Abstract
Human phospholipid transfer protein (PLTP) mediates the transfer of phospholipids among atheroprotective high-density lipoproteins (HDL) and atherogenic low-density lipoproteins (LDL) by an unknown mechanism. Delineating this mechanism would represent the first step towards understanding PLTP-mediated lipid transfers, which may be important for treating lipoprotein abnormalities and cardiovascular disease. Here, using various electron microscopy techniques, PLTP is revealed to have a banana-shaped structure similar to cholesteryl ester transfer protein (CETP). We provide evidence that PLTP penetrates into the HDL and LDL surfaces, respectively, and then forms a ternary complex with HDL and LDL. Insights into the interaction of PLTP with lipoproteins at the molecular level provide a basis to understand the PLTP-dependent lipid transfer mechanisms for dyslipidemia treatment.
Keywords: PLTP, phospholipid transfer protein, PLTP bound to HDL, PLTP bound to liposome, electron microscopy, HDL, liposome
Introduction
Phospholipid transfer protein (PLTP) mediates the phospholipid transfer among lipoproteins, including high-density lipoproteins (HDL), low-density lipoproteins (LDL), intermediate density lipoproteins (IDL), very low-density lipoproteins (VLDL) and chylomicrons.[1, 2] Additionally, PLTP has been reported to have a unique function in remodeling HDL.[3] Since it is well known that the HDL cholesterol level is closely related to atherosclerosis and coronary artery disease (CAD), [4, 5] exploring the connection between PLTP and CAD is necessary. Some studies suggest that high PLTP activity may be a risk factor for CVD based on the increased PLTP activity reported in CAD patients and the inverse correlation between PLTP expression and the HDL level.[6–9] However, there are also opposite results, indicating that low PLTP activity is a marker for peripheral arterial disease (PAD)[10] and PLTP deficiency causes accumulation of cholesterol in the circulatory system and accelerates the development of atherosclerosis.[11] These controversial results about PLTP function in developing CAD motivated our study of the mechanism of PLTP-mediated lipid transfer among lipoproteins at the molecular level.
PLTP is a plasma glycoprotein with a molecular mass of ~81 kDa.[12] As a family member of the lipopolysaccharide (LPS)-binding/lipid transfer proteins, PLTP shares approximately 20% sequence identity with cholesteryl ester transfer protein (CETP) and bactericidal permeability increasing protein (BPI).[13, 14] Due to the similar roles of PLTP and CETP as lipid transfer vehicles, many biochemical experiments have been performed comparing their similarities and differences.[13, 15] Unlike CETP, which primarily mediates net lipid transfer, including the exchange of cholesterol ester (CE) and triglycerides (TG) from HDL to triglyceride-rich lipoproteins such as LDL, [16] PLTP is mainly responsible for promoting the transfer of phospholipids from lipid-rich lipoproteins to HDL.[17] Both PLTP and CETP enable the remodeling of the HDL size, even without the involvement of another lipoprotein species.[18, 19] Moreover, incubation experiments in vitro have predicted that both PLTP and CETP could form a ternary complex with two HDL particles, creating a large unstable fusion intermediate, which would finally result in either three smaller particles or remain as an enlarged fusion particle.[19, 20] However, electron microscopy experiments did not favor this hypothesis, at least for CETP, due to the absence of observation of the ternary HDL-CETP-HDL complexes.[21–24] Whether the ternary complex of HDL-PLTP-HDL could be observed by electron microscopy remains a question.
Although the structure of PLTP is still unavailable, homology models have been constructed based on the X-ray structure of human BPI.[25–27] PLTP was predicted to have a banana-shaped structure with a long narrow tunnel connecting two distal end openings and two lipid binding pockets, [25] which is very similar to the structure of CETP.[25] However, the Ω1 flap on the C-terminal end of CETP[28] end adopts a very different conformation to that in PLTP. Moreover, two α-helixes along the PLTP tunnel are relatively short compared to those of CETP, potentially affecting the structural flexibility of PLTP. Considering that PLTP and CETP have different functions and activities during lipid transfer, [15] it is necessary to investigate whether PLTP adopts a similar binding conformation with HDL as that of CETP, and whether it shares a similar lipid transfer mechanism to CETP, such as the shuttle mechanism[21, 29] or the tunnel model.[30]
Difficulties in studying the structure-based PLTP-mediated lipid transfer and HDL remodeling mechanisms lie in lipoprotein structure heterogeneity, [31–33] which is caused by the variety of lipoprotein compositions, such as difference in lipid and protein components among or within each species of lipoproteins. Moreover, the physical properties of the lipid components result in lipoprotein structural softness and dynamics, especially for HDL.[34–36] A particle-by-particle structural study is required to examine lipoprotein-PLTP interactions. Here, we used our reported and validated optimized negative-staining electron microscopy (OpNS EM) protocol, [34, 37] rapidly settling the structure and eliminating the major artifacts[22, 38–40] without losing high contrast or structure details, [34, 37, 41] to examine the example. We then used individual-particle electron tomography (IPET)[42] and single-particle analysis (SPA) techniques[43] to study the three-dimensional (3D) structure of PLTP and its interaction with lipoproteins and the liposome. As a comparison, CETP was also used for the incubation with different lipoproteins.
Results
Structure of PLTP
We first examined a PLTP sample using the optimized OpNS-EM methodology.[19, 27] Both the surveyed micrographs (Fig. 1A) and the selected particle views (Fig. 1B) revealed a boomerang- or banana-shaped structure of PLTP, as predicted by homology modeling, [25] with dimensions of ~12.4 ± 1.9 nm × ~3.8 ± 0.6 nm. These measurements excluded aggregated particles, which may related to the exposure of PLTP surface hydrophobic residues[44] to the solvent. As a comparison, a sample of mutated PLTP (M159E), [45] which lacks lipid transfer activity, [5] was also examined (Supplementary Fig. 1A). Isolated particles of wild-type PLTP were windowed (Fig. 1B) and the particle images were submitted to reference-free classification and averaging.[38] The result showed that the particle shape and size are highly similar to the homology model (Fig. 1C) and some detailed features could be distinguished, such as the concave-shaped surface, the tapper N-terminal β-barrel domain and globular C-terminal β-barrel domain (Fig. 1C and D).
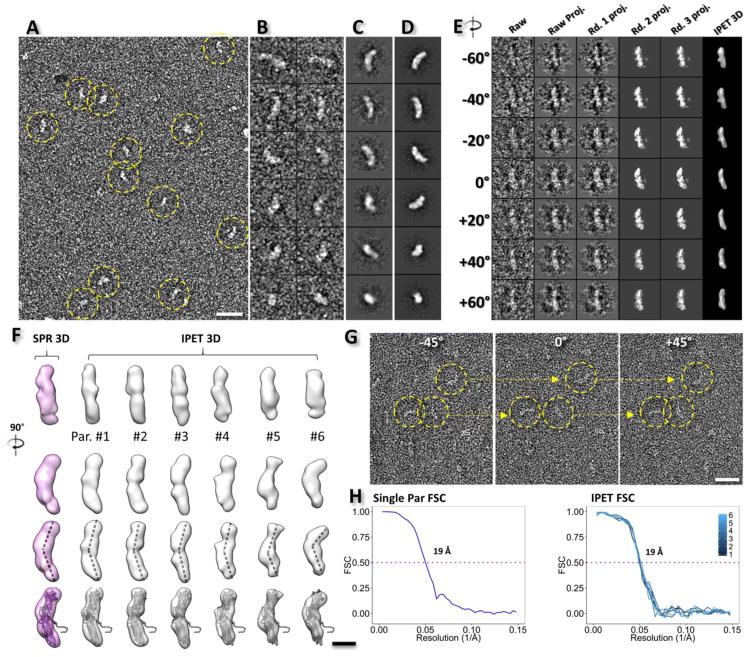
Fig. 1. OpNS EM images and 3D reconstruction of PLTP particles.
A) Survey view of the sample of PLTP particles (yellow dashed circles). B) Representative raw PLTP particles. C) Selected reference-free class averages. D) Selected projections of the single particle 3D reconstruction (average from a thousand particles). E) Ab-initio 3D reconstruction of an individual PLTP particle by individual particle electron tomography (IPET). The particle was imaged by electron tomography (ET, tilt angles ranging from −60° to 60° in steps of 2°). Seven representative tilt views of a targeted PLTP particle present the step-by-step process of IPET 3D reconstruction shown in columns 2 to 5. The final IPET 3D reconstruction is displayed in the last column. F) The comparison of the single particle 3D reconstructions (column 1, pink model) and the IPET 3D reconstructions (columns 2 to 7, gray models). Six representative IPET 3D reconstructions from six targeted PLTP particles are displayed, with different curvatures indicated by docking of the homology model into each density map (row 4). G) Three representative tilt views of the single-axis tilt series of PLTP particles. H) The resolutions of the single particle 3D reconstruction and the resolution of the IPET 3D reconstruction are estimated by FSC between two models built from odd- and even-numbered views, respectively. Scale Bars: A, 25 nm; F, 5 nm; G, 20 nm. The box sizes are 29 nm for B, C, D and E.
A further examination of the 3D structure and dynamics of the PLTP particles were conducted by electron tomography (ET). The targeted particles were imaged from a series of tilt angles ranging from −60° to 60° in steps of 2°. The images of each individual PLTP particle were extracted from the tilted series after contrast transfer function (CTF) correction, and then the particle tilt series were submitted for 3D individual-particle electron tomography (IPET) reconstruction[42] (Fig. 1E). Representative IPET 3D reconstructions (Fig. 1F, right six columns) and tilted views of representative particles (Fig. 1G) confirmed the banana-shaped structure of PLTP, with similar dimensions to the homology model[25]. The analysis of the curvature of IPET-reconstructed PLTP particles showed a noticeable variation in the angle between the two center axes of the N- and C-terminal β-barrel domains (with a mean of 140.63° ± 10.61°) (Fig. 1F, third row), which has been predicted by molecular dynamics simulations in CETP.[46] The resolutions of the IPET 3D maps were ~19 Å based on the intra-Fourier shell correlation (intra-FSC) analysis [42] (Fig. 1H, right panel).
