Abstract
Background
Staphylococcal food poisoning (SFP) is one of the most common food-borne diseases in the world. Pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST) and spa typing methods were used to characterize Staphylococcus aureus isolates from food surveillance during 2013–2015 in southwest China, and Staphylococcal cassette chromosome mec (SCCmec) typing was used for methicillin-resistant S. aureus (MRSA). Isolates were also examined for their antibiotic resistance and carriage of virulence genes.
Results
Isolation rate of S. aureus was 2.60% during the three years’ surveillance and 29.50% of them were MRSA. All the S. aureus had hla genes (100%), 14.34% of the strains had tst, and 16.73% had PVL. 163 PFGE-SmaI patterns, 41 ST types and 36 spa types were obtained for all the S. aureus. Among them, ST6-t701 (13.15%), ST7-t091 (12.75%), ST59-t437 (9.96%) and ST5-t002 (7.57%) were the prevalent genotypes. Most of MRSA in this study belonged to SCCmec IV and V, accounted for 74.32% and 20.27% respectively. ST6-SCCmec IV-t701 (36.50%) was the most prevalent clone among isolates from food, followed by ST59-SCCmec V-t437 (20.30%), ST5-SCCmec IV-t002 (12.20%) and ST59-SCCmec IV-t437 (12.20%). Some strains had the identical PFGE patterns, ST and spa types with isolates from patients.
Conclusions
S. aureus isolated from food in southwest China displayed heterogeneity. Isolates had the same genotype profiles with isolates from patients, indicating high homology.
Electronic supplementary material
The online version of this article (10.1186/s12866-018-1239-z) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus aureus, Food surveillance, Southwest China
Background
Food-borne diseases (FBD) represent major public health concern worldwide [1, 2]. Staphylococcal food poisoning (SFP) is one of the most common FBD in the world due to the ingestion of staphylococcal enterotoxins (SEs) that are produced by enterotoxigenic strains [3]. In the United States, SFP is the major concern of public health and it is one of the most common causes of reported FBD, causing an estimated 241,000 illnesses per year, imposing a great economic burden [4, 5]. In China, 371 outbreaks of bacterial FBDs have been reported from 2008 to 2010, involving 20,062 cases and 41 deaths [6]. Furthermore, 94 outbreaks of SFP were reported from 2003 to 2007 according to the National Monitoring Network with 2223 individual patients and 1186 hospitalizations. During these SFP outbreaks, Staphylococcus aureus was the fifth most isolated pathogenic bacteria followed by Vibrio parahaemolyticus, Bacillus cereus, Bacillus proteus, and Salmonella [7]. Since 2009, Food safety law of People’s Republic of China was enacted, and S. aureus was one of the bacteria that must be tested according to this law. Therefore, the systemic surveillance of S. aureus for food products is very important.
S. aureus is a commensal and opportunistic pathogen that can cause wide spectrum of infections, from superficial skin infections to severe and invasive disease [8]. The toxigenic S. aureus strains associated with SFP can express one or more of a family of genes coded for heat-stable enterotoxins. These genes showed high sequence homology in their structure and functions [1]. While, other virulence genes such as toxic shock syndrome toxin 1 (TSST-1), exfoliative toxins and cytolytic toxins (leukocidin and hemolysins) can also cause related diseases [9]. At present, the major molecular typing methods for S. aureus were pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), spa typing and Staphylococcal cassette chromosome mec (SCCmec) typing [10–13]. PFGE was considered as the golden method for S. aureus typing, showing a high discriminatory power. MLST was suitable for analyzing the evolutionary relation and clone complex of the strains. SCCmec developed by International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements, (IWG-SCC: www.sccmec.org) was mainly used for genotyping of MRSA [14].
Yunnan is located in southwest China, including 16 cities. The lifestyle and eating habits are specific, and there is no systemic surveillance program for S. aureus in foods at present. Therefore this study was the first analyzing the characteristics of S. aureus isolates from food surveillance between 2013 and 2015 in this province.
Results
Isolation of S. aureus
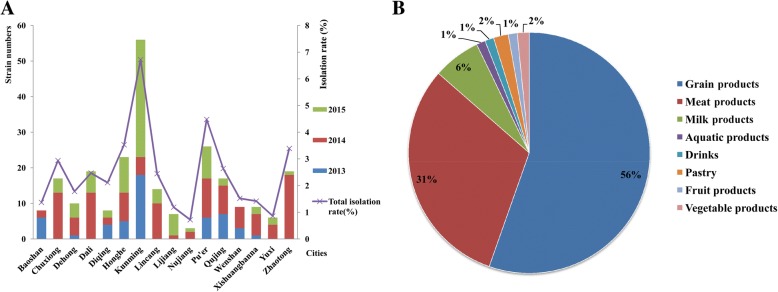
The isolation rate of S. aureus was 2.60% (251/9646) for three years in Yunnan province. The highest isolation rate was 6.72% (56/833) in Kunming city followed by Pu’er 4.48% (26/581), Honghe 3.53% (23/651), Zhaotong 3.39% (19/561), Chuxiong 2.94% (17/579) and Qujing 2.64% (17/645). The lowest isolation rates were 0.72% (3/419) and 0.86% (6/699) observed in Nujiang and Yuxi respectively (Fig. 1a).
