Abstract
Protein G can be a valuable binding agent for antibodies and immunoglobulins in methods such as immunosensors, chromatographic-based immunoassays, and immunoaffinity chromatography. This report used the method of peak decay analysis along with frontal analysis and zonal elution studies to characterize the binding, elution and regeneration properties of affinity microcolumns that contained immobilized protein G. Frontal analysis was employed with rabbit immunoglobulin G (IgG) to characterize the binding capacity of these affinity microcolumns. Zonal elution experiments looking at the retained peaks for small injections of labeled rabbit IgG were used to optimize the column regeneration conditions. Peak decay analysis was then used to look at the effects of flow rate and elution pH on the release of several types of IgG from the protein G microcolumns. This approach made it possible to obtain detailed information on the use and behavior of such columns, as could be used in future work to optimize the capture or analysis of IgG and antibodies by such devices. The same approach and tools that were used in this report could also be adapted for work with affinity columns that make use of other supports, binding agents or targets.
Keywords: protein G, immunoglobulin G, peak decay method, frontal analysis, dissociation rate, affinity microcolumn
1. Introduction
Supports that contain immobilized or adsorbed antibodies have been popular for many years in analytical techniques for measuring various targets [1-10]. Examples of flow-based methods in which such supports have been used have included some types of immunosensors, chromatographic immunoassays, immunoaffinity chromatography and even some approaches for studying biological interactions [1-5]. Some attractive features of these methods are the high affinity and specificity with which antibodies can bind to their complementary targets and the ability of these agents to be employed with various types of labels and detection formats [7].
A chromatographic method that employs antibodies with supports such as HPLC-grade silica or monoliths is often referred to as high-performance immunoaffinity chromatography (HPIAC) [4,9-12]. One common approach for placing antibodies onto these supports is by using covalent immobilization; however, this method can result in some loss of an antibody’s activity through multi-site attachment or incorrect orientation of the antibody [8,12]. An alternative approach that avoids or minimizes these effects is to instead use biospecific adsorption of the antibody to a secondary immobilized binding agent such as protein G [12]. Protein G is a protein found in the cell walls of group G Streptococci bacteria that can bind tightly to the constant region of many types of immunoglobulins and antibodies [8,13-14]. This feature, plus the ability to release the bound antibodies through a decrease in pH and to later apply a fresh batch of antibodies, has made protein G and related immunoglobulin-binding proteins useful as tools for the capture, analysis or utilization of antibodies in chromatographic systems [8,9,15-18].
The development and optimization of methods based on protein G supports ideally requires information on such factors as the retention and elution properties of these materials when they are employed with antibodies (or, in the more general sense, immunoglobulins). Methods that have been used to study the rates of these or other biological interactions in chromatographic systems have included the split-peak method and various methods based on peak fitting or band-broadening measurements [19-22]. This report will examine the use of a technique known as the peak decay method [23,24] to study the dissociation of various types of immunoglobulins from immobilized protein G. In this technique, a small pulse of an analyte (e.g., an antibody/immunoglobulin) is injected onto an affinity column that contains the binding agent of interest. A mobile phase is then introduced onto the column under conditions that disrupt binding of the analyte with the immobilized agent and prevents re-association of the analyte with the column. This type of dissociation can often be produced by changing the mobile phase pH or by adding a displacement agent to the mobile phase. As the analyte is released from the column, it produces a decay curve that can be used to determine the dissociation rate constant for the analyte from the immobilized agent under the given elution conditions [23,24]. Although this method has been used in previous work to examine antibody-antigen interactions [4], it has not been used in prior work to study the protein G and its interactions with antibodies/immunoglobulins.
This report will examine the extension and use of the peak decay method to study the elution and dissociation kinetics of various types of immunoglobulin G (IgG) from protein G that has been immobilized onto HPLC-grade silica and placed into affinity microcolumns (i.e., columns containing an immobilized binding agent and with volumes in the low-to-mid microliter range) [22]. The general principles of this method will be discussed, along with various practical factors to consider in the use of this technique. This method will then be employed to examine the elution of various types of IgG from protein G microcolumns, thus providing new fundamental information on these interactions. The effect of the elution flow rate and pH will also be considered with the goal of improving the characterization and optimization of these microcolumns for future use in antibody- or immunoassay-related applications.
2. Experimental
2.1. Materials
The 3-glycidoxypropyltrimethoxysilane, periodic acid, sodium cyanoborohydride and sodium borohydride were from Sigma-Aldrich (St. Louis, MO, USA). The following types of IgG were also obtained from Sigma–Aldrich: rabbit IgG (> 95% pure), goat IgG (>95%), human IgG (>95%), and mouse IgG (> 95%). The protein G (recombinant, albumin binding domains removed) was purchased from Pierce (Rockford, IL, USA). The amount of immobilized protein on each support was determined in triplicate by using a bicinchoninic acid (BCA) protein assay, which was conducted by using reagents that were also obtained from Pierce. These immunoglobulins were labeled with N–hydroxysuccinimide (NHS) ester-activated fluorescein from Pierce. Other sources of protein G and IgG, or related binding agents and targets, can also be used for the peak decay method, as well as other types of fluorescent or chemical labels. All running buffers and aqueous solutions that were used in this report were prepared using deionized water, as was obtained in the following examples by using an EMD MILLI-Q water purification system using 0.2 ¼m GNWP nylon filters, both of which were from Millipore (Billerica, MA, USA).
