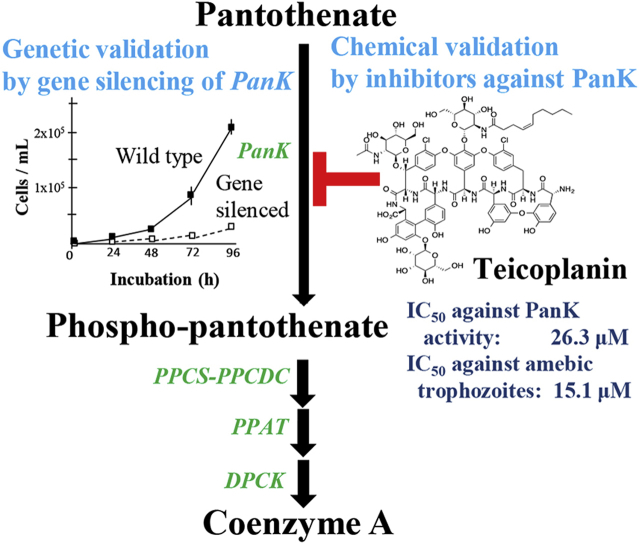
Abstract
The Coenzyme A (CoA), as a cofactor involved in >100 metabolic reactions, is essential to the basic biochemistry of life. Here, we investigated the CoA biosynthetic pathway of Entamoeba histolytica (E. histolytica), an enteric protozoan parasite responsible for human amebiasis. We identified four key enzymes involved in the CoA pathway: pantothenate kinase (PanK, EC 2.7.1.33), bifunctional phosphopantothenate-cysteine ligase/decarboxylase (PPCS-PPCDC), phosphopantetheine adenylyltransferase (PPAT) and dephospho-CoA kinase (DPCK). Cytosolic enzyme PanK, was selected for further biochemical, genetic, and phylogenetic characterization. Since E. histolytica PanK (EhPanK) is physiologically important and sufficiently divergent from its human orthologs, this enzyme represents an attractive target for the development of novel anti-amebic chemotherapies. Epigenetic gene silencing of PanK resulted in a significant reduction of PanK activity, intracellular CoA concentrations, and growth retardation in vitro, reinforcing the importance of this gene in E. histolytica. Furthermore, we screened the Kitasato Natural Products Library for inhibitors of recombinant EhPanK, and identified 14 such compounds. One compound demonstrated moderate inhibition of PanK activity and cell growth at a low concentration, as well as differential toxicity towards E. histolytica and human cells.
Keywords: Entamoeba histolytica, Amebiasis, Coenzyme A, Pantothenate kinase, Gene silencing, Drug development
Graphical abstract
Highlights
-
•
Pantothenate kinase (PanK) is essential in Entamoeba histolytica parasite.
-
•
PanK gene silencing caused a significant growth defect, CoA level, and PanK activity.
-
•
PanK represents an attractive target for new drug development.
-
•
Potential inhibitors have been found against E. histolytica PanK.
1. Introduction
Coenzyme A (CoA) is an essential cofactor in all living organisms as an acyl group carrier and carbonyl-activating group involved in more than 100 cellular reactions (Begley et al., 2001); it is estimated to be a cofactor used in 9% of identified enzymatic reactions (Strauss, 2010). CoA biosynthesis is considered to be an essential and universal pathway of the majority of prokaryotes and eukaryotes (Leonardi et al., 2005b). In general, CoA participates in fatty acid metabolism, the tricarboxylic acid cycle and numerous other intermediary metabolic reactions (Abiko, 1975). CoA is synthesized from pantothenate (vitamin B5), cysteine, and ATP (Jackowski, 1996; Leonardi et al., 2005b). Most of the eukaryotes are unable to synthesize pantothenic acid and thus rely on an external supply.
Entamoeba histolytica is the protozoan agent responsible for human amebiasis, an infectious disease causing dysentery and amebic liver abscesses, responsible for 100,000 deaths annually throughout the world. It represents the third most common parasitic cause of death, after malaria and schistosomiasis (Stanley, 2003; Ali and Nozaki, 2007; Ralston and Petri, 2011). The elaborate pathogenesis of this parasite is well documented (Espinosa-Cantellano and Martínez-Palomo, 2000; Nozaki and Bhattacharya, 2015). Metronidazole has, for decades, been the most effective drug in the treatment of amebiasis despite its known side effects and low efficacy against asymptomatic cyst carriers. Moreover, resistance, virulence and host immune response to metronidazole treatment in amebiasis have been reported in some countries (Griffin, 1973; Pittman and Pittmann, 1974; Johnson, 1993; Koch et al., 1997; Ali and Nozaki, 2007). The molecular target of metronidazole has been well described as a key metabolic enzyme, pyruvate: ferredoxin oxidoreductase, which is involved in acetyl CoA production from pyruvate as part of central energy metabolism. Identification and characterization of novel drug targets unique to E. histolytica are therefore needed to design better therapeutics against amebiasis.
Here we investigate the CoA biosynthetic pathway of E. histoltyica. We identified four enzymes, including one bifunctional enzyme, involved in this pathway. We further characterized one of these enzymes, pantothenate kinase (PanK, EC 2.7.1.33), with biochemical and reverse genetic approaches. Moreover, we identified E. histolytica PanK inhibitors by screening the Kitasato Natural Products Library against EhPanK recombinant enzyme. Taken together, we demonstrate that the CoA biosynthetic pathway, in general, and PanK, specifically, represents a rational and novel drug target against amebiasis.
2. Materials and methods
2.1. Organisms, cultivation, and chemicals
Trophozoites of E. histolytica clonal strains HM-1: IMSS cl 6 and G3 (Bracha et al., 2006) were maintained axenically in Diamond's BI-S-33 medium at 35.5 °C as described previously (Diamond et al., 1978). Trophozoites were continuously maintained in mid-log phase after inoculation of one-thirtieth to one-twelfth of the total culture volume. Escherichia coli BL21 (DE3) strain was obtained from Invitrogen (Carlsbad, CA, USA). Magnesium-free ATP was procured from DiscoverX (Fremont, CA, USA). Ni2+-NTA agarose was procured from Novagen (Darmstadt, Germany). Lipofectamine and geneticin (G418) were procured from Invitrogen. Chemicals to evaluate metals in PanK activity assay were procured from Wako (Tokyo, Japan). All other chemicals of analytical grade were procured from Sigma-Aldrich (Tokyo, Japan) unless otherwise stated.
2.2. Production of PanK gene-silenced strain
In order to construct a plasmid for epigenetic gene silencing of E. histolytica (Bracha et al., 2006; Zhang et al., 2011) PanK (EhPanK), a fragment corresponding to a 430 bp 5′ open reading frame of EhPanK gene was amplified by PCR from cDNA using the following primer set (sense primer, 5′-CAGAGGCCTATGTCTCAACCATCCCATTCT-3′ and antisense primer, 5′-AATGAGCTCTCTGAAGATTACCAATCCCATAAA-3′). These oligonucleotides contained StuI and SacI restriction sites (shown in bold). The amplified product was digested with StuI and SacI, and ligated into the StuI and SacI double digested psAP2-Gunma construct (Husain et al., 2011) to synthesize a EhPanK gene silencing plasmid. The G3 strain trophozoites were transformed with an empty vector as a control and the silencing plasmid was transfected by liposome-mediated transfection as previously described (Nozaki et al., 1999). Transformants were initially selected in the presence of 1 μg/mL geneticin gradually increased to 10 μg/mL.