Fig. 1.
The isolation results of S. aureus in this study. a: The isolation rates and strain distributions; b: The isolation sources of the strains
A total of 251 S. aureus strains were isolated from food, 56% (139/251) from grain products, 31% (78/251) from meat products and 6% (16/251) from dairy products, these strains accounted for 93% of all the isolates (Fig. 1b). The distribution of strains from different foods had statistical significance among the 16 cities (H = 39.55, p = 0.001).
Antibiotic resistance
Seventy four S. aureus strains (29.50%) were MRSA exhibiting resistance to OXA, carrying the mecA gene. The resistance rate against to the other antibiotic was: 71.10% (180/251) to SXT, 51.00% (128/251) to EM, 46.60% (117/251) to TET, 44.20% (111/251) to CIP, and 37.80% (95/251) to CM. All isolates were sensitive to VAN (Table 1 and Additional file 1). Comparison of antibiotic resistance results between cities and sources, we found that only OXA (H = 30.52, p = 0.010) and GEN (H = 30.63, p = 0.010) had significance in 16 cities, while MXF (H = 29.46, p = 0.000) and TET (H = 33.63, p = 0.000) had statistical difference depending on food sources.
Table 1.
The antibiotics resistant results in this study
| Antibiotics | MICs | Strain numbers | Percent (%) |
|---|---|---|---|
| PCN | ≤0.12 | 177 | 70.50% |
| – | – | – | |
| ≥0.25 | 74 | 29.50% | |
| OXA | ≤2 | 177 | 70.50% |
| – | – | – | |
| ≥4 | 74 | 29.50% | |
| MXF | ≤0.5 | 231 | 92.00% |
| 1 | 1 | 0.40% | |
| ≥2 | 19 | 7.60% | |
| SXT | ≤2/38 | 71 | 28.30% |
| – | – | – | |
| ≥4/76 | 180 | 71.70% | |
| LEV | ≤1 | 78 | 31.10% |
| 2 | 155 | 61.80% | |
| ≥4 | 18 | 7.20% | |
| EM | ≤0.5 | 118 | 47.00% |
| 1–4 | 5 | 2.00% | |
| ≥8 | 128 | 51.00% | |
| LZD | ≤4 | 154 | 61.35% |
| – | – | – | |
| ≥8 | 97 | 38.65% | |
| TET | ≤4 | 134 | 53.40% |
| 8 | 0 | 0 | |
| ≥16 | 117 | 46.60% | |
| CIP | ≤1 | 8 | 3.20% |
| 2 | 132 | 52.60% | |
| ≥4 | 111 | 44.20% | |
| CM | ≤0.5 | 133 | 53.00% |
| 1–2 | 23 | 9.20% | |
| ≥4 | 95 | 37.80% | |
| GEN | ≤4 | 208 | 82.90% |
| 8 | 6 | 2.40% | |
| ≥16 | 37 | 14.70% | |
| RIF | ≤1 | 248 | 98.80% |
| 2 | 0 | 0 | |
| ≥4 | 3 | 1.20% | |
| VAN | ≤2 | 251 | 100% |
| 4–8 | 0 | 0 | |
| ≥16 | 0 | 0 |
Virulence genes
All the S. aureus in this study had hla genes, the positive rate of other enterotoxin genes were sea 30.86% (77/251), seb 49.00% (123/251), sec 17.28% (43/251), sed 57.77% (145/251), seg 19.12% (48/251), seh 11.55% (29/251), sei 34.26% (86/251), sej 15.54% (39/251), sen 14.34% (36/251), sem 17.93% (45/251) and seo 12.75% (32/251). Thirty six strains had toxic shock syndrome toxin gene tst (14.34%), and the positive rate of exfoliative toxin genes was 21.91% (55/251) for eta and 12.35% (31/251) for etb respectively. Forty two isolates (16.73%) had leucocytic toxin gene lukS-PV-lukF-PV (PVL), the positive rate of other virulence genes were hlb 96.30% (242/251), hld 90.12% (226/251), hlg 87.65% (220/251), hlg-2 85.19% (214/251) and edin 35.46% (89/251). The virulence genes results had no statistical significance with source or distributions of the bacteria.