2.2. Apparatus
Many standard HPLC systems can be adapted for use in the peak decay measurements. The HPLC system that was used in this particular study was a Jasco 2000 system (Easton, MD, USA) that contained a DG-2080-53 three-solvent degasser, three PU-2080 isocratic pumps, an AS-2057 autosampler equipped with a 100 μL sample loop (operated in the partial loop injection mode), a UV-2075 absorbance detector, and a FP-2020 fluorescence detector. Two Advantage PF six-port switching valves (Rheodyne, Cotati, CA) were used for alternating passage of an IgG solution and acidic or neutral buffer solutions through the microcolumns during the frontal analysis studies. The system components were controlled by a Jasco LC-Net II/ADC system and a Jasco ChromNav chromatography data system. The breakthrough times for the frontal analysis data and elution profiles were examined by using PeakFit 4.12 (SeaSolve Software, San Jose, CA).
All binding studies were carried out at room temperature (25° C) in this report. Work at other temperatures can also be conducted by using an on-line column heater or a circulating water bath and column jacket for temperature control [1]. The elution of unlabeled IgG was monitored at 280 nm. Detection of fluorescein-labeled IgG was monitored by using an excitation wavelength of 494 nm and an emission wavelength of 518 nm.
Purification of the labeled IgG was performed by using Zeba spin columns (7 kDa MW cutoff, 0.7–4 mL sample capacity) from Pierce, along with a 5702RH temperature-controlled centrifuge from Eppendorf (New York, NY, USA) and a fixed-angle centrifuge rotor from VWR (West Chester, PA, USA). The microcolumns were packed using an HPLC slurry packing system from ChromTech (Apple Valley, MN, USA); however, other column packing systems can also be used for such work.
2.3. Antibody labeling
IgG was labelled with NHS-fluorescein, according to the manufacturer’s instructions. In this process, the initial protein solution was prepared by dissolving 5 mg IgG in 5 mL of pH 8.5, 0.10 M potassium phosphate buffer (i.e., giving a 1.0 mg/mL IgG solution). A 1 mg portion of NHS-fluorescein was dissolved in 100 μL dimethylformamide, and 25 μL of this NHS-fluorescein solution was added to 5 mL of the pH 8.5 IgG solution, resulting in a reaction mixture that contained 0.05 mg NHS-fluorescein per mg IgG. This mixture was allowed to shake for 1 h in the dark at room temperature.
Zeba spin columns were utilized to purify and separate the labeled IgG from any unreacted NHS-fluorescein. Prior to use, each spin column was washed three times with pH 7.4, 0.067 M potassium phosphate buffer for buffer exchange. The labeled IgG solution was then loaded into two spin columns and centrifuged at 1000 × g for 2 min. The labeled IgG solutions that remained in the spin columns were then pooled for further use. The label/protein ratio and concentration of the final labeled IgG solution were determined by making absorbance measurements at 494 and 280 nm, according to the manufacturer’s directions. The final labeled IgG solutions had the following measured concentrations: rabbit IgG, 0.76 mg/mL (5.4 μM); mouse IgG, 0.81 mg/mL (5.1 μM); goat IgG, 0.77 mg/mL (5.2 μM); and human IgG, 0.92 mg/mL (6.1 μM). The sample solutions contained 3–6 (average, 5) moles of label per mol of IgG. The labeled IgG solutions were stored at 4°C in pH 7.4, 0.067 M potassium phosphate buffer when not in use. These labeled IgG conjugates and were stable for up to 2 weeks when protected from light and stored under these conditions [25].
2.4. Support and microcolumn preparation
The method that is described in this article can be employed with a variety of supports and immobilization schemes. The specific examples that are described here were all conducted using Nucleosil Si-1000-7 silica (7 μm particle size, 1000 Å pore size) that was purchased from Macherey-Nagel (Duren, Germany). A pore size of 1000Å was chosen for this support to allow sufficient room for the immobilization of protein G and the later binding of this agent to antibodies. Although silica with a smaller pore size (e.g., 50–500Å) could be employed to make protein G supports, and it has been shown that supports with the larger pore sizes that were used here allow for maximum binding (on a mol-per-mol basis) of IgG-class antibodies to immobilized protein G [26].
This silica was first converted into a diol-bonded form to provide a material that had low non-specific binding for most biological compounds but that still could be further modified for the immobilization of a binding agent such as protein G [12,27]. This material was made as described previously [27], by mixing 1.00 g of bare silica with 4.0 mL of pH 5.5, 0.10 M sodium acetate buffer. This mixture was sonicated under vacuum for at least 5 min to remove any air bubbles that were trapped inside the pores of the support and to allow all the support’s surface to be available for immobilization. This mixture was then combined with 50 μL of 3-glycidoxypropyltrimethoxysilane and shaken in a water bath at 90°C for approximately 5 h. The silica was then washed four times with water and refluxed in a diluted solution of sulfuric acid (pH 3.0) for 1 h. The resulting product was washed with water, dried in a vacuum oven at 50°C, and stored in a desiccator until further use.
