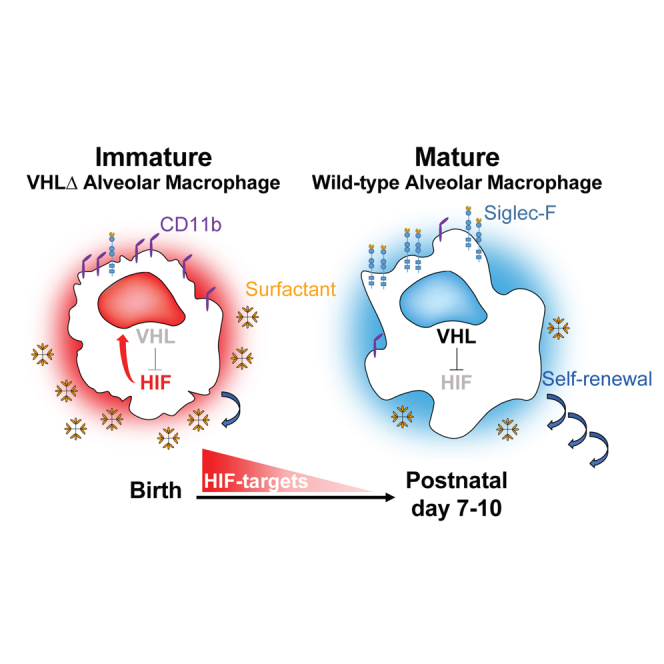
Summary
The rapid transit from hypoxia to normoxia in the lung that follows the first breath in newborn mice coincides with alveolar macrophage (AM) differentiation. However, whether sensing of oxygen affects AM maturation and function has not been previously explored. We have generated mice whose AMs show a deficient ability to sense oxygen after birth by deleting Vhl, a negative regulator of HIF transcription factors, in the CD11c compartment (CD11cΔVhl mice). VHL-deficient AMs show an immature-like phenotype and an impaired self-renewal capacity in vivo that persists upon culture ex vivo. VHL-deficient phenotype is intrinsic in AMs derived from monocyte precursors in mixed bone marrow chimeras. Moreover, unlike control Vhlfl/fl, AMs from CD11cΔVhl mice do not reverse pulmonary alveolar proteinosis when transplanted into Csf2rb−/− mice, demonstrating that VHL contributes to AM-mediated surfactant clearance. Thus, our results suggest that optimal AM terminal differentiation, self-renewal, and homeostatic function requires their intact oxygen-sensing capacity.
Keywords: alveolar macrophage, oxygen, VHL, HIF, self-renewal, differentiation, surfactant, lung, glycolysis, lipid metabolism
Graphical Abstract
Highlights
-
•
Hypoxia and glycolysis signatures are downregulated during alveolar macrophage maturation
-
•
VHL-deficient alveolar macrophages induce HIF-target genes and are more glycolytic
-
•
VHL-deficient alveolar macrophages show an intrinsic immature-like phenotype
-
•
VHL promotes alveolar macrophage self-renewal and surfactant clearance capacity
Izquierdo et al. show that the absence of VHL in lung alveolar macrophages prevents their terminal maturation. VHL-deficient alveolar macrophages induce HIF-target gene expression and are partially desensitized to oxygen. Moreover, they have a decreased self-renewal and surfactant clearance capacity, suggesting that oxygen sensing contributes to alveolar macrophage functional maturation.
Introduction
Tissue resident macrophages are essential for tissue homeostasis and are in turn modulated by tissue factors. Alveolar macrophages (AMs) show a high capacity for self-maintenance by local proliferation (Sieweke and Allen, 2013), with little repopulation by blood-circulating monocytes in the steady state (Guilliams et al., 2013, Hashimoto et al., 2013). Mature AMs regulate surfactant levels in the lung (Baker et al., 2010, Robb et al., 1995, Stanley et al., 1994). Factors including granulocyte-macrophage colony-stimulating factor (GM-CSF) (Csf2−/−), GM-CSF receptor (Csf2rb−/−) (Robb et al., 1995, Stanley et al., 1994), PPAR-γ (Schneider et al., 2014), and mammalian target of rapamycin (mTOR) (Sinclair et al., 2017) are required for AM development and function.
Initial lung development occurs in a relatively hypoxic environment, beginning in mice on embryonic day 9 (E9) (Lee et al., 2001). Fetal monocytes colonize the developing lung at approximately E12.5 and E16.5 and only differentiate into mature AMs after birth (Guilliams et al., 2013). Epithelial cell-derived GM-CSF is essential for the early commitment of AM precursors (Guilliams et al., 2013, Schneider et al., 2014). However, it remains unknown whether AM adaptation to increased oxygen concentration in the postnatal lung influences AM maturation and function.
We hypothesized that sensing the increased oxygen concentration just after birth may be important for postnatal AM maturation. Hypoxia-inducible transcription factors (HIFs) are the master regulators of the transcriptional response to low oxygen tensions, and HIF depletion affects macrophage physiology (Imtiyaz et al., 2010, Li et al., 2018, Peyssonnaux et al., 2005). Under normoxia, HIF-α subunits are hydroxylated and bind to the Von Hippel-Lindau tumor suppressor protein (VHL), which is part of an E3 ubiquitin ligase complex targeting HIF-α subunits for proteasomal degradation. Therefore, Vhl deletion induces normoxic expression of HIF target genes (Cramer et al., 2003, Peyssonnaux et al., 2005). Here, we deleted Vhl in the CD11c+ compartment (CD11cΔVhl) to desensitize AMs to oxygen after birth. Vhl deletion induced the expression of HIF target genes and altered the AM metabolic profile. VHL-deficient AMs showed an immature-like phenotype and a decreased self-renewal capacity that were cell intrinsic, as demonstrated using mixed bone marrow (BM) chimeras. In addition, VHL contributed to AM-mediated surfactant clearance in a model of alveolar proteinosis. Our results therefore support the notion that intact oxygen-sensing capacity is required for AM terminal differentiation, self-renewal, and function.