PFGE
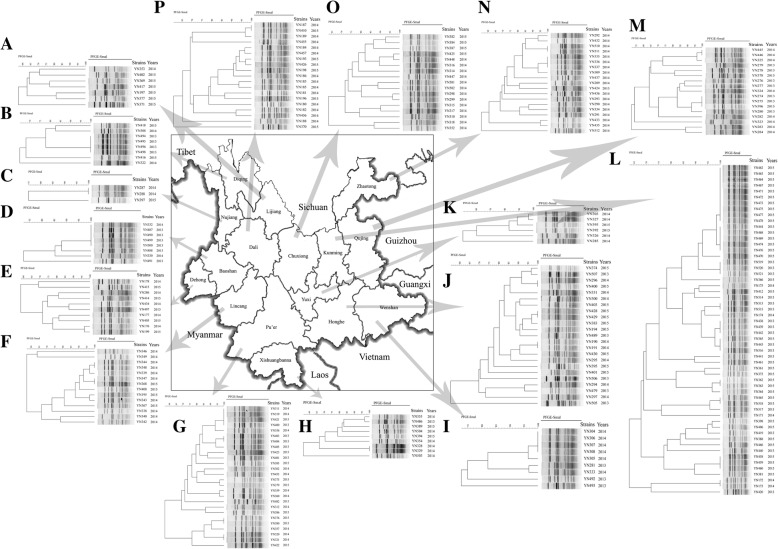
PFGE examination revealed 163 PFGE-SmaI patterns among all the S. aureus isolates. The isolates showed great heterogenous PFGE patterns distribution, especially for Lijiang, Nujiang, Dehong, Lincang, Pu’er, Xishuangbanna, Honghe, Yuxi, Dali, Chuxiong, Zhaotong and Qujing cities, almost no identical PFGE patterns were shown in these cities (Fig. 2a, c, e, f, g, h, j, k, m, n, o and p). However, some strains showed identical patterns, such as YN494, YN495, YN496 and YN498 in Diqing (Fig. 2b); YN332, YN487, YN490, YN499 and YN500 in Baoshan (Fig. 2d.); YN304, YN306, YN307 and YN308 in Wenshan (Fig. 2i); YN462, YN463, YN464, YN467, YN471, YN472, YN473, YN474, YN477 and YN478 in Kunming (Fig. 2l). Specifically, most of these strains were isolated from rice noodles and rice rolls.
Fig. 2.
PFGE cluster results of 251 S.aureus in southwest China. a: Lijiang; b: Diqing; c: Nujiang; d: Baoshan; e: Dehong; f: Lincang; g: Pu’er; h: Xishuangbanna; i: Wenshan; j: Honghe; k: Yuxi; l: Kunming; m: Qujing; n: Zhaotong; o: Chuxiong; p: Dali
MLST
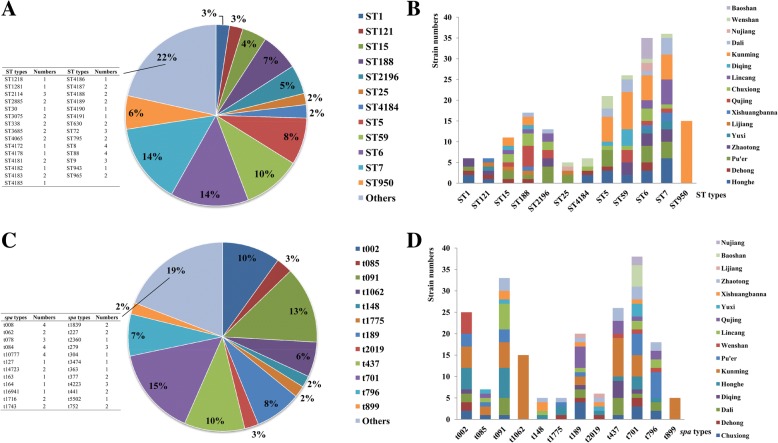
A total of 41 ST types were obtained; among them ST4172, ST4178, ST4181 to ST4191 were isolated for the first time in China (Additional file 1). ST7 (36/251, 14%), ST6 (35/251, 14%), ST59 (26/251, 10%) and ST5 (21/251, 8%) were the most common ST types distributed in Yunnan province, accounted for 46% of all the S. aureus (Fig. 3a). The majority of the ST types were discrete distributed in 16 cities, and each city had different diversified ST types, except the ST950 in Kunming (Fig. 3b).
Fig. 3.
MLST and spa typing results of S. aureus in this study. a: The ST types distributions; b: The ST types distribution in 16 cities; c: The spa types distributions; d: The spa types distribution in 16 cities
Spa typing
A total of 36 spa types were obtained for the 251 S. aureus isolates (Additional file 1); t701 (38/251, 15%), t091 (33/251, 13%), t437 (26/251, 10%) and t002 (25/251, 10%) were the prevalent spa types, accounted for 48% (Fig. 3c). The spa types of isolates also discrete distributed in 16 cities, and each city had different diversified spa types, except t1062 and t899 in Kunming (Fig. 3d).