The protein G that was used in this work was immobilized by using the Schiff base method [28-30], although other methods for coupling proteins to supports such as silica could also have been utilized [12,29]. In this approach, also known as reductive amination, the diol-bonded silica was oxidized to give an aldehyde-activated form by mixing 100 mg of the silica with 100 mg of periodic acid in 2 mL of a 90% acetic acid solution [5,27-28]. This mixture was allowed to shake at room temperature for 2 h. The support was next washed three times with water, followed by additional washes with pH 6.0, 0.10 M potassium phosphate buffer. The aldehyde-activated silica was then mixed with 2.5 mL of a 1.2 mg/mL protein G solution (i.e., 3.0 mg protein G) that had been prepared in pH 6.0, 0.10 M potassium phosphate buffer and that included 250 mg sodium cyanoborohydride as a mild reducing agent (Note: this reducing agent is mild enough to not reduce the aldehyde groups on the activated silica or the disulfide bridges of a protein but will reduce a Schiff base). This mixture was allowed to shake for three days at 4°C. At the end of this immobilization step, the support was washed four times with water, three times with pH 8.0, 0.10 M potassium phosphate buffer, and shaken for 90 min in the presence of 2.5 mL of a 2 mg/mL solution of sodium borohydride in pH 8.0, 0.10 M potassium phosphate buffer, which was used to remove any remaining aldehyde groups on the silica. The final support was washed thoroughly with water and pH 7.4, 0.067 M potassium phosphate buffer and stored in the same buffer at 4°C until use. A separate control support was prepared using the same initial starting material but without the addition of any protein G during the immobilization step.
Stainless steel microcolumns in Delrin housing and with PEEK-lined frits were downward slurry packed with the protein G silica or the control support at packing pressures of approximately 3000-4000 psi and using a neutral buffer as the packing solution (e.g., a pH 7.0–7.4 phosphate buffer). The column size that was used in this study was 5 mm × 2.1 mm I.D. These microcolumns were stored in a neutral pH phosphate buffer at 4°C when not in use. The types of protein G microcolumns that were used in this study have been found to be stable for up to one year and over approximately 200 application and elution cycles [6].
2.5. Chromatographic studies
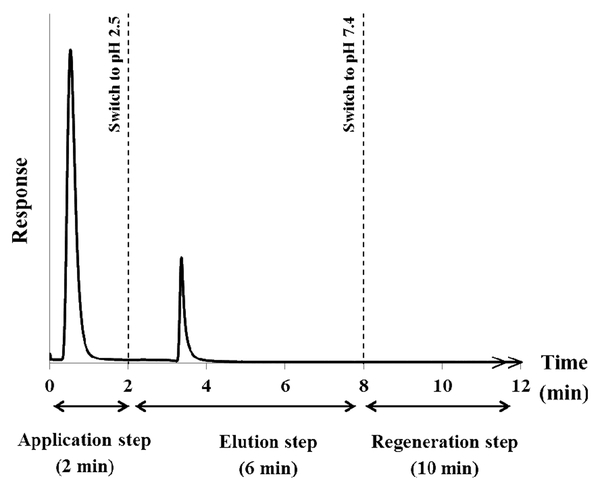
The chromatogram in Figure 1 shows the sequence that was used in this study to apply the IgG samples, elute the retained IgG, and regenerate a microcolumn that contained protein G. This format is based on the on/off elution scheme that is common in HPIAC and other types of affinity chromatography [2-5]. The first step in this scheme involved application of the target analyte (e.g., IgG) to the column. In this study, this involved injecting 20 μL of 0.20 mg/mL labeled IgG onto the protein G microcolumn at 0.50 mL/min and using pH 7.4, 0.0670 M potassium phosphate buffer as the application buffer. It was found for the type of Support and microcolumn that were used in this report that only 2 min was needed to completely elute any non-retained components (e.g., excess labeled IgG) and for the signal of the detector to return to the baseline level.
Figure 1.
A typical chromatogram obtained for the application and elution of rabbit IgG from a 5 mm × 2.1 mm I.D. column containing immobilized protein G. Details on the experimental conditions and flow rates are described in the text.
The second step in the process consisted of the elution and dissociation of the bound IgG from the immobilized protein G. This step occurred when an elution buffer was passed through the microcolumn. In this case, the mobile phase was switched to a lower pH elution buffer (e.g., pH 2.5, 0.067 M phosphate buffer at 0.50 mL/min) to release the bound IgG while the elution profile for this released analyte was monitored. This type general step elution mode was used throughout this report. However, other ways to elute a bound target, such as through the addition of a competing agent that will displace the target from the column, could also have been employed for this purpose [2].