To validate the significance of the IPET 3D reconstruction, the single-particle 3D reconstruction method was used to classify and average a total of ~7,000 particle images.[43] In this process, the IPET 3D map was used as an initial model after low-pass filtration to 80 Å to avoid potential initial model bias. The single-particle 3D reconstruction at a resolution of ~19 Å confirmed the banana-shaped structure of PLTP (~129 Å × 36 Å), which is very similar to the IPET 3D reconstruction and the homology model (Fig. 1F, first column and last row). Fitting the homology model into the envelopes of the single particle 3D reconstruction showed a similar quality to that fitted to the IPET 3D reconstruction, suggesting similar resolution between the IPET and the single particle 3D reconstructions (Fig. 1H, left panel).
Binary conformation of the PLTP-HDL3 complex
To examine how PLTP interacts with HDL3, PLTP was incubated with HDL3 at molar ratios of ~3:1, and then examined by OpNS EM. The survey EM images showed that protruding features were observed on the spherical-shaped HDL3 particles (with an HDL3 diameter of ~12.28 ± 1.91 nm) (Fig. 2B), in which no more than two PLTP molecules could be observed on a single HDL3 particle (Supplementary Fig. 1G). In comparison, the HDL-PLTPM159E sample only had 1.88 ± 0.89% of the HDL3 particles that were observed with protruding features, which is significant lower than the percentage of bound HDL3 in the HDL-wild-type PLTP sample (25.06 ± 4.19%). This lower percentage of binding to HDL3 may explain the ineffective lipid transfer activity [5] of PLTPM159E. Interestingly, much less aggregation of the PLTP particles was observed compared with the sample of PLTP alone (Fig. 2A), which implies that the disassembling of the aggregates may occur through the absorption of PLTP onto HDL3 particles. To analyze the detailed structure of PLTP-bound HDL3, complex particle images were submitted to reference-free 2D classification and averaging (Fig. 2C and D). The class averages showed that the distal end of PLTP inserts into the HDL3 particles, with ~8.09 ± 1.41 nm remaining outside the HDL3 surface. The averaged PLTP width is ~3.44 ± 0.51 nm and the binding conformation is similar to the garlic-shaped structure of CETP binding to HDL3.[31] This result suggests that the hydrophobic N-terminal β-barrel domain of PLTP interacts with HDL3.
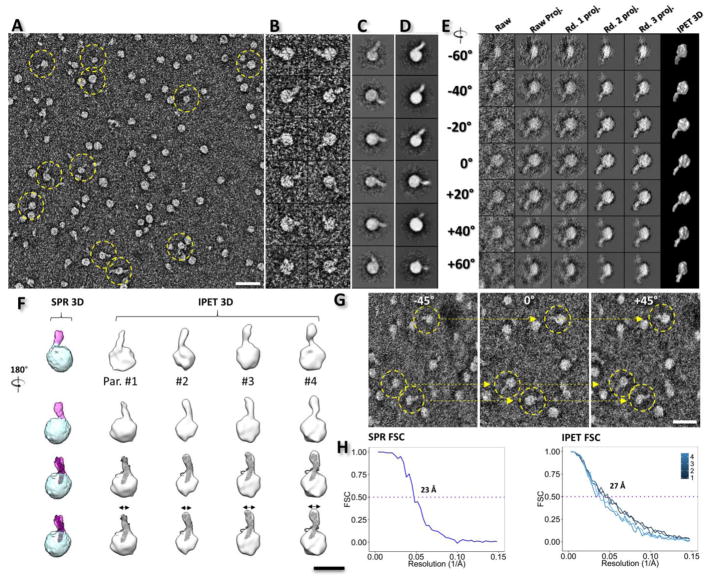
Fig. 2. OpNS EM images and 3D reconstruction of the PLTP-HDL3 complexes.
A) Survey view of PLTP-HDL3 complexes, indicated by yellow dashed circles. B) Representative particle images of PLTP-HDL3 complexes. C) The selected reference-free class averages. D) Selected projections of the single particle 3D reconstruction. E) Ab-initio 3D reconstruction of an individual PLTP-HDL3 complex by IPET. The complex was imaged by ET tilting from −62° to 67° in steps of 1.5°. Seven representative tilting views (column 1) were compared to the projections from the intermediate 3D reconstruction (columns 2 to 5) at corresponding angles. The final IPET 3D reconstruction is displayed in the last column. F) The comparison of the 3D reconstructions by the single particle averaging method (column 1, a quasi-spherical shape density and a single protrusion were labeled with cyan and pink colors, respectively) and IPET (column 2 to 7, gray models). Four representative IPET 3D reconstructions of PLTP-HDL3 complexes were docked with the homology model of PLTP (row 3). G) Three representative views of the single-axis tilt series of PLTP-HDL3 complexes. H) Single particle 3D averaging and IPET 3D reconstruction resolution were estimated by Fourier-shell correlation (FSC) between two models built from odd- and even-numbered views, respectively. Scale Bars: A, 40 nm; F, 15 nm; G, 30 nm. All the box sizes are 37 nm for B, C, D and E.
To further examine the structure of PLTP-bound HDL3 in 3D, PLTP-HDL3 complexes were also imaged using OpNS ET (Fig. 2E). A series of tilt views of the images (ranging from −62° to +67° in steps of 1.5°) confirmed the garlic shaped structure (Fig. 2G), in which a banana-shaped PLTP molecule is attached to a spherical HDL3 particle, as observed in 2D images. By using IPET, 3D density maps were reconstructed from the tilted images of individual complexes. Representative complex particles showed resolutions of ~27 Å (indicated by intra-FSC analysis) (Fig. 2H, right, and Fig. 2F, right four columns). The density contour level of these 3D maps was defined as close to a molecular weight of ~330 kDa, which confirmed the garlic-shaped 3D structure of the complex. Additionally, we detected a variety of HDL3-PLTP complexes, especially in terms of the size of the HDL3 particle (10.82 ± 1.43 nm) and the width of the free end of PLTP (3.39 ± 0.64 nm; Fig. 2F, last row).
To evaluate the statistical significance of the IPET 3D structures, the single-particle 3D reconstruction method[38] was also used to classify and average a total of ~32,000 complexes of PLTP-HDL3 (Fig. 2F). As an initial model, the IPET 3D maps were low-pass filtered to 80 Å to avoid potential bias. Due to the heterogeneity of HDL3, only a small portion of complexes (~4,700 particles) with relatively homogeneous HDL3 diameters were selected for 3D reconstruction (Fig. 2H, left panel). The single-particle 3D reconstruction at a resolution of ~23 Å again confirmed a garlic-shaped structure, with a quasi-spherical HDL3 (with dimensions of ~11.5×11.6×11.6 nm) carrying a rod-shaped PLTP protrusion (with a length of ~8.0 nm and a width of ~3.8 nm, at a contour level corresponding to the molecular volume of the complex). This spherical HDL3 contains a low-density core and a relatively high-density outer shell, whereas the density of the PLTP protrusion is even higher than the outer shell. The angle between the inserted PLTP and the tangent plane of the HDL3 surface is ~63°, which is similar to that of CETP-bound HDL.[31]
To determine the orientation of PLTP in binding to HDL3, two structural analysis methods in Chimera[47] were used to analyze the PLTP homology model fitting into the protruded density on the HDL3 surface (Fig. 2F, third row). Unfortunately, these analyses were insensitive to distinguish which distal end penetrates the HDL3 surface. In detail, i) by fitting each of the β-barrel domains into the rod-shaped density portion, the average map fit value showed a similar score (~4.65 vs. ~4.69; and ~22.4% vs. ~18.2% atoms outside the contour for the C-terminal and the N-terminal fitting, respectively); ii) by computing the correlation values between the protrusion density and 14-Å-resolution density maps generated from the above two fittings, respectively, the R-value results of ~0.95 vs. ~0.94 were insufficient to distinguish which was better. However, considering the distal end of the N-terminal β-barrel domain contains a hydrophobic lipid pocket similar to CETP, and the CETP N-terminal β-barrel domain penetrates into the HDL3 surface[21, 31] due to hydrophobic interactions[22], we hypothesize that the N-terminal pocket opening penetrates the HDL3 surface.
Binary conformation of PLTP interacting with LDL, VLDL or Liposome
To examine the interactions between PLTP and other lipoproteins, such as LDL, VLDL, and liposome, the lipoprotein samples were incubated with or without the presence of PLTP, and then imaged using OpNS EM. Without PLTP, the spherical shaped particles of LDL (diameter ~200–300 Å), VLDL (diameter ~370–600 Å) and liposome (diameter ~200–1000 Å) showed no protrusions (Supplementary Fig. 1D, E and F). However, in the presence of PLTP, rod-shaped PLTP molecules were observed on the globular surface of LDL, VLDL and liposome (Fig. 3A, B and C). Approximately 7.3% of the LDL particles, ~4.3% of the VLDL particles and ~13% of the liposomes showed one or two protrusions (Fig. 2A and B, and Supplementary Fig. 1H). The angle between the inserted PLTP and the tangent plane of the LDL and VLDL surface is difficult to measure due to the large difference in diameters between LDL/VLDL and PLTP, and the much floppier 3D structure of LDL/VLDL compared with HDL3. The percentages of particles bound to PLTP were significantly higher for HDL3 than for LDL and VLDL, suggesting that PLTP has a higher binding affinity for HDL3 than for LDL/VLDL/liposome.
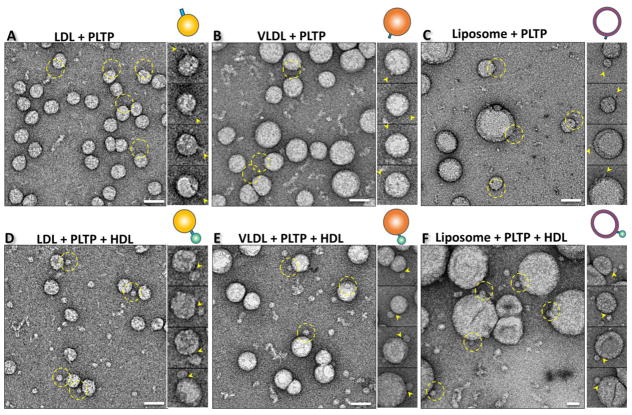
Fig. 3. Structure of PLTP bound to LDL, VLDL and liposome by OpNS EM.
