Abstract
The Teladorsagia circumcincta P-glycoprotein-9 (Tci-pgp-9) gene has previously been implicated in multiple-anthelmintic resistance in this parasite. Here we further characterise genetic diversity in Tci-pgp-9 and its possible role in ivermectin (IVM) and multi-drug resistance using two UK field isolates of T. circumcincta, one susceptible to anthelmintics (MTci2) and the other resistant to most available anthelmintics including IVM (MTci5). A comparison of full-length Tci-pgp-9 cDNA transcripts from the MTci2 and MTci5 isolates (∼3.8 kb in both cases) indicated that they shared 95.6% and 99.5% identity at the nucleotide and amino acid levels, respectively. Nine non-synonymous SNPs were found in the MTci5 sequences relative to their MTci2 counterparts. Twelve genomic sequence variants of the first internucleotide binding domain of Tci-pgp-9 were identified and up to 10 of these were present in some individual worms, strongly supporting previous evidence that amplification of this gene has occurred in T. circumcincta. On average, fewer distinct sequence variants of Tci-pgp-9 were present in individual worms of the MTci5 isolate than in those of the MTci2 isolate. A further reduction in the number of sequence variants was observed in individuals derived from an IVM-treated sub-population of MTci5. These findings suggest that Tci-pgp-9 was under purifying selection in the face of IVM treatment in T. circumcincta, with some sequence variants being selected against.
Keywords: Teladorsagia circumcincta, Anthelmintic resistance, P-glycoprotein, Ivermectin
Graphical abstract
Highlights
-
•
Tci-pgp-9 cDNA sequences from the MTci2 and MTci5 isolates shared 95.6% identity.
-
•
Comparison of Tci-pgp-9 sequences from MTci5 and MTci2 showed 9 non-synonymous SNPs.
-
•
Multiple Tci-pgp-9-IBDA sequence variants occurred in the majority of individual larvae.
-
•
Tci-pgp-9 appears to be under purifying selection after anthelmintics are applied.
1. Introduction
The control of parasitic nematodes of ruminants, such as Teladorsagia circumcincta, currently relies heavily on the use of anthelmintic drugs, and this has led to widespread selection for drug resistance. Anthelmintic resistance in T. circumcincta has been documented in most countries where this parasite occurs (Kaplan, 2004) and it has been reported in UK field populations against three of the five major classes of anthelmintics. Resistance to single and multiple classes of anthelmintic has frequently been diagnosed in populations of this species, with ‘triple-resistance’ (i.e. resistant to benzimidazoles (BZs), imidothiazoles and macrocyclic lactones (MLs)) being reported in T. circumcincta isolates from sheep in Europe more than a decade ago (Sargison et al., 2001). Although as yet there have been no reported field cases in the UK of resistance to the amino-acetonitrile derivative class of anthelmintics, which includes Monepantel, monepantel-resistance has been observed in T. circumcincta under experimental conditions (Bartley et al., 2015). Disturbingly, monepantel-resistance has been reported in field populations of T. circumcincta (and Trichostronglus colubriformis) from sheep and goats in New Zealand (Leathwick et al., 2013, Scott et al., 2013) and in Haemonchus contortus in sheep in Australia (Sales and Love, 2016) and in the Netherlands (Van den Brom et al., 2015).
Anthelmintic resistance in most cases is accepted as a pre-adaptive phenomenon (Jackson and Coop, 2000) in which the genes responsible for resistance already exist at a low frequency within a population but become dominant under drug selection. Anthelmintic resistance has been attributed to several genetic factors including qualitative and/or quantitative changes in putative drug targets (e.g. glutamate-gated chloride channels and β-tubulin), and members of the adenosine triphosphate (ATP)-binding cassette (ABC)-transporter superfamily (P-glycoproteins (Pgps), multi-drug resistant proteins and half-transporters) have also been implicated (Xu et al., 1998, Prichard and Roulet, 2007, James and Davey, 2009, Dupuy et al., 2010). Pgps have been implicated in the molecular basis of IVM-resistance in H. contortus (Xu et al., 1998, Molento and Prichard, 2001), and polymorphisms in certain alleles have the potential to improve drug efflux from the cell, thereby changing the drug distribution within the parasite's tissues and preventing anthelmintics reaching their site of action (Wolstenholme et al., 2004, Prichard and Roulet, 2007). Studies in which IVM or moxidectin (MOX) were co-administered with Pgp-inhibitors (such as verapamil) have shown increased efficacy of these drugs against MOX-resistant H. contortus (Xu et al., 1998, Molento et al., 2004, Bartley et al., 2009). Functional recombinant expression of Cylicocyclus elongatus pgp-9 in a Saccharomyces cerevisiae strain that was deficient in endogenous ABC-transported demonstrated that MLs are substrates for pgp-9 in this equine parasite (Kaschny et al., 2015). Moreover, recent studies have linked polymorphisms in Pgps with a multi-drug-resistant phenotype in T. circumcincta (Bisset, 2007, Dicker et al., 2011a, Dicker et al., 2011b), H. contortus (Blackhall et al., 1998, Williamson et al., 2011), Cooperia oncophora (Demeler et al., 2013), Parascaris equorum (Janssen et al., 2013) and Onchocerca volvulus (Bourguinat et al., 2008). Increased expression of Pgp genes has also been associated with IVM resistance in T. circumcincta (Dicker et al., 2011b, Raza et al., 2016, Choi et al., 2017) H. contortus (Xu et al., 1998), C. oncophora (Areskog et al., 2013, De Graef et al., 2013) and C. elegans (James and Davey, 2009).
One specific Pgp gene, Tci-pgp-9, has been associated with multi-drug resistant phenotypes in two independent studies which demonstrated an increase in expression and polymorphisms in Tci-pgp-9 from multiple-anthelmintic resistant T. circumcincta (Bisset, 2007, Dicker, 2010). Bisset (2007) and subsequently Choi et al. (2017) have reported gene amplification, alternative splicing and four non-synonymous, exonic SNPs in Tci-pgp-9 when comparing a multiple-anthelmintic resistant strain with its anthelmintic-susceptible “near-isogenic” sister strain in New Zealand (NZ). Using field isolates from the UK, Dicker (2010) showed that there was a higher level of constitutive expression of Tci-pgp-9 in the anthelmintic resistant isolate (MTci5) in all life-cycle stages, relative to an unrelated susceptible isolate (MTci2). In a subsequent study conducted on an IVM-exposed population of MTci5, evidence for inducible Tci-pgp-9 expression was also obtained (Dicker et al., 2011b). Significantly higher levels of transcription of pgp-9.1 and pgp-9.2 have similarly been shown in comparisons of anthelmintic susceptible and resistant isolates of H. contortus following in vitro exposure to IVM and LEV (Raza et al., 2016).
The aim of the present study was to further characterise Tci-pgp-9 and its putative role in IVM resistance in T. circumcincta. Specifically, we wished to sequence similar regions of the gene in susceptible and resistant isolates from the UK to determine whether there was an association of drug resistance with any specific sequence variants, and to develop a genotyping assay and determine the frequencies of the different sequence variants in susceptible and resistant isolates, and in the resistant isolate after further selection with IVM.