The final step for the scheme in Figure 1 was the regeneration of the immobilized binding agent and the microcolumn. After the IgG had eluted, the mobile phase was switched back to the pH 7.4, 0.067 M phosphate buffer that had been used for sample application. This buffer was passed through the microcolumn for 10 min at 0.50 mL/min to give essentially full regeneration and equilibration of the microcolumn prior to the injection of another IgG sample (see Section 3.2)
The frontal analysis studies with the protein G supports were carried out using rabbit IgG as the model analyte. The following series of events were used during these studies. First, the protein G microcolumn was equilibrated with the pH 7.4, 0.067 M phosphate buffer for 1 min at 0.5 mL/min. The mobile phase was then switched to a solution that contained 0.05 mg/mL of rabbit IgG and that was applied at 0.10 mL/min until a breakthrough curve with a level plateau was achieved (i.e., at approximately 30 min). The mobile phase was next switched to a pH 2.5, 0.067 M phosphate buffer to elute the bound rabbit IgG, with this buffer being passed through the microcolumn for 10 min at 0.5 mL/min. Finally, the microcolumn was regenerated and equilibrated for the next run by applying pH 7.4, 0.067 M phosphate buffer at 0.5 mL/min for 10 min. The central location of each breakthrough curve was determined by using a Savitzky-Golay first derivative algorithm for smoothing, followed by fitting of the first derivative to an exponentially-modified Gaussian curve. A correction for any non-specific binding of the IgG to the support was made by subtracting the breakthrough times of the control microcolumn from the breakthrough times obtained on the protein G microcolumn at the same flow rate and using the same concentration for the applied IgG [7,34].
3. Results and discussion
3.1. Initial characterization of protein G supports and microcolumns
The protein G supports that were made and used in this report were found through protein assays to contain 6.1 (± 0.4) mg protein G/g silica or 0.19 (± 0.01) μmol protein G/g silica. This gave these supports an average surface coverage of 0.24 (± 0.02) mg protein G/m2 silica, or 0.01 (± 0.005) μmol protein G/m2 silica. These results agreed with those of a previous study in which protein G was immobilized to similar silica-based supports [26, 31].
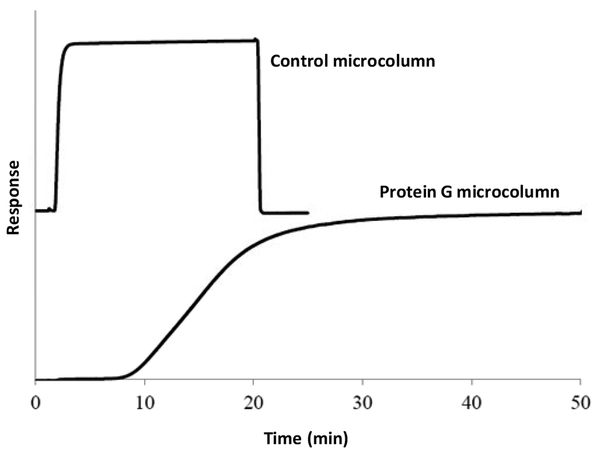
Frontal analysis was used to estimate the binding capacity of the protein G supports [1,2,26,32]. Figure 2 shows some typical results that were obtained when using a 0.05 mg/mL of rabbit IgG and that was applied at 0.10 mL/min. The protein G supports had an apparent binding capacity of 8.7 (± 1.0) mg of IgG/g silica under these conditions. This corresponded to a binding capacity of 0.067 (± 0.020) mg IgG for a 2.1 mm i.d. × 5 mm protein G microcolumn. The specific activity of the immobilized protein G was found by calculating the ratio of the moles of bound IgG per mole of immobilized protein G. In this study, the specific activity for these microcolumns was around 31%. This result was in good agreement with previous studies that have reported specific activities of 30–47% for high capacity protein G supports based on porous silica [26].
Figure 2.
Typical frontal analysis curves obtained for the binding of rabbit IgG to a protein G microcolumn. These curves were obtained for a solution of 0.05 mg/mL rabbit IgG that was applied to 2.1 mm I.D. × 5 mm protein G microcolumn or control microcolumn at pH 7.4 and 0.10 mL/min.
Frontal analysis measurements can also be used to examine the stability of the protein G microcolumns and the effectiveness of the column regeneration conditions (e.g., as shown in Figure 1). This has previously been done by making repeated measurements of the binding capacity for rabbit IgG over the course of up to 50 application and elution cycles. In work using a similar protein G support to that employed in this current study, it was found that a variation in the binding capacity of less than one 1% (e.g., 0.3-0.9%) occurred from one application/elution cycle to the next during the course of these experiments [6].
3.2. Microcolumn regeneration
Further studies based on zonal elution were conducted to determine the minimum time that could be used for regeneration time of the protein G microcolumns. In these studies, 20 μL of 0.20 mg/mL labeled rabbit IgG (i.e., an amount less than 10% of the total binding capacity) was injected onto a protein G microcolumn at 0.50 mL/min in the presence of pH 7.4, 0.067 M phosphate buffer. A pH 2.5, 0.067 M phosphate buffer was then passed through the microcolumn at 0.50 mL/min to elute the retained IgG, and the peak size for the retained and released amount of labeled IgG was determined. After this retained target had been completely eluted, the mobile phase was switched back to the pH 7.4 application buffer, and the microcolumn was regenerated for times ranging from 20 min down to 1 min at 0.50 mL/min prior to sample injection. The amount of retained and released labeled IgG was then again measured during the elution step and compared to the peak that was obtained when using a regeneration time of 20 min. The process was then repeated.