Survey OpNS EM images (left panel), representative particle images (right panel) and the corresponding particle cartoon (top right panel) of the binary complex after incubating PLTP with A) LDL, B) VLDL, or C) liposome and the ternary complex after incubating HDL3 and PLTP with D) LDL, E) VLDL, or F) liposome at 37°C for 1 minute. The PLTP binding positions on each type of substrate were labeled with yellow dashed circles and yellow arrows. Particle window size: A, 44 nm; B, 60 nm; C, 120 nm D, 44 nm; E, 74 nm; F, 96 nm. All scale bars: A–F, 40 nm.
Ternary conformation of PLTP interacting with HDL3 and LDL, VLDL or liposome simultaneously
To examine how PLTP interacts with atheroprotective and atherogenic lipoproteins simultaneously, samples of the co-incubation of LDL with HDL3 in the presence of PLTP were imaged using OpNS EM. The micrographs showed a small amount of LDL particles forming a ternary complex via the bridging of two HDL3 particles through a PLTP molecule (Fig. 3D). The portion of bridges are ~25 Å, which is markedly shorter than the length of PLTP alone (~129 Å) or the length of the PLTP protrusion on the HDL3 surface (~80 Å), suggesting that both distal ends of PLTP penetrated into the corresponding surface of LDL and HDL3 simultaneously. When repeating the above experiments by co-incubating with VLDL or liposome, a similar phenomenon was observed, in which a rod-shaped PLTP density (with a length of ~30 Å) bridging a large spherically shaped VLDL or liposome with a small spherically shaped HDL3 particle (Fig. 3E and F). Interestingly, the percentages of PLTP-bound LDL, VLDL and liposome particles were about doubled in forming the ternary complexes, i.e., ~16.6%, ~11.9%, and ~18.2%, respectively. A similar phenomenon was also previously observed for CETP-lipoprotein incubation [23], which suggests that CETP and PLTP share a similar binding mechanism. The interaction of the N-terminal β-barrel domain with HDL3 may trigger a conformational change in the C-terminal β-barrel domain to increase the binding affinity to other lipoproteins.
PLTP function in HDL3 remodeling
To examine the differences in PLTP-mediated lipid transfer efficiency among HDLs themselves, between HDL3 and other lipid-rich particles or merely fusing the HDL3 particles, the above samples, HDL-PLTP-LDL/VLDL/liposome (with a molar ratio of PLTP, HDL3 and LDL/VLDL/liposome of 9:3:1) and PLTP-HDL3 (molar ratio of 3:1) were further incubated separately for up to 24–48 hours at 37°C. Samples without HDL3 (LDL/VLDL-PLTP) or PLTP (HDL3-liposome) were used as controls. The molar ratios of lipoproteins were estimated based on the protein concentration of apoA-I and apo-B in HDL and LDL/VLDL, in which it is assumed that the spherical HDL contains three copies of ApoA-I and LDL/VLDL contains one copy of ApoB-100. The molar ratios were further validated by EM images, in which the lipoprotein samples were mixed under different ratios and dilution conditions for TEM examination. The average number of particles per unit area on the carbon film was calculated to confirm the molar ratio in the original samples.
Since the amount of lipid transfer cannot be measured by OpNS EM, the change in the HDL3 diameter was used as an alternative to indirectly reflect the lipid transfer activity from lipid-rich particles to HDL3. The observation of HDL size change in this simplified model can also be affected by the PLTP-mediated fusion process.[48] At certain point, pre-β particles are released, and these particles are prone to fuse thereby increasing particle diameter. Therefore, the increase in particle size is not a perfect surrogate for PLTP transfer activity.
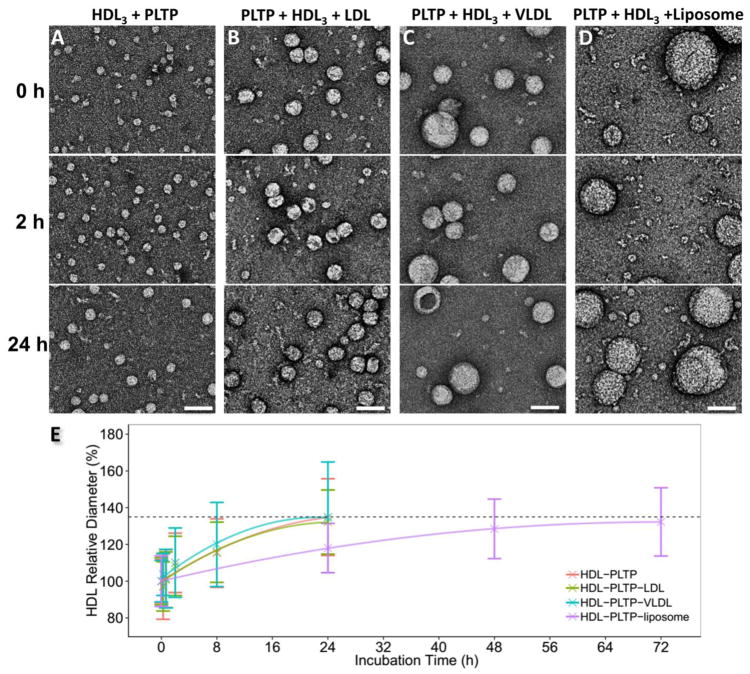
Starting with a relatively simple case, in which only HDL3 and PLTP were incubated at a molar ratio of 1:3, OpNS EM images showed an increased HDL3 size with incubation time (Fig. 4A, Supplementary Fig. 2A and B). In brief, although the HDL3 size remains stable at 15 min and 40 min, a significantly increased HDL3 size (~110%) was observed after a two-hour incubation (Fig. 4E), and it grows continually until 8 hours with its diameter increased to ~135% compared with the initial conditions. Meanwhile, the number of HDL3 particles showed an observable decrease. After 8 hours, the growth started to slow down. This may be due to the lack of free volume, which will not allow more lipids to be loaded onto the surface of a large HDL3 particle. The largest particle at 24 hours was approximately 4 times larger in diameter than normal HDL3. In these experiments, the ternary complex of one PLTP bridging two HDL3 particles was not observed.
Fig. 4. PLTP-induced HDL3 remodeling in the samples of HDL3-PLTP, HDL3-PLTP-LDL, HDL3-PLTP-VLDL and HDL3-PLTP-liposome.
A) OpNS EM images of the sample of HDL3, B) HDL3 and LDL, C) HDL3 and VLDL, and D) HDL3 and liposome were incubated with PLTP at 37°C for an incubation time of 0 h, 2 hrs and 24 hrs (for HDL3-PLTP, HDL3-PLTP-LDL and HDL3-PLTP-VLDL) and 0 h, 24 hrs and 72 hrs (for HDL3-PLTP-liposome). E) The statistical distribution of HDL3 size against the incubation time. Approximately 300–500 HDL3 particles were assessed for each category. The relative mean diameter measurement of HDL3 starting at 0 min (~12.50 nm) is set as 100%. The particle diameters were measured based on the geometric mean of two diameters: the longest diameter and its perpendicular diameter. All scale bars: 50 nm.
As a control, LDL and PLTP were also incubated together at a molar ratio of 1:3. The particle size measurement from OpNS EM images showed that the LDL size remains at 100% (~25.5 nm) over the whole incubation time up to 24 hours, suggesting that PLTP does not cause any significant size variation in the LDL particle (Supplementary Fig. 2C and D).
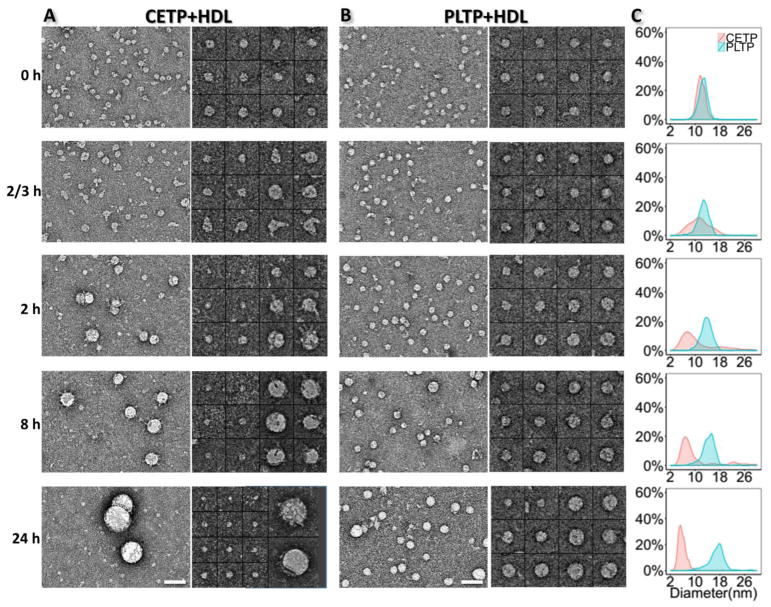
Comparison of PLTP and CETP functions in HDL3 remodeling
To further understand the functional specificity of PLTP in remodeling the HDL3 size, we repeated the above experiments using CETP to compare with PLTP (Fig. 5). The statistical analyses of the HDL3 size from the OpNS images showed that: i) PLTP-mediated HDL3 remodeling was much slower than that by CETP. The peak HDL3 size shifted away from its original position at 8 hours for PLTP, while it took only 2 hours for CETP to produce a noticeable HDL3 size change; ii) PLTP-mediated remodeling led to a general increase in the HDL3 size, which results in a more homogenous population. Small particles are rarely observed in the images of the PLTP-HDL3 samples. In comparison, the density of the HDL3 size peak decreased dramatically after 40 minutes for CETP but not PLTP, and the peak split at 8 hours, indicating that CETP redistributed the HDL3 population and most of the particles centered on a smaller size. iii) Though both PLTP and CETP can cause the formation of large HDL3 particles, the HDL3 particles enlarged by PLTP have a much “smoother” surface, while CETP-induced large HDL3 particles are generally attached to multiple CETPs, indicating differences in the mechanisms of HDL3 remodeling by PLTP and CETP.
Fig. 5. Comparison of the HDL3 remodeling process by PLTP and CETP.