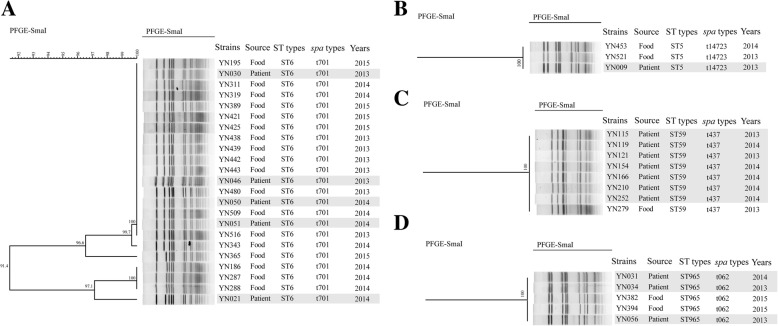
Together analysis MLST with spa typing, ST6-t701 (33/251, 13.15%), ST7-t091 (32/251, 12.75%), ST59-t437 (25/251, 9.96%) and ST5-t002 (19/251, 7.57%) were the most types for all the isolates, accounted for 43.43%. These strains were discrete distributed in different cities, no dominant type was found. The comparison of S. aureus isolates from patients and food was also performed; the results showed that some strains from patients had identical PFGE patterns, ST and spa types with food isolates, indicated high homology relationship. These strains were specifically for ST6-t701 (Fig. 4a), ST5-t14723 (Fig. 4b), ST59-t437 (Fig. 4c) and ST965-t062 (Fig. 4d). Among them, ST6-t701 was the prevalent type, since five strains of patients from our database showed identical patterns with food strains and the majority of food strains were isolated from rice noodles and rice rolls.
Fig. 4.
PFGE cluster results for patients strains with food strains. a: ST6-t701 type; b: ST5-t14723 type; c: ST59-t437 type; d: ST965-t062 type. The grey areas of figure indicated the S. aureus isolated from patients
SCCmec typing of MRSA
SCCmec typing was performed on 74 MRSA isolates in this study. Among them, type IV and V were found between all these strains. 74.32% of all MRSA were type IV (55/74), 20.27% were type V (15/74), and four isolates could not be assigned (Table 2). Combined analysis of MLST, spa and SCCmec types for MRSA indicated ST6-SCCmec IV-t701 (36.50%) was the most prevalent clone among MRSA isolates from food, followed by ST59-SCCmec V-t437 (20.30%), ST5-SCCmec IV-t002 (12.20%) and ST59-SCCmec IV-t437 (12.20%).
Table 2.
Molecular types of MRSA isolates from food in this study
| MLST | spa type | SCCmec | Numbers | ||
|---|---|---|---|---|---|
| IV | V | NA | |||
| ST1 | t1775 | 4(5.40%) | – | – | 4 |
| ST5 | t002 | 9(12.20%) | – | – | 9 |
| ST6 | t701 | 27(36.50%) | – | – | 27 |
| ST7 | t091 | 4(5.40%) | – | 4(5.40%) | 8 |
| ST59 | t437 | 9(12.20%) | 15(20.30%) | – | 24 |
| ST88 | t10777 | 2(2.60%) | – | – | 2 |
Discussion
SEs produced by toxigenic strains are predisposing causes of SFP, among them, SEA is the most common ones. In this study, sea, seb, sec and sed were the most found toxin genes, 14.34% of the strains had tst, and 16.73% had PVL. Some similar results could be found from other Chinese studies, for example, Chao et al. [15] analyzed 568 isolates from different sources for the prevalence of SEs genes and performed spa typing. Approximately 54.40% of the isolates from different sources harbored one or more SE genes. Seven genes, sea, seb, seg, seo, sem, seq, and sel were more frequently detected. In Li et al. study, [16] the most detected toxin genes were seu, lukED, hla, hlb and hld, followed by hlg, hlgv, lukPV, sea, see, etd, seb, sec, sed, sej, and sek. Although S. aureus strains in this study were isolated from food surveillance and not from SFP outbreak or patients, the high carriage rates of virulence genes indicated high risk for SFP in case of improper food handling and compromised good hygienic practices during food production and storage.