The peak areas and amplitudes of the retained peaks that were measured during these experiments had relative standard deviations that ranged from ± 0.1 to ± 2%. It was found that the use of a regeneration times longer than 5 min did not pro<zduce any significant changes in the peak areas and peak amplitudes. In addition, only small changes in the retained peak areas and amplitudes (i.e., changes of less than 2%) were seen when comparing the results acquired at a regeneration of at least 5 min versus a 20 min regeneration time. Based on these results, a regeneration time of 10 min at 0.50 mL/min was selected for use in all later studies with the protein G microcolumns as a compromise between the regeneration time and peak reproducibility. However, the data from these experiments also indicated that shorter regeneration times can be used for such work in many cases.
3.3. Target elution and dissociation rates from protein G microcolumns
The elution and dissociation of IgG from the protein G microcolumns was next characterized by using the peak decay method. In this method, the following equation describes the elution profile that will occur when a step change is made that suddenly allows a retained target to be released from a binding agent on a chromatographic support [22-24].
| (1) |
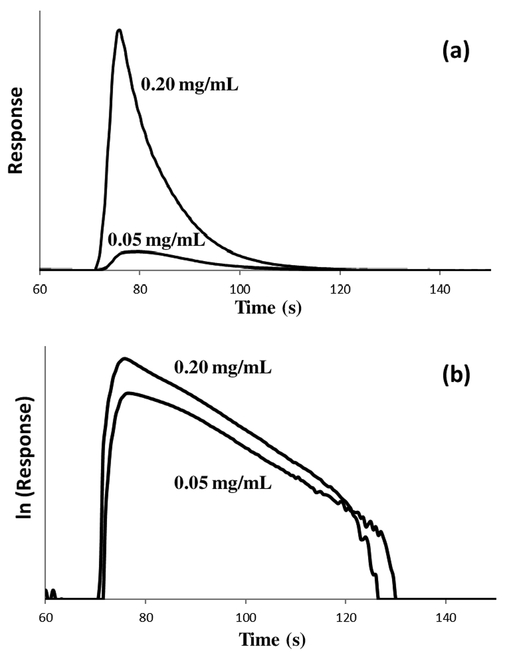
In eqn. (1), mAe represents the moles of the analyte or target that is eluting at time t, and mA0 represents the moles of analyte that were initially bound to the column. The reverse rate constant (k−1) describes the movement of analyte from the stagnant mobile phase to the flowing mobile phase in the column, and kd is the dissociation rate constant for release of the analyte or target from the immobilized binding agent [22-24]. Figure 3(a) shows some typical elution profiles that were generated for goat IgG when using a step change in pH to release this target from a protein G microcolumn. Similar elution profiles were obtained for the other types of IgG that were examined later in this report.
Figure 3.
(a) Elution profiles and (b) natural logarithm of the elution profiles obtained following the application of 20 μL of 0.05 or 0.20 mg/mL labeled goat IgG onto a 5 mm × 2.1 mm I.D. protein G microcolumn. These elution profiles were obtained using a pH 2.5 elution buffer that was passed through the microcolumn at 0.50 mL/min.
The relationship in eqn. (1) converts into the following expression if experimental conditions are present during the peak decay method that make dissociation of the target from the immobilized binding agent the rate-limiting step in release of the target from the column (i.e., kd < k−1) [22-24].
| (2) |
Eqn. (2) indicates that a plot of the natural logarithm of the elution profile versus time in this situation should result in a linear response with a slope that is equal to −kd [22-24]. This value can then be easily used directly to find the dissociation rate constant, kd, for the interaction. Figure 3(b) shows some examples of these natural logarithm elution profiles and linear responses, as obtained for goat IgG on a protein G microcolumn. Similar plots were seen for the other types of IgG that were examined in this study.