2.3. Reverse transcriptase PCR
RNA was extracted from approximately 1 × 106 trophozoites of EhPanK gene silenced (PanK gs) and control transformant strains using TRIzol reagent (Ambion, Life Technologies) as previously described (Chomczynski and Mackey, 1995). DNase treatment was performed using DNase I (Invitrogen) to exclude genomic DNA. RNA quantity was determined by measuring the absorbance at 260 nm with a NanoDrop ND-1000 UV–Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Approximately one μg total RNA was used for cDNA synthesis using First-Stand cDNA Synthesis (Superscript® III, Invitrogen) with reverse transcriptase and oligo (dT) primers according to manufacturer's instructions. The cDNA product was diluted 10-fold and PCR reactions were carried out in 50 μl, using the primer pair (Sense primer, 5′-ATGTCTCAACCATCCCATTC-3′ and antisense primer, 5′-TTACATTAGTTCTTCTTCATACTC-3′). The PCR conditions were: 98 °C, 10 s; 55 °C, 1 min; and 72 °C, 1 min; 20–25 cycles. The PCR products obtained were resolved by agarose gel electrophoresis.
2.4. Quantitative real-time (qRT) PCR
Relative levels of steady-state mRNA of the following genes were measured using qRT-PCR: PanK (EHI_183060), bifunctional phosphopantothenate-cysteine ligase/decarboxylase (PPCS-PPCDC, EHI_164490), phosphopantetheine adenylyltransferase (PPAT, EHI_006680), dephospho CoA kinase 1 and 2 (DPCK1, EHI_040840; and DPCK2, EHI_155780), and RNA polymerase II gene (EHI_056690) as a control. Each 20 μL reaction contained 10 μL 2X Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA), 0.6 μL each of 10 μM sense and antisense primers, 5 μL 10x diluted cDNA, and nuclease-free water. PCR was performed using the StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with the following cycling conditions: enzyme activation at 95 °C for 20 s, followed by 40 cycles of denaturation at 95 °C for 3 s and annealing-extension at 60 °C for 30 s. All reactions were carried out in triplicate, including cDNA-minus controls. The amount of the steady-state mRNA of each target gene was determined by the ΔCt method with RNA polymerase II as a reference gene (Livak and Schmittgen, 2001). The mRNA expression level of each gene in the transformant was expressed relative to that in the control transfected with psAP2.
2.5. Production of whole lysates from E. histolytica trophozoites
Approximately 1 × 106 trophozoites were harvested 48 h after initiation of culture and washed with 2% glucose in 1X phosphate buffer saline (PBS) three times. Cells were counted and resuspended in 500 μL homogenization buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 50 mM NaCl) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 0.5 mg/mL E-64 (Peptide Institute, Osaka, Japan). Cells were disrupted mechanically by a Dounce homogenizer and kept on ice for 30 min with intermittent vortexing followed by centrifugation at 500 × g for 30 min at 4 °C for removing the insoluble cellular debris. The supernatant, representing total cell lysate, was carefully collected. Protein concentrations were spectrophotometrically determined by the Bradford method using bovine serum albumin as a standard as previously described (Bradford, 1976).
2.6. Enzyme assays and quantitation of CoA in cell lysates
PanK and DPCK activities in the cell lysate were measured with coupled assays using ADP Hunter™ Plus Assay kit (DiscoverX, US) according to the manufacturer's instructions. Briefly, fluorescence intensities were continuously measured to estimate the formation of resorufin at 37 °C by excitation at 530 nm and emission at 590 nm in a 25 μL reaction mixture [10 mM MgCl2, 15 mM HEPES, 20 mM NaCl, 1 mM EGTA, 0.02% tween 20, 0.1 mg/mL β-globulin, 2 mM pantothenate or 2 mM dephospho CoA for PanK or DPCK, respectively, 0.1 mM ATP, 2 μL of cell lysate (∼5 μg protein)]. Kinetic data were estimated by curve fitting with the Michaelis–Menten equation using GraphPad Prism (GraphPad Software Inc., San Diego, USA). This experiment was repeated three times in triplicate with proteins isolated from two independent extractions, and kinetic values are presented as the means ± S.E. for three independent kinetic assays.
Concentrations of CoA in the cell lysate were measured using the CoA assay kit (BioVision, CA, USA) according to manufacturer's instructions. CoA at 0.05–1 nmole was used to produce a standard curve to determine the amounts of CoA in lysates. Experiment was conducted in triplicate, and repeated three times on three different days.
2.7. Monitoring of growth kinetics
Trophozoite cultures were continuously maintained in mid-log phase as described previously, and placed on ice for 5 min to detach cells from the glass surface. Cells were collected by centrifugation at 500 × g for 5 min at room temperature. After discarding the spent medium, the pellet was re-suspended in 1 mL of BI-S-33 medium. Cell densities were estimated on a haemocytometer. Approximately 10,000 trophozoites were inoculated in 6 mL fresh BI-S-33 medium. Cultures were examined every 24 h for 5 days.
2.8. Genome-wide survey of enzymes involved in CoA biosynthesis in the E. histolytica genome
Putative genes encoding PanK, PPCS-PPCDC, PPAT, and DPCK were identified in the genome of HM-1:IMSS in the E. histolytica genome database (AmoebaDB, http://amoebadb.org/amoeba/) using the blastp online search tool (protein-protein BLAST). Human or bacterial orthologs (Human PanK1α/NP_683878.1, E. coli bifunctional CoaBC/WP_089667520.1, Human bifunctional COASY/AAL50813.1) retrieved from non-redundant protein sequences (nr) database of National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) were used as query sequences. Pathway analysis was conducted using Kyoto Encyclopedia of Genes and Genomes database (KEGG, https://www.genome.jp/kegg). The steady-state mRNA level of each gene in the trophozoite stage was examined using our previous array data as an independent experiment in both HM1: IMSS cl6 (Penuliar et al., 2012, 2015) and G3 strains (Nakada-Tsukui et al., 2012; Furukawa et al., 2012, 2013). Then using Entamoeba invadens orthologs, we compared the mRNA expression levels of these genes during various timepoints during the encystation stage (De Cádiz et al., 2013).
2.9. Phylogenetic analysis of PanK
Putative orthologs of PanK from a variety of organisms were retrieved by a blastp search of non-redundant protein sequences (nr) database of National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) using the EhPanK sequence (XP_001913460) as a query. To comprehensively retrieve all orthologs from representative organisms in the variety of major taxa, we carried out a blastp search against each taxonomic group as shown in Table S1. In each blastp analysis, a list of selected sequences was made with an E-value less than 1 × 10−10 in pairwise alignments with EhPanK. Based on the lists for all taxonomic groups in Table S1, we selected 81 sequences as a final data set. The Muscle program (Edgar, 2004) in SeaView software package version 4.6.1 (Gouy et al., 2010) was used for sequence alignment. Then 145 unambiguously aligned positions were selected by manual inspection and used for phylogenetic analyses.