A) OpNS EM images (left panel) and selected particles (right panel) of CETP incubated with HDL3 samples after an incubation of 0 min, 15 min, 40 min, 2 hrs, 8 hrs and 24 hrs at 37°C. B) The OpNS EM images and selected particles of PLTP incubated with HDL3 with the same time series and temperature. C) The statistical analysis of the HDL3 size distribution at each time point. A different peak shift direction can be observed in the two groups. The molar ratios of HDL3 vs. CETP/PLTP were both set at 1:3. Scale bars: A and B, 50 nm. Box sizes A and B, 37 nm. For 24 hrs, the box sizes are 28 nm and 55 nm, respectively.
PLTP-mediated lipid transfer between HDL3 and LDL/VLDL/Liposome
To study the PLTP-mediated lipid transfer function in the presence of both atheroprotective and atherogenic lipoproteins simultaneously, we incubated LDL, HDL3 and PLTP together at a molar ratio of 1:3:9 with different incubation times. OpNS EM images showed that the mean diameter of the HDL3 increased with incubation time for the HDL3-PLTP-LDL sample (Fig. 4B). In brief, although the HDL3 size (~10.3 nm in the beginning) was not significantly changed after incubation for 15 min and 40 min, the mean diameter increased to ~108% of the initial diameter after 2 hours (Fig. 4B and E, Supplementary Fig. 3A and B) and further increased to ~132% after 24 hours. This performance was similar to the increase in the HDL3 size in the absence of LDL, suggesting that LDL is not a major factor that influences the rate of HDL3 size growth. Similarly, we compared the HDL3-PLTP-LDL incubation to the previously reported incubation of HDL3-CETP-LDL.[23] In the incubation with CETP, the HDL3 size peak did not diverge as shown in the CETP-HDL3 incubation (Fig. 5A and C). However, in the presence of LDL, the larger “spiky” particles (the particles attached to multiple PLTPs) disappeared and the HDL3 particle size continued declining (Supplementary Fig. 4). This significant difference between PLTP and CETP indicates that CETP-mediated HDL3 remodeling is more dependent on other species of lipoproteins. This activity is consistent with the observation that a higher percentage of LDL/VLDL particles showed bound CETPs[23] compared to bind PLTPs (Fig. 3A and B).
As a parallel comparison experiment, VLDL instead of LDL was used to repeat the above experiments (HDL-PLTP-VLDL) (Fig. 4C and E, Supplementary Fig. 3C and D). The OpNS EM images showed that the HDL3 sizes increased to ~110% and ~134% at 2 hours and 24 hours, respectively. The fitted curve was similar to that of HDL3-PLTP and HDL3-PLTP-LDL, as described above (Fig. 4E). These results confirm that the rate of PLTP-mediated HDL3 growth is also not significantly altered by VLDL.
Similar to the above, liposome instead of LDL/VLDL was used to repeat the incubation experiments (HDL3-PLTP-liposome). Liposomes are well-defined phospholipid donors to HDL3, designed to measure the phospholipid transfer activity by radioactive labeling.[49, 50] The samples of liposome particles were incubated with HDL3 particles with or without PLTP at a molar ratio of 1:3:9 (liposome:HDL3:PLTP) or 1:3 (liposome:HDL3), respectively. The control samples without PLTP showed that the HDL3 particles retained a constant size during long-term incubation (Supplementary Fig. 5A). In the presence of PLTP, the mean diameter of the HDL3 particles increased throughout the incubation time (Fig. 4D and E). However, the speed was slower than for LDL/VLDL. For example, after 24 hours, the particle size only increases to ~120%, similar to the size increase in the VLDL/LDL-PLTP-HDL3 experiment in 8 hours. To achieve a similar final HDL3 particle size of ~135%, as seen in the VLDL/LDL-PLTP-HDL3 experiments at 24 hours, the liposome-PLTP-HD3L sample incubation had to be extended to 72 hours (~132% of the initial size) (Fig. 4D, Supplementary Fig. 5B).
Based on the evidence from the above incubation experiment that the HDL3 size increase rate is similar in both the HDL3-PLTP and the HDL3-PLTP-LDL/VLDL groups, we can conclude that PLTP-induced HDL3 self-remodeling has greater effect on the HDL3 size compared with lipid transfer from other lipoprotein species to HDL3. This is in agreement with the observation that liposome slows down the PLTP-mediated HDL3 remodeling. Given that more PLTP bound to liposome than to LDL and VLDL (~13% for liposome vs. 7.3% for LDL and 4.3% for VLDL bound to PLTPs, plus each liposome can bind more PLTPs due to its large surface area), a lower percentage of PLTP participated in the HDL3 self-remodeling, although it may increase the formation of ternary complexes for lipid transfer. However, the effect of the latter is much weaker than the former.
Discussion
The structure of PLTP determined by the OpNS IPET method
Since the discovery of PLTP and its cloning in 1994, [10] numerous questions about its 3D structure and the nature of its interactions with lipids and lipoproteins have remained unanswered because of the technical difficulties associated with the crystallization of PLTP and the direct observation of its interactions with lipoprotein particles. Using our OpNS protocol, [34, 37] the IPET approach, [42] and a conventional single-particle reconstruction approach, we visualized the 3D molecular structure of PLTP and its interactions with various lipoproteins. The observed 3D structure of the PLTP molecule revealed by the studies reported here remarkably resembles the homology model based on the BPI crystal, [25–27] including the previously putative model of an allegedly disordered portion of the C-terminal tail-end sequence of PLTP, not seen in any of the other molecular family members. These studies confirm the two β barrel-like domains containing the N- and C-terminal lipid binding pockets (and their relative positions), the curvature of the banana-shaped molecule, and the presence of a channel through the entire length of the molecule.
In addition to individual PLTP molecules, we also observed PLTP molecular aggregates. These aggregates may be the inactive form of the PLTP, which represents up to 70% of all PLTP in human blood, and have an average size distribution from 340 to over 600 kDa.[51–53] Interestingly, a PLTP mutant that is inactive in lipid transfer, PLTPM159E, formed more aggregates than wild-type PLTP and had even fewer interactions with HDL, supporting the idea that some of the lipid transfer inactivity may be associated with self-aggregation of PLTP molecules and their inability to bind lipoproteins.
The concepts of active and inactive PLTP refer to PLTP found in the blood. Recombinant PLTP is not bound to lipoproteins (the presumed basis of active PLTP in the blood. The self-aggregated forms in the rPLTP preparation are rare and contain no other proteins, such as those found in the inactive PLTP complexes in the blood. Therefore, it is not possible to compare rPLTP with the so-called active or inactive forms of PLTP in the blood. Even if some inactive form of PLTP is present, it is unlikely that it would introduce a bias in our observations.
The PLTP-HDL complex is the basic functional unit for lipid transfer activity
The observation of a clear rod-shaped protrusion adopting a vertical conformation relative to the HDL3 surface is direct evidence that PLTP is part of the binary complex binding to the HDL3 surface. No PLTP molecule was found with its concave surface docking to the convex surface of HDL3, or with its two distal ends bridging two HDL3 particles, together forming a typical “dumbbell” shaped ternary complex. This observation of the convex surface of PLTP binding to HDL3 implies that that PLTP binding to HDL3 particles is directional. The observation of fewer PLTPM159E-HDL3 complexes suggests that mutation may disable this HDL3 binding function of PLTP either directly (by changing the PLTP structure) or indirectly (by enhancing PLTP aggregation). Overall, fitting the PLTP model into the binary complex showed consistency between the structure and the density map. However, the free C-terminal globular end of the PLTP was larger than expected. Since one function of PLTP is the delivery of phospholipid molecules from the HDL3, this extra electron density may be related to accumulated phospholipid molecules around the surface of the PLTP C-terminus, as we observed a similarly enlarged PLTP protrusion in the 2D images (Fig. 2).
Our data suggest that the portion of PLTP that penetrates into the HDL3 surface represents ~35% of the molecule, most likely representing the N-terminus of PLTP. The free end of PLTP seems more likely to be the globular and curved C-terminus of the PLTP molecule. These findings suggest that, under physiological conditions, the distal PLTP N-terminal end penetrates into or through the phospholipid portion of the HDL3 surface. The enlarged, rounded C-terminus may be associated with the initiation of the lipid transfer activity. Considering that the thickness of the monolayer of the lipid shell of phospholipids is ~28 Å, and the inserted portion of the PLTP molecule is markedly larger (~50 Å, Fig. 2F), the PLTP N-terminus could reach through the phospholipid layer and deep into the cholesteryl ester core of HDL3. PLTP penetrates particles at an angle, which differs for the N- vs. the C-terminal. Therefore, although the length of PLTP molecule immersed into the HDL particle is approximately 40–50Å, the actual depth PLTP molecule reaches within the particle is shorter. We assume that the position of the N-terminal pocket requires insertion into HDL at the specified length and angle for the lipid-binding pocket to become accessible.
Whether the HDL surface apolipoproteins especially apoA-I involved in the interaction with PLTP is still unclear. One possibility is that PLTP directly interacts with the N-terminal of apoA-I HDL2 Other evidence showed that increasing the amount of apoA-II, which displaces apoA-I from HDL particles, decreases PLTP activity, and hinders the increase in HDL particle size.2 Another possibility is that, similar to CETP, the surface lipid mediates the PLTP bound to lipoprotein and liposome,3 in which smaller size of HDL particles showed more curvature and more hydrophobicity resulting more binding CETPs. Considering the apoA-II can increase HDL size particles,4,5 reducing the surface hydrophobicity and further resulting less bind PLTPs as the case of CETP.3 Unfortunately, our approach and resolution are not able to distinguish the mechanism.
PLTP can interact with HDL3, LDL, VLDL and PL-liposomes both separately and simultaneously in binary complexes. However, PLTP has higher affinity for binding HDL3 than apoB-containing lipoprotein particles. We presume that PLTP binds to the apoA-I and apoB-containing particles using the N-terminal of PLTP when binding only to a single particle. Whether LDL/VLDL-bound PLTP is released from the complex in the presence of HDL3 due to the affinity differences is unclear. More experiments are required to test this possibility, such as immune-EM experiment to identify the orientation of PLTP binding by designing antibodies against the distal ends of PLTP N- and C-terminal β-barrel domains.