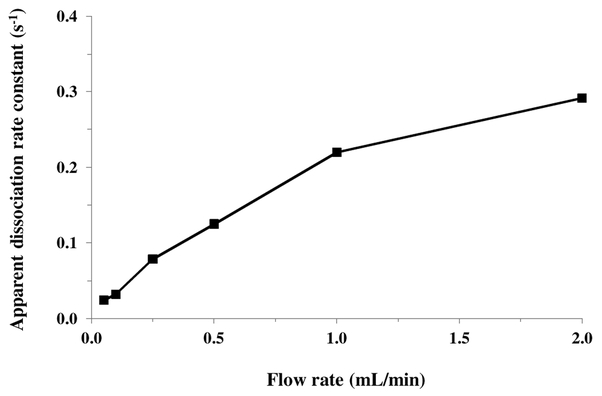
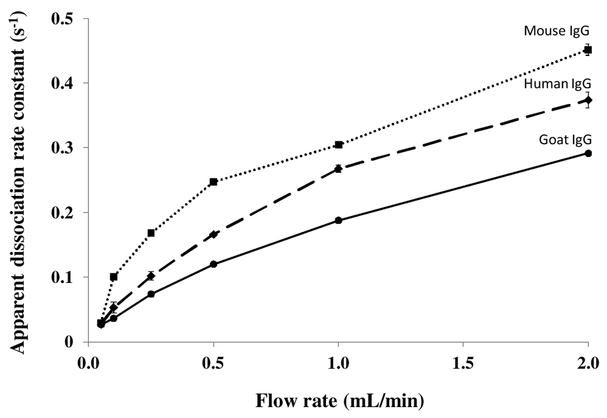
According to eqn. (1), the flow rate can affect the apparent slope of a logarithmic elution profile if mobile phase mass transfer still plays some role in determining the net rate of target dissociation [24]. This effect is illustrated in Figure 4 by the fact that the slopes for such plots represented by their apparent dissociation constants) tended to increase slightly and level off at higher flow rates when goat IgG was eluted from a protein G microcolumn. A similar effect has been noted in use of the peak decay method with other systems [18]. Ideally, values obtained at high flow rates should be used in peak decay studies to avoid or minimize these flow rate effects [23,24]. However, work at lower or intermediate flow rates, as used in this study, is also useful in characterizing the elution behaviour of such a system under typical operating conditions [18]. Work under these latter conditions provides apparent dissociation rate constant values that reflect the net contribution of both the inherent rate of target dissociation and mass transfer effects [18,23,24].
Figure 4.
Apparent dissociation rate constants measured by the peak decay method for rabbit IgG as a function of flow rate. These results were obtained using a 5 mm × 2.1 mm I.D. protein G microcolumn and a pH 2.5 elution buffer that was passed through the microcolumn at 0.05–2.0 mL/min.
Plots like those obtained for rabbit IgG in Figure 4 were also acquired for goat, mouse and human IgG on protein G microcolumns by using the peak decay method over elution flow rates that ranged from 0.05 to 2 mL/min (see Figure 5). In each case, the slopes for the logarithmic decay profiles and their apparent dissociation rate constants showed a steady increase in value when going from low flow rates to moderate flow rates (i.e., 0.05-0.5 mL/min), with the values then levelling off at higher flow rates (i.e., 1-2 mL/min). The results that were obtained over elution flow rates from 0.5 to 2.0 mL/min and at an elution pH of 2.5 are summarized in Table 1. The precision of the apparent dissociation constants rates that were measured for the protein G microcolumns under these conditions ranged from ± 0.9 to ± 6.5%. Although the overall dissociation rates for all the types of IgG that were examined showed a similar change with flow rate, the apparent dissociation rate constants differed that were obtained for these targets. In some cases, a difference in kd as large as 1.5-fold was obtained between IgG from two different species. For instance, the fastest rate of dissociation in Table 1 was obtained for mouse IgG, followed by human IgG and then rabbit and goat IgG.
Figure 5.
Apparent dissociation rate constants measured by the peak decay method for goat (●), human (♦) and mouse (■) IgG as a function of elution flow rate. These results were obtained using a 5 mm × 2.1 mm I.D. protein G microcolumn and a pH 2.5 elution buffer that was passed through the microcolumn at 0.05–2.0 mL/min. The results obtained for rabbit IgG were given in Figure 4. The error bars represent ± 1 standard error of the mean (n = 3).
Table 1.
Dissociation rate constants measured for various types of IgG with protein G at several flow ratesa
| Type of IgG | Flow rate and apparent dissociation rate constant, kd (s−1) | ||
|---|---|---|---|
| 0.50 mL/min | 1.0 mL/min | 2.0 mL/min | |
| Rabbit | 0.125 (± 0.004) | 0.220(± 0.002) | 0.292 (± 0.005) |
| Goat | 0.120 (± 0.002) | 0.188(± 0.002) | 0.291 (± 0.003) |
| Human | 0.166 (± 0.001) | 0.268(± 0.005) | 0.374 (± 0.012) |
| Mouse | 0.247 (± 0.003) | 0.305(± 0.004) | 0.452 (± 0.009) |
The kd values were measured at pH 2.5 and room temperature. The values in parentheses represent ± 1 S.D.
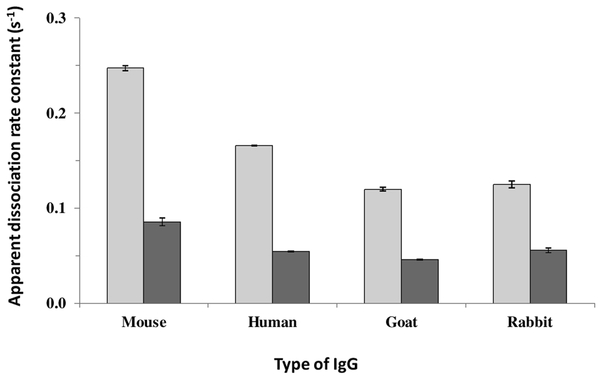
It was possible to apply this approach based on peak decay analysis to determine how the apparent dissociation rate constants for each type of IgG changed with the pH of the elution buffer. Figure 6 summarizes the results that were obtained in going from an elution pH of 2.5 to 3.0 and when using a flow rate of 0.50 mL/min for elution for all the tested types of IgG. All the IgG samples decreased in the value of their apparent dissociation rate constants when going from an elution pH of 2.5 to 3.0. For example, the kd for human IgG decreased by 3.1-fold in going from an elution pH of 2.5 to pH 3.0. The kd values that were obtained under these conditions are similar to those observed at pH 2.5 for the dissociation of small target compounds from HPIAC columns containing immobilized antibodies for these targets [4]. In addition the kd values shown for rabbit IgG at pH 2.5 to 3.0 agree with a general estimate of 10−1 s−1 that would be expected from prior kinetic studies that have been conducted between rabbit IgG and protein G at pH 7.0 and 25°C [35], the observed decrease in binding strength for this interaction that has been measured in going from pH 7 to pH 2.8 [36].