2. Materials & methods
2.1. Parasite isolates
Two field isolates of T. circumcincta from the UK were used: MTci2, an isolate with verified susceptibility to IVM, levamisole (LEV) and BZs (Skuce et al., 2010; D. Bartley, Pers. Comm.), and MTci5, a multiple-anthelmintic resistance isolate from a Scottish lowland sheep farm which was first described by Sargison et al. (2001) and has since been shown to be resistant to BZs, LEV and IVM (Bartley et al., 2004, Bartley et al., 2005). A separate population of MTci5, representing progeny from adult survivors of IVM treatment, was generated for the current study by treating an experimentally infected donor lamb with IVM at the manufacturer's recommended dose rate (0.2 mg/kg). Faecal samples were collected from day-21 post-treatment, eggs were recovered and cultured to third-stage larvae (L3). This IVM-screened population is, henceforth, referred to as MTci5 Post-Treatment (MTci5PT).
In addition, sequences from two near-isogenic lines of T. circumcincta generated in New Zealand, one susceptible to the available anthelmintic classes and the other resistant to multiple-anthelmintic classes (Bisset, 2007, Choi et al., 2017), were used in the present study for comparative purposes.
2.2. Generation of full-length Tci-pgp-9 cDNA sequence from UK isolates
The primers used for first strand cDNA synthesis of Tci-pgp-9, and subsequent PCR amplification, are listed in the Supplementary Table S1. RNA for this work was extracted from pools of L3 from the respective MTci2 and MTci5 isolates and approximately 100 ng were reverse transcribed to cDNA using rapid amplification of cDNA ends (RACE) to generate the sequences of the 5′ and 3′ untranslated regions (UTR). A cDNA fragment at the 5′ end of Tci-pgp-9 was generated using the antisense primer, Tci-pgp-9_A1, and the sense primer Tci-pgp-9_5′_F1, which incorporated part of the spliced leader-1 sequence (Krause and Hirsh, 1987). To amplify the 3′ region, a 3′ RACE approach was used replacing the Oligo(dT)20 primer with the Tci-pgp-9_3′RACE_dT primer in the SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen) kit, following the manufacturer's protocol, which provided a binding site for the adapter-specific nested primer, Tci-pgp-9_3′RACE.
The full-length Tci-pgp-9 transcript was amplified using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen) in a final volume of 50 μl (High Fidelity PCR Buffer, 0.2 μM of dNTP mix, 2 mM MgSO4, 1 μM of each primer, 1 unit of Platinum® Taq High Fidelity, 1 μl template cDNA), under the following conditions: initial denaturing at 94 °C for 2 min followed by 35 three-step cycles of 94 °C for 30 s, 55 °C for 30 s, 68 °C for 4 min with a final extension step of 68 °C for 5 min. The primer pair Tci-pgp-9_5′_F2 and Tci-pgp-9_R2 was used for the MTci2 isolate, and Tci-pgp-9_5′_F1 and Tci-pgp-9_R1 was used for the MTci5 isolate. Amplicons of the expected size (∼3.8 kb) were ligated into pGEM®-T Easy Vector (Promega) and transformed into chemically competent JM109 E. coli (Promega), following the manufacturer's protocols. Three clones derived from each UK isolate were sequenced in both sense and antisense directions.
2.3. Sequence variant-specific PCR
Crude gDNA lysates were generated from individual L3 lysed in 10 μl of a 3% solution of recombinant PCR grade proteinase K (Roche) in PCRDirect lysis reagent (Tail) (Viagen Biotech). Lysis was achieved by incubating at 55 °C for 16 h, followed by incubation at 90 °C for 1 h to denature the proteinase K.
Fragments representing the first (“N-terminal”) internucleotide binding domain of the Tci-pgp-9 gene (Tci-pgp-9-IBDA) were amplified from gDNA lysates prepared from 84 individual L3 randomly selected from each of the MTci2, MTci5, and MTci5PT isolates. A nested PCR was used, with the initial reaction using the primer pair Tci-pgp-9_IBDAGF3B and Tci-pgp-9_IBDAGR2 (Table S1). The use of degenerate primers increased the likelihood of amplifying all potential sequence variants present in these isolates. End-point PCR was performed with each 10 μl reaction containing Taq buffer, 2.5 mM MgCl2, dNTP mix (4 μM each), 0.5 units of Platinum® Taq (all Invitrogen), 1 μM each primer and 1 μl template gDNA (crude lysate). Reactions were carried out using the following thermocycling program: 94 °C for 8 min, followed by 35 three-step cycles of 94 °C for 10 s, 50 °C for 20 s and 68 °C for 40 s, followed by a final elongation step of 68 °C for 7 min.
A similar Platinum® Taq nested-PCR reaction was undertaken using 1 μl of a 50-fold dilution of the primary reactions as template with the primer pair, Tci-pgp-9_IBDAGF4 and Tci-pgp-9_IBDAGR3. Successful PCR amplification and evidence of amplicon length variants present in each T. circumcincta larva were assessed using a QIAxcel Advance System (QIAgen) using the QIAxcel Screening Kit (QIAgen), and analysed using the QIAxcel ScreenGel 1.0.2 software (QIAgen). Tci-pgp-9-IBDA PCR products from 37 individuals (10 from MTci2, 11 from MTci5, and 16 from MTci5PT) were selected for sequencing to account for each of the different sized amplicons observed. These were ligated into pCR® 4-TOPO® Vector (Invitrogen), transfected into One Shot® TOP10 Electrocomp™ Cells (Invitrogen), plasmids purified and the amplicon inserts sequenced.
