channel clustering domain on the axon initial segment of mammalian neurons. J Comp Neurol 522:2594–2608. 10.1002/cne.23551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossov VL, Sivaguru M, Huff J, Luby K, Kanakaraju K, Gaskins HR (2018) Airyscan super-resolution microscopy of mitochondrial morphology and dynamics in living tumor cells. Microsc Res Tech 81:115–128. 10.1002/jemt.22968 [DOI] [PubMed] [Google Scholar]
- Kumagai K, Kawano-Kawada M, Hanada K (2014) Phosphoregulation of the ceramide transport protein CERT at serine 315 in the interaction with VAMP-associated protein (VAP) for inter-organelle trafficking of ceramide in mammalian cells. J Biol Chem 289:10748–10760. 10.1074/jbc.M113.528380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Jan LY (2006) The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci 7:548–562. 10.1038/nrn1938 [DOI] [PubMed] [Google Scholar]
- Lee HC, Wang JM, Swartz KJ (2003) Interaction between extracellular hanatoxin and the resting conformation of the voltage-sensor paddle in kv channels. Neuron 40:527–536. 10.1016/S0896-6273(03)00636-6 [DOI] [PubMed] [Google Scholar]
- Lim ST, Antonucci DE, Scannevin RH, Trimmer JS (2000) A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons. Neuron 25:385–397. 10.1016/S0896-6273(00)80902-2 [DOI] [PubMed] [Google Scholar]
- Liu PW, Bean BP (2014) Kv2 channel regulation of action potential repolarization and firing patterns in superior cervical ganglion neurons and hippocampal CA1 pyramidal neurons. J Neurosci 34:4991–5002. 10.1523/JNEUROSCI.1925-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomant AJ, Fairbanks G (1976) Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J Mol Biol 104:243–261. 10.1016/0022-2836(76)90011-5 [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Lenn NJ, Trimmer JS (1995) Differential spatiotemporal expression of K+ channel polypeptides in rat hippocampal neurons developing in situ and in vitro. J Neurosci 15:3840–3851. 10.1523/JNEUROSCI.15-05-03840.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandikian D, Bocksteins E, Parajuli LK, Bishop HI, Cerda O, Shigemoto R, Trimmer JS (2014) Cell type-specific spatial and functional coupling between mammalian brain Kv2.1 K(+) channels and ryanodine receptors. J Comp Neurol 522:3555–3574. 10.1002/cne.23641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning CF, Bundros AM, Trimmer JS (2012) Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS One 7:e38313. 10.1371/journal.pone.0038313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, Romoli M, Parrini E, Costa C, Mei D, Mari F, Parmeggiani L, Procopio E, Metitieri T, Cellini E, Virdò S, De Vita D, Gentile M, Prontera P, Calabresi P, Guerrini R (2017) Clinical features and outcome of 6 new patients carrying de novo KCNB1 gene mutations. Neurol Genet 3:e206. 10.1212/NXG.0000000000000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune BT, Tang W, Lu J, Eaglesham JB, Thorne L, Mayer AE, Condiff E, Nice TJ, Goodfellow I, Krezel AM, Virgin HW (2017) Noroviruses co-opt the function of host proteins VAPA and VAPB for replication via a phenylalanine-phenylalanine-acidic-tract-motif mimic in nonstructural viral protein NS1/2. MBio 8: pii: e00668–17. 10.1128/mBio.00668-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middaugh CR, Vanin EF, Ji TH (1983) Chemical cross-linking of cell membranes. Mol Cell Biochem 50:115–141. [DOI] [PubMed] [Google Scholar]
- Min SW, Chang WP, Südhof TC (2007) E-syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci U S A 104:3823–3828. 10.1073/pnas.0611725104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, Anderson AE, Trimmer JS (2004) Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci 7:711–718. 10.1038/nn1260 [DOI] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Trimmer JS (2005a) Kv2.1: a voltage-gated K+ channel critical to dynamic control of neuronal excitability. Neurotoxicology 26:743–752. 10.1016/j.neuro.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Misonou H, Mohapatra DP, Menegola M, Trimmer JS (2005b) Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci 25:11184–11193. 10.1523/JNEUROSCI.3370-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misonou H, Menegola M, Mohapatra DP, Guy LK, Park KS, Trimmer JS (2006) Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci 26:13505–13514. 10.1523/JNEUROSCI.3970-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia F, Zuccolo E, Soda T, Tanzi F, Guerra G, Mapelli L, Lodola F, D'Angelo E (2015) Stim and orai proteins in neuronal Ca(2+) signaling and excitability. Front Cell Neurosci 9:153. 10.3389/fncel.2015.