Figure 6.
Apparent dissociation rate constants measured at 0.50 mL/min for various types of IgG on a protein G microcolumn when using an elution buffer with a pH of 2.5 (light bars) or 3.0 (darker bars). The error bars represent ± 1 standard error of the mean (n = 3).
4. Conclusions
The report examined the binding and elution of various types of IgG from protein G microcolumns. Frontal analysis was used to determine the binding capacity of these columns, while zonal elution was used to examine the regeneration of these columns. The peak decay method was then employed with this system to study the apparent dissociation rates for IgG from several species and as a function of elution flow rate or pH. The apparent dissociation rate constants obtained at low-to-moderate flow rates had values that were affected by both the inherent target dissociation rate and mass transfer effects. However, the use of higher flow rates gave conditions that helped to minimize mass transfer effects, thus providing more accurate estimates of the dissociation rate constants [18]. Although similar dissociation behavior was seen for the various types of IgG, there were differences in the apparent dissociation rates constants that were as large as 1.5-fold for IgG from two different species. It was also possible to use this approach to determine how the apparent dissociation rate constants changed with a different pH elution buffer. This is the first time that the dissociation rates of protein G in these interactions have been examined under these elution conditions and with various types of IgG. These results should be useful in the future use and optimization of protein G columns for IgG analysis, capture and purification. These results also demonstrate how the peak decay method and approaches based on frontal analysis or zonal elution can be used together to study target–protein binding and dissociation for systems like IgG and protein G.
Highlights.
Antibody binding and dissociation from protein G microcolumns were examined
Frontal analysis was used to characterize the binding capacity of the microcolumns
Zonal elution was utilized to optimize the microcolumn regeneration conditions
The peak decay method was used to measure dissociation rates during elution
Immunoglobulin G from several species were compared in the elution studies
Acknowledgements
This work was supported by the National Institutes of Health under Grant R01 DK069629. J.A. Anguizola also acknowledges the National System of Investigators (SNI) for funding during the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hage DS, Bian M, Burks R, Karle E, Ohnmacht C, Wa C, Bioaffinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 101–126. [Google Scholar]
- [2].Phillips TM, High performance immunoaffinity chromatography. An introduction, LC Mag. 3 (1985) 962–972. [Google Scholar]
- [3].Weller MG, Immunochromatographic techniques—a critical review, Fresenius J. Anal. Chem 366 (2000) 635–645. [DOI] [PubMed] [Google Scholar]
- [4].Nelson MA, Moser A, Hage DS, Biointeraction analysis by high-performance affinity chromatography: kinetic studies of immobilized antibodies, J. Chromatogr. B 878 (2010) 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pfaunmiller EL, Moser A, Hage DS, Biointeraction analysis of immobilized antibodies and related agents by high-performance immunoaffinity chromatography, Methods, 56 (2012) 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pfaunmiller EL, Anguizola JA, Milanuk ML, Carter N, Hage DS, Use of protein G microcolumns in chromatographic immunoassays: a comparison of competitive binding formats, J. Chromatogr. B 1021 (2016) 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hage DS, Xuan H, Nelson MA, Application and elution in affinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 79–97. [Google Scholar]
- [8].Hage DS, Phillips TM, Immunoaffinity chromatography, in: Hage DS (Ed.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 127–172. [Google Scholar]
- [9].Moser AC, Hage DS, Chromatographic immunoassays, in: Hage DS (Ed.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 789–836. [Google Scholar]
- [10].Hage DS, Anguizola JA, Li R, Matsuda R, Papastavros E, Pfaunmiller E, Sobansky M, Zheng X, Affinity chromatography, in: Fanali DS, Haddad PR, Poole CF, Schoenmakers P, Lloyd D (Eds.), Liquid Chromatography: Applications, Elsevier, Massachusetts, 2013, pp. 1–23. [Google Scholar]
- [11].Moser AC, Hage DS, Immunoaffinity chromatography: an introduction to applications and recent developments, Bioanalysis 2 (2010) 769–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim HS, Hage DS Immobilization methods for affinity chromatography, in: Hage DS (Eds.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 35–78. [Google Scholar]