Results and Discussion
Downregulation of Hypoxia and Glycolysis during Postnatal AM Maturation
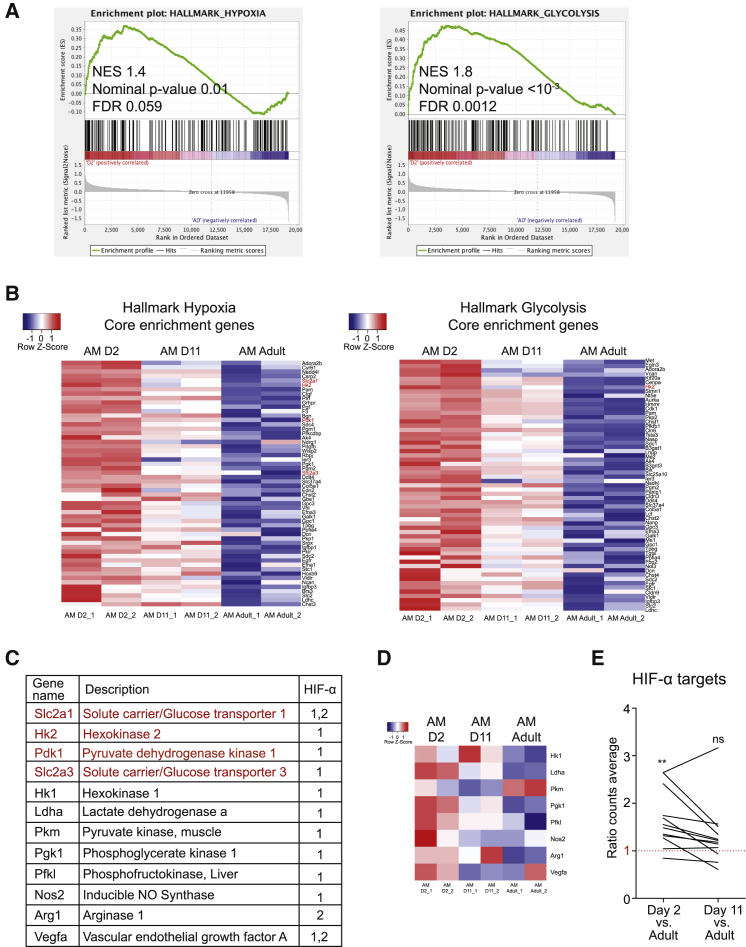
To explore whether oxygen sensing by AMs could be linked to their maturation, we analyzed a publicly available microarray data (Schneider et al., 2014) containing the transcriptional profile of AMs from pups at postnatal days 2 and 11 and from adult mice that were 8–12 weeks old. Gene set enrichment analysis (GSEA) was performed on the gene expression profiles using the hallmark collection of the Molecular Signatures Database (MSigDB). Results showed significant enrichment of both hypoxia and glycolysis gene sets in immature AMs from 2-day-old pups compared with mature AMs from adults (Figure 1A). In addition, the expression of core genes from these two enriched hallmark gene sets was gradually downregulated during AM maturation (Figure 1B), suggesting postnatal adaptation of AMs to oxygen. Core genes from both gene sets included well-known HIF targets such as Slc2a1, Hk2, Pdk1, and Slc2a3 (Figures 1B and 1C [genes marked in red]). Analysis of other classical HIF target genes in most cases revealed the same expression dynamics (Figures 1C and 1D). The mean expression of all HIF target genes was thus significantly higher in immature AMs (day 2) than in mature AMs from adult mice, whereas no significant changes were found between postnatal day 11 and adulthood (Figure 1E), correlating with the end of AM terminal differentiation at approximately postnatal day 7 (Guilliams et al., 2013). These results suggest that downregulation of HIF activity in AMs upon exposure to high oxygen tension after birth may be important for AM terminal differentiation, concurring with the HIF-1-independent role of mTOR in AM development after birth (Sinclair et al., 2017).
Figure 1.
Downregulation of Hypoxia and Glycolysis during Postnatal AM Maturation
(A) Enrichment plots from GSEA of postnatal day 2 (D2) compared with adult (AD) AMs using the hallmark gene set collection from the Molecular Signatures Database (MSigDB).
(B) mRNA expression (normalized log2, robust multiarray average [RMA] counts) of core genes from hypoxia and glycolysis gene sets in lung AMs from postnatal day 2 (D2), D11, and adult mice. Red and blue represent overexpressed and underexpressed genes, respectively.
(C) Table depicting some classic HIF target genes. Genes marked in red are present in Figure 1B.
(D) mRNA expression (as seen in Figure 1B) of HIF target genes not included in the hallmark gene sets indicated in Figure 1B (marked in black); color intensity defined as seen in Figure 1B.
(E) Expression of HIF target genes from Figure 1C in D2 and D11 AMs. Values represented as the ratio of the mean expression in AMs from D2 or D11 mice into the mean expression in AMs from adult mice. ns, not significant; ∗∗p < 0.01 by column statistics analysis, hypothetical value = 1.