Comparing the binding percentages of LDL, VLDL or liposome to PLTP after introducing HDL3 into the solution, it is clear that the percentage of particles bound to PLTP increased (~16.6%, ~11.9%, and ~18.2%, respectively), which implies an allosteric regulation of PLTP after binding to HDL3. The coexistence of ternary complexes of HDL3–PLTP–LDL, HDL3–PLTP–VLDL and HDL3–PLTP–liposome is consistent with the mechanistic model of lipid transfer through a tunnel within the PLTP molecule. However, the low percentage of these complex particles implies that the interaction might be an instantaneous process or at least not stable. The observation of the ternary complex does not exclude the coexistence of a shuttle mechanism. Other experimental approaches, such as asymmetrical flow field–flow fractionation (AsFlFFF) showed that part of 35S-labeled PLTP initially binding with small unilamellar vesicles migrate into HDL population after 45 min incubation [57], These data are in agreement with a higher likelihood of observing the PLTP-HDL3 binary complex than the PLTP-LDL/VLDL/liposome binary complex, suggesting that PLTP molecules generally prefer to attach to the HDL3 surface.
The LDL sample (d = 1.006–1.069 g/ml, apoB 64.9 mg/dL) may include a small fraction of IDL particles and/or VLDL remnants (1.006 <d< 1.019 g/ml). Considering the LDL were isolated from healthy individuals with low triglyceride levels, our experience suggests that less than 5% of the β-migration particles would constitute remnants. Furthermore, based on the particle diameters we did not observe other distinguishable population of particles by EM (Supplementary Fig. 1D). However, we cannot exclude the possibility that a small fraction of IDL interact with PLTP.
Comparing the HDL3 remodeling dynamics between PLTP and CETP, generalizing the HDL3 self-remodeling mechanism
PLTP and CETP both belong to a family of lipid transfer proteins. Here we list the similarities and differences after comparing the current PLTP study with an earlier CETP study.[21, 22] The similarities include: i) PLTP and CETP both show a banana-shaped structure; ii) Both PLTP and CETP can insert into the surfaces of HDL3, LDL, VLDL and liposome by using one distal end of a β-barrel domain, forming a binary complex; iii) Both PLTP and CETP can form a ternary complex between different classes of lipoproteins (such as HDL3 vs. LDL; HDL3 vs. VLDL and HDL3 vs. liposome). However, neither PLTP nor CETP can form a ternary complex within the same class of lipoproteins. The differences include: i) One of the major known functions of PLTP is to regulate the HDL3 size and composition.[58] We observed marked differences in the dynamics and the form of HDL3 modifications by PLTP and CETP. The comparison experiment of PLTP/CETP-induced HDL3 self-remodeling showed that the PLTP-induced HDL3 remodeling speed is much slower in terms of the HDL3 size change compared to CETP (see the detailed discussion in the next section). In addition, the final product of the incubation with CETP included two populations of HDL3 particles, ~6 nm smaller particles and large “spikey” particles, which were not observed in the incubation with PLTP; ii) Comparing the incubation of HDL3-PLTP, HDL3-PLTP-LDL, and HDL3-PTLP-VLDL, we noticed that the HDL3 size change did not show a significant difference with or without LDL/VLDL (Fig. 4A–C). This suggests that PLTP-mediated HDL3 self-remodeling is the major function of PLTP in remodeling HDL3. However, in the same experiment CETP showed that the addition of LDL to an HDL3-CETP mixture can limit the generation of large HDL3 particles (Supplementary Fig. 4), which suggests that CETP-mediated HDL3 remodeling is more dependent on the lipid transfer between HDL3 and LDL comparing to PLTP; iii) In our studies of CETP, [21, 22] we reported that more than five CETP molecules can bind to a single plasma HDL2 particle. In contrast, no more than two PLTP molecules bound to HDL3. It is plausible that the binding of PLTP to HDL3, like that of CETP, is a result of a hydrophobic interaction between PLTP and HDL.[22] However, the N-terminus of PLTP is known to interact with and bind to apoA-I, [54] the main apolipoprotein in HDL3 particles. It is, therefore, possible that protein-protein interactions together with hydrophobic interactions determine the number of PLTP molecules bound to HDL3.
Hypothesized models for the mediation of HDL3 self-remodeling by CETP and PLTP
Our observations are insufficient to provide a full picture of how CETP and PLTP coordinate the transport of lipids and transformation between HDL3 and other types of lipoproteins. However, for a relatively simple system of HDL3-PLTP/CETP, the above comparisons provide some useful hints to understand the HDL3 self-remodeling mechanisms by PLTP/CETP.
Following the previous study of CETP-mediated CE/TG transfer between HDL3 and LDL, [21] two mechanistic models of HDL3 self-remodeling by CETP and PLTP can be proposed: i) the tunnel model, [29] where PLTP/CETP acts as a bridge between two HDL3 particles, facilitating the lipid transfer; and ii) the shuttle model, [30] in which PLTP/CETP acts as a vehicle that transfers lipids from one HDL3 particle to another HDL3 through the aqueous phase. Both models were plausible given the known functions of PLTP/CETP. However, in the tunnel model, we should observe one PLTP/CETP bridge between two HDLs to form a “dumbbell” structure. Unfortunately, the absence of observation of these ternary complexes while imaging thousands of complexes does not support the tunnel mechanism. For a shuttle model, the shrinkage of part of the HDL3 population must be followed by the enlargement of the remaining population. We observed a large variation in the HDL3 size caused by PLTP and CETP, in which the HDL3 sizes were increased by PLTP, while both large and small size HDLs were generated by CETP. However, it is difficult to explain how PLTP/CETP could directionally transfer lipids among the same species of HDLs, and more than half of the CETP naturally appeared on the surface of HDL3, which is less likely to support the shuttle model.
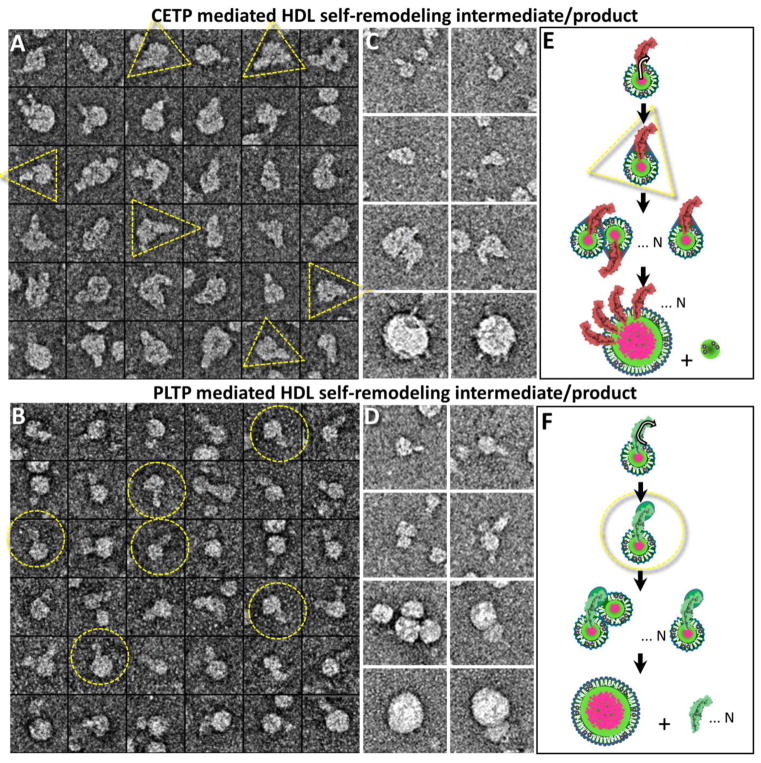
Absent of full support for either mechanism forced us to consider of another possibility. As a small functional unit, the HDL3-PLTP/CETP complex must interact with other HDLs to generate the observed size variation. If this interaction is not caused by a PLTP/CETP bridge or shuttle, then it must be induced by HDL3 itself. Since HDL3 alone does not show a size variation after a sufficiently long incubation time (Supplementary Fig. 1C), the HDL3 variation in the presence of PLTP/CETP must be caused by PLTP/CETP mechanisms. These detailed mechanisms about how PLTP/CETP remodeled the HDL3 size were unknown. To study these mechanisms, we carefully examined the images. In addition to the above observations that large particles bound to many CETPs but rarely bound to PLTP, and that small globular particles (likely the small particles) appeared in the background of the CETP-HDL3 samples but rod-shaped particles (likely PLTP particles) appeared in the background of the PLTP-HDL3 samples, we also unexpectedly found a “triangular-shaped” interaction between CETP and HDL3 in many complexes of CETP-HDL3, especially in the initial stage of incubation (Fig. 6A). These “triangular-shaped” interactions did not clearly appear in the complexes of PLTP-HDL3. In contrast, large HDL3 surfaces were relatively smooth in the presence of PLTP. Moreover, a small bulb was found in the far distal end of PLTP in many PLTP-HDL3 complexes (Fig. 6B), which was not clearly seen in the CETP-HDL3 complexes. These differences may reflect different mechanisms for PLTP and CETP.
Fig. 6. Hypothesized mechanism of PLTP/CETP-mediated HDL3 self-remodeling.
A) Selected particle views of CETP-bound HDL3 showed a “triangular” shaped complex at the initial stage of CETP-HDL3 incubation. B) Selected particle views of PLTP-bound HDL3 showed an “enlarged free tapper end” of PLTP at the initial stage of PLTP-HDL3 incubation. The yellow dashed circle and triangle mark out the free terminal end of PLTP and the CETP-HDL3 complex, respectively. C and D) Hypothesized mechanism of CETP- and PLTP-mediated HDL3 self-remodeling. Selected particles of the HDL3-CETP/PLTP complexes shown in the left panel. Schematic of PLTP remodeling of HDL3 through three stages: i) the formation of an unstable HDL3-CETP/PLTP complex, ii) HDL3-CETP/PLTP complex merging and iii) re-stabilizing, which results in different appearances of enlarged HDL3 due to the amount of lipid transfer protein binding. CETP, PLTP, phospholipid, neutral lipids and ApoA-I molecules are displayed in magenta, green, white, pink and black, respectively. Box size: A and B, 37 nm; C and D, 62 nm.