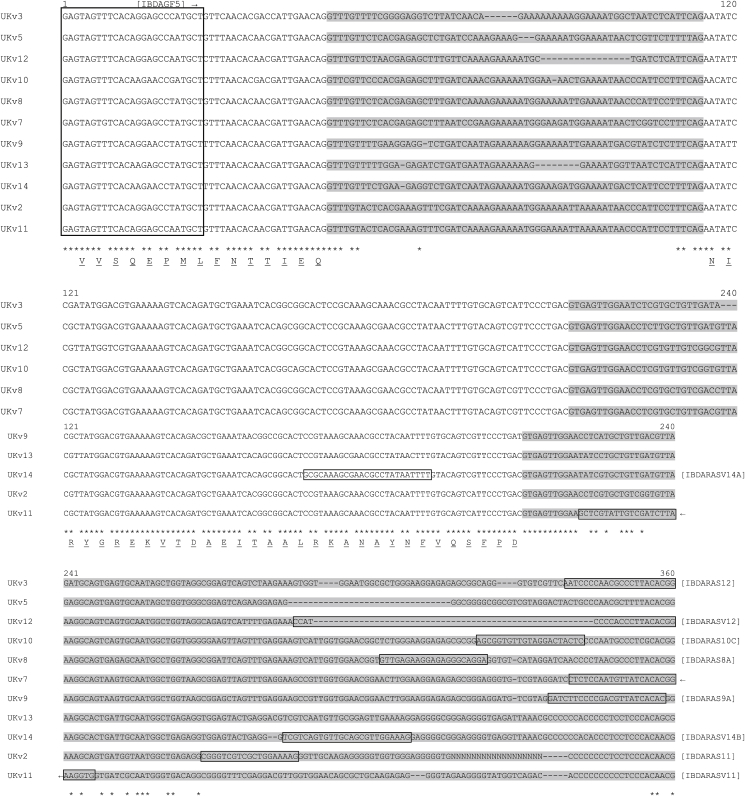
The DNA sequences generated from the initial Tci-pgp-9-IBDA PCR products were aligned with 9 sequence variants (numbered NZv1-3, NZv5-10) that had previously been identified from NZ isolates of T. circumcincta (Bisset, 2007). Sequence variant-specific primers (Supplementary Table S1) were designed to target polymorphic regions within the introns of Tci-pgp-9-IBDA and were used to differentiate among variants (Fig. 3). Each sequence variant-specific antisense primer was paired with a degenerate (‘non variant-specific’) sense primer, Tci-pgp-9_IBDAGF5, located near the 5′ end of the Tci-pgp-9-IBDA fragment initially amplified with the primer pair Tci-pgp-9_IBDAGF3B and Tci-pgp-9_IBDAGR2. A touchdown and nested PCR strategy was adopted for the sequence variant-specific PCR reactions where the template used was a 50-fold dilution of the PCR products amplified using the primers, Tci-pgp-9_IBDAGF3B and Tci-pgp-9_IBDAGR2. The touchdown PCR thermocycling conditions were: 94 °C for 8 min, then 12 three-step cycles with decreasing annealing temperature by 0.5 °C per cycle, starting at 94 °C for 10 s, at 62 °C for 20 s and 68 °C for 30 s, each cycle; this was followed by 24 three-step cycles of 94 °C for 10 s, 56 °C for 20 s and 68 °C for 30 s with a final extension step of 68 °C for 5 min. PCR products were analysed using the QIAxcel Advanced System and the QIAxcel DNA Screening Kit (QIAgen).
Fig. 3.
Sequence Variants Within the Tci-pgp-9-IBDA Domain of UK strains MTci2 and MTci5
Clustal Omega multiple alignment of partial gDNA sequences of Tci-pgp-9 sequence variants showing introns (shaded) and predicted amino acid translation (underlined). The locations of sequence variant-specific primers (boxed) designed to distinguish between sequence variants are shown and the primers used are listed in Table S1.
2.4. Statistical analyses
Sequence data were assembled and analysed using SeqMan Pro™ software from DNASTAR® Lasergene® v15 with default parameters. Molecular phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA) v7.0.26 software (Kumar et al., 2016) to select the best fit substitution model. The Maximum Likelihood method based on the Kimura 2-parameter model (Kimura, 1980) was used to construct a bootstrap consensus tree inferred from 2000 replicates (Felsenstein, 1985) to show the relationships between the sequence variants of Tci-pgp-9-IBDA, found in the UK and NZ strains of T. circumcincta, respectively. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.6377)). Sequence variants identified from the UK strains were numbered according to their relationships with sequence variants identified from the NZ resistant and susceptible isolates (Bisset, 2007). Those that differed by ≥ 10% were recorded as new variants and numbered accordingly.
Presence or absence of each sequence variant in DNA from each of the individual larvae tested was recorded. The data were treated as multi-locus binary data, in which each sequence variant was considered to have two possible states (absent = 0, present = 1). One-wayanalysis of variance (ANOVA) for the number of sequence variants per individual larva, by population, and subsequent Tukey pairwise comparisons and Kruskal-Wallis tests were conducted using Minitab17 statistical software.
3. Results
3.1. Generation of full-length Tci-pgp-9 cDNA sequences
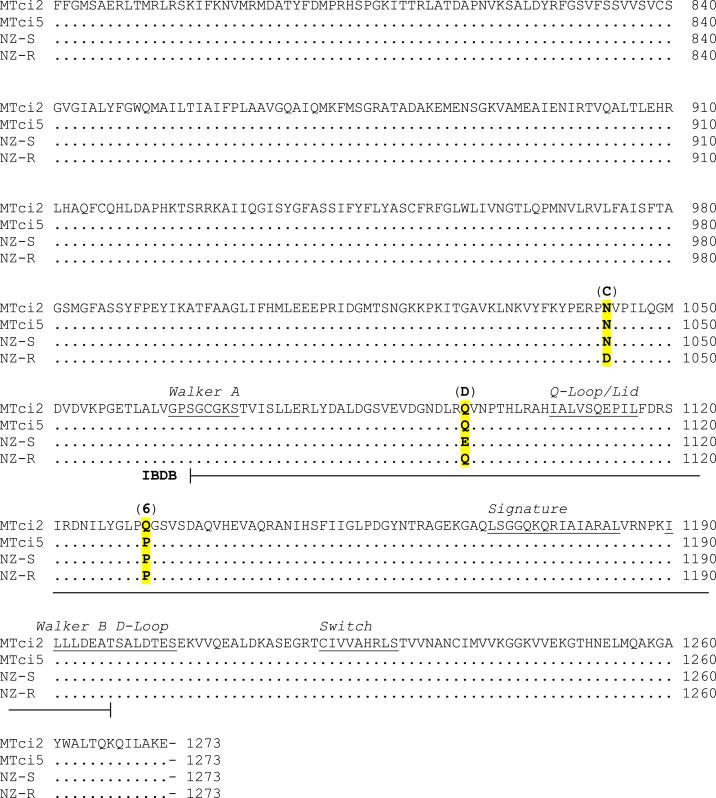
Full-length Tci-pgp-9 cDNAs (3.8 kb) were successfully amplified from pools of L3 using primer pairs Tci-pgp-9_5′_F2 and Tci-pgp-9_R2 for the MTci2 isolate and Tci-pgp-9_5′_F1 and Tci-pgp-9_R1 for the MTci5 isolate. Sequences were generated in both the sense and antisense directions from each of three clones per isolate, and assembled to form contiguous sequences for each UK field isolate and comparisons of the Tci-pgp-9 gene sequences from MTci2 and MTci5 conducted. Common motifs shared by members of the ABC-transporter superfamily were identified by comparing with ABC-transporter sequence motifs characterised in Brugia malayi (Ardelli et al., 2010) and annotated in the contiguous sequences (Fig. 1), thus verifying that the sequenced transcript was that of a P-glycoprotein.
Fig. 1.
Amino Acid Sequence of Tci-pgp-9
Amino acid sequences from the UK isolates MTci2 and MTci5, aligned with sequence information derived from two near-isogenic strains of T. circumcincta from NZ. The first of the NZ strains (NZ-S) was known to be susceptible to all available anthelmintics while the second (NZ-R), which shared a largely common genetic background, was multiple-anthelmintic resistant. Sequence motifs are underlined and annotated above the sequence. The positions of inter-nucleotide binding domains A and B (IBDA and IBDB) are underlined and annotated below the sequences. Highlighted in yellow are six residue substitutions identified in the UK isolates (numbered 1–6), and four previously identified residue substitutions in NZ isogenic strains (annotated A-D).