Lack of VHL Results in AM-Immature Phenotype and Modified Metabolic Profile
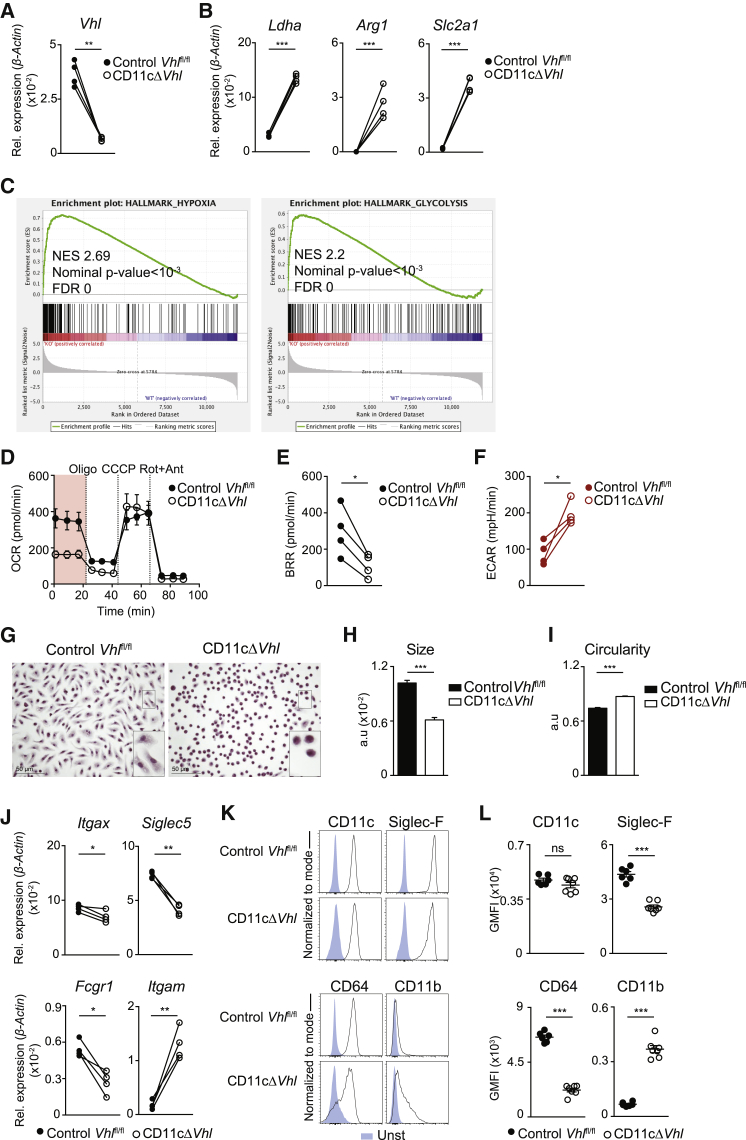
To investigate the biological significance of oxygen-sensing modulation in AMs after birth, we generated a genetic system to prevent HIF degradation in response to high postnatal oxygen pressures by deleting Vhl in the CD11c lineage (CD11c-Cre Vhlfl/fl mice). Compared to the LysM(Lyz2)-Cre driver, the CD11c-Cre driver specifically targets postnatal AMs (Guilliams et al., 2013). Vhl deletion leads to an increased glycolytic activity of macrophages under basal conditions (Cramer et al., 2003). Sorted bronchoalveolar lavage (BAL) AMs from CD11c-Cre+ Vhlfl/fl (CD11cΔVhl) mice showed decreased Vhl mRNA levels relative to AMs from CD11c-Cre− Vhlfl/fl littermates (control Vhlfl/fl) (Figure 2A). Vhl deletion in AMs resulted in the increased expression of HIF-1 targets such as Ldha (lactate dehydrogenase) (Semenza et al., 1996), HIF-2 targets such as Arg1 (arginase 1) (Takeda et al., 2010), and common HIF-1 and HIF-2 target genes such as Slc2a1 (glucose transporter 1) (Chen et al., 2001) (Figure 2B). To explore the functional consequences of Vhl deletion in AMs, we conducted an RNA sequencing (RNA-seq) analysis to compare the transcriptome of BAL AMs isolated from CD11cΔVhl and control Vhlfl/fl mice. GSEA showed significant enrichment of hypoxia and glycolysis gene sets from the hallmark collection in the absence of VHL (Figure 2C), resembling the comparison of day 2 postnatal versus adult AMs (Figure 1A) and indicating that, at the transcriptional level, the absence of VHL affects the ability of AMs to detect oxygen. To address this influence functionally, we determined the real-time metabolic profile of AMs from BAL. The absence of VHL in AMs significantly decreased their basal respiratory rate (BRR) (Figures 2D and 2E) while increasing their basal extracellular acidification rate (Figure 2F). These results indicate that the deletion of VHL in AMs induces the expression of HIF-1 and HIF-2 target genes and alters their basal metabolic profile.
Figure 2.
Deletion of VHL Alters AM Metabolic Profile and Phenotype
(A and B) Mean expression of Vhl (A), Ldha, Arg1, and Slc2a1 (B) by qPCR in sorted BAL AMs from the indicated genotypes. Data represent four pools of mice corresponding to four independent experiments. n = 4–5 mice per genotype. ∗∗p < 0.01, ∗∗∗p < 0.001 by ratio paired Student’s t test.
(C) Enrichment plots from GSEA analysis of CD11cΔVhl (knockout [KO]), compared with control Vhlfl/fl (wild-type [WT]) AMs using the hallmark gene set collections as seen in Figure 1B.
(D) Oxygen-consumption rate (OCR) by BAL AMs in the presence of glucose, glutamine, and pyruvate sequentially treated with oligomycin (Oligo), carbonyl cyanide m-chlorophenylhydrazone (CCCP) and rotenone+antimycin (Rot+Ant). Data are means ± SDs and are representative of four independent experiments. n = 6–7 mice per genotype.
(E and F) Basal respiratory rate (BRR) (E) and basal extracellular acidification rate (ECAR) (F) of BAL AMs. Data represent four independent experiments. n = 6–7 mice per genotype. ∗p < 0.05 by ratio paired Student’s t test.
(G–I) Morphological parameters of BAL AMs from each genotype after 5-day culture in complete medium. Representative images of H&E staining (error bars, 50 μm) (G), mean AM size ± SEM (H), and mean AM circularity ± SEM (I). Data are representative of two independent experiments. n = 4–5 mice per genotype. ∗∗∗p < 0.001, by unpaired Student’s t test.
(J) Expression of Itgax, Siglec5, Fcgr1, and Itgam by qPCR as in (A) and (B). ∗p < 0.05, ∗∗p < 0.01 by ratio paired Student’s t test.
(K) Representative flow cytometry histograms for CD11c, Siglec-F, CD64, and CD11b (black line) expression compared with unstained samples (blue) in BAL AMs from the indicated genotypes.
(L) Normalized geometric mean fluorescence intensity (GMFI) for markers from Figure 2K. Data shown as means ± SEMs and representative of three experiments. n = 6–7 per genotype. ∗∗∗p < 0.001 by unpaired Student’s t test.
See also Figure S1.