We hypothesize that the HDL3 remodeling mechanism of CETP mainly involves the transfer of lipids from the HDL3 core to the HDL3 surface through the central lipid pore to the concave surface of CETP, which leads to the formation of a triangle shape (Fig. 6C, first two rows). When more CETP particles insert into the HDL3 surface, more imperfections may appear on the outer phospholipid surface of HDL3, causing more hydrophobic interactions and a greater opportunity to fuse with other HDL3-CETP complexes to form large particles. The number of CETP on the large HDL surface shall keep increasing as more HDL3-CETP complexes are incorporated. However, the balancing process of the HDL3 surface pressure may result in apolipoprotein separation from the merged HDL3 (Fig. 6C, last two rows). In comparison to CETP, PLTP has a lower binding number to HDLs, resulting the presence of more PLTP in the sample background (Fig. 6D, last two rows). We hypothesize that the PLTP mechanism of HDL3 remodeling mainly involves the transfer of phospholipids from the HDL3 surface to the distal end of PLTP (Fig. 6D, first two rows). The transfer of lipids to the outside of HDL3 by PLTP would also cause the exposure of more hydrophobic components, destabilizing the original HDL3 structure. The merging of several HDL3-PLTP complexes may cause the release of the bound PLTP molecules. The HDL3 remodeling also includes the release of surface apoA-I-PL pre-β molecular complexes from the HDL3 particles and ensuing further particle fusion.[48]
As a result, the HDL3 particle transformation step induced by the bound PLTP/CETP may be the prerequisite for the initiation of HDL3 particle merging.[22, 59] This explains why the HDL3 size exhibits almost no change within the first 40 minutes for PLTP and 15 minutes for CETP (Fig. 5). The capture of the intermediate stage of PLTP/CETP-mediated destabilization of the HDL3 particle provides direct evidence to support the proposed particle merging model for HDL3 self-remodeling.
In summary, the EM 3D structure of PLTP supports the homology model of PLTP. PLTP has a similar interaction with HDL and LDL to that of CETP. However, the PLTP activity in HDL remodeling is slower than that of CETP and occurs via a different mechanism. The EM images of the interaction of PLTP with lipoproteins provide the first picture for understanding PLTP-mediated lipid transfer, relevant for treating dyslipidemia.
Methods
Synthesis and isolation of PLTP and lipoprotein
PLTP (~1.625 mg/ml) was isolated using the previously reported procedure.[60, 61] Briefly, a PLTP His-tagged construct was expressed in mammalian cells, isolated from the concentrated conditioned media, purified, and stored at −80°C until use. The purity of the isolated PLTP was evaluated by mass spectrometry, and no significant contaminants or other proteins were found in the isolated material. Wild-type plasma HDL3 was isolated from fresh human plasma using ultracentrifugation as previously described.[62] It contained 4.28 mg/ml protein, 2.39 mg/ml CE and 1.03 mg/ml TG. LDL (d = 1.006–1.069 g/ml, apoB 64.9 mg/dL) and VLDL (d < 1.006 g/ml, apoB 24.5 mg/dL) were isolated in the Krauss laboratory by sequential flotation of plasma from fasted, healthy male volunteers and further purified by ultracentrifugation.[63] Liposome vesicle samples were produced by Encapsula NanoSciences (Brentwood, TN). The sample contained 1 mg/ml 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC, from Avanti Polar lipids) with a peak vesicle size of ~50 nm in a buffer containing 20 mM Tris-Cl, 154 mM NaCl, at pH 7.4.
EM specimen preparation by the optimized NS (OpNS)
Conventional cryo-EM is often used for protein structural studies under physiological conditions since it avoids the potential artifacts induced by fixatives and stains.[37] Still, cryo-EM studies of PLTP are challenging; small molecules (<150 kDa) are difficult to image or reconstruct using the cryo-EM single-particle approach because of low contrast.[64] Thus, we studied human PLTP using the OpNS protocol.[34, 37] Our OpNS protocol, refined from the conventional negative-staining protocol, eliminates rouleaux artifacts of lipoprotein particles and has been statistically validated as a way to determine lipoprotein particle shapes and sizes.[34, 37]
Both the PLTP and PLTP-lipoprotein/liposome complex samples were prepared with the optimized NS protocol (OpNS).[37] The PLTP was either directly diluted 1000 times (final concentration 1.625 μg/ml) with Dulbecco’s phosphate-buffered saline (DPBS: 2.7 mM KCl, 1.46 mM KH2PO4, 136.9 mM NaCl, and 8.1 mM Na2HPO4; Invitrogen) buffer for PLTP Single Particle and IPET 3D reconstruction or co-incubated with HDL3 at a molar ratio of 3:1 for PLTP-HDL3 complex 3D reconstruction. LDL, VLDL and liposome were also co-incubated with PLTP and HDL3 at molar ratios of 9:3:1 (PLTP:HDL3:LDL/VLDL/liposome) with their original buffer at 37°C for studying the PLTP-mediated lipid transfer activity among lipoproteins/liposome. To examine complexes at different time intervals, samples from the original incubation solution were fast fixed at 0 min, 15 min, 40 min, 2 hrs, 8 hrs, and 24 hrs on a carbon grid following the OpNS protocol. In brief, an aliquot (~3 μl) was placed on a thin-carbon-coated 200 mesh copper grid (Cu-200CN, Pacific Grid-Tech, San Francisco, CA) that had been glow-discharged. After ~1 min, the excess solution was blotted with filter paper, followed by a procedure of washing and staining as described.[34, 37] Three drops of 1% (w/v) uranyl formate (UF) negative stain on parafilm were then applied successively before being nitrogen-air-dried at room temperature. Since the UF solutions are light sensitive and unstable, this operation was performed in the dark.[34, 37]
Electron microscopy data acquisition and image pre-processing
The OpNS micrographs were acquired at room temperature under a defocus of ~0.6 um on a Gatan UltraScan 4 K × 4 K CCD equipped on a Zeiss Libra 120 Plus transmission electron microscope (Carl Zeiss NTS GmbH, Oberkochen, Germany). TEM was operated under a high-tension of 120 kV, energy filtering of 20 eV and 4 magnification range of 31.5 K to 80 K, in which each pixel of the micrographs corresponded to 3.68 to 1.48 Å, respectively. A total of ~230 micrographs were collected for the single particle reconstruction for the PLTP and PLTP-HDL3 complex and ~10 micrographs were collected from each sample condition for the HDL-PLTP-LDL/VLDL/liposome incubation. The defocus of each micrograph was determined by fitting the contrast transfer function (CTF) parameters with its power spectrum using ctffind3 in the FREALIGN software package.[65] The phase of each micrograph was corrected by a Wiener filter with the SPIDER software package.[66]. Approximately 200–500 particles from each ternary mixture sample at different time points were windowed and selected by the boxer software in the EMAN software package.[43] These particles are submitted for Gaussian low-pass filtering before statistical analysis.
Electron tomography data collection and image pre-processing
Electron tomography (ET) data of the PLTP and PLTP-HDL3 specimens were acquired under a less than 2 μm defocus with a high-sensitivity 4,096 × 4,096 pixel Gatan Ultrascan CCD camera at 80,000x magnification and with the same Zeiss Libra 120 TEM (each pixel of the micrograph corresponds to 1.48 Å). The specimens mounted on a Gatan 626 high-tilt room-temperature holder were tilted at angles ranging from −60° to 60° in steps of 2° for the PLTP specimen and −62° to 67° in steps of 1.5° for the PLTP-HDL3 specimen. The total illumination electron dose was ~200 e−/Å2. The tilt series of tomographic data were controlled and imaged by manual operation with the Gatan tomography software (Zeiss Libra 120 TEM) and automated tomography software preinstalled in the microscopes.[67] Collected micrographs were initially aligned together following the procedure of the IMOD software package.[68] The tilt series of each particle image in windows of 220 × 220 pixels (PLTP) and 256 × 256 pixels (PLTP-HDL3) were semi-automatically tracked and selected by the IPET software. The defocus of the small particle image area of each tilt micrograph was examined by fitting the CTF parameters with its power spectrum by ctffind3 in the FREALIGN software package.[69] The CTF was then corrected by TOMOCTF.[70]
Individual particle electron tomography (IPET) 3D reconstruction
The 3D density maps of an individual PLTP and PLTP-HDL3 complex were reconstructed by the IPET method.[42] In brief, a small image area containing only a single PLTP particle and PLTP-HDL3 complex particle were windowed from each tilted whole-micrograph after CTF correction. An ab-initio model was generated by directly back-projecting these small images into a 3D map. The map was then refined via three rounds of refinement loops (including more than a hundred iterations) by the focused electron tomography reconstruction (FETR) algorithm.[42] In FETR, an automatically generated dynamic Gaussian low-pass filter and an automatically generated soft-mask were applied to both the references and tilted images to achieve the final 3D reconstruction. To analyze tomographic 3D reconstructions, the center-refined raw ET images were split into two groups based on having an odd- or even-numbered index in the order of tilt angles.[42] The frequency at which the intra-FSC curve falls to a value of 0.5 showed that the resolutions of the reconstruction density maps of the PLTP and PLTP-HDL3 particles are in the range of ~19–27 Å. A 3D IPET density map from the IPET reconstruction was low pass filtered to 80 Å, which prepared for the initial model for single particle reconstruction.
Single particle 3D reconstruction of PLTP and PLTP-HDL3 particles
Approximately 7,000 isolated PLTP particles and ~5,000 HDL3-PLTP complexes (from an initial pool of ~32,000 particles) were extracted from the micrographs by using a window of 196 × 196 and 256 × 256 pixel images, respectively, using the e2boxer.py program in EMAN2.[71] The CTF-corrected images of the particles were submitted for reference-free class averaging and approximately 300 class averages were generated for both the PLTP and the PLTP-HDL3 complex using refind2d.py in EMAN.[43] To prevent bias from a given initial model for single particle 3D reconstruction, the IPET 3D reconstructions of the PLTP and the PLTP-HDL3 complex was filtered to 80 Å and then used as the initial models.