The phylogenic data matrices were subjected to analysis with the IQTREE program (Nguyen et al., 2015) to select appropriate models for amino acid sequence evolution. The LG + Γ4 model was found to be the best for analysis. Maximum likelihood (ML) analysis implemented in the RAxML program version 7.2.6 (Stamatakis, 2006) was used to infer the ML tree by applying LG + Γ4 model. In the bootstrap analysis, heuristic tree search was performed with a rapid bootstrap algorithm option (-f) for 100 bootstrap replicates. Bootstrap proportion (BP) values greater than 50 were indicated on the corresponding internal branches of the ML tree drawn with FigTree program Version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
2.10. Production of E. histolytica transformant line to express Myc-tagged PanK
The EhPanK gene was amplified from cDNA using Ex Taq DNA polymerase (Takara) using the following primer set (sense primer, 5′- GCCCCGGGATGTCTCAACCATCCCATTC -3′ and antisense, 5′- GCCTCGAG TTACATTAGTTCTTCTTCATACTC-3′), restriction sites are indicated in bold. An amplified fragment was ligated into SmaI and XhoI double digested pEhEx-Myc expression vector (Nakada-Tsukui et al., 2012) to produce pEhExMyc-PanK. The plasmid was introduced into trophozoites of E. histolytica HM-1:IMSS cl6 (Diamond et al., 1978) by liposome-mediated transfection (Nozaki et al., 1999). Transformants were selected and maintained as described previously.
2.11. Cell fractionation and immunoblot analysis
Trophozoites of the amoeba transformant expressing Myc-EhPanK and the mock transformant, transfected with pEhEx-Myc, were washed three times with PBS containing 2% glucose. After resuspension in homogenization buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 50 mM NaCl and 0.5 mg/mL E-64 protease inhibitor), cells were disrupted mechanically by a Dounce homogenizer on the ice, centrifuged at 500 × g for 5 min, and the supernatant was collected to remove unbroken cells. The supernatant fraction was centrifuged at 5000 × g for 10 min to isolate pellet and supernatant fractions. The 5000 × g supernatant fraction was further centrifuged at 100,000 × g for 60 min to produce a 100,000 × g supernatant and pellet fractions. The pellets at each step were further washed twice with homogenization buffer and re-centrifuged at 100,000 × g for 10 min to minimize carryover. Immunoblot analysis was performed using the fractions and anti-Myc mouse monoclonal antibody. Anti-CPBF1 (cysteine protease binding family protein 1) and anti-CS1 (cysteine synthase 1) rabbit antisera were used as organelle membrane and cytosolic markers, respectively.
2.12. Production of EhPanK recombinant protein
The plasmid was constructed as previously described (Sambrook and Russell, 2001). DNA fragment was amplified from E. histolytica cDNA. Primers used were sense, 5′- GCCGGGATCCATGTCTCAACCATCCCATTC -3′ and antisense, 5′- GCCG GTCGACTTACATTAGTTCTTCTTCATACTC-3’. Bold letters indicate BamHI and SalI restriction sites. PCR was performed with primeSTAR HS DNA polymerase (Takara) and the following parameters: initial incubation at 98 °C for 30 s; followed by the 30 cycles of denaturation at 98 °C for 10 s; annealing at 55 °C for 30 s; and elongation at 72 °C for 1 min; and a final extension at 72 °C for 7 min. The PCR fragment was digested with BamHI and SalI, purified with Wizard® SV Gel and PCR Clean-up System (Promega). The fragment was cloned into BamHI and SalI double digested pCOLD1™ histidine-tag vector (Takara) to finally produce pCOLD1-EhPanK. The nucleotide sequence of the engineered plasmid was verified by sequencing.
pCOLD1-EhPanK was introduced into E. coli BL21(DE3) cells via heat shock at 42 °C for 1 min. E. coli BL21 (DE3) harboring pCOLD1-EhPanK was grown at 37 °C in 100 mL of Luria Bertani medium (Invitrogen) in the presence of 100 μg/mL ampicillin (Nacalai Tesque). The overnight culture was used to inoculate 500 mL of fresh medium, and the culture was further continued at 37 °C with shaking at 180 rpm. When A600 absorbance reached 0.8, then 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) was added, and cultivation was continued for another 24 h at 15 °C. E. coli cells from the induced culture were harvested by centrifugation at 5000 × g for 20 min at 4 °C. The cell pellet was washed with PBS, pH 7.4, re-suspended in 20 mL of the lysis buffer (50 mM Tris HCl, pH 8.0, 300 mM NaCl, and 10 mM imidazole) containing 0.1% Triton x 100 (v/v), 100 μg/mL lysozyme, and 1 mM PMSF, and incubated at room temperature for 30 min. Then the mixture was sonicated on ice and centrifuged at 25,000 × g for 15 min at 4 °C. The supernatant was mixed with 1.2 mL of 50% Ni2+-NTA His-bind slurry, incubated for 1 h at 4 °C with mild shaking. The recombinant enzyme-bound resin in a column was washed three times with buffer A (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 0.1% Triton X-100, v/v) containing 10–50 mM of imidazole. Bound protein was eluted with buffer A containing 100–300 mM imidazole. After the integrity and the purity of recombinant protein were confirmed with 12% SDS-PAGE analysis, followed by Coomassie Brilliant Blue staining, the sample was dialyzed against a 300-fold volume of 50 mM Tris-HCl, 150 mM NaCl, pH 8.0 containing 10% glycerol (v/v) and the Complete Mini protease inhibitor cocktail (Roche, Mannheim, Germany) for 18 h at 4 °C. Pure enzyme was stored at −80 °C with 20% glycerol in small aliquots until use.
2.13. Screening of natural compounds for EhPanK inhibitors
We screened 244 compounds from the Kitasato Natural Products Library for inhibition of EhPanK activity. Initially, each compound was dissolved in 50% DMSO and water at a final concentration of 1 mg/mL. Enzymatic reactions were carried out on a black 384-well microtiter plate with a 20 μL reaction mixture composed of 19 μl enzyme mix (100 μM pantothenate, 100 μM ATP, 50 ng of EhPanK recombinant enzyme in kinase buffer (see above) and 1 μl of the individual compounds (final concentration 50 μg/mL or 25–200 μM) at 37 °C for 2 h. After the kinase reaction, ADP was measured using ADP Hunter™ Plus kinase assay kit as described previously. Inhibition values were measured in triplicate.
Compounds that showed more than 50% inhibition at <400 μM were re-tested to confirm that they did not inhibit the enzyme in the coupled assay (pyruvate kinase and pyruvate oxidase) in ADP Hunter™ Plus kinase assay kit. Inhibition of the enzymes used in the coupled assay was examined in the reaction mixture (10 mM MgCl2, 15 mM HEPES, 20 mM NaCl, 1 mM EGTA, 0.02% tween 20, 0.1 mg/mL β-globulin, 60 μM pantothenate, 50 μM ATP, 10–50 μM ADP, and 25–200 μM of selected active compounds). No significant change was observed in a range of ADP and inhibitor concentrations, suggesting no inhibition of coupled enzymes by the EhPanK inhibitors. Compounds that did not inhibit enzymes used in the coupled assay were selected as hits for further evaluation, including dose-response, determination of concentration needed for 50% inhibition (IC50) of PanK activity as well as both E. histolytica and human cell growth.