Altered AM terminal differentiation can lead to the development of AM-like cells with a distorted phenotype (Schneider et al., 2014). We found that VHL-deficient AMs from BAL were smaller and unable to acquire the fusiform morphology of control Vhlfl/fl AMs after culture ex vivo for 5 days, instead maintaining a rounded shape that is similar to that of precursor cells or monocytes (Figures 2G–2I) (Guilliams et al., 2013). Because altered maturation can affect AM phenotype (Schneider et al., 2014), we measured the expression of canonical surface markers on BAL AMs ex vivo. VHL-deficient AMs slightly reduced mRNA for Itgax (CD11c) without affecting membrane receptor expression (Figures 2J–2L). Notably, VHL-deficient AMs showed decreased mRNA and protein for Siglec5 (Siglec-F) and Fcgr1a (CD64), whereas Itgam (CD11b) was increased (Figures 2J–2L). This phenotype is linked to an immature AM state (Guilliams et al., 2013, Schneider et al., 2014).
Although access to the alveolar niche is restricted in steady-state conditions, blood monocytes can access the lung alveoli and differentiate into AMs under specific conditions (Hashimoto et al., 2013). Thus, the immature-like phenotype of VHL-deficient AMs could be either cell intrinsic or caused by increased infiltration of monocytes in CD11cΔVhl mice. To investigate this, we performed surgical parabiosis of CD45.1 wild-type (WT) mice with either CD45.2 control Vhlfl/fl or CD11cΔVhl mice (Figure S1A). After 30 days, analysis of the peripheral blood in CD45.2 parabionts revealed equivalent frequencies of CD45.1+ neutrophils and Ly6Chi monocytes, found at the same level as those exchanged after longer protocols of parabiosis in the steady state (Hashimoto et al., 2013) (Figures S1B and S1C). Lung CD45.1+ Ly6Chi monocytes and BAL AMs frequencies were similar in both genotypes (Figures S1D and S1E). These results suggest that the immature-like phenotype of VHL-deficient AMs is caused by a cell-autonomous effect and is not a consequence of differential lung infiltration by monocytes.
Cell-Cycle Regulation and Lipid Handling Are Altered in VHL-Deficient AMs
RNA-seq identified 2,531 differentially expressed genes in CD11cΔVhl AMs relative to control Vhlfl/fl and control CD11cCre+ AMs (Figure S2A). Functional analysis of these genes with Ingenuity Pathway Analysis software revealed several pathways altered by the absence of VHL in AMs (Figure S2B). We validated the expression of selected genes in pathways related to relevant features of AM physiology, such as cell cycle and liver X receptor/retinoid X receptor (LXR/RXR) activation, the latter being implicated in intracellular lipid sensing (Figures S2C and S2D). The absence of VHL in AMs resulted in the reduced expression of genes involved in cell-cycle progression, such as cyclin B1 (Ccnb1), cyclin B2 (Ccnb2), CDK1 (Cdk1), CD25c (Cd25c), and PLK3 (Plk3) (Figure S2C [top]) and cell-cycle control, including BRCA1 (Brca1), CHEK1 (Chek1), and CHEK2 (Chek2), while the cell-cycle master regulators p53 (Tp53) and p21 (Cdkn1a) displayed similar expression levels (Figure S2C [bottom]). Among those genes associated with the LXR/RXR activation pathway, upregulated transcript expression of LXRα (Nr1h3) and LXRβ (Nr1h2) genes (Figure S2D) was confirmed. In accordance with this, we confirmed that there was an increased expression of genes involved in cholesterol efflux, such as the transporters ABCG1 (Abcg1) and apolipoprotein E (ApoE) and the lipoprotein remodeling enzyme PLTP (Pltp) (Figure S2D). These results suggest that crucial distinctive features of AMs, such as proliferation and lipid handling, are affected by the absence of VHL.
Functional VHL Is Required for AM Self-Renewal Capacity
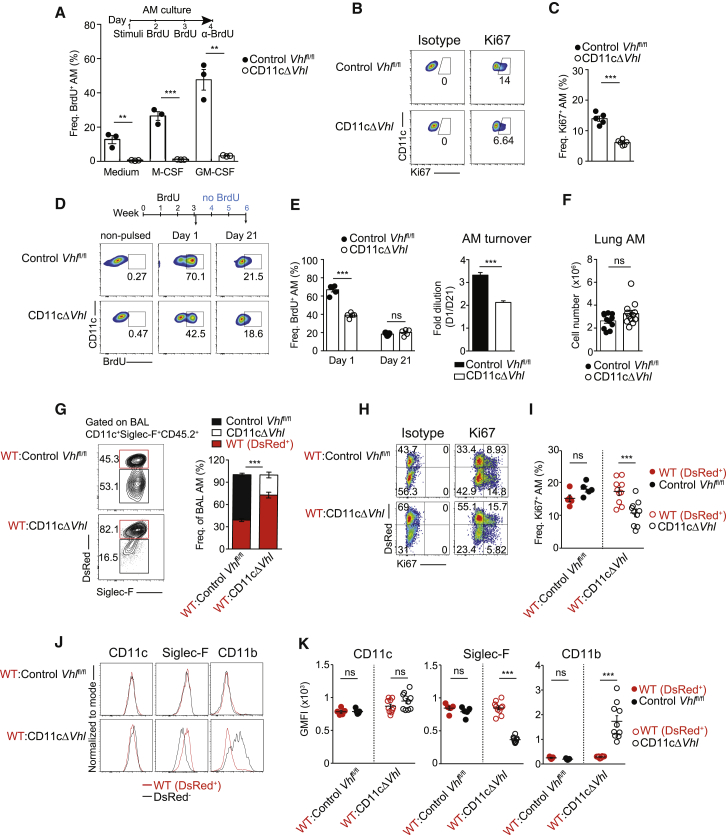
We next investigated whether the absence of VHL affected the self-renewal capacity of AMs. We measured bromodeoxyuridine (BrdU) incorporation in BAL AMs cultured in vitro in the absence or presence of macrophage colony-stimulating factor (M-CSF) and GM-CSF, tissue-derived cytokines that contribute to the maintenance and proliferation of mature AMs (Hashimoto et al., 2013). VHL-deficient AMs proliferated significantly less than did control Vhlfl/fl AMs both in the absence and presence of these cytokines (Figure 3A).
Figure 3.