According to the 0.5 Fourier shell correlation criterion, [72] the final resolutions of the asymmetric reconstructions of the PLTP and the PLTP-HDL3 complex were 19 Å and 23 Å, respectively. The contour level of the 81 kDa PLTP was estimated from the average density of protein of 1.22 g cm−3. [73]
Statistical analyses of PLTP binding to lipoprotein particles
To harvest a sufficient number of isolated lipoprotein/PLTP particles for statistical analysis, 4–5 images (containing 300–500 particles) were collected from each sample at a given time mentioned above. For the statistic of the percentage of HDL3 binding to PLTPs, the number of bound PLTPs was counted by accumulating the number of observed rod-shaped protrusions on the edge of the sphere. This number could be slightly different by including the undetectable CETPs that were located behind or in front of the lipoprotein particles. As we previously calculated, [22] the probability (℘) (i.e., the ratio of the PLTP visible area vs. the overall sphere area) is , where d is the lipoprotein diameter and l is the PLTP protrusion length. Since the sizes of HDLs were small, this correction was not significant. The particle diameter and the PLTP protrusion width and length were determined by measuring the diameters in two orthogonal directions, as previously described.[22] In brief, the geometric mean of the two perpendicular diameters was used to represent the particle diameter and the PLTP protrusion geometry.
Supplementary Material
Highlights.
TEM imaging the interaction between phospholipid transfer protein and lipoproteins
Acknowledgments
We thank Drs. Drs. Douglas and Ronald Krauss for providing the HDL3 and LDL samples. This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (R01HL115153, R01GM104427, and P01HL030086). Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
AUTHOR CONTRIBUTIONS
This project was initiated and designed by SV and GR. MZ, DGJ and RK prepared the materials. MZ conducted the experiments and acquired the OpNS data. MZ processed the NS data and computed the statistics. MZ and XZ reconstructed the 3D IPET. MZ drafted the initial manuscript, which was revised by JJA, SV, DGJ, RK and GR.
Conflict Interest
Authors have no conflict interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rao R, Albers JJ, Wolfbauer G, Pownall HJ. Molecular and macromolecular specificity of human plasma phospholipid transfer protein. Biochemistry. 1997;36:3645–3653. doi: 10.1021/bi962776b. [DOI] [PubMed] [Google Scholar]
- 2.Tall AR, Krumholz S, Olivecrona T, Deckelbaum RJ. Plasma phospholipid transfer protein enhances transfer and exchange of phospholipids between very low density lipoproteins and high density lipoproteins during lipolysis. Journal of lipid research. 1985;26:842–851. [PubMed] [Google Scholar]
- 3.Jauhiainen M, Metso J, Pahlman R, Blomqvist S, van Tol A, Ehnholm C. Human plasma phospholipid transfer protein causes high density lipoprotein conversion. The Journal of biological chemistry. 1993;268:4032–4036. [PubMed] [Google Scholar]
- 4.Asztalos BF, Study HDLAT. High-density lipoprotein metabolism and progression of atherosclerosis: new insights from the HDL Atherosclerosis Treatment Study. Current opinion in cardiology. 2004;19:385–391. doi: 10.1097/01.hco.0000126979.41946.7e. [DOI] [PubMed] [Google Scholar]
- 5.Oram JF, Wolfbauer G, Tang C, Davidson WS, Albers JJ. An amphipathic helical region of the N-terminal barrel of phospholipid transfer protein is critical for ABCA1-dependent cholesterol efflux. The Journal of biological chemistry. 2008;283:11541–11549. doi: 10.1074/jbc.M800117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlitt A, Bickel C, Thumma P, Blankenberg S, Rupprecht HJ, Meyer J, Jiang XC. High plasma phospholipid transfer protein levels as a risk factor for coronary artery disease. Arterioscl Throm Vas. 2003;23:1857–1862. doi: 10.1161/01.ATV.0000094433.98445.7F. [DOI] [PubMed] [Google Scholar]
- 7.Bickel C, Thumma P, Blankenberg S, Rupprecht HJ, Schlitt A, Meyer J, Jiang XC. High plasma phospholipid transfer protein (PLTP) levels as a risk factor for coronary artery disease. Circulation. 2002;106:45–45. doi: 10.1161/01.ATV.0000094433.98445.7F. [DOI] [PubMed] [Google Scholar]
- 8.Chen XY, Sun AJ, Mansoor A, Zou YZ, Ge JB, Lazar JM, Jiang XC. Plasma PLTP activity is inversely associated with HDL-C levels. Nutr Metab. 2009;6 doi: 10.1186/1743-7075-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlitt A, Blankenberg S, Bickel C, Lackner KJ, Heine GH, Buerke M, Werdan K, Maegdefessel L, Raaz U, Rupprecht HJ, Munzel T, Jiang XC. PLTP activity is a risk factor for subsequent cardiovascular events in CAD patients under statin therapy: the AtheroGene Study. Journal of lipid research. 2009;50:723–729. doi: 10.1194/jlr.M800414-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schgoer W, Mueller T, Jauhiainen M, Wehinger A, Gander R, Tancevski I, Salzmann K, Eller P, Ritsch A, Haltmayer M, Ehnholm C, Patsch JR, Foeger B. Low phospholipid transfer protein (PLTP) is a risk factor for peripheral atherosclerosis. Atherosclerosis. 2008;196:219–226. doi: 10.1016/j.atherosclerosis.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 11.Liu RJ, Hojjati MR, Devlin CM, Hansen IH, Jiang XC. Macrophage phospholipid transfer protein deficiency and ApoE secretion - Impact on mouse plasma cholesterol levels and atherosclerosis. Arterioscl Throm Vas. 2007;27:190–196. doi: 10.1161/01.ATV.0000249721.96666.e5. [DOI] [PubMed] [Google Scholar]
- 12.Day JR, Albers JJ, Lofton-Day CE, Gilbert TL, Ching AF, Grant FJ, O’Hara PJ, Marcovina SM, Adolphson JL. Complete cDNA encoding human phospholipid transfer protein from human endothelial cells. The Journal of biological chemistry. 1994;269:9388–9391. [PubMed] [Google Scholar]
- 13.Masson D, Jiang XC, Lagrost L, Tall AR. The role of plasma lipid transfer proteins in lipoprotein metabolism and atherogenesis. Journal of lipid research. 2009;50(Suppl):S201–206. doi: 10.1194/jlr.R800061-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beamer LJ. Structure of human BPI (bactericidal/permeability-increasing protein) and implications for related proteins. Biochemical Society transactions. 2003;31:791–794. doi: 10.1042/bst0310791. [DOI] [PubMed] [Google Scholar]
- 15.Kawano K, Qin SC, Lin M, Tall AR, Jiang XC. Cholesteryl ester transfer protein and phospholipid transfer protein have nonoverlapping functions in vivo. Journal of Biological Chemistry. 2000;275:29477–29481. doi: 10.1074/jbc.M003523200. [DOI] [PubMed] [Google Scholar]
- 16.Morton RE, Izem L. Modification of CETP function by changing its substrate preference: a new paradigm for CETP drug design. Journal of lipid research. 2015;56:612–619. doi: 10.1194/jlr.M056333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang XC, Bruce C, Mar J, Lin M, Ji Y, Francone OL, Tall AR. Targeted mutation of plasma phospholipid transfer protein gene markedly reduces high-density lipoprotein levels. The Journal of clinical investigation. 1999;103:907–914. doi: 10.1172/JCI5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rye KA, Hime NJ, Barter PJ. Evidence that cholesteryl ester transfer protein-mediated reductions in reconstituted high density lipoprotein size involve particle fusion. Journal of Biological Chemistry. 1997;272:3953–3960. doi: 10.1074/jbc.272.7.3953. [DOI] [PubMed] [Google Scholar]
- 19.Settasatian N, Duong M, Curtiss LK, Ehnholm C, Jauhiainen M, Huuskonen J, Rye KA. The mechanism of the remodeling of high density lipoproteins by phospholipid transfer protein. The Journal of biological chemistry. 2001;276:26898–26905. doi: 10.1074/jbc.M010708200. [DOI] [PubMed] [Google Scholar]
- 20.Rye KA, Hime NJ, Barter PJ. Evidence that cholesteryl ester transfer protein-mediated reductions in reconstituted high density lipoprotein size involve particle fusion. The Journal of biological chemistry. 1997;272:3953–3960. doi: 10.1074/jbc.272.7.3953. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Yan F, Zhang S, Lei D, Charles MA, Cavigiolio G, Oda M, Krauss RM, Weisgraber KH, Rye KA, Pownall HJ, Qiu X, Ren G. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nature chemical biology. 2012;8:342–349. doi: 10.1038/nchembio.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Charles R, Tong H, Zhang L, Patel M, Wang F, Rames MJ, Ren A, Rye KA, Qiu X, Johns DG, Charles MA, Ren G. HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. Scientific reports. 2015;5:8741. doi: 10.1038/srep08741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Lei D, Peng B, Yang M, Zhang L, Charles MA, Rye KA, Krauss RM, Johns DG, Ren G. Assessing the mechanisms of cholesteryl ester transfer protein inhibitors. Biochimica et biophysica acta. 2017;1862:1606–1617. doi: 10.1016/j.bbalip.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauer ME, Graff-Meyer A, Rufer AC, Maugeais C, von der Mark E, Matile H, D’Arcy B, Magg C, Ringler P, Muller SA, Scherer S, Dernick G, Thoma R, Hennig M, Niesor EJ, Stahlberg H. Cholesteryl ester transfer between lipoproteins does not require a ternary tunnel complex with CETP. J Struct Biol. 2016;194:191–198. doi: 10.1016/j.jsb.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Huuskonen J, Wohlfahrt G, Jauhiainen M, Ehnholm C, Teleman O, Olkkonen VM. Structure and phospholipid transfer activity of human PLTP: analysis by molecular modeling and site-directed mutagenesis. Journal of lipid research. 1999;40:1123–1130. [PubMed] [Google Scholar]
- 26.Bruce C, Beamer LJ, Tall AR. The implications of the structure of the bactericidal/permeability-increasing protein on the lipid-transfer function of the cholesteryl ester transfer protein. Curr Opin Struc Biol. 1998;8:426–434. doi: 10.1016/s0959-440x(98)80118-8. [DOI] [PubMed] [Google Scholar]
- 27.Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu X, Mistry A, Ammirati MJ, Chrunyk BA, Clark RW, Cong Y, Culp JS, Danley DE, Freeman TB, Geoghegan KF, Griffor MC, Hawrylik SJ, Hayward CM, Hensley P, Hoth LR, Karam GA, Lira ME, Lloyd DB, McGrath KM, Stutzman-Engwall KJ, Subashi AK, Subashi TA, Thompson JF, Wang IK, Zhao H, Seddon AP. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nature structural & molecular biology. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 29.Ihm J, Quinn DM, Busch SJ, Chataing B, Harmony JA. Kinetics of plasma protein-catalyzed exchange of phosphatidylcholine and cholesteryl ester between plasma lipoproteins. Journal of lipid research. 1982;23:1328–1341. [PubMed] [Google Scholar]