At the nucleotide level, the full-length Tci-pgp-9 cDNAs from the MTci2 and MTci5 isolates shared 95.6% identity (identical at 3651/3822 bases) [Accession Numbers: MTci2 - LT724249 and MTci5 – LT724250] while the amino acid sequences deduced from them shared 99.5% identity (1267/1273 residues). These sequences are likely to represent two of several different sequence variants of Tci-pgp-9 subsequently found in these two isolates. In each case, those cloned were assumed to be the most abundant of the Tci-pgp-9 variants present in the isolate from which they were derived. Nine point-mutations not present in the transcript derived from the MTci2 isolate were identified in that derived from the MTci5 isolate, and these resulted in six residue changes: V28E, A662T, T663A, A664T, R697S, and Q1131P, respectively (Fig. 1). None of the non-synonymous SNPs that were identified were located in any of the conserved sequence motifs. Of the 162 synonymous SNPs present, 12 were located in each of the sequence motifs in the first Tci-pgp-9-internucleotide binding domain (IBDA) and two were found in the Walker A motif within the second Tci-pgp-9-internucleotide binding domain (IBDB). The remaining 148 were distributed throughout the sequence.
Comparisons were made between amino acid sequences deduced from the full-length Tci-pgp-9 cDNA clones from the UK isolates of T. circumcincta and those deduced from the Tci-pgp-9 sequences from anthelmintic susceptible and multiple-resistant strains of NZ origin (Bisset, 2007; and unpublished data, S. A. Bisset). The amino acid sequences of the susceptible UK field-derived MTci2 and NZ inbred susceptible (NZ-S) strains shared 99.6% identity (identical at 1268/1273 residues) and the UK and NZ resistant (NZ-R) strains shared 99.9% identity (identical at 1272/1273 residues). Clones derived from the MTci2 and MTci5 isolates both displayed similar sequences to those derived from the anthelmintic resistant strain from NZ at three of the four non-synonymous SNPs at residues 79, 86 and 1097 (annotated A, B and D, respectively, in Fig. 1). At the fourth non-synonymous SNP, residue 1043 (annotated C in Fig. 1), the UK MTci2 and MTci5 isolates shared the ‘susceptible genotype’ displayed in the NZ near-isogenic strains. The four non-synonymous SNPs that were identified in the resistant NZ strain but absent from the susceptible NZ strain were all absent in the UK isolates of T. circumcincta.
3.2. Amplifying and sequencing Tci-pgp-9-IBDA variants
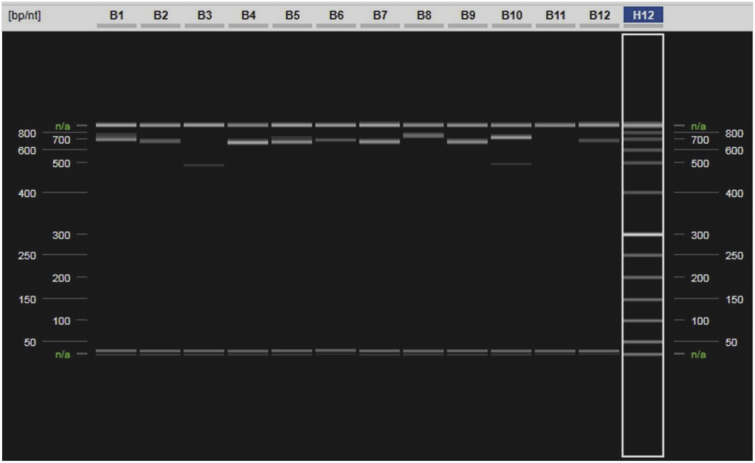
To identify genomic sequence variants of Tci-pgp-9-IBDA, a nested PCR strategy and degenerate primers were used. PCR products generated varied in size, both within and between individuals (Fig. 2). A panel of amplicons of varying size from individual larvae were selected for sequencing and the data aligned with nine distinct sequence variants from the NZ resistant and susceptible isolates. When classifying sequence variants it was considered impractical to take into account every apparent SNP. In most cases, groupings were based on blocks of nucleotide substitutions, insertions or deletions within introns, which were repeatedly associated with particular sequence variants (Fig. 3).
Fig. 2.
Secondary Nested PCR Products from MTci2 (individuals 1–12)
Virtual gel output from the QIAxcel Advanced System showing the secondary nested-PCR amplicons generated using primer pair Tci-pgp-9_IBDAGF4 and Tci-pgp-9_IBDAGR3 from a selection of individuals of the MTci5 strain. The alignment markers, 15 bp and 1 kb are shown in each lane along with PCR products of differing sizes, ranging from ∼500 to 800 bp. The QX DNA Size Marker 50–800 bp (v2.0) is highlighted in lane H12.
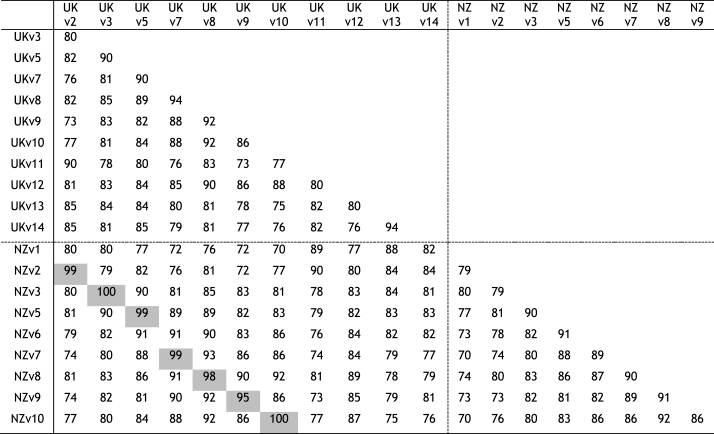
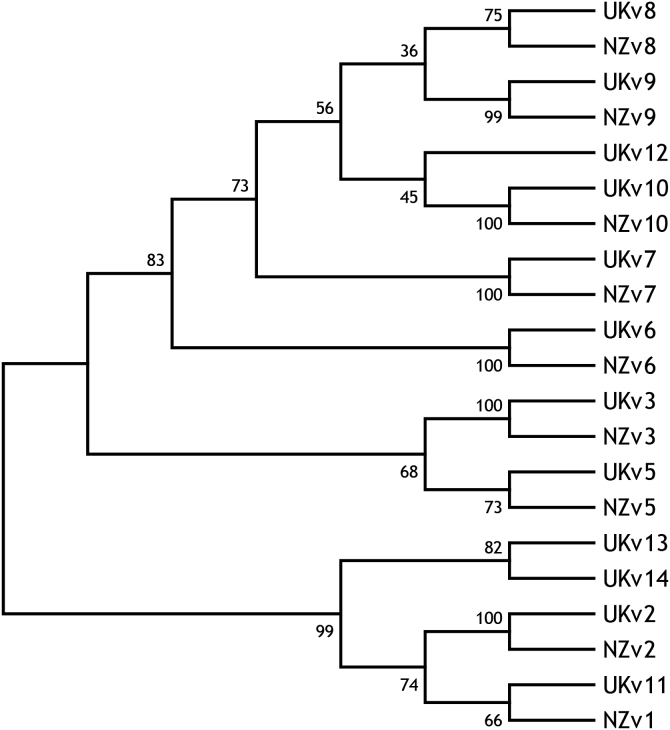
Eight of the ten Tci-pgp-9-IBDA sequence variants identified in NZ T. circumcincta strains (Bisset, 2007) were also found in the UK strains. These sequence variants aligned closely with NZ variants (NZv) sharing 95–100% identity (Table 1), and were annotated accordingly. Four sequence variants identified in the UK strains were more divergent from the known NZ sequence variants, and were assigned the numbers 11–14. A phylogenetic tree was constructed to show relationships (Fig. 4) between the sequence variants found in UK and NZ strains of T. circumcincta, supporting the high levels of identity observed within this amplified gDNA fragment of Tci-pgp-9-IBDA. Sequence variants NZv1 and NZv4 were not observed in the UK strains, although UKv11 was found to be most closely related to NZv1, sharing 89% identity. Sequence data for a sequence variant that most closely resembled the NZv6 was generated from a single larva belonging to the MTci5 isolate. However, it was not encountered again in larvae from these UK isolates, suggesting it was relatively rare.