Cell-Intrinsic VHL Requirement for AM Self-Renewal and Terminal Maturation
(A) In vitro BrdU assay of BAL AMs in complete RPMI (medium) or stimulated with recombinant mouse (rM)-CSF or recombinant granulocyte-macrophage (rGM)-CSF as indicated. Data are means ± SEMs and are representative of three experiments. n = 15–18 mice of each genotype. ∗∗p < 0.01, ∗∗∗p < 0.001, by unpaired Student’s t test.
(B and C) Ki67 staining in ex vivo BAL AMs. Representative flow cytometry plots (B) and frequencies of Ki67+ cells (C). Data are means ± SEMs and are representative of three experiments. n = 5 mice per genotype. ∗∗∗p < 0.001 by unpaired Student’s t test.
(D and E) Mice were pulsed with BrdU for 3 weeks (1 mg intraperitoneally [i.p.] daily), and BrdU incorporation into BAL AMs was assessed 1 and 21 days after the last pulse. Representative flow cytometry plots (D); frequencies of BrdU+ AMs (left) and AM turnover (right) calculated as the ratio of frequencies of BrdU+ AMs at day 1 versus day 21 (E). Data are means ± SEMs and are representative of three experiments. n = 5 mice per genotype. ∗∗∗p < 0.001; ns, not significant, by unpaired Student’s t test.
(F) Total lung AM number. Data are means ± SEMs and show a pool of two experiments representative of four performed. n = 5–6 per genotype. ∗∗∗p < 0.001; ns, not significant, by unpaired Student’s t test.
(G) Flow cytometry plots and frequencies of DsRed+ (WT) and DsRed− (Control Vhlfl/fl or CD11cΔVhl) AMs (normalized to blood Ly6Chi monocyte frequency) from CD45.2+ AM fraction of BM chimeras.
(H and I) Staining of Ki67 in ex vivo BAL AMs of the indicated genotypes from BM chimeras. Representative flow cytometry plots (H) and frequencies of Ki67+ BAL AMs (I).
(J and K) Representative flow cytometry histograms for AM selected markers (red line for WT DsRed+ AMs or black line for Control Vhlfl/fl or CD11cΔVhl AMs) in BAL AMs from the indicated BM chimera (J). Normalized geometric mean fluorescence intensity (GMFI) for AM selected markers (K).
(G, I, and K) Data shown as means ± SEMs and are representative of three (G) or two (I) performed. n = 5–10 per group. ∗∗∗p < 0.001; ns, not significant, by unpaired Student’s t test.
See also Figures S2 and S3.
To further investigate the potential role of VHL in AM self-renewal, we analyzed BAL AMs ex vivo. These experiments revealed significantly reduced Ki67 staining in VHL-deficient AMs than in control Vhlfl/fl AMs (Figures 3B and 3C), suggesting impaired proliferation. We next tracked self-renewal in vivo by injecting BrdU intraperitoneally every day for 3 consecutive weeks. One day after the last pulse, VHL-deficient AMs from BAL showed lower BrdU incorporation than control Vhlfl/fl AMs (Figures 3D and 3E), which is indicative of a decreased replication rate. In contrast, 21 days after the last pulse, BrdU labeling was similar in both genotypes (Figures 3D and 3E), which translated into a lower fold dilution of the incorporated BrdU in the absence of VHL (Figure 3F [right]). Despite the reduced turnover in VHL-deficient AMs, total lung AM numbers were similar in both genotypes (Figure 3F), suggesting that VHL-deficient AMs have a longer lifespan. Consistent with this, previous results showed increased primary macrophage survival under hypoxia (Roiniotis et al., 2009) and reduced apoptosis in Phd3−/− BM-macrophages (Swain et al., 2014).
AMs are replaced by BM-derived blood monocytes in lethal irradiation chimeras (Hashimoto et al., 2013, Yona et al., 2013). Therefore, we next asked whether Vhl deletion could also affect the replenishment of AMs after irradiation-induced cytoablation. We generated chimeric mice by intravenous adoptive transfer of BM cells from WT CD45.2+ DsRed mice (DsRed+) mixed at a 1:1 ratio with BM cells from either DsRed− CD45.2+ CD11cΔVhl mice (WT:CD11cΔVhl) or DsRed− CD45.2+ control Vhlfl/fl (WT:control Vhlfl/fl) into lethally irradiated CD45.1+ WT recipient mice (Figures 3G–3K). After 47 days, the frequency of AMs was analyzed. Most BAL and lung AMs (after BAL) were of donor origin (CD45.2+) in both types of chimeras at this time point (Figures S3A and S3B) (Hashimoto et al., 2013). Notably, the frequency of CD11cΔVhl AMs in the BAL (Figure 3G) and the lung (Figure S3C) of chimeric mice was significantly lower compared with control Vhlfl/fl AMs, recapitulating the suboptimal reconstitution of the alveolar space by prolyl hydroxylase 3 (PHD3)-deficient AMs (Tavernier et al., 2017). This result correlated with a significantly decreased proliferative capacity of BAL CD11cΔVhl AMs compared with WT AMs from co-transferred BM chimeras (Figures 3H and 3I). As a control, no differences were found in the potential of reconstitution of lung eosinophils from the different donors (Figure S3D). Moreover, CD11cΔVhl AMs showed an immature-like phenotype compared with WT AM counterparts (Figures 3J and 3K). Monocytic AM precursors only differentiate into mature AMs once they reach the lung (Guilliams et al., 2013, Schneider et al., 2014, van de Laar et al., 2016). Therefore, these results suggest that the absence of VHL driven by the CD11c promoter during AM differentiation intrinsically prevents their terminal phenotypic maturation and restrains their self-renewal capacity, decreasing their potential to reconstitute the alveolar niche in a context of competition with WT AMs. Because AM precursors come from oxygen-rich arterial blood, this result also supports the notion that VHL-mediated oxygen sensing is required but not sufficient for AM terminal differentiation.
VHL Contributes to Surfactant Handling by AMs
Mice deficient for Csf2 (GM-CSF) or Csf2r or with Pparg deletion in AMs show impaired AM maturation, resulting in low numbers of lung immature AMs that accumulate lipids intracellularly (Robb et al., 1995, Schneider et al., 2014, Stanley et al., 1994). Immature AM-like cells from patients with surfactant accumulation and Csf2−/− mice, as well as Pparg-deficient AMs, show an induction of LXR signaling (Baker et al., 2010, Thomassen et al., 2007). These findings correlate with the LXR pathway activation predicted by our RNA-seq analysis (Figure S2B) and with the increased expression of LXR-target genes implicated in lipid efflux in VHL-deficient AMs (Figure S2D).