Several studies showed that high levels of MRSA were found on US and European farms and commercially-distributed meats [17–19]. The prevalence of MRSA isolated from raw retail meat products was reported, ranging from less than 1% in Asia [20] to 11.9% in the Netherlands [21], with intermediate prevalence in other studies [22, 23]. A recent study found that 45% of pork and 63% of beef products were positive for S. aureus in the United States [24]. However, other studies reported low isolation rates of MRSA in foods. El-Bayomi et al. [25] analyzed 110 samples from chicken products, and 30 samples were positive for S. aureus. Examination of virulence-associated factors revealed that 31.3% of isolates harbored the PVL gene, whereas 10.4% were positive for the sea and sed genes each. Thapaliya et al. [26] investigated S. aureus and MRSA in commercially-distributed raw meat products. The prevalence of S. aureus on meat samples was 27.8%. Higher prevalence of MRSA was found in meat samples. Among the S. aureus isolates, 18 were PVL-positive and 41 for the mecA gene. Al-Ashmawy et al. [27] analyzed the MRSA isolated from raw milk and dairy products in Egypt, and the average isolation rate of MRSA was 53% among all milk and dairy products, with 35, 50, 40, 65 and 75% in yogurt samples, ice cream, Kareish cheese, Damietta cheese, and raw milk, respectively. In our study, 29.50% of isolates were MRSA, being a higher isolation rate to the reported in the above study. ST6-SCCmec IV-t701 was the most prevalent clone among MRSA isolates from food, followed by ST59-SCCmec V-t437, ST5-SCCmec IV-t002 and ST59-SCCmec IV-t437. Most of these strains were from grains and meat products. Grains are the favorite food for Chinese population; it can be processed into a variety of products, such as steamed bread, noodles, bean jelly, rice noodles and rice rolls etc. Among them, rice noodles, rice rolls are the most favorite foods in southwest China, people lived there eat these foods almost every day, and they eat them raw and cold. The comparison between food and patients’ isolates, where a high homology was observed for ST6-t701 types isolated from rice noodles and rice rolls, indicated the possible circulation of those types between patients and foods. Therefore, the origin and distribution of MRSA or S. aureus are different between countries probably due to their specific eating habits and the resources of food.
Some studies performed in Europe indicated that ST398 was the most common MRSA type in retail meat, which might be a potential transmission factor of ST398 from farm to the community [28]. Yan et al. [29] analyzed 52 S. aureus isolates from 11 outbreaks in Shenzhen, China. They found that ST6 was the major ST type, and constituted 63.5% of all the strains in seven outbreaks. The second most commonly found ST type was ST943, constituted 23.1% of all the isolates from three outbreaks. The dominant spa types were t701, t091 and t2360, accounting 67.3% of the isolates. Li et al. [16] investigated the molecular epidemiology of S. aureus isolates from seven SFP outbreaks between 2006 and 2013 in Xi’an, northwest China. The results showed that ST6-t701 (71.4%) was the dominant clone, followed by ST5-t002 (14.3%) and ST59-t172 (14.3%). The antibiotic resistance results showed that 71.4% of the strains were resistant to trimethoprim and penicillin, 28.6% resistant to erythromycin, and 14.3% to ampicillin, clindamycin, and tetracycline. All the isolates were sensitive to chloramphenicol, cefoxitin, oxacillin, amikacin, and vancomycin. Furthermore, studies referred to MRSA circulated in the China hospitals revealed that ST59-SCCmec IV-t437 was the most common epidemic clone [30, 31]. All these researches mentioned above indicated that ST6-SCCmec IV-t701 and ST59-SCCmec IV-t437 were predominant MRSA genotypes for patients. In this study, ST6-t701, ST7-t091, ST59-t437 and ST5-t002 were the prevalent types of the isolates, accounted for 43.43%. ST6-SCCmec IV-t701, ST59-SCCmec V-t437, ST5-SCCmec IV-t002 and ST59-SCCmec IV-t437 were the most prevalence MRSA clones. The strains showed great heterogenous distributions characteristics; however, ST6-t701 (or ST6-SCCmec IV-t701) was the dominant type in southwest China, because of their homology with strains isolated from patients and the aforementioned outbreaks.
This study was the first systemic S. aureus food surveillance in southwest China, in which the molecular characteristics of strains were determined. However, there were several limitations in this study. Firstly, the study was conducted in urban regions that probably showed a poor representation of the potential S. aureus food surveillance. Secondly, the surveillance time was only for three years, which might not fully reflect the characteristics of local strains. Therefore, further research including urban, rural, and other food sources might be done to comprehensively evaluate the prevalence and features of S. aureus.
Conclusions
Two hundred fifty-one S. aureus for food surveillance were analyzed from 2013 to 2015 in southwest China. Isolation rate of S. aureus was 2.60% during the three years’ surveillance and 29.50% of them were MRSA. 163 PFGE-SmaI patterns were obtained for all the strains and the isolates displayed great heterogenous distributions. ST6-t701 (13.15%), ST7-t091 (12.75%), ST59-t437 (9.96%) and ST5-t002 (7.57%) were the most types for all the isolates, scattered distributed in different cities. ST6-SCCmec IV-t701 was the most prevalent clone among MRSA isolates from food, followed by ST59-SCCmec V-t437, ST5-SCCmec IV-t002 and ST59-SCCmec IV-t437. Some strains had the identical PFGE patterns, ST and spa types with isolates from patients. S. aureus isolated from food in southwest China displayed heterogeneity. Isolates had the same genotype profiles with isolates from patients, indicating high homology. Future research including urban, rural, and other food sources might be done to comprehensively evaluate the prevalence and features of S. aureus in southwest China.