2.14. Measurement of anti-amebic activity of EhPanK inhibitor compounds
Approximately 1 × 104 trophozoites of E. histolytica clonal strain HM-1:IMSS cl6 in 200 μl of BI-S-33 medium were dispensed into each well of a 96-well plate and incubated at 35.5 °C for 2 h. Medium was then removed and replaced with 200 μl of BI-S-33 medium that contained various concentrations of each compound. The final concentrations of test compounds are 50–600 μM for cephaloridine, hygromycin A, erythromycin A, and fluorescamine; 5–200 μM for kasugamycin, gardimycin, rosamycin, streptomycin, tirandamycin A, neomycin, and teicoplanin; 0.1–10 μM for O-methylnanaomycin A, echinomycin, and trichostatin. The plate was incubated under anaerobic conditions at 35.5 °C for 48 h. After the medium was removed, 100 μl of pre-warmed Opti-MEM I (Life Technologies, Grand Islands, NY, USA) containing one-tenth volume of WST-1 (Roche, Mannheim, Germany) was added. The viability of trophozoites was estimated by measuring absorbance at 450 nm by SpectraMax® Paradigm® (Molecular Devices, CA, USA). Metronidazole was used as positive control at final concentrations of 0.1–10 μM and 0.5% DMSO as negative control. Each assay was performed in triplicate. IC50 values were calculated by least squares curve fitting of the dose inhibition curves using GraphPad Prism (GraphPad Software Inc., San Diego, USA).
2.15. Evaluation of cytotoxicity against MRC-5 cells
Human fibroblast cells, MRC-5, were cultured on 96-well flat bottom plates at a density of 1.5 × 104 cells/well with 100 μl of MEM medium (Life Technologie) containing 10% fetal bovine serum (Hana-nesco Bio, Tokyo, Japan) and 1% penicillin-streptomycin (Life Technologies). Cells were incubated at 37 °C with 5% CO2 for 48 h. Approximately 100 μl of MEM medium containing 5 μl of each compound dissolved in 50% DMSO in water were added to each well. After the plates were incubated for 48 h, approximately 10 μl of WST-8 (Dojindo, Kumamoto, Japan) was added to each well and the plate was incubated at 37 °C with 5% CO2 for 2 h. The absorbance was measured at 450 nm by spectrophotometer (SH-9000Lab, Corona Electric, Ibaraki, Japan). Growth of MRC-5 cells was measured in duplicate. Staurosporine (Kitasato Natural Products Library) was used as a positive (cytotoxic) control in a concentration range of 0.5 pM to 0.1 nM IC50 values were calculated as described previously.
3. Results
3.1. Identification of four enzymes involved in CoA biosynthesis in E. histolytica
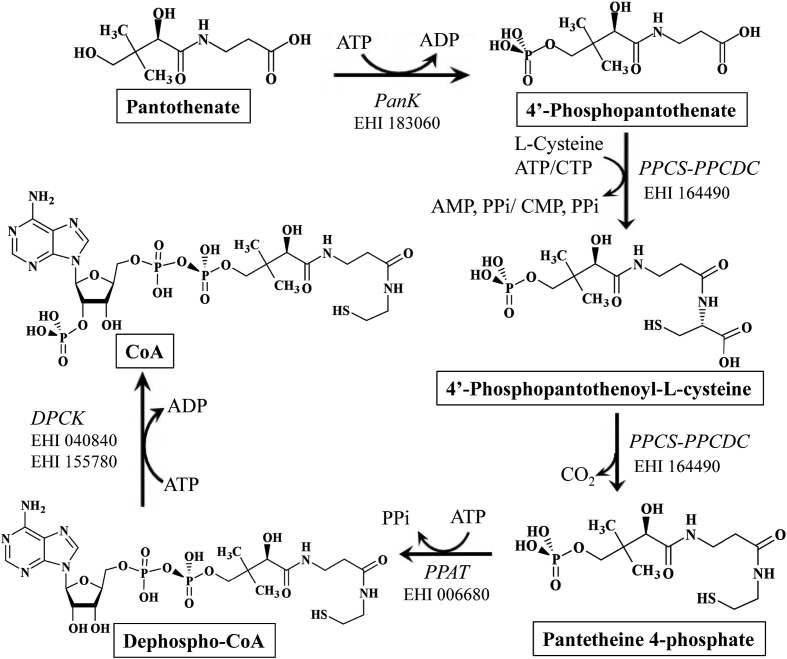
We identified all putative genes involved in CoA biosynthesis in the reference genome of E. histolytica: HM-1:IMSS (Fig. 1). Four genes encoding sequential pathway enzymes were identified: pantothenate kinase (EC 2.7.1.33, PanK), bifunctional phosphopantothenate-cysteine ligase/decarboxylase (EC 6.3.2.5/EC 4.1.1.36), phosphopantetheine adenylyltransferase (EC 2.7.7.3), and dephospho CoA kinase (EC 2.7.1.24). We identified a single putative PanK gene in each strain (e.g., EHI_183060 in HM-1:IMSS and EHI7A_012650 in HM-1:IMSS-A) in Amoeba Genomics Resources database, AmoebaDB (http://amoebadb.org). The nucleotide sequences of the genes in different strains are 100% identical. Therefore, E. histolytica seems to have only one PanK gene.
Fig. 1.
Coenzyme A biosynthetic pathway in E. histolytica. ID numbers of individual enzymes in AmoebaDB are also shown. PanK, pantothenate kinase; PPCS-PPCDC, bifunctional phosphopantothenoylcysteine synthetase and phosphopantothenoylcysteine decarboxylase; PPAT, phosphopantetheine adenylyltransferase; and DPCK, dephosphocoenzyme A kinase.
Our previous transcriptome data verified that all four genes are expressed at low to moderate levels (Fig. S1A). The level of mRNA expression relative to RNA polymerase for PanK, PPCS-PPCDC, PPAT, DPCK1, and DPCK2 in trophozoites of the E. histolytica clonal strain HM1: IMSS cl6 (Penuliar et al., 2015, 2012) and G3 (Nakada-Tsukui et al., 2012; Furukawa et al., 2012, 2013) strain were 1.2–1.8, 0.5–0.6, 0.3–0.4, 0.04–0.05 and 0.3–0.4, respectively. In this research, we describe biochemical and genetic characterization of PanK; investigation of other enzymes involved in CoA biosynthesis will be described elsewhere. Our previous transcriptomic analysis of E. invadens (De Cádiz et al., 2013), as a model for encystation (Donaldson et al., 1975; Kojimoto et al., 2001; Stanley, 2003; Chia et al., 2009), showed that genes encoding these four enzymes were expressed during encystation, but the expression profiles of individual enzymes largely differed (Fig. S1B). The expression profiles of PPCS-PPCDC and DPCK1 were quite peculiar with expression peaking at certain time points (2 h and 24 h, respectively); this subject was, however, not investigated as part of this study.