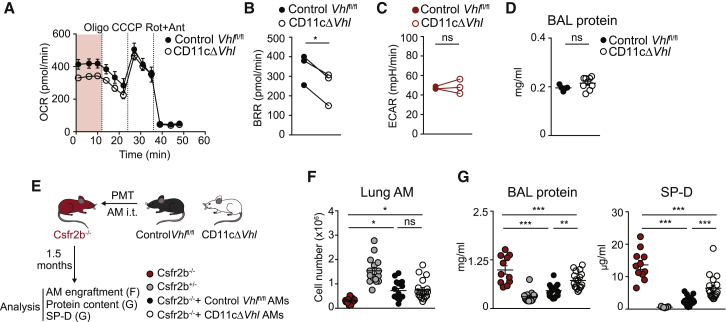
To quantify the lipid β-oxidation capacity of AMs, we measured the oxygen-consumption rate of BAL AMs in the presence of palmytoyl-coenzyme A (CoA) as a carbon source (Figure 4A). The basal respiration rate was significantly lower in AMs lacking VHL, indicating a reduced lipid oxidation capacity (Figure 4B), whereas the basal extracellular acidification rate was unaltered (Figure 4C). Unlike other models of impaired AM function, 3-month-old CD11cΔVhl mice did not show an increased accumulation of total protein in BAL supernatants (Figure 4D).
Figure 4.
VHL Contributes to Surfactant Handling by AMs
(A) OCR by BAL AMs in the presence of palmitoyl-CoA treated as seen in Figure 2D. Data are means ± SDs and are representative of three independent experiments. n = 6–7 mice of each genotype.
(B and C) Basal respiratory rate (BRR) (B) and basal ECAR (C) of BAL AMs. Data represent three independent experiments. n = 6–7 mice per genotype. ∗p < 0.05; ns, not significant, by ratio paired Student’s t test.
(D) Protein concentration in BAL from 3-month-old mice. Data are means ± SEMs and representative of three independent experiments. n = 5–9 mice per genotype. ns, not significant, by unpaired Student’s t test.
(E–G) 5 × 104 AMs from control Vhlfl/fl or CD11cΔVhl AMs were transferred intratracheally (i.t.) to Csfr2b−/− mice. Scheme of the experiment (E). After 1.5 months, lung AM numbers (F) and BAL concentrations of total protein (G [left]) and SP-D (G [right]) were determined. Data represented as means ± SEMs of three pooled independent experiments. Additional control mice (Csfr2b−/− and Csfr2b+/−) were added. n = 10–18 per group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant, by one-way ANOVA and Bonferroni post hoc test. Only some relevant comparisons indicated.
See also Figure S2.
To determine whether the decreased ability to oxidize lipids of VHL-deficient AMs affected their therapeutic potential to degrade surfactant excess in vivo, we transplanted a limited number of AMs from BALs of CD11cΔVhl or control Vhlfl/fl mice into Csf2rb−/− mice, which lack mature AMs and spontaneously develop lung proteinosis (Figures 4E–4G) (Robb et al., 1995). As a marker of disease severity, we measured total protein and surfactant protein D concentrations in BAL supernatants at 1.5 months after pulmonary macrophage transplantation (Figure 4E), a time point at which transferred WT AMs had removed surfactant nearly at the levels found in healthy mice (Figure 4G) (Happle et al., 2014, Suzuki et al., 2014). Consistent with our earlier results (Figure 3F), AM engraftment in the lung was similar in both genotypes (Figure 4F). Transplanted AMs from CD11cΔVhl mice were less efficient than control Vhlfl/fl mice at reversing surfactant accumulation in BAL from Csf2rb−/− recipient mice BAL (Figure 4G). These results indicate that VHL contributes to the ability of AMs to oxidize lipids and to clear pulmonary surfactant in vivo.
In conclusion, we investigated whether AM adaptation to increased oxygen concentrations in the postnatal lung is relevant for AM function. Deletion of Vhl in AMs induced the expression of HIF target genes and altered their metabolic profile, leading to partial desensitization of AMs to oxygen. VHL-deficient AMs resembled immature-like AMs in terms of morphology and expression of maturation markers and have a limited self-renewal potential and altered transcriptional signature. Experiments of parabiosis and BM chimeras indicated that this is a cell-autonomous phenotype. These results suggest that besides lung tissue-specific factors, oxygen sensing is also needed for AM terminal differentiation. Functionally, VHL-deficient AMs showed defective therapeutic capacity to clear surfactant excess in vivo. Our data therefore suggest that an intact oxygen-sensing machinery is required for AM terminal differentiation, self-renewal, and functional capacity.
Experimental Procedures
Animals
Mice were bred at CNIC under specific pathogen-free conditions. C57BL/6J-Crl mice (Charles River), Csf2rb−/− mice (Robb et al., 1995), and Vhlfl/fl mice (Menendez-Montes et al., 2016) mated with CD11c-Cre BAC transgenic mice (Caton et al., 2007) were used. B6/SJL (Ptprca Pepcb/BoyJ) mice expressing the CD45.1 allele and DsRed (Actb-DsRed.T3) mice were both acquired from the Jackson Laboratory. All of the animal procedures were approved by the Animal Ethics Committee of the CNIC, the Madrid Autonomous University Ethics Committee, and the Community of Madrid Authority, and were compliant with EU Directives 2010/63/EU and 2007/526/EC and with Spanish law under Real Decreto 1201/2005.
Parabiosis
Parabiosis was performed by surgical attachment of the skin of two mice, which promoted the formation of microvasculature at the site of inflammation. CD11c-Cre+ Vhlfl/fl and CD11c-Cre− Vhlfl/fl littermates were joined with B6/SJL, expressing the CD45.1 congenic marker to facilitate cell tracking after 1 month of parabiosis.