Methods
Sample collection and isolation of S. aureus
The collection of samples used in this study was complied with national regulation on risk monitoring and management of food safety guidelines, and permitted by Center for Food Safety Risk Assessment of Yunnan Province. A total of 9646 food samples were collected from 16 cities in Yunnan province, from 2013 to 2015. The types of foods included grain products, meat products, milk products, fishery products, drinks, pastry, fruit products and vegetable products etc. The food samples were processed based on S. aureus isolation methods for foods of national standards in China (GB/T 4789.10–2010). We used qualitative method to isolate the strains. Each sample was aseptically weighed in an analytical balance, twenty-five grams were placed into a sterile plastic bag and 225 mL of buffered peptone water (Oxoid, UK) was added, then homogenized in a Stomacher Bagmixer 400 W (Interscience, France) for one minute. The subsequent enrichments and isolation process were according to the method described by Puah et al. [32]. All the isolated S. aureus were identified by Vitek Compact 2 (bioMérieux, France). S. aureus isolated from patients from hospital surveillance of Yunnan province were used for comparison to the food isolates.
Antibiotic resistance test
Minimum inhibitory concentrations (MICs) for 13 antibiotics was determined through broth micro-dilution method using customized microtiter plates (Sensititre, UK) according to the manufacturers’ instructions: Penicillin (PCN), Oxacillin (OXA), Gentamicin (GEN), Ciprofloxacin (CIP), Levofloxacin (LEV), Moxifloxacin (MXF), Erythromycin (EM), Clindamycin (CM), Linezolid (LZD), Vancomycin (VAN), Tetracycline (TET), Rifampicin (RIF) and Trimethoprim/Sulfamethoxazole (SXT). The tests were performed and interpreted in accordance to the Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-S25, 2015); S. aureus ATCC 29213 was used as quality control. The mecA gene was detected by PCR, as described by Velasco et al. [33].
Virulence genes detection
The genomic DNA of the bacteria was extracted using Bacterial Genomic Extraction Kit (Tiangen, Beijing) followed the manufacturer’s instructions. The virulence genes were detected based on previous study [34]. The enterotoxin, toxic shock syndrome toxin, exfoliative toxin, leucocytic toxin, hemolysin and epidermic cell differential inhibition gene were all amplified (MyCycler, Bio-Rad). Taq premix (TaKaRa, Japan) was used, and the amplification procedures were 94 °C 5 min followed by 30 cycles: 94 °C 15 s, 55 °C 30s, 72 °C 30s, and finally 72 °C 10 min. Electrophoresis of the amplified products was performed in 1.5% agarose, and detected by Gel Imaging instrument (Bio-Rad, USA).
Pulsed-field gel electrophoresis
PFGE was performed according to Coombs et al. [35], and each plug was digested with 50 U SmaI (TaKaRa, Japan) at 37 °C for 4 h. CHEF-Mapper (Bio-Rad, USA) was used for electrophoresis, and the pulse time ranged from 4 s to 40 s for 19 h. The gel was stained with Gel Red (Biotium) and visualized using a gel imaging system (Bio-Rad, Gel DocXR). PFGE patterns were analyzed using BioNumerics version 6.6, and a dendrogram was constructed using the Dice coefficient and un-weighted pair group methods with the arithmetic mean algorithm (UPGMA).
Multilocus sequence typing
MLST was performed according to MLST database (http://saureus.mlst.net) and Enright et al. method [12]. The amplification products were sent for bidirectional sequencing by TaKaRa, Japan; and the sequences were analyzed using DNAStar and MEGA 4.0 software. All the sequences were submitted to MLST database for identifying their allele and ST assignments.
Spa typing
The genomic DNA of the bacteria was extracted as described above. PCR amplification and sequencing primers were used according to Harmsen et al. method [36]. The amplification products were sent for bidirectional sequencing by TaKaRa, Japan, and the sequences were submitted to the database for their spa type assignments (http://spatyper.fortinbras.us/).
SCCmec typing
SCCmec typing was performed by discriminating the mec complex and the cassette chromosome recombinase (ccr) genes complex as described previously [37], which was based on a set of multiplex PCR methods. SCCmec types were assigned according to the combination of the mec class and ccr type. MRSA strains which could not be assigned were defined as non-typable (NA).