Table 1.
Percentage identity of Tci-pgp-9-IBDA sequence variants.
The percentage identity between the Tci-pgp-9-IBDA sequence variants from the UK and NZ strains of T. circumcincta were calculated from the number of base substitutions per site from between sequences. Analyses were conducted in MEGA7 (Kumar et al., 2016), using the Kimura 2-parameter model (Kimura, 1980). Fewer than 5% alignment gaps, missing data and ambiguous bases were allowed at any position. The sequence variants with the identity >95% (shaded) formed the basis for the numbered classification of the corresponding UK sequence variants. Note that UKv11-UKv14 had no corresponding NZ variant with >90% identity.
Fig. 4.
Phylogenetic Relationships of Tci-pgp-9-IBDA Sequence Variants
Molecular phylogenetic analyses were conducted using MEGA7 to select the best fit substitution model. The Maximum Likelihood method based on the Kimura 2-parameter model was used to construct a bootstrap consensus tree inferred from 2000 replicates to show the relationships between the sequence variants of Tci-pgp-9-IBDA, found in the UK and NZ strains of T. circumcincta, respectively. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.6377)).
3.3. Sequence variant-specific genotyping
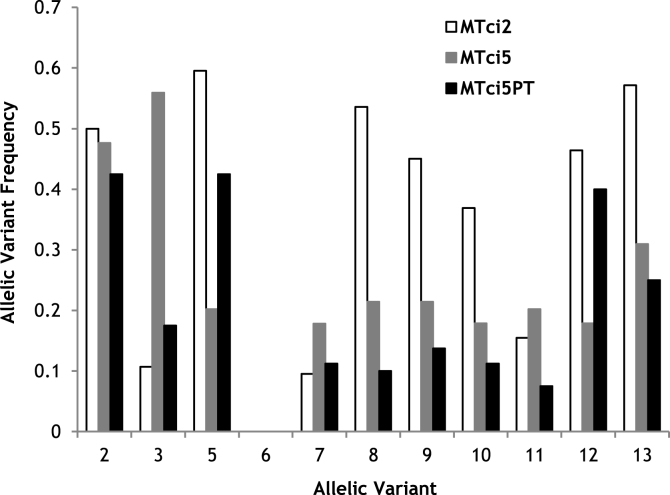
A series of sequence variant-specific primers were designed to distinguish between the Tci-pgp-9-IBDA sequence variant(s) present in individual larvae (Fig. 3). These primers targeted unique areas of polymorphism in each sequence variant (GenBank accession numbers listed in Table S2), which in most cases ensured that they did not cross-react with other sequence variants. However, as can be seen in Fig. 3, the sequences generated from the sequence variant ‘UKv14’ of Tci-pgp-9-IBDA possessed relatively few polymorphisms that were unique to this variant, limiting the sites available for the placement of a sequence variant-specific primer. As a result, the primers designed to identify the presence of this variant, Tci-pgp-9_IBDARASV14A and Tci-pgp-9_IBDARASV14B, proved to be unsuitable due to problems with cross-reactivity with other variants, therefore their use was discontinued. Using sequence variant-specific PCR reactions, the presence or absence of each sequence variant was determined in 84 individual larvae from each of the MTci2 and MTci5 isolates and in 80 individual larvae from the MTci5PT isolate. The frequencies of each of the sequence variants in each of the isolates are shown in Fig. 5. As is clear from this figure, column percentage totals in all cases exceed 200%, the total that might be expected if every individual larva was heterozygous and generated two distinct genotypes. The results thus suggest the involvement of gene amplification.
Fig. 5.
Frequency of Tci-pgp-9-IBDA Sequence Variants in the MTci2 and MTci5 Isolates
Frequency of Tci-pgp-9-IBDA sequence variants identified from the MTci2 (white), MTci5 (grey) and MTci5PT (black) isolates of T. circumcincta. Note that column percentages for each isolate add up to >200% because many individual larva yielded multiple genotypes.
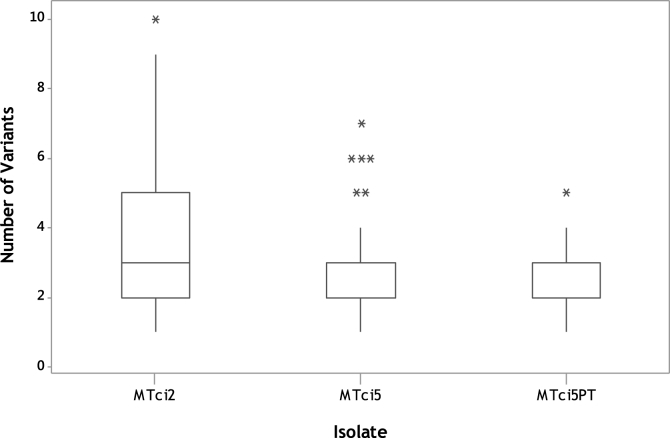
Frequently more than one sequence variant occurred in an individual larva, as indicated by more than one PCR product, for example in lanes B1, B5 and B10 in Fig. 2. Indeed, the results of the sequence variant-specific PCR revealed that up to ten different Tci-pgp-9-IBDA sequence variants occurred in some individual larvae belonging to the MTci2 isolate, while individual larvae belonging to the MTci5 isolate possessed up to 7 different variants, and larvae from the MTci5PT isolate possessed up to 5 different variants (p < 0.001) (Fig. 6). The one way-ANOVA method was used to compare the average number of sequence variants in larvae from each isolate, and showed that MTci2 had a significantly higher mean of 3.8 sequence variants, compared with 2.7 in the MTci5 isolate and 2.2 in the MTci5PT isolate (p < 0.001). A Tukey pairwise comparison between the isolates indicated the MTci2 isolate was significantly different from the MTci5 and MTci5PT isolates.