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra DP, Trimmer JS (2006) The Kv2.1 C terminus can autonomously transfer Kv2.1-like phosphorylation-dependent localization, voltage-dependent gating, and muscarinic modulation to diverse Kv channels. J Neurosci 26:685–695. 10.1523/JNEUROSCI.4620-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shi G, Scannevin RH, Trimmer JS (1997) Phosphorylation of the Kv2.1 K+ channel alters voltage-dependent activation. Mol Pharmacol 52:821–828. 10.1124/mol.52.5.821 [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Trimmer JS (1999) Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons. J Neurosci 19:1728–1735. 10.1523/JNEUROSCI.19-05-01728.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP (2016) VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta 1861:952–961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Nishi M, Sakagami H, Komazaki S, Kondo H, Takeshima H (2003) Coexpression of junctophilin type 3 and type 4 in brain. Brain Res Mol Brain Res 118:102–110. 10.1016/S0169-328X(03)00341-3 [DOI] [PubMed] [Google Scholar]
- Nusser Z. (2012) Differential subcellular distribution of ion channels and the diversity of neuronal function. Curr Opin Neurobiol 22:366–371. 10.1016/j.conb.2011.10.006 [DOI] [PubMed] [Google Scholar]
- O'Connell KM, Rolig AS, Whitesell JD, Tamkun MM (2006) Kv2.1 potassium channels are retained within dynamic cell surface microdomains that are defined by a perimeter fence. J Neurosci 26:9609–9618. 10.1523/JNEUROSCI.1825-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio S, Chevaleyre V, Brann DH, Murray KD, Piskorowski RA, Trimmer JS (2017) Heterogeneity in Kv2 channel expression shapes action potential characteristics and firing patterns in CA1 versus CA2 hippocampal pyramidal neurons. eNeuro 4: pii: ENEURO.0267–17.2017. 10.1523/ENEURO.0267-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Mohapatra DP, Misonou H, Trimmer JS (2006) Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science 313:976–979. 10.1126/science.1124254 [DOI] [PubMed] [Google Scholar]
- Peltola MA, Kuja-Panula J, Lauri SE, Taira T, Rauvala H (2011) AMIGO is an auxiliary subunit of the Kv2.1 potassium channel. EMBO Rep 12:1293–1299. 10.1038/embor.2011.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Proskauer CC, Kaiserman-Abramof IR (1968) The small pyramidal neuron of the rat cerebral cortex: the axon hillock and initial segment. J Cell Biol 39:604–619. 10.1083/jcb.39.3.604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS (2015) Store-operated calcium channels. Physiol Rev 95:1383–1436. 10.1152/physrev.00020.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman PT, He K, Hartnett KA, Jefferson BS, Hu L, Rosenberg PA, Levitan ES, Aizenman E (2007) Apoptotic surge of potassium currents is mediated by p38 phosphorylation of Kv2.1. Proc Natl Acad Sci U S A 104:3568–3573. 10.1073/pnas.0610159104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J. (1962) Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol 13:405–421. 10.1083/jcb.13.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P (2017) Endoplasmic reticulum-plasma membrane contact sites. Annu Rev Biochem 86:659–684. 10.1146/annurev-biochem-061516-044932 [DOI] [PubMed] [Google Scholar]
- Saitsu H, Akita T, Tohyama J, Goldberg-Stern H, Kobayashi Y, Cohen R, Kato M, Ohba C, Miyatake S, Tsurusaki Y, Nakashima M, Miyake N, Fukuda A, Matsumoto N (2015) De novo KCNB1 mutations in infantile epilepsy inhibit repetitive neuronal firing. Sci Rep 5:15199. 10.1038/srep15199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ponce D, DeFelipe J, Garrido JJ, Muñoz A (2011) In vitro maturation of the cisternal organelle in the hippocampal neuron's axon initial segment. Mol Cell Neurosci 48:104–116. 10.1016/j.mcn.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Sarmiere PD, Weigle CM, Tamkun MM (2008) The Kv2.1 K+ channel targets to the axon initial segment of hippocampal and cortical neurons in culture and in situ. BMC Neurosci 9:112. 10.1186/1471-2202-9-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev SV, Woodroofe CC, Sabat G, Adams CM, Klaubert D, Wood K, Urh M (2013) Mass spectrometry compatible surfactant for optimized in-gel protein digestion. Anal Chem 85:907–914. 10.1021/ac302423t [DOI] [PubMed] [Google Scholar]
- Scannevin RH, Murakoshi H, Rhodes KJ, Trimmer JS (1996) Identification of a cytoplasmic domain important in the polarized expression and clustering of the Kv2.1 K+ channel. J Cell Biol 135:1619–1632. 10.1083/jcb.135.6.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A, Del Turco D, Deller T, Gutzmann A, Schultz C, Engelhardt M (2017) Structural plasticity of synaptopodin in the axon initial segment during visual cortex development. Cereb Cortex 27:4662–4675. 10.1093/cercor/bhx208 [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL (2002) Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16:869–871. 10.1096/fj.01-0995fje [DOI] [PubMed] [Google Scholar]