- 30.Barter PJ, Jones ME. Kinetic studies of the transfer of esterified cholesterol between human plasma low and high density lipoproteins. Journal of lipid research. 1980;21:238–249. [PubMed] [Google Scholar]
- 31.Ren G, Rudenko G, Ludtke SJ, Deisenhofer J, Chiu W, Pownall HJ. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1059–1064. doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y, Kuang YL, Lei D, Zhai X, Zhang M, Krauss RM, Ren G. Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography1. Journal of lipid research. 2016;57:1879–1888. doi: 10.1194/jlr.M070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Antwerpen R, La Belle M, Navratilova E, Krauss RM. Structural heterogeneity of apoB-containing serum lipoproteins visualized using cryo-electron microscopy. Journal of lipid research. 1999;40:1827–1836. [PubMed] [Google Scholar]
- 34.Zhang L, Song J, Cavigiolio G, Ishida BY, Zhang S, Kane JP, Weisgraber KH, Oda MN, Rye KA, Pownall HJ, Ren G. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. Journal of lipid research. 2011;52:175–184. doi: 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B, Ren X, Neville T, Jerome WG, Hoyt DW, Sparks D, Ren G, Wang J. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein Sci. 2009;18:921–935. doi: 10.1002/pro.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Song J, Newhouse Y, Zhang S, Weisgraber KH, Ren G. An optimized negative-staining protocol of electron microscopy for apoE4 POPC lipoprotein. Journal of lipid research. 2010;51:1228–1236. doi: 10.1194/jlr.D002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Tong H, Garewal M, Ren G. Optimized negative-staining electron microscopy for lipoprotein studies. Biochimica et biophysica acta. 2013;1830:2150–2159. doi: 10.1016/j.bbagen.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong H, Zhang L, Kaspar A, Rames MJ, Huang L, Woodnutt G, Ren G. Peptide-conjugation induced conformational changes in human IgG1 observed by optimized negative-staining and individual-particle electron tomography. Sci Rep-Uk. 2013;3:1089. doi: 10.1038/srep01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Lei DS, Smith JM, Zhang M, Tong HM, Zhang X, Lu ZY, Liu JK, Alivisatos AP, Ren G. Three-dimensional structural dynamics and fluctuations of DNA-nanogold conjugates by individual-particle electron tomography. Nature communications. 2016;7 doi: 10.1038/ncomms11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rames M, Yu Y, Ren G. Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy. Journal of visualized experiments: JoVE. 2014:e51087. doi: 10.3791/51087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Ren G. IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. PloS one. 2012;7:e30249. doi: 10.1371/journal.pone.0030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Desrumaux C, Labeur C, Verhee A, Tavernier J, Vandekerckhove J, Rosseneu M, Peelman F. A hydrophobic cluster at the surface of the human plasma phospholipid transfer protein is critical for activity on high density lipoproteins. Journal of Biological Chemistry. 2001;276:5908–5915. doi: 10.1074/jbc.M008420200. [DOI] [PubMed] [Google Scholar]
- 45.Oram JF, Wolfbauer G, Tang C, Davidson WS, Albers JJ. An amphipathic helical region of the N-terminal barrel of phospholipid transfer protein is critical for ABCA1-dependent cholesterol efflux. Journal of Biological Chemistry. 2008;283:11541–11549. doi: 10.1074/jbc.M800117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chirasani VR, Revanasiddappa PD, Senapati S. Structural Plasticity of Cholesteryl Ester Transfer Protein Assists the Lipid Transfer Activity. The Journal of biological chemistry. 2016;291:19462–19473. doi: 10.1074/jbc.M116.744623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Korhonen A, Jauhiainen M, Ehnholm C, Kovanen PT, Ala-Korpela M. Remodeling of HDL by phospholipid transfer protein: demonstration of particle fusion by 1H NMR spectroscopy. Biochemical and biophysical research communications. 1998;249:910–916. doi: 10.1006/bbrc.1998.9162. [DOI] [PubMed] [Google Scholar]
- 49.Damen J, Regts J, Scherphof G. Transfer of [14C]phosphatidylcholine between liposomes and human plasma high density lipoprotein. Partial purification of a transfer-stimulating plasma factor using a rapid transfer assay. Biochimica et biophysica acta. 1982;712:444–452. doi: 10.1016/0005-2760(82)90271-5. [DOI] [PubMed] [Google Scholar]
- 50.Cheung MC, Wolfbauer G, Albers JJ. Plasma phospholipid mass transfer rate: relationship to plasma phospholipid and cholesteryl ester transfer activities and lipid parameters. Biochimica et biophysica acta. 1996;1303:103–110. doi: 10.1016/0005-2760(96)00082-3. [DOI] [PubMed] [Google Scholar]
- 51.Oka T, Kujiraoka T, Ito M, Egashira T, Takahashi S, Nanjee MN, Miller NE, Metso J, Olkkonen VM, Ehnholm C, Jauhiainen M, Hattori H. Distribution of phospholipid transfer protein in human plasma: presence of two forms of phospholipid transfer protein, one catalytically active and the other inactive. Journal of lipid research. 2000;41:1651–1657. [PubMed] [Google Scholar]
- 52.Janis MT, Siggins S, Tahvanainen E, Vikstedt R, Silander K, Metso J, Aromaa A, Taskinen MR, Olkkonen VM, Jauhiainen M, Ehnholm C. Active and low-active forms of serum phospholipid transfer protein in a normal Finnish population sample. Journal of lipid research. 2004;45:2303–2309. doi: 10.1194/jlr.M400250-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Cheung MC, Wolfbauer G, Albers JJ. Different phospholipid transfer protein complexes contribute to the variation in plasma PLTP specific activity. Biochimica et biophysica acta. 2011;1811:343–347. doi: 10.1016/j.bbalip.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pussinen PJ, Jauhiainen M, Metso J, Pyle LE, Marcel YL, Fidge NH, Ehnholm C. Binding of phospholipid transfer protein (PLTP) to apolipoproteins A-I and A-II: location of a PLTP binding domain in the amino terminal region of apoA-I. Journal of lipid research. 1998;39:152–161. [PubMed] [Google Scholar]
- 55.Boucher J, Ramsamy TA, Braschi S, Sahoo D, Neville TA, Sparks DL. Apolipoprotein A-II regulates HDL stability and affects hepatic lipase association and activity. Journal of lipid research. 2004;45:849–858. doi: 10.1194/jlr.M300431-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Weng W, Brandenburg NA, Zhong S, Halkias J, Wu L, Jiang XC, Tall A, Breslow JL. ApoA-II maintains HDL levels in part by inhibition of hepatic lipase. Studies In apoA-II and hepatic lipase double knockout mice. Journal of lipid research. 1999;40:1064–1070. [PubMed] [Google Scholar]
- 57.Setala NL, Holopainen JM, Metso J, Wiedmer SK, Yohannes G, Kinnunen PK, Ehnholm C, Jauhiainen M. Interfacial and lipid transfer properties of human phospholipid transfer protein: implications for the transfer mechanism of phospholipids. Biochemistry. 2007;46:1312–1319. doi: 10.1021/bi0621866. [DOI] [PubMed] [Google Scholar]
- 58.Albers JJ, Vuletic S, Cheung MC. Role of plasma phospholipid transfer protein in lipid and lipoprotein metabolism. Biochimica et biophysica acta. 2012;1821:345–357. doi: 10.1016/j.bbalip.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu YD, Kuang YL, Lei DS, Zhai XB, Zhang M, Krauss RM, Ren G. Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo-electron tomography. Journal of lipid research. 2016;57:1879–1888. doi: 10.1194/jlr.M070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albers JJ, Wolfbauer G, Cheung MC, Day JR, Ching AF, Lok S, Tu AY. Functional expression of human and mouse plasma phospholipid transfer protein: effect of recombinant and plasma PLTP on HDL subspecies. Biochimica et biophysica acta. 1995;1258:27–34. doi: 10.1016/0005-2760(95)00091-p. [DOI] [PubMed] [Google Scholar]
- 61.Albers JJ, Day JR, Wolfbauer G, Kennedy H, Vuletic S, Cheung MC. Impact of site-specific N-glycosylation on cellular secretion, activity and specific activity of the plasma phospholipid transfer protein. Biochimica et biophysica acta. 2011;1814:908–911. doi: 10.1016/j.bbapap.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han S, Flattery AM, McLaren D, Raubertas R, Lee SH, Mendoza V, Rosa R, Geoghagen N, Castro-Perez JM, Roddy TP, Forrest G, Johns D, Hubbard BK, Li J. Comparison of lipoprotein separation and lipid analysis methodologies for human and cynomolgus monkey plasma samples. Journal of cardiovascular translational research. 2012;5:75–83. doi: 10.1007/s12265-011-9340-9. [DOI] [PubMed] [Google Scholar]
- 63.Caulfield MP, Li S, Lee G, Blanche PJ, Salameh WA, Benner WH, Reitz RE, Krauss RM. Direct determination of lipoprotein particle sizes and concentrations by ion mobility analysis. Clinical chemistry. 2008;54:1307–1316. doi: 10.1373/clinchem.2007.100586. [DOI] [PubMed] [Google Scholar]
- 64.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biological procedures online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. Journal of structural biology. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of structural biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Li H, Zhang L, Rames M, Zhang M, Yu Y, Peng B, Celis CD, Xu A, Zou Q, Yang X, Chen X, Ren G. Fully Mechanically Controlled Automated Electron Microscopic Tomography. Scientific reports. 2016;6:29231. doi: 10.1038/srep29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of Structural Biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 69.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Fernandez JJ, Li S, Crowther RA. CTF determination and correction in electron cryotomography. Ultramicroscopy. 2006;106:587–596. doi: 10.1016/j.ultramic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 71.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 72.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 73.Andersson KM, Hovmoller S. The protein content in crystals and packing coefficients in different space groups. Acta crystallographica Section D, Biological crystallography. 2000;56:789–790. doi: 10.1107/s0907444900005163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.