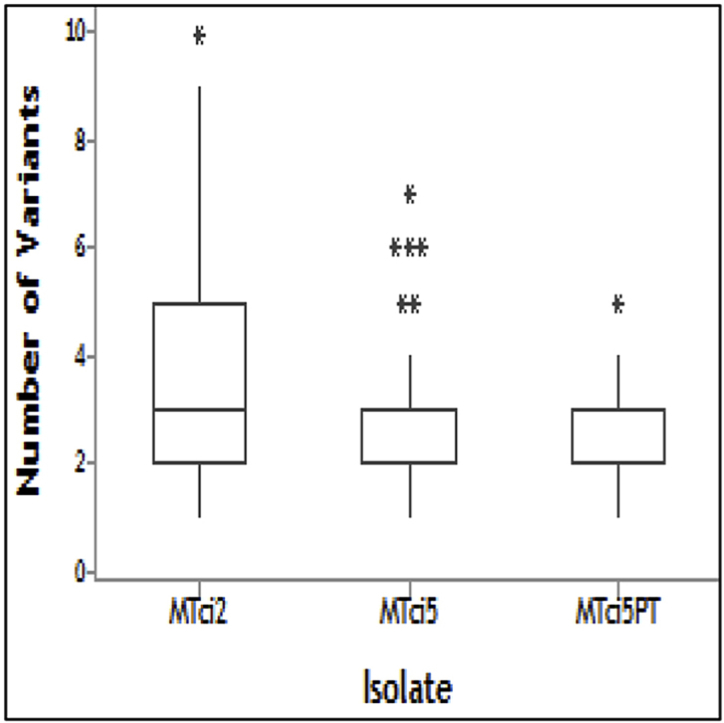
Fig. 6.
Number of Tci-pgp-9-IBDA Sequence Variants Identified in Individual Larvae
Larvae from the MTci2 (84), MTci5 (84) and MTci5PT (80) were screened for the presence of the 13 different sequence variants. The number or sequence variants per individual was plotted and a Kruskal-Wallis test was conducted to compare the median values: MTci2 = 3, MTci5 = 3 and MTci5PT = 2 (p < 0.001).
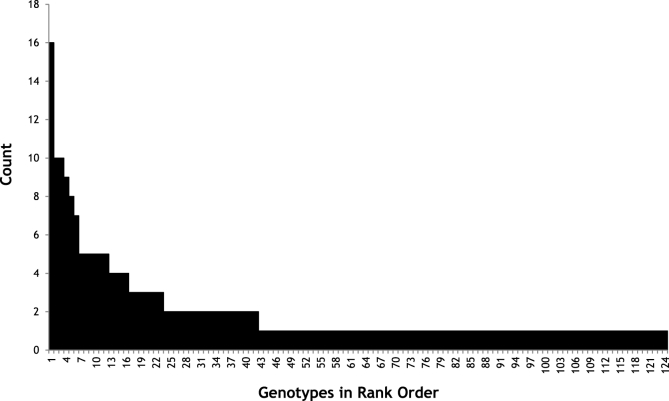
The combination of sequence variants that each larva possessed was allocated a genotype number (Table S3). A total of 124 different genotypes were identified across the three worm isolates. The majority of these were unique in the sense that they occurred in just a single larva. Only five of them occurred in 8 or more individuals (Fig. 7). Because the Tci-pgp-9 copy number in individual larvae was not known it was not possible to determine which of them carried multiple copies of any particular sequence variant.
Fig. 7.
Frequency Distribution of Tci-pgp-9 Genotypes Over All Populations Combined
Different combinations of sequence variants that occurred in the larvae were assigned a “genotype”. The genotypes displayed by larvae from the 3 UK strains studied (MTci2, MTi5 and MTci5PT) were combined to show that the majority of genotypes were unique and seen in only a single larva, with just 5 genotypes being seen in 8 or more individual larvae.
4. Discussion
Full-length (3.8 kb) cDNA transcripts of Tci-pgp-9 from anthelmintic susceptible and multi-drug resistant UK isolates of T. circumcincta were sequenced and compared to Tci-pgp-9 cDNA sequences derived from two near-isogenic strains of T. circumcincta from NZ (Bisset, 2007; and unpublished data, S. A. Bisset). Comparisons of the Tci-pgp-9 transcripts amplified from MTci2 and MTci5 identified 171 polymorphic loci, of which 9 nucleotide substitutions resulted in 6 amino acid substitutions. Four substitutions at codons 662–664 and 697 were found to occur in sequences from both the NZ and UK resistant isolates. Tci-pgp-9 sequences derived from the anthelmintic susceptible strains, MTci2 and NZ-S, both displayed Ala-Thr-Ala at amino acids 662–664 and arginine at amino acid 697, whilst those amplified from MTci5 resistant strain displayed Thr-Ala-Thr the 662–664 residues and serine at amino acid 697. The significance of these geographically distinct resistant isolates sharing non-synonymous SNPs at these positions in Tci-pgp-9 remains to be determined. We do not yet know whether similar polymorphisms in Tci-pgp-9 are associated with resistance to IVM in other isolates of T. circumcincta. Furthermore, these polymorphisms may not be isolate-specific, but rather genotype-specific. For example, arginine and threonine were found to be present at residues 79 and 86 in sequence variant 1 which was the most common genotype in the NZ inbred susceptible strain but was not in the other sequence variants; similarly Ala-Thr-Ala and Arg at residues 662–664 and 697, respectively, were only present in sequence variants 1 and 2 while Thr-Ala-Thr and Ser at the same residues was seen in sequence variants 3, 6, 9 and 10 which were most commonly found in the NZ multiple resistant strain (Bisset, 2007; and unpublished data, S. A. Bisset).
As a possible mechanism of resistance, the distribution of non-synonymous SNPs observed throughout the Tci-pgp-9 gene might influence the tertiary structure of the gene, perhaps altering drug binding (Gottesman and Pastan, 1993). Additionally, the synonymous SNPs may cause changes in substrate specificity, possibly by changing the timing of co-translational folding and thus the conformation of the Pgp molecule (Buss and Callaghan, 2008, Kimchi-Sarfaty et al., 2007). Changes in the codon sequence, and therefore codon usage, have been linked to the secondary structure of the proteins they encode. Some residues show a higher probability of being buried in the centre of the protein molecule and others are more likely to be exposed on the surfaces of the folded protein (Saunders and Deane, 2010). Synonymous codons encoded by different nucleotides are not used with equal frequency (Tao and Dafu, 1998) resulting in the incorporation of “rarer” codons, the availability of which may impede translation speed. Structural information that is linked to translation speed (Saunders and Deane, 2010), might influence translation rate and subsequently protein folding (Kimchi-Sarfaty et al., 2007). Such changes could alter Pgp tertiary structure and potentially substrate specificity and drug binding, therefore, the influence of both synonymous and non-synonymous SNPs on the rate of drug efflux warrants further investigation in multi-drug resistant isolates of T. circumcincta.
Bisset (2007) reported the occurrence of alternative splicing that resulted in a deletion of 45 residues in the N-terminal transmembrane region of Tci-pgp-9 in multiple resistant T. circumcincta. An alternatively spliced Pgp with a deletion of a similar size and location, has also been reported to occur at a high level in two independently derived multidrug resistant Chinese hamster lung cell lines, while being absent in a drug sensitive Chinese hamster lung cell line (Devine et al., 1991). As only full-length Tci-pgp-9 cDNA PCR products were cloned and sequenced in the present study, we are unable to confirm whether a similar alternatively spliced Tci-pgp-9 gene occurs in these UK isolates of T. circumcincta.