- Song MY, Hong C, Bae SH, So I, Park KS (2012) Dynamic modulation of the Kv2.1 channel by SRC-dependent tyrosine phosphorylation. J Proteome Res 11:1018–1026. 10.1021/pr200770v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speca DJ, Ogata G, Mandikian D, Bishop HI, Wiler SW, Eum K, Wenzel HJ, Doisy ET, Matt L, Campi KL, Golub MS, Nerbonne JM, Hell JW, Trainor BC, Sack JT, Schwartzkroin PA, Trimmer JS (2014) Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav 13:394–408. 10.1111/gbb.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N, McBain C (2007) Structural and functional properties of hippocampal neurons. In: The hippocampus book (Andersen P, Morris R, Amaral D, Bliss T, O'Keefe J, eds), pp 133–201. New York, NY: OUP. [Google Scholar]
- Sun XH, Protasi F, Takahashi M, Takeshima H, Ferguson DG, Franzini-Armstrong C (1995) Molecular architecture of membranes involved in excitation-contraction coupling of cardiac muscle. J Cell Biol 129:659–671. 10.1083/jcb.129.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz KJ. (2004) Towards a structural view of gating in potassium channels. Nat Rev Neurosci 5:905–916. 10.1038/nrn1559 [DOI] [PubMed] [Google Scholar]
- Takeshima H, Hoshijima M, Song LS (2015) Ca(2)(+) microdomains organized by junctophilins. Cell Calcium 58:349–356. 10.1016/j.ceca.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun MM, O'Connell KM, Rolig AS (2007) A cytoskeletal-based perimeter fence selectively corrals a sub-population of cell surface Kv2.1 channels. J Cell Sci 120:2413–2423. 10.1242/jcs.007351 [DOI] [PubMed] [Google Scholar]
- Thiffault I, Speca DJ, Austin DC, Cobb MM, Eum KS, Safina NP, Grote L, Farrow EG, Miller N, Soden S, Kingsmore SF, Trimmer JS, Saunders CJ, Sack JT (2015) A novel epileptic encephalopathy mutation in KCNB1 disrupts Kv2.1 ion selectivity, expression, and localization. J Gen Physiol 146:399–410. 10.1085/jgp.201511444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Bersell K, Jorge BS, Bjork RL Jr, Friedman JR, Bloss CS, Cohen J, Gupta S, Naidu S, Vanoye CG, George AL Jr, Kearney JA (2014) De novo KCNB1 mutations in epileptic encephalopathy. Ann Neurol 76:529–540. 10.1002/ana.24263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. (1991) Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc Natl Acad Sci U S A 88:10764–10768. 10.1073/pnas.88.23.10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS. (2015) Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron 85:238–256. 10.1016/j.neuron.2014.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimmer JS, Misonou H (2015) Phosphorylation of voltage-gated ion channels. In: Handbook of ion channels (Zheng J, Trudeau MC, eds), pp 531–544. Boca Raton, FL: CRC. [Google Scholar]
- Trimmer JS, Trowbridge IS, Vacquier VD (1985) Monoclonal antibody to a membrane glycoprotein inhibits the acrosome reaction and associated Ca2+ and H+ fluxes of sea urchin sperm. Cell 40:697–703. 10.1016/0092-8674(85)90218-1 [DOI] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS (2008) Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev 88:1407–1447. 10.1152/physrev.00002.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDongen AM, Frech GC, Drewe JA, Joho RH, Brown AM (1990) Alteration and restoration of K+ channel function by deletions at the N- and C-termini. Neuron 5:433–443. 10.1016/0896-6273(90)90082-Q [DOI] [PubMed] [Google Scholar]
- Wu C, Ivanova E, Zhang Y, Pan ZH (2013) rAAV-mediated subcellular targeting of optogenetic tools in retinal ganglion cells in vivo. PLoS One 8:e66332. 10.1371/journal.pone.0066332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS (2006) Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol 174:803–813. 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, Hess HF, De Camilli P (2017) Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci U S A 114:E4859–E4867. 10.1073/pnas.1701078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SP, Kerchner GA (1998) Endogenous voltage-gated potassium channels in human embryonic kidney (HEK293) cells. J Neurosci Res 52:612–617. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rasband MN (2016) Cytoskeletal control of axon domain assembly and function. Curr Opin Neurobiol 39:116–121. 10.1016/j.conb.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437:902–905. 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]