Statistical analysis
Data analysis was performed by IBM SPSS software (version 19.0 for Windows, Armonk, NY). Non-parameter analysis was performed using a Kruskal-Wallis H test in SPSS statistics package. Two-sided significance was assessed for all variables, applying a cut-off value of P<0.05.
Additional file
The bacterial information, antibiotic resistance and molecular typing results for all the strains used in this study. (XLS 121 kb)
Acknowledgements
We sent our manuscript to American Journal Experts (www.aje.com) for English language revisions.
Funding
This work was supported by Yunnan Science and Technology Talent and Platform Program (2017HC019) and National Sci-Tech key project (2017ZX10103010). The funders have no role in the design, collection, analysis, interpretation of data and in writing the manuscript of the study.
Availability of data and materials
All data generated or analysed during this study are included in this published article (Additional file 1).
Abbreviations
- MLST
Multilocus sequence typing
- MRSA
Methicillin-resistant S. aureus
- PFGE
Pulsed field gel electrophoresis
- SEs
Enterotoxins
- SFP
Staphylococcal food poisoning
Authors’ contributions
WX, ZY and WG conceived and designed the experiments; FL, ZM, LF and YG performed the experiments; CL performed the statistical analysis; FL and XF wrote the paper. JD gave a critical reading and important suggestions of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Feng Liao, Email: lf_0305@163.com.
Wenpeng Gu, Phone: +86087163632033, Email: gu_02788@163.com.
Zushun Yang, Email: yncdcjck@163.com.
Zhishuo Mo, Email: vbstone@126.com.
Lu Fan, Email: 273886578@qq.com.
Yidan Guo, Email: 369402316@qq.com.
Xiaoqing Fu, Email: yncdcfxq@163.com.
Wen Xu, Email: yncdcxuwen@sohu.com.
Chaoqun Li, Email: 78430359@qq.com.
Jiejie Dai, Email: djj@imbcams.com.cn.
References
- 1.Balaban N, Rasooly A. Staphylococcal enterotoxins. Int J Food Microbiol. 2000;61(1):1–10. doi: 10.1016/S0168-1605(00)00377-9. [DOI] [PubMed] [Google Scholar]
- 2.Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003;2(1):63–76. [PubMed] [Google Scholar]
- 3.Hennekinne JA, De Buyser ML, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev. 2012;36(4):815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 4.Murray RJ. Recognition and management of Staphylococcus aureus toxin-mediated disease. Intern Med J. 2005;35(Suppl 2):S106–S119. doi: 10.1111/j.1444-0903.2005.00984.x. [DOI] [PubMed] [Google Scholar]
- 5.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam HM, Remais J, Fung MC, Xu L, Sun SS. Food supply and food safety issues in China. Lancet. 2013;381(9882):2044–2053. doi: 10.1016/S0140-6736(13)60776-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Duan H, Zhang W, Li JW. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol Med Microbiol. 2007;51(1):8–13. doi: 10.1111/j.1574-695X.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 8.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 9.Argudin MA, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2010;2(7):1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babouee B, Frei R, Schultheiss E, Widmer AF, Goldenberger D. Comparison of the DiversiLab repetitive element PCR system with spa typing and pulsed-field gel electrophoresis for clonal characterization of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2011;49(4):1549–1555. doi: 10.1128/JCM.02254-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y, Kong F, Wang Q, Tong Z, Sintchenko V, Zeng X, Gilbert GL. Comparison of single- and multilocus sequence typing and toxin gene profiling for characterization of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45(10):3302–3308. doi: 10.1128/JCM.01082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185(11):3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53(12):4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao G, Bao G, Cao Y, Yan W, Wang Y, Zhang X, Zhou L, Wu Y. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int J Food Microbiol. 2015;211:142–147. doi: 10.1016/j.ijfoodmicro.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Wu S, Luo W, Su Y, Luan Y, Wang X. Staphylococcus aureus ST6-t701 isolates from food-poisoning outbreaks (2006-2013) in Xi'an, China. Foodborne Pathog Dis. 2015;12(3):203–206. doi: 10.1089/fpd.2014.1850. [DOI] [PubMed] [Google Scholar]