BM Chimeras
B6/SJL mice expressing the CD45.1 congenic marker were lethally irradiated (12 Gy) and were reconstituted intravenously (i.v.) with 5 × 106 cells of a 1:1 mixture of CD45.2+ WT DsRed+ BM cells and CD45.2+ CD11cΔVhl BM cells, or CD45.2+ WT DsRed+ BM cells and CD45.2+ control Vhlfl/fl BM cells. Mice were analyzed 47 days after reconstitution.
AM Isolation
Mice were sacrificed with a lethal dose of pentobarbital (Dolethal, Vetoquinol). Ten BALs per mouse were performed with a blunt fill needle (18G × 1.5; 1.2 × 40 mm, BD Safety Products), with 1 mL of PBS, 2.5 mM EDTA, and 2% fetal bovine serum (FBS, HyClone) (flow cytometry buffer) at 37°C. Samples were kept on ice until further processing. Red blood cell lysis was performed at room temperature (RT) for 3 min (RBC Lysis Buffer, Sigma-Aldrich). AMs were counted with a Neubauer chamber and cultured in RPMI 1640 (GIBCO) supplemented with 10% FBS plus 100 U/mL penicillin and 2 mM l-glutamine (complete RPMI) at 37°C and 5% CO2. Additional information can be found in the Supplemental Experimental Procedures.
RNA Isolation
Cell lysis was performed with buffer RLT (QIAGEN) containing 10 μm/mL β-mercaptoethanol, and RNA was isolated with the RNeasy Plus Mini Kit (QIAGEN). RNA concentration and integrity were determined with an Agilent 2100 Bioanalyzer (Caliper Life Science). Samples with RNA integrity values >8 were further processed. cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Additional information can be found in the Supplemental Experimental Procedures.
Metabolic Measurements
Real-time oxygen-consumption rate (OCR) and extracellular acidification rate (ECAR) in AMs were determined with an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). BAL AMs were pooled from 6–7 mice per genotype. After red blood cell lysis, AMs were plated and incubated at 37°C without CO2 for 30 min before the Seahorse assay was run. Additional information can be found in the Supplemental Experimental Procedures.
BrdU Proliferation In Vivo
Four groups of mice (two groups of each genotype) were injected daily with 1 mg BrdU (Sigma-Aldrich) for 2 weeks. One day and 21 days after the last injection, two groups (one of each genotype) were sacrificed, and BAL AMs were analyzed for BrdU incorporation (Hashimoto et al., 2013). The ratio of frequencies of BrdU+ AMs at days 21 and 1 was calculated as a measurement of BrdU dilution for 21 days.
Pulmonary Macrophage Transplantation
Csf2rb−/− mice were pulmonary transplanted with 5 × 104 BAL AM pools of control Vhlfl/fl and CD11cΔVhl mice. Receptor mice were anesthetized with ketamine (Imalgene, Merial) and xylazine (Rompun, Bayer). Then, a small incision was made to partially expose the trachea and animals were intubated orotracheally with an i.v. catheter (22G, 0.9 × 25 mm; BD Insyte). Cells were inoculated with a pipette in 30 μL of PBS. The viability of AMs was >90% after the last transplantation. Six weeks after transplantation, BAL was performed with 1 mL of PBS, centrifuged at 1,700 rpm for 5 min at RT, and supernatant was used to measure total protein and surfactant protein D (SP-D) concentrations. Additional information can be found in the Supplemental Experimental Procedures.
Flow Cytometry
Stainings were performed at 4°C with the appropriate antibody (Ab) cocktail in cold flow cytometry buffer. Samples were processed with a Spectral Analyzer flow cytometer (SP6800, Sony), and data were analyzed with FlowJo software (Tree Star). Additional information can be found in the Supplemental Experimental Procedures.
Bioinformatics Analysis
GEO: GSE60249 Array Data
Array data (CEL files) (Schneider et al., 2014) were downloaded from GEO and imported into R software with functions provided by the packages GEOquery and oligo. RMA (affy package) was used for data processing, normalization, and calculation of log2 transformed expression values. The array annotation database mogene11sttranscriptcluster.db was used to map expression values with their associated Ensembl gene IDs by selecting for each gene the transcript cluster with the highest median value across all of the samples. Additional information can be found in the Supplemental Experimental Procedures.
RNA-Seq
BAL AMs were pooled from 5–7 mice per genotype and further purified by positive selection with anti-CD11c-microbeads (Miltenyi Biotec), following the manufacturer’s instructions. A total of three pools per genotype were used for RNA-seq. Additional information can be found in the Supplemental Experimental Procedures.
Statistical Analysis
Statistical comparisons were made with Prism version 7 (GraphPad Software, La Jolla, CA). Statistical tests are detailed in the figure legends. p values: ns, not significant; ∗p < 0.05; ∗∗p < 0.01; and ∗∗∗p < 0.001.
Acknowledgments
We are grateful to the members of the David Sancho lab for discussions and critical reading of the manuscript. We are grateful to Andrés Hidalgo for providing DsRed mice, to Stefanie K. Wculek for helping with BM transplantation, and to Natalia Pietrosemoli for support in the in silico analysis. We thank the CNIC facilities and personnel for technical support and S. Bartlett for editorial assistance. H.M.I. was funded by the Spanish Ministerio de Ciencia, Innovación y Universidades (MICINN; BES-2011-044928). Work in the David Sancho laboratory is funded by the CNIC and grant SAF2016-79040-R from MICINN, Agencia Estatal de Investigación, and Fondo Europeo de Desarrollo Regional (FEDER); B2017/BMD-3733 Immunothercan-CM from Comunidad de Madrid; RD16/0015/0018-REEM from FIS-Instituto de Salud Carlos III, MICINN, and FEDER; Acteria Foundation; Constantes y Vitales prize (Atresmedia); La Marató de TV3 Foundation (201723); the European Commission (635122-PROCROP H2020); and the European Research Council (ERC-2016-Consolidator Grant 725091). The CNIC is supported by the MICINN and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Author Contributions
Conceptualization, H.M.I. and D.S.; Methodology, H.M.I. and D.S.; Investigation, H.M.I., P.B., R.C.-G., E.P., M.E., S.M.-C., I.S., and L.C.; Formal Analysis, H.M.I., M.-J.G., D.J.-C., and D.S.; Resources, R.C.-G., S.M.-C., I.S., S.M.-P., and M.G.; Writing – Original Draft, H.M.I. and D.S.; Writing – Review & Editing, all authors; Supervision, Project Administration, and Funding Acquisition, D.S.