3.2. Features of EhPanK and its encoded protein
The 1219-bp E. histolytica PanK gene contains two small introns, and is predicted to encode a 403 a.a. polypeptide with a molecular mass of 45.2 kDa and a pI value of 5.2. EhPanK showed 99, 96, 83, and 70% sequence identity to PanK from E. nuttalli, E. dispar, E. moshkovskii, and E. invadens, respectively. In general, PanK is classified into three groups, type I through III, as determined by primary sequence analysis and kinetic properties (Song and Jackowski, 1993; Rock et al., 2002; Yang et al., 2006). E. histolytica PanK is classified as PanK type II, and shows a low level (∼35%) of positional identity to the A. thaliana and human orthologs, both of which also belong to type II (Fig. S2 and S3). When compared to human PanK, Entamoeba PanK shows the highest similarity to human PanK4 (35%), whereas similarity to other human PanK isotypes (PanK1α, PanK1β, PanK2, and PanK3) is ∼32%. All amino acid residues were inferred to be involved in pantothenate and ATP catalysis and substrate binding include the typical ATP-binding motif have been reported in human PanK1 and PanK3 (Bum et al., 2007). Some conserved glycine residues in the ATP binding motif (Gly19 and Gly321 in human PanK3; Gly59 and Gly351 in E. histolytica) are indispensable for activity. Mutations of this amino acid have been demonstrated to abolish enzymatic activity because ATP is no longer able to properly bind (Bum et al., 2007). Furthermore, it is understood that phosphorylation is proceeded by an ordered sequential mechanism, with ATP binding preceding pantothenate binding (Leonardi et al., 2005a).
3.3. Phylogenetic analysis of EhPanK
We determined the phylogenetic relationship between 81 putative PanK orthologs collected from bacteria and eukaryotes (Table S1, Fig. S2). The majority of eukaryotes possess the PanK gene. Mammals (Opisthokonta) and land plants (Viridiplantae) apparently possess twice or more two genes. For instance, four PanK genes were identified from Homo sapiens, PanK1α and PanK1β (Rock et al., 2002), PanK2 (annotated as a mitochondrial precursor protein) (Hörtnagel et al., 2003; Kotzbauer et al., 2005), and PanK3 (Zhang et al., 2005). Archaea and most bacterial groups do not possess PanK homologs that show similarity to EhPanK with the E-value less than 10−10. PanK homologs were identified, however in Firmicutes, specifically Bacilli. During sequence alignment, only the ‘fumble’ domain (PFAM:PFO03630) could be aligned among all 81 sequences, although insertions and deletions were needed for proper alignment due to a high degree of divergence. PanKs from Euglenozoa have an ∼1000-a.a. long N-terminal extension, which contains an exonuclease-endonuclease-phosphatase (EEP) domain in the 300 a.a.-long N-terminal region and a 550 a.a.-long adenylate forming (AF) domain. Some of PanK from Viridiplantae have a 300 a.a.-long C-terminal extension, annotated as DUF89 (Domain of Unknown Function 89) (Huang et al., 2016). PanK from other organisms including the genus Entamoeba basically contain only a ‘fumble’ domain; neither the EEP/AF nor DUF89 domain is present in PanK from other organisms except that some possess an N-terminal extension that appears to be an organelle targeting sequence.
Based on 145 aligned positions from the ‘fumble’ domain, an optimal ML tree was created with bootstrap proportion (BP) support values (Fig. S2). Eukaryotes and Firmicutes were separated clearly with a 100% BP support value. The Eukaryota clade had monophylies of several taxonomic groups reconstructed, but BP values were generally low except for those of Viridiplantae monophyly (88%) and Haptophyceae monophyly (97%). Four Entamoeba species are monophyletic with 100% BP support, and the Entamoeba clade shares a sister group position to the clade of Euglenozoa. However, since BP support values for deep branching patterns were very low, monophyly of Amoebozoa including the Entamoeba clade cannot be ruled out. Although this analysis could not precisely infer the phylogenetic position of the Entamoeba PanK, it is apparently of a eukaryotic origin.
3.4. Expression and purification of recombinant PanK
A soluble EhPanK recombinant protein with a 2.6 kDa histidine tag at the amino terminus was successfully produced using the pCOLD I E. coli expression system and purified to homogeneity (>95% as evaluated with Coomassie Brilliant Blue stained SDS-PAGE gel) (Fig. S4A, Table S2). Immunoblot analysis of the purified recombinant protein using His-Tag antibody confirmed the absence of truncation (Fig. S4B). The molecular mass of the purified protein under reducing conditions was consistent with the predicted molecular mass 45.2 kDa excluding the histidine tag. The specific activity of the purified enzyme was estimated to be 1.5 μmol/min/mg when assayed under the standard conditions described in the “Materials and methods” section. EhPanK is robust and catalytically active in broad pH range with maximum activity obtained at pH 6 and 37 °C (Fig. S5A).
3.5. Kinetic properties and phosphoryl donor specificities of EhPanK, and effects of metal ions on EhPanK
Table 1 summarizes the apparent Km, Vmax, and Kcat values for EhPanK using pantothenate and ATP as substrates. EhPanK exhibited hyperbolic saturation kinetics when assayed over the substrate range of 4–256 μM for pantothenate in the presence of 25–100 μM ATP (Fig. S5B) and 1–100 μM ATP in the presence of 8–128 μM pantothenate (Fig. S5C). The apparent Km value for pantothenate at saturating ATP concentrations was 53.2 ± 7.1 μM, and the Km value for ATP at saturating pantothenate concentrations was 41.4 ± 3.9 μM.
Table 1.
Kinetic parameters of E. histolytica pantothenate kinase.
| Substrate | Km (μM) | Vmax (μmole/min/mg) | Kcat (min−1) | Kcat/Km (min−1μM−1) |
|---|---|---|---|---|
| Pantothenate | 53.2 ± 7.1 | 2.79 ± 0.1 | 126.8 ± 4.5 | 2.38 ± 0.08 |
| ATP | 41.4 ± 3.9 | 2.84 ± 0.2 | 129.1 ± 9.2 | 3.11 ± 0.22 |
Mean ± SEM of three replicates are shown.
EhPanK utilized various nucleoside triphosphates, ATP, CTP, GTP, UTP, dATP, and polyphosphates, as well as deoxynucleotides as phosphoryl donors, with ATP being the best phosphate donor (Table 2). EhPanK showed an absolute requirement for a free bivalent metal cofactor, with Mg2+ as the preferred cation (Table 3).
Table 2.
Phosphoryl donor specificity of E. histolytica pantothenate kinase.
| Phosphoryl donora | Relative activity (%) | Phosphoryl donora | Relative activity (%) |
|---|---|---|---|
| ATP | 100.0 ± 4.1 | CTP | 1.4 ± 0.2 |
| GTP | 78.8 ± 4.9 | Metaphosphate | ND |
| dATP | 19.3 ± 4.3 | Hexametaphosphate | ND |
| Tripolyphosphate | 16.9 ± 0.8 | Trimetaphosphate | ND |
| UTP | 16.6 ± 1.2 | Glucose-6-phosphate | ND |
| Polyphosphate | 16.2 ± 2.5 | Phosphoenolpyruvate | ND |
| TTP | 5.4 ± 1.9 | None | ND |
Assays were performed as described in Materials and methods, in the presence 15 mM HEPES, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL β−globulins and 0.2 mM pantothenate. Reactions were conducted at 37 °C at pH 6.
The activity is shown in percentage (%) relative to that toward ATP.
ND, not detected.
Mean ± SEM of three replicates are shown.
The final concentration used was 100 μM.
Table 3.