Considerable variation in the gDNA sequence of the N-terminal internucleotide binding domain, Tci-pgp-9-IBDA, was identified in individuals from UK isolates of T. circumcincta. The sequences generated (Fig. 3) were compared with 9 sequence variants previously identified in the same Tci-pgp-9-IBDA region isolated from NZ strains of T. circumcincta (Bisset, 2007). The sequence variants in over 80 randomly selected larvae from each of MTci2, MTci5 and MTci5PT isolates of T. circumcincta were classified into 12 distinct “sequence variants”. Eight of the nine sequence variants seen in the NZ strains were also present in the UK isolates, with NZv1 the only exception. The level of identity shared between the UK and NZ Tci-pgp-9-IBDA sequence variants (95–100%), suggests a high degree of conservation of these sequence variants in these two geographically diverse T. circumcincta isolates. Despite this however, the results showed a high degree of sequence diversity in the introns, as well as numerous synonymous SNPs in the exons of Tci-pgp-9-IBDA. It is conceivable that polymorphisms elsewhere in the Tci-pgp-9 cDNA sequence may be linked to the sequence variants identified in the intronic regions in the Tci-pgp-9-IBDA domain, although this remains to be proven. The possibility of having an allele that is capable of diminishing the toxic effects of a drug is greater in genetically diverse species, such as the trichostrongyloid nematodes, H. contortus (Beech et al., 1994) and T. circumcincta (Blackhall et al., 1998).
Based on the amplification of sequence variant-specific PCR products from individual worms, comparisons of the observed frequency of sequence variants were conducted. The majority (78.6%) of individuals in the anthelmintic susceptible isolate, MTci2, carried between 1 and 4 sequence variants, while the remaining 21.4% carried >5 sequence variants of Tci-pgp-9. The maximum number of sequence variants observed in an individual MTci2 larva was 10, suggesting at least 5 heterozygous copies of the Tci-pgp-9 gene were amplified by the sequence variant-specific PCR. Possessing up to ten different variants may provide some individuals with a panel of Tci-pgp-9 sequence variants which potentially offers a fitness advantage over other individuals. A reduction in the number of sequence variants in individuals of the anthelmintic resistant isolate (MTci5) was observed, with 92.9% of individuals carrying ≤ 4 variants and the remaining 7.1% of larvae possessing 5–7 sequence variants. The MTci5 population was not derived from MTci2 so we cannot definitively ascribe changes in population structure to selection, however, the results are in line with the possibility that purifying selection might have eliminated sequence variants that were not advantageous under exposure to anthelmintics. The MTci5 isolate exhibits a resistance phenotype to BZs and LEV, thus differences in genotype between MTci2 and MTci5 might also be due to selection with anthelmintics other than IVM. The further reduction in the number of sequence variants observed in the MTci5PT isolate (which had survived in vivo exposure to IVM), provides support for this possibility as 79 of the 80 larvae from this population which were genotyped displayed ≤ 4 variants with just one individual possessing 5 sequence variants.
In summary, the present study revealed 1) 4 non-synonymous nucleotide substitutions (residues 662–664 and 697) in Tci-pgp-9 sequence variants shared by the multiple-resistant UK and NZ isolates but which were not observed in their drug-susceptible counterparts; 2) that multiple sequence variants of Tci-pgp-9 were present in the majority of individuals, consistent with previous evidence that amplification of this gene has occurred in T. circumcincta; 3) that there was a reduction in the diversity of Tci-pgp-9 sequence variants in individuals of the anthelmintic resistant MTci5 isolate relative to those in the anthelmintic susceptible MTci2 isolate and a further reduction in the number of different variants present in individuals derived from an IVM treated population of MTci5, suggesting that IVM treatment had applied purifying selection pressure.
Acknowledgements
The authors would like to thank Dave Bartley, Alison Morrison and Leigh Devin of Moredun Research Institute (UK), and Charlotte Bouchet and Jacqui Knight at AgResearch (NZ), for the provision of parasite material and technical assistance throughout the study. This work was funded by Moredun Research Institute (UK) and AgResearch (NZ).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.01.004.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ardelli B.F., Stitt L.E., Tompkins J.B. Inventory and analysis of ATP-binding cassette (ABC) systems in Brugia malayi. Parasitology. 2010;137(8):1195–1212. doi: 10.1017/S0031182010000120. [DOI] [PubMed] [Google Scholar]
- Areskog M., Engström A., Tallkvist J., von Samson-Himmelstjerna G., Höglund J. PGP expression in Cooperia onchophora before and after ivermectin selection. Parasitol. Res. 2013;112(8):3005–3012. doi: 10.1007/s00436-013-3473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley D.J., Jackson F., Jackson E., Sargison N. Characterisation of two triple resistant field isolates of Teladorsagia from Scottish lowland sheep farms. Vet. Parasitol. 2004;123(3–4):189–199. doi: 10.1016/j.vetpar.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., Jackson E., Sargison N., Jackson F. Further characterisation of a triple resistant field isolate of Teladorsagia from a Scottish lowland sheep farm. Vet. Parasitol. 2005;134(3–4):261–266. doi: 10.1016/j.vetpar.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., McAllister H., Bartley Y., Dupuy J., Ménez C., Alvinerie M., Jackson F., Lespine A. P-glycoprotein interfering agents potentiate ivermectin susceptibility in ivermectin sensitive and resistant isolates of Teladorsagia circumcincta and Haemonchus contortus. Parasitology. 2009;136(09):1081–1088. doi: 10.1017/S0031182009990345. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., Devin L., Nath M., Morrison A.A. Selection and characterisation of monepantel resistance in Teladorsagia circumcincta isolates. Int. J. Parasitol. Drugs Drug Resist. 2015;5(2):69–76. doi: 10.1016/j.ijpddr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech R.N., Prichard R.K., Scott M.E. Genetic variability of the beta-tubulin genes in benzimidazole- susceptible and -resistant strains of Haemonchus contortus. Genetics. 1994;138(1):103–110. doi: 10.1093/genetics/138.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisset S.A. Flinders University of South Australia; 2007. The Genetic Basis of Multiple-anthelmintic Resistance in Teladorsagia circumcincta, a Gastrointestinal nematode Parasite of Sheep and Goats; p. 273. PhD Thesis. [Google Scholar]
- Blackhall W.J., Pouliot J.F., Prichard R.K., Beech R.N. Haemonchus contortus: selection at a glutamate-gated chloride channel gene in ivermectin- and moxidectin-selected strains. Exp. Parasitol. 1998;90(1):42–48. doi: 10.1006/expr.1998.4316. [DOI] [PubMed] [Google Scholar]
- Bourguinat C., Ardelli B.F., Pion S.D.S., Kamgno J., Gardon J., Duke B.O.L., Boussinesq M., Prichard R.K. P-glycoprotein-like protein, a possible genetic marker for ivermectin resistance selection in Onchocerca volvulus. Mol. Biochem. Parasitol. 2008;158(2):101–111. doi: 10.1016/j.molbiopara.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Buss D., Callaghan A. Interaction of pesticides with p-glycoprotein and other ABC proteins: a survey of the possible importance to insecticide, herbicide and fungicide resistance. Pestic. Biochem. Physiol. 2008;90(3):141–153. [Google Scholar]