Declaration of Interests
The authors declare no competing interests.
Published: August 14, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.07.034.
Data and Software Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE108975.
Supplemental Information
References
- Baker A.D., Malur A., Barna B.P., Ghosh S., Kavuru M.S., Malur A.G., Thomassen M.J. Targeted PPARgamma deficiency in alveolar macrophages disrupts surfactant catabolism. J. Lipid Res. 2010;51:1325–1331. doi: 10.1194/jlr.M001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Pore N., Behrooz A., Ismail-Beigi F., Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- Cramer T., Yamanishi Y., Clausen B.E., Förster I., Pawlinski R., Mackman N., Haase V.H., Jaenisch R., Corr M., Nizet V. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., Lambrecht B.N. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happle C., Lachmann N., Škuljec J., Wetzke M., Ackermann M., Brennig S., Mucci A., Jirmo A.C., Groos S., Mirenska A. Pulmonary transplantation of macrophage progenitors as effective and long-lasting therapy for hereditary pulmonary alveolar proteinosis. Sci. Transl. Med. 2014;6:250ra113. doi: 10.1126/scitranslmed.3009750. [DOI] [PubMed] [Google Scholar]
- Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiyaz H.Z., Williams E.P., Hickey M.M., Patel S.A., Durham A.C., Yuan L.J., Hammond R., Gimotty P.A., Keith B., Simon M.C. Hypoxia-inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J. Clin. Invest. 2010;120:2699–2714. doi: 10.1172/JCI39506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Jeong C.H., Koo S.Y., Son M.J., Song H.S., Bae S.K., Raleigh J.A., Chung H.Y., Yoo M.A., Kim K.W. Determination of hypoxic region by hypoxia marker in developing mouse embryos in vivo: a possible signal for vessel development. Dev. Dyn. 2001;220:175–186. doi: 10.1002/1097-0177(20010201)220:2<175::AID-DVDY1101>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Li C., Wang Y., Li Y., Yu Q., Jin X., Wang X., Jia A., Hu Y., Han L., Wang J. HIF1α-dependent glycolysis promotes macrophage functional activities in protecting against bacterial and fungal infection. Sci. Rep. 2018;8:3603. doi: 10.1038/s41598-018-22039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Montes I., Escobar B., Palacios B., Gómez M.J., Izquierdo-Garcia J.L., Flores L., Jiménez-Borreguero L.J., Aragones J., Ruiz-Cabello J., Torres M., Martin-Puig S. Myocardial VHL-HIF signaling controls an embryonic metabolic switch essential for cardiac maturation. Dev. Cell. 2016;39:724–739. doi: 10.1016/j.devcel.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C., Datta V., Cramer T., Doedens A., Theodorakis E.A., Gallo R.L., Hurtado-Ziola N., Nizet V., Johnson R.S. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L., Drinkwater C.C., Metcalf D., Li R., Köntgen F., Nicola N.A., Begley C.G. Hematopoietic and lung abnormalities in mice with a null mutation of the common beta subunit of the receptors for granulocyte-macrophage colony-stimulating factor and interleukins 3 and 5. Proc. Natl. Acad. Sci. USA. 1995;92:9565–9569. doi: 10.1073/pnas.92.21.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiniotis J., Dinh H., Masendycz P., Turner A., Elsegood C.L., Scholz G.M., Hamilton J.A. Hypoxia prolongs monocyte/macrophage survival and enhanced glycolysis is associated with their maturation under aerobic conditions. J. Immunol. 2009;182:7974–7981. doi: 10.4049/jimmunol.0804216. [DOI] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- Semenza G.L., Jiang B.H., Leung S.W., Passantino R., Concordet J.P., Maire P., Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Sieweke M.H., Allen J.E. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- Sinclair C., Bommakanti G., Gardinassi L., Loebbermann J., Johnson M.J., Hakimpour P., Hagan T., Benitez L., Todor A., Machiah D. mTOR regulates metabolic adaptation of APCs in the lung and controls the outcome of allergic inflammation. Science. 2017;357:1014–1021. doi: 10.1126/science.aaj2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E., Lieschke G.J., Grail D., Metcalf D., Hodgson G., Gall J.A., Maher D.W., Cebon J., Sinickas V., Dunn A.R. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc. Natl. Acad. Sci. USA. 1994;91:5592–5596. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Arumugam P., Sakagami T., Lachmann N., Chalk C., Sallese A., Abe S., Trapnell C., Carey B., Moritz T. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain L., Wottawa M., Hillemann A., Beneke A., Odagiri H., Terada K., Endo M., Oike Y., Farhat K., Katschinski D.M. Prolyl-4-hydroxylase domain 3 (PHD3) is a critical terminator for cell survival of macrophages under stress conditions. J. Leukoc. Biol. 2014;96:365–375. doi: 10.1189/jlb.2HI1013-533R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., O’Dea E.L., Doedens A., Kim J.W., Weidemann A., Stockmann C., Asagiri M., Simon M.C., Hoffmann A., Johnson R.S. Differential activation and antagonistic function of HIF-alpha isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010;24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier S.J., Vanlangenakker N., Vetters J., Carmeliet P., Janssens S., Lambrecht B.N. Opposing regulation and roles for PHD3 in lung dendritic cells and alveolar macrophages. J. Leukoc. Biol. 2017;102:1115–1126. doi: 10.1189/jlb.3A0916-405R. [DOI] [PubMed] [Google Scholar]
- Thomassen M.J., Barna B.P., Malur A.G., Bonfield T.L., Farver C.F., Malur A., Dalrymple H., Kavuru M.S., Febbraio M. ABCG1 is deficient in alveolar macrophages of GM-CSF knockout mice and patients with pulmonary alveolar proteinosis. J. Lipid Res. 2007;48:2762–2768. doi: 10.1194/jlr.P700022-JLR200. [DOI] [PubMed] [Google Scholar]
- van de Laar L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B.N., Guilliams M. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.