Effect of metal ions on the activity of E. histolytica pantothenate kinase.
| Metala | Relative activity (%) |
|---|---|
| MgCl2 | 100.0 ± 0.0 |
| CuCl2 | 84.3 ± 4.7 |
| MnCl2 | 83.2 ± 2.0 |
| CoCl2 | 74.2 ± 6.1 |
| FeCl2 | 72.8 ± 1.7 |
| LiCl2 | 34.5 ± 1.8 |
| CaCl2 | 17.0 ± 3.0 |
| ZnCl2 | 3.9 ± 0.7 |
| NiCl2 | ND |
| NaCl | ND |
| KCl | ND |
| None | ND |
Assays were performed as described in Materials and methods, in the presence of 100 mM ATP, 15 mM HEPES, 20 mM NaCl, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL β−globulins and 0.2 mM pantothenate. Reactions were conducted at 37 °C at pH 6.0.
The activity is shown in percentage (%) relative to that toward MgCl2. ND, not detected.
Mean ± SEM of three replicates are shown.
The cation final concentration used was 5 mM.
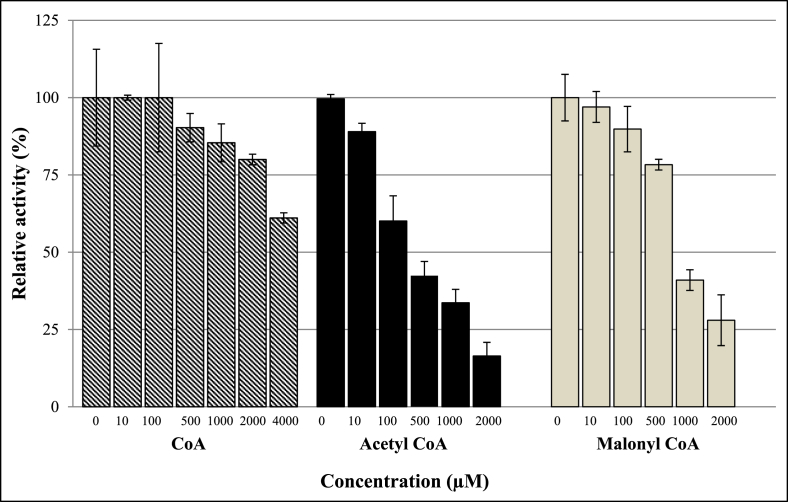
3.6. Regulation of EhPanK by CoA, acetyl CoA, and malonyl CoA
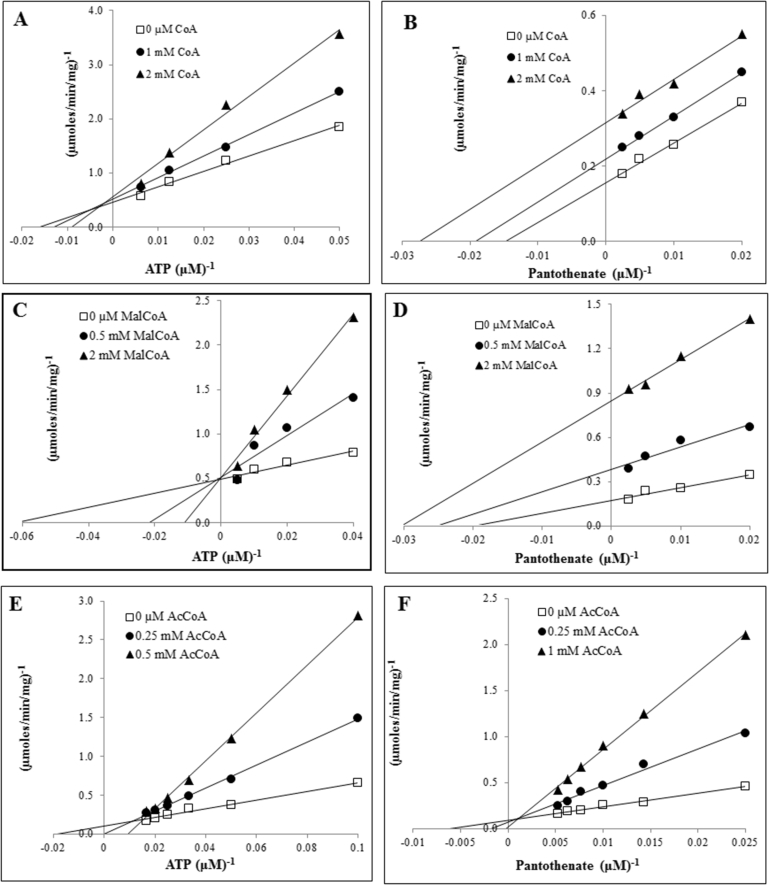
PanK from other organisms was reported to be negatively regulated by allosteric inhibition with CoA, acetyl CoA, and malonyl CoA (Calder et al., 1999; Takagi et al., 2010; Vallari et al., 1987). Similarly, EhPanK was also inhibited by CoA and its derivatives, although relatively higher concentrations were needed for inhibition compared with PanK from other organisms (Brand and Strauss, 2005; Calder et al., 1999) (Fig. 2). Double-reciprocal plots revealed the mechanisms of inhibition by CoA and malonyl CoA are competitive with ATP, while inhibition by CoA and malonyl CoA are uncompetitive or non-competitive with pantothenate, respectively. The mechanisms of inhibition by acetyl CoA were not clear but seemed to be the mixed type with respect to ATP and pantothenate (Fig. 3).
Fig. 2.
Effect of CoA, acetyl CoA, and malonyl CoA on the pantothenate kinase activities. Relative activities of EhPanK in the presence of various concentrations of inhibitors to those without inhibitors are shown. The assay was performed in 10 mM MgCl2, 15 mM HEPES, 20 mM NaCl, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL γ−globulins, 0.2 mM pantothenate, and 100 μM ATP with various concentrations of CoA, acetyl CoA, or malonyl CoA at 37 °C at pH 6. The assays were carried out three times independently, and the results are shown as means ± SEM of triplicates.
Fig. 3.
Double-reciprocal plots of the recombinant EhPanK in the presence of CoA (A and B), malonyl CoA (C and D), or acetyl CoA (E and F). The enzymatic activities were determined with various concentrations of ATP and 0.2 mM pantothenate (A, C, and E) or various concentrations of pantothenate and 100 μM ATP (B, D, and F), in the presence of three concentrations of inhibitors (0, 0.5, and 2 mM). Data are shown in means ± SEM of triplicate analyses.
3.7. Cellular localization of EhPanK
To examine the localization of EhPanK, we performed fractionation and immunoblot analysis of PanK using E. histolytica transformant-expressing Myc-tagged EhPanK. We demonstrated that EhPanK is exclusively present in the 100,000 × g supernatant fraction corresponding the cytosol (Fig. S6).