- Choi Y., Bisset S.A., Doyle S.R., Hallsworth-Pepin K., Martin J., Grant W.N., Mitreva M. Genomic introgression mapping of field-derived multiple-resistance in the nematode parasite Teladorsagia circumcincta. PLoS Genet. 2017;13(6) doi: 10.1371/journal.pgen.1006857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graef J., Demeler J., Skuce P., Mitreva M., von Samson-Himmelstjerna G., Vercruysse J., Claerebout E., Geldhof P. Gene expression analysis of ABC transporters in a resistant Cooperia oncophora isolate following in vivo and in vitro exposure to macrocyclic lactones. Parasitology. 2013;140(4):499–508. doi: 10.1017/S0031182012001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler J., Küttler U., Al Gusbi S., Ramünke S., De Graef J., Kerboeuf D., Geldhof P., Pomroy W.E., von Samson-Himmelstjerna G. Potential contribution of P-glycoproteins to macrocyclic lactone resistance in the cattle parasitic nematode Cooperia onchophora. Mol. Biochem. Parasitol. 2013;118(1):10–19. doi: 10.1016/j.molbiopara.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Devine S.E., Hussain A., Davide J.P., Merla P.W. Full length and alternatively spliced pgp1 transcripts in multidrug-resistance Chinese hamster cells. J. Biol. Chem. 1991;266(7):4545–4555. [PubMed] [Google Scholar]
- Dicker A.J. University of Glasgow; 2010. Comparative Gene Expression Studies of Anthelmintic Resistance in the Parasitic nematode, Teladorsagia circumcincta. PhD Thesis. [Google Scholar]
- Dicker A.J., Nath M., Yaga R., Nisbet A.J., Lainson F.A., Gilleard J.S., Skuce P.J. Teladorsagia circumcincta: the transcriptomic response of a multi-drug-resistant isolate to ivermectin exposure in vitro. Exp. Parasitol. 2011;127(2):351–356. doi: 10.1016/j.exppara.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Dicker A.J., Nisbet A.J., Skuce P.J. Gene expression changes in a P-glycoprotein (Tci-pgp-9) putatively associated with ivermectin resistance in Teladorsagia circumcincta. Int. J. Parasitol. 2011;41(9):935–942. doi: 10.1016/j.ijpara.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Dupuy J., Alvinerie M., Ménez C., Lespine A. Interaction of anthelmintic drugs with P-glycoprotein in recombinant LLC-PK1-mdr1a cells. Chem. Biol. Interact. 2010;186(3):280–286. doi: 10.1016/j.cbi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gottesman M.M., Pastan I. Biochemistry of multidrug-resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Jackson F., Coop R.L. The development of anthelmintic resistance in sheep nematodes. Parasitology. 2000;120(07):95–107. doi: 10.1017/s0031182099005740. [DOI] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39(2):213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Janssen I.J.I., Krϋcken J., Demeler D., Basiaga M., Kornaś S., von Samson-Himmelstjerna G. Genetic variants and increased expression of Parascaris equorum P-glycoprotein-11 in populations with decreased ivermectin susceptibility. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20(10):477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kaschny M., Demeler J., Janssen I.J.I., Kuzmina T.A., Besognet B., Kanellos T., Kerboeuf D., von Samson-Himmelstjerna G., Krücken J. Macrocyclic lactones differ in interaction with recombinant P-glycoprotein 9 of the parasitic nematode Cylicocylus elongatus and ketoconazole in a yeast growth assay. PLoS Pathog. 2015;11(4) doi: 10.1371/journal.ppat.1004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Krause M., Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49(6):753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathwick D., Miller C., McMurty L. The 24th International Conference of the World Association for the Advancement of Veterinary Parasitology. World Association for the Advancement of Veterinary Parasitology; 2013. Resistance to monepantel in two nematode species in goats. [Google Scholar]
- Molento M.B., Prichard R.K. Effect of multidrug resistance modulators on the activity if ivermectin and moxidectin against selected strains of Haemonchus contortus infective larvae. Pesqui. Vet. Bras. 2001;21:117–121. [Google Scholar]
- Molento M.B., Lifschitz A., Sallovitz J., Lanusse C., Prichard R. Influence of verapamil on the pharmacokinetics of the antiparasitic drugs ivermectin and moxidectin in sheep. Parasitol. Res. 2004;92(2):121–127. doi: 10.1007/s00436-003-1022-3. [DOI] [PubMed] [Google Scholar]
- Prichard R.K., Roulet A. ABC transporters and β-tubulin in macrocyclic lactone resistance: prospects for marker development. Parasitology. 2007;134(Pt 8):1123–1132. doi: 10.1017/S0031182007000091. [DOI] [PubMed] [Google Scholar]
- Raza A., Kopp S.R., Bagnall N.H., Jabbar A., Kotze A.C. Effects of in vitro exposure to ivermectin and levamisole on the expression patterns of ABC transporters in Haemonchus contortus larvae. Int. J. Parasitol. Drugs Drug Resist. 2016;6:103–115. doi: 10.1016/j.ijpddr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales N., Love S. Resistance of Haemonchus sp. To monepantel and reduced efficacy of a derquantel/abamectin combination confirmed in sheep in NSW. Australia. Vet. Parasitol. 2016;228:193–196. doi: 10.1016/j.vetpar.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Sargison N., Scott P., Jackson F. Multiple anthelmintic resistance in sheep. Vet. Rec. 2001;149(25):778–779. [PubMed] [Google Scholar]
- Saunders R., Deane C.M. Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res. 2010;38(19):6719–6728. doi: 10.1093/nar/gkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Pomroy W.E., Kenyon P.R., Smith G., Adlington B., Moss A. Lack of efficacy of monepantel against Teladorsagia circumcincta and Trichostrongylus colubriformis. Vet. Parasitol. 2013;198:166–171. doi: 10.1016/j.vetpar.2013.07.037. [DOI] [PubMed] [Google Scholar]
- Skuce P., Stenhouse L., Jackson F., Hypša V., Gilleard J. Benzimidazole resistance allele haplotype diversity in United Kingdom isolates of Teladorsagia circumcincta supports a hypothesis of multiple origins of resistance by recurrent mutation. Int. J. Parasitol. 2010;40(11):1247–1255. doi: 10.1016/j.ijpara.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Tao X., Dafu D. The relationship between synonymous codon usage and protein structure. FEBS Lett. 1998;434(1):93–96. doi: 10.1016/s0014-5793(98)00955-7. [DOI] [PubMed] [Google Scholar]
- Van den Brom R., Moll L., Kappert C., Vellema P. Haemonchus contortus resistance to monepantel in sheep. Vet. Parasitol. 2015;209(3–4):278–280. doi: 10.1016/j.vetpar.2015.02.026. [DOI] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180(2):99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wolstenholme A.J., Fairweather I., Prichard R., von Samson-Himmelstjerna G., Sangster N.C. Drug resistance in veterinary helminths. Trends Parasitol. 2004;20(10):469–476. doi: 10.1016/j.pt.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Xu M., Molento M., Blackhall W., Ribeiro P., Beech R., Prichard R. Ivermectin resistance in nematodes may be caused by alteration of P-glycoprotein homolog. Mol. Biochem. Parasitol. 1998;91(2):327–335. doi: 10.1016/s0166-6851(97)00215-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.