- [13].Björck L, Kronvall G, Purification and some properties of streptococcal protein G, a novel IgG-binding reagent, J. Immunol 133 (1984) 969–974. [PubMed] [Google Scholar]
- [14].Akerström B, Björck L, A physicochemical study of protein G, a molecule with unique immunoglobulin G-binding properties, J. Biol. Chem 261 (1986) 10240–10247. [PubMed] [Google Scholar]
- [15].Riggin A, Regnier FE and Sportsman JR, Quantification of antibodies to human growth hormone by high-performance protein G affinity chromatography with fluorescence detection, Anal. Chem 63 (1991) 468–474. [DOI] [PubMed] [Google Scholar]
- [16].de Frutos M, Paliwal SK and Regnier FE, Liquid chromatography based enzyme-amplified immunological assays in fused-silica capillaries at the zeptomole level, Anal. Chem 65 (1993) 2159–2163. [DOI] [PubMed] [Google Scholar]
- [17].Shen H, Aspinwall CA and Kennedy RT, Dual microcolumn immunoassay applied to determination of insulin secretion from single islets of Langerhans and insulin in serum, J. Chromatogr. B: Biomed. Sci. Appl 689 (1997) 295–303. [DOI] [PubMed] [Google Scholar]
- [18].Holtzapple CK, Pishko EJ and Stanker LH, Separation and quantification of two fluoroquinolones in serum by on-line high-performance immunoaffinity chromatography, Anal. Chem 72 (2000) 4148–4153. [DOI] [PubMed] [Google Scholar]
- [19].Schiel JE, Ohnmacht CM, Hage DS, Measurement of drug-protein dissociation rates by high-performance affinity chromatography and peak profiling, Anal. Chem 81 (2009) 4320–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tong Z, Schiel JE, Papastavros E, Ohnmacht CM, Smith QR, Hage DS, Kinetic studies of drug-protein interactions by using peak profiling and high-performance affinity chromatography: examination of multi-site interactions of drugs with human serum albumin columns, J. Chromatogr. A 1218 (2011) 2065–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee W-C, Chuang C-Y, Performance of pH elution in high-performance affinity chromatography of proteins using non-porous silica, J. Chromatogr. A 721 (1996) 31–39. [Google Scholar]
- [22].Schiel JE, Hage DS, Kinetic studies of biological interactions by affinity chromatography, J. Sep. Sci 32 (2009) 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yoo MJ, Hage DS, Use of peak decay analysis and affinity microcolumns containing silica monoliths for rapid determination of drug-protein dissociation rates, J. Chromatogr. A 1218 (2011) 2072–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Moore RM, Walters RR, Peak-decay method for the measurement of dissociation rate constants by high-performance affinity chromatography, J. Chromatogr 384 (1987) 91–103. [Google Scholar]
- [25].Clarke W, Hage DS, Development of sandwich HPLC microcolumns for analyte adsorption on the millisecond time scale, Anal. Chem 73 (2001) 1366–1373. [DOI] [PubMed] [Google Scholar]
- [26].Jackson AJ, Karle EM, Hage DS, Preparation of high-capacity supports containing protein G immobilized to porous silica, Anal. Biochem 15 (2010) 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ruhn P, Garver S, Hage DS, Development of dihydrazide-activated silica supports for high-performance affinity chromatography, J. Chromatogr. A 669 (1994) 9–19. [DOI] [PubMed] [Google Scholar]
- [28].Walters RR, High-performance affinity chromatography. pore-size effects, J. Chrom. A, 249 (1982) 19–28. [Google Scholar]
- [29].Hermanson GT, Mallia AK, Smith PK, Immobilized Affinity Ligand Techniques, Academic Press, Boca Raton, 1992. [Google Scholar]
- [30].Larsson PO, High-performance liquid affinity chromatography, Methods Enzymol. 104 (1984) 212–223. [DOI] [PubMed] [Google Scholar]
- [31].Walker JM, The bicinchoninic acid (BCA) assay for protein quantitation, Methods Mol. Biol 32 (1994) 5–8. [DOI] [PubMed] [Google Scholar]
- [32].Zheng X, Li Z, Beeram S, Matsuda R, Pfaunmiller EL, Podariu M, White CJ II, Carter DS Hage, Analysis of biomolecular interactions using affinity microcolumns review, J. Chromatogr. B 968 (2014) 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schiel JE, Tong Z, Z, Hage DS, Development of a flow-based ultrafast immunoextraction and reverse displacement immunoassay: analysis of free drug fractions, Anal. Chem 83 (2011) 9384–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hage DS, Chen J Quantitative affinity chromatography: practical aspects, in: Hage DS (Eds.), Handbook of Affinity Chromatography, 2nd edn., Taylor & Francis, New York, 2006, pp. 595–628. [Google Scholar]
- [35].Rollag JG, Hage DS Non-linear elution effects in split-peak chromatography. II Role of ligand heterogeneity in solute binding to columns with adsorption-limited kinetics, J. Chromatogr. A 795 (1998) 185–198. [DOI] [PubMed] [Google Scholar]
- [36].Akerstrom B, Bjorck L A physiochemical study of protein G, a molecule with unique immunoglobulin G-binding properties, J. Biol. Chem 261 (1986) 10240–10247. [PubMed] [Google Scholar]