3.8. Effects of EhPanK gene silencing on growth, cellular CoA levels, and gene expression of the CoA biosynthetic pathway
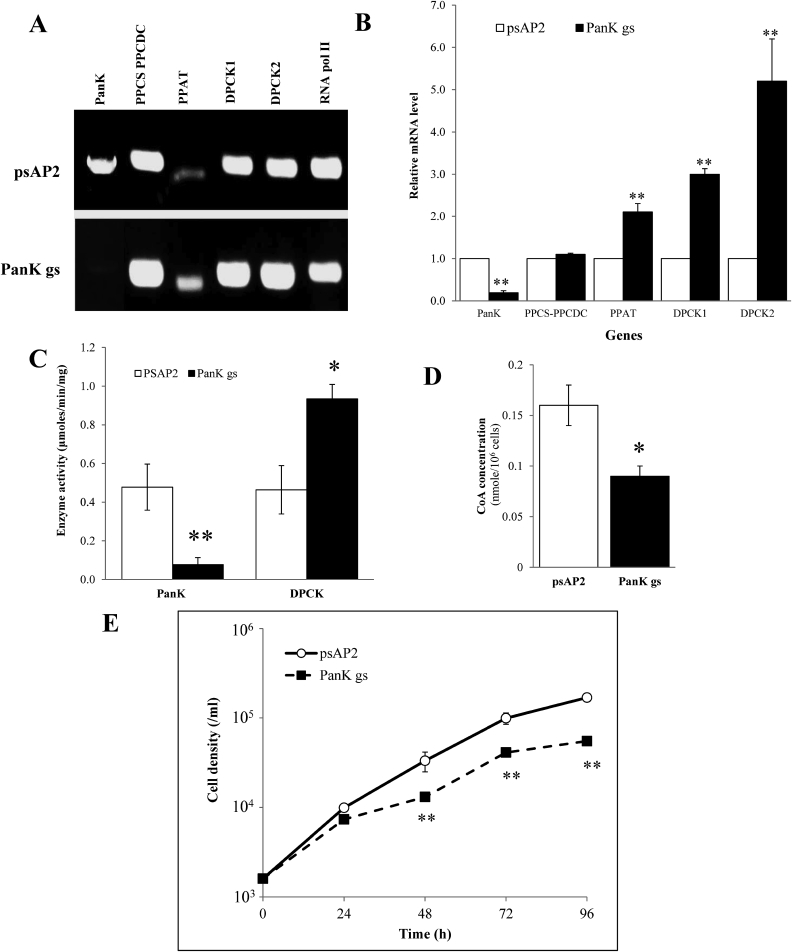
To investigate the physiological importance and essentiality of PanK in E. histolytica, we created and examined an E. histolytica strain where EhPanK was silenced by antisense small RNA-mediated transcriptional gene silencing (Mirelman et al., 2008); EhPanK gene expression was successfully silenced (Fig. 4A). The level of silencing was estimated to be approximately 85% by qRT-PCR measurement. Interestingly, the steady-state transcript of the genes encoding other enzymes involved in CoA biosynthesis were up-regulated 1.2 ± 0.03, 2.1 ± 0.2, 3 ± 0.1, and 5.2 ± 1.0-fold for PPCS-PPCDC, PPAT, DPCK1, and DPCK2, respectively (Fig. 4B).
Fig. 4.
Analyses of EhPanK gene silenced strain. A) RT-PCR analysis of EhPanK gene transcript from E. histolytica transformants. “psAP” indicates control strain transfected with psAP-2-Gunma empty vector, and “PanK gs” indicates EhPanK gene silenced strain. B) Relative levels of gene transcripts encoding all four enzymes involved in CoA biosynthesis by qRT-PCR analysis in psAP and Pan Kgs transformants. qRT-PCR data were normalized against RNA polymerase II, and are shown in percentage relative to the transcript level of each gene in psAP control. C) PanK and DPCK activity in cell lysates of psAP and PanKgs transformants. D) CoA concentration in cell lysates. For B-D, data are shown in mean ± SEM of three biological replicates. Statistical comparison is made by Student's t-test (*P < 0.05, **P < 0.01). E) Growth kinetic of E. histolytica transformants during 96 h incubation in BI-S-33 medium. Data are shown in mean ± SEM of three replicates. Data from one representative experiment of three conducted independently are shown. Statistical comparison is made by Student's t-test (*P < 0.05, **P < 0.01).
In order to determine whether the protein level also increased, we measured PanK and DPCK activity in cell lysates. PanK activity decreased by approximately 81% in the EhPanK-silenced strain compared to the control. In contrast, DPCK activity increased 2.1 fold (Fig. 4C). CoA concentration decreased by approximately 40% in EhPanK-silenced strain compared to the control (Fig. 4D). Growth kinetic analysis also showed that in EhPanK-silenced strain showed remarkable growth retardation as the cell density of EhPanK-silenced strain decreased to approximately 30–52% of the control at 48–96 h, respectively (Fig. 4E, Fig. S7A, Fig. S7B).
3.9. Identification of EhPanK inhibitors from Kitasato Natural Products Library
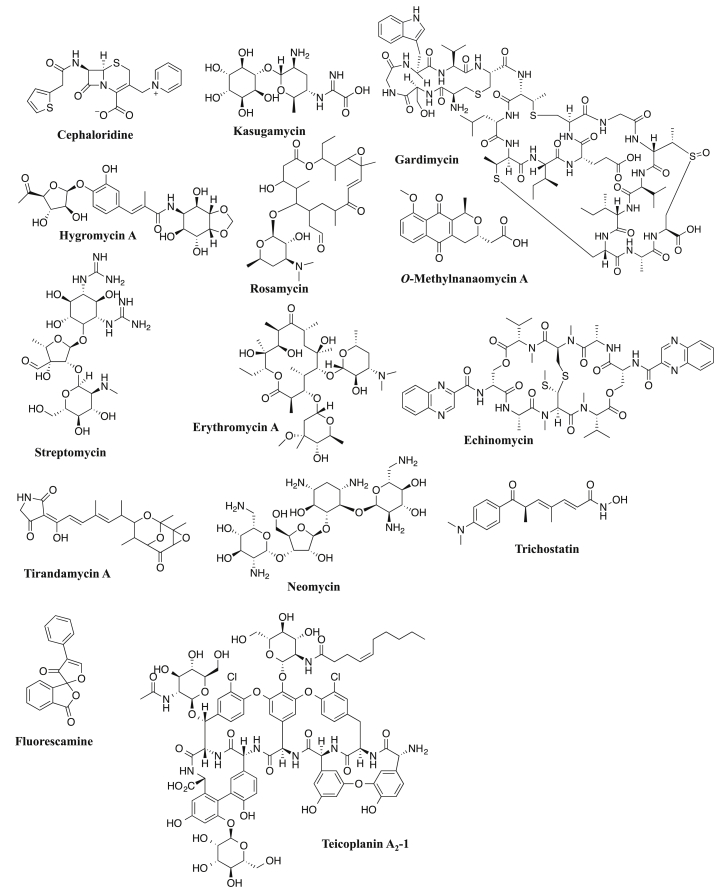
Based on the significant biological role of EhPanK, inhibitors of this enzyme should be considered to possess properties needed for novel anti-amoebic agents. Therefore, we conducted enzyme-based screening of the Kitasato Natural Products Library composed of 244 structurally elucidated compounds using recombinant EhPanK to identify potential PanK inhibitors. We identified 14 compounds (Fig. 5) that demonstrated an IC50 value of <400 μM (Table 4, Fig. S8). Among these compounds, Teicoplanin had the best inhibitory activity against EhPanK with an IC50 value of 26.3 ± 2.6 μM. All EhPanK inhibitors also inhibited E. histolytica trophozoites although the IC50 values of most of the identified EhPanK inhibitors were high. Six compounds, O-methylnanaomycin A, Echinomycin, Tirandamycin A, Neomycin, Trichostatin, and Teicoplanin, showed the IC50 values of <20 μM against E. histolytica trophozoites, but the IC50 values of O-methylnanaomycin A, Echinomycin, Tirandamycin A, Neomycin, and Trichostatin against E. histolytica trophozoites were lower than their IC50 values against EhPanK activity, suggesting that the target of these compounds for growth inhibition are unlikely to be through PanK inhibition. In contrast, Teicoplanin demonstrated comparable IC50 values against PanK activity and E. histolytica growth (Table 5).