- 17.Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128(3–4):298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Monaco M, Pedroni P, Sanchini A, Bonomini A, Indelicato A, Pantosti A. Livestock-associated methicillin-resistant Staphylococcus aureus responsible for human colonization and infection in an area of Italy with high density of pig farming. BMC Infect Dis. 2013;13:258. doi: 10.1186/1471-2334-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien AM, Hanson BM, Farina SA, Wu JY, Simmering JE, Wardyn SE, Forshey BM, Kulick ME, Wallinga DB, Smith TC. MRSA in conventional and alternative retail pork products. PLoS One. 2012;7(1):e30092. doi: 10.1371/journal.pone.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon NH, Park KT, Jung WK, Youn HY, Lee Y, Kim SH, Bae W, Lim JY, Kim JY, Kim JM, et al. Characteristics of methicillin resistant Staphylococcus aureus isolated from chicken meat and hospitalized dogs in Korea and their epidemiological relatedness. Vet Microbiol. 2006;117(2–4):304–312. doi: 10.1016/j.vetmic.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 21.de Boer E, Zwartkruis-Nahuis JT, Wit B, Huijsdens XW, de Neeling AJ, Bosch T, van Oosterom RA, Vila A, Heuvelink AE. Prevalence of methicillin-resistant Staphylococcus aureus in meat. Int J Food Microbiol. 2009;134(1–2):52–56. doi: 10.1016/j.ijfoodmicro.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 22.van Loo IH, Diederen BM, Savelkoul PH, Woudenberg JH, Roosendaal R, van Belkum A, Lemmens-den Toom N, Verhulst C, van Keulen PH, Kluytmans JA. Methicillin-resistant Staphylococcus aureus in meat products, the Netherlands. Emerg Infect Dis. 2007;13(11):1753–1755. doi: 10.3201/eid1311.070358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozano C, Lopez M, Gomez-Sanz E, Ruiz-Larrea F, Torres C, Zarazaga M. Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. J Antimicrob Chemother. 2009;64(6):1325–1326. doi: 10.1093/jac/dkp378. [DOI] [PubMed] [Google Scholar]
- 24.Jackson CR, Davis JA, Barrett JB. Prevalence and characterization of methicillin-resistant Staphylococcus aureus isolates from retail meat and humans in Georgia. J Clin Microbiol. 2013;51(4):1199–1207. doi: 10.1128/JCM.03166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Bayomi RM, Ahmed HA, Awadallah MA, Mohsen RA, Abd El-Ghafar AE, Abdelrahman MA. Occurrence, virulence factors, antimicrobial resistance, and genotyping of Staphylococcus aureus strains isolated from chicken products and humans. Vector Borne Zoonotic Dis. 2016;16(3):157–164. doi: 10.1089/vbz.2015.1891. [DOI] [PubMed] [Google Scholar]
- 26.Thapaliya D, Forshey BM, Kadariya J, Quick MK, Farina S, A OB, Nair R, Nworie A, Hanson B, Kates A, et al. Prevalence and molecular characterization of Staphylococcus aureus in commercially available meat over a one-year period in Iowa, USA. Food Microbiol. 2017;65:122–129. doi: 10.1016/j.fm.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Al-Ashmawy MA, Sallam KI, Abd-Elghany SM, Elhadidy M, Tamura T. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathog Dis. 2016;13(3):156–162. doi: 10.1089/fpd.2015.2038. [DOI] [PubMed] [Google Scholar]
- 28.Fessler AT, Kadlec K, Hassel M, Hauschild T, Eidam C, Ehricht R, Monecke S, Schwarz S. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl Environ Microbiol. 2011;77(20):7151–7157. doi: 10.1128/AEM.00561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan X, Wang B, Tao X, Hu Q, Cui Z, Zhang J, Lin Y, You Y, Shi X, Grundmann H. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl Environ Microbiol. 2012;78(18):6637–6642. doi: 10.1128/AEM.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Z, Gu FF, Guo XK, Ni YX, He P, Han LZ. Antimicrobial resistance and molecular characterization of Staphylococcus aureus causing childhood pneumonia in shanghai. Front Microbiol. 2017;8:455. doi: 10.3389/fmicb.2017.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Sun S, Yang C, Chen H, Yin Y, Li H, Zhao C, Wang H. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Front Microbiol. 2018;9:332. doi: 10.3389/fmicb.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puah SM, Chua KH, Tan JA. Virulence factors and antibiotic susceptibility of Staphylococcus aureus isolates in ready-to-eat foods: detection of S. Aureus contamination and a high prevalence of virulence genes. Int J Environ Res Public Health. 2016;13(2):199. doi: 10.3390/ijerph13020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasco V, Sherwood JS, Rojas-Garcia PP, Logue CM. Multiplex real-time PCR for detection of Staphylococcus aureus, mecA and Panton-valentine Leukocidin (PVL) genes from selective enrichments from animals and retail meat. PLoS One. 2014;9(5):e97617. doi: 10.1371/journal.pone.0097617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70(2):631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coombs GW, Monecke S, Ehricht R, Slickers P, Pearson JC, Tan HL, Christiansen KJ, O'Brien FG. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob Agents Chemother. 2010;54(5):1914–1921. doi: 10.1128/AAC.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The bacterial information, antibiotic resistance and molecular typing results for all the strains used in this study. (XLS 121 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article (Additional file 1).