Fig. 5.
Structures of EhPanK inhibitors identified in this study.
Table 4.
Inhibitory activities of EhPanK inhibitors from Kitasato Natural Products Library.
| Compound | IC50 (μM) |
|---|---|
| Cephaloridine | 229.9 ± 10.1 |
| Kasugamycin | 254.1 ± 4.7 |
| Gardimycin | 49.2 ± 4.8 |
| Hygromycin A | 193.7 ± 17.8 |
| O-methylnanaomycin A | 283.8 ± 28.7 |
| Rosamycin | 165.7 ± 2.0 |
| Streptomycin | 155.4 ± 10.1 |
| Erythromycin A | 131.3 ± 6.0 |
| Echinomycin | 77.8 ± 4.4 |
| Tirandamycin A | 228.8 ± 3.4 |
| Neomycin | 147.2 ± 6.8 |
| Fluorescamine | 346.4 ± 14.7 |
| Trichostatin | 309.9 ± 14.9 |
| Teicoplanin | 26.3 ± 2.6 |
Mean ± SEM of three replicates are shown.
Table 5.
In vitro anti-amebic activity and cytotoxicity against MRC-5 cells of EhPanK inhibitors.
| Compound | IC50 (μM) |
|
|---|---|---|
| E. histolytica trophozoite | Normal human cell (MRC-5) | |
| Cephaloridine | 245.78 ± 34.92 | 48.20 ± 3.61 |
| Kasugamycin | 66.16 ± 13.68 | 87.10 ± 7.22 |
| Gardimycin | 34.40 ± 3.16 | >52.9 |
| Hygromycin A | 142.22 ± 16.34 | >196 |
| O-methylnanaomycin A | 1.68 ± 0.05 | 18.98 ± 0.49 |
| Rosamycin | 56.34 ± 11.72 | 34.40 ± 2.71 |
| Streptomycin | 77.53 ± 1.52 | >172 |
| Erythromycin A | 155.88 ± 20.57 | >136 |
| Echinomycin | 0.26 ± 0.04 | <0.38 |
| Tirandamycin A | 13.71 ± 4.85 | 143.88 ± 3.80 |
| Neomycin | 10.58 ± 1.12 | >163 |
| Fluorescamine | 167.90 ± 20.07 | 287.77 ± 17.84 |
| Trichostatin | 0.29 ± 0.04 | 2.62 ± 0.49 |
| Teicoplanin | 15.08 ± 2.25 | >53.3 |
| Metronidazole | 0.66 ± 0.05 | >100 |
Mean ± SEM of three replicates are shown.
4. Discussion
4.1. Identification of CoA biosynthesis as rational drug target
CoA is an essential and ubiquitous cofactor functioning as an acyl carrier as well as a carbonyl activating group for many metabolic reactions. In the present study, we identified four enzymes that are involved in CoA synthesis in E. histolytica (Fig. 1) and characterized the first rate-limiting enzyme in this process, PanK. We have provided evidence for the pivotal role of this enzyme in cell proliferation by gene silencing.
Since all genes involved in CoA biosynthesis are transcribed in all amoeba life cycle stages, this pathway is the good target not only for killing the pathogenic trophozoites but inhibiting stage conversion and thus amoeba transmission. The mechanism coordinating gene expression regulation among the five genes involved in E. histolytica CoA biosynthesis remains elusive. However, based on our previous transcriptomic analysis of E. invadens, a related reptilian amoeba species that also causes an invasive disease and has been used as a model system for E. histolytica (Donaldson et al., 1975; Kojimoto et al., 2001; Chia et al., 2009) during encystation (De Cádiz et al., 2013), we found that PanK is transcribed in both trophozoite and encystation life cycle stages at comparable levels, suggesting that PanK is required in both stages.
A EhPanK gene-silenced strain was successfully established, after producing a strain in which EhPanK gene expression was ∼85% repressed. Four failed attempts to create of such a cell line (data not shown) suggest the essentiality of this gene. The low levels of EhPanK in our experiment can keep the cells viable, consistent with the previous publication in Mycobacterium tuberculosis PanK (Reddy et al., 2014). Interestingly, EhPanK gene silencing resulted in the simultaneous transcriptional up-regulation of genes for downstream CoA pathway enzymes. Remarkably, such genes, DPCK1 and DPCK2, were up-regulated 3- and 5-fold, respectively. This observation is similar to that in experimentation with Plasmodium yoelii, where other genes in the CoA biosynthetic pathway were up-regulated when PanK2 gene was knocked out (Hart et al., 2016). Despite the apparent compensatory upregulation of downstream enzyme genes, CoA concentration was significantly reduced, which is most likely a direct cause of decreased ATP generation and retarded growth.
In order to validate EhPanK as the drug target, there should be notable biochemical, structural, and genetic differences in their human homologs. Such characteristics should, in theory, minimize toxic side effects of pharmacological inhibition of human enzymes via amebiasis chemotherapy. E. histolytica has one PanK enzyme whereas Homo sapiens has five PanK isoforms encoded by four genes. EhPanK is most closely related to human PanK4 with 35% amino acid identity, but phylogenetic analyses suggest a distant relationship between EhPanK and its human counterparts.
While PanK is indispensable for the optimal growth of E. histolytica trophozoites, but in some eukaryotes, including human, PanK was shown non-essential. It has been demonstrated that human cells have the ability to hydrolyze exogenous CoA to 4′-phosphopantetheine by ectonucleotide pyrophosphatase, and then incorporate 4′-phosphopantetheine and enzymatically convert it back to CoA by the bifunctional enzyme CoA synthase (Srinivasan et al., 2015). In Plasmodium, it was reported that PanK is nonessential in blood stage parasite development, but essential in the hepatic stage, as well as oocyst and sporozoite formation in the mosquito (Hart et al., 2016; Srivastava et al., 2016). In M. tuberculosis, CoaA was proven to be an essential enzyme (Awasthy et al., 2010), but was also shown in in vitro and in vivo studies to be a suboptimal target for antimycobacterial drug development because expression of CoaA gene needs to be repressed to obtain sufficient cidal effects by inhibitors (Reddy et al., 2014). On the other hand, CoaBC, a bifunctional enzyme in the CoA biosynthesis, has proven to be essential, but its suitability as drug target needs to be chemically validated in future (Evans et al., 2016). Inhibition of PanK in human cells may be tolerated, which also supports the premise that PanK is a reasonable chemotherapeutic target against pathogens for humans.
4.2. Hit discovery of EhPanK inhibitors
Historically, natural products, especially secondary metabolites, have provided various drugs against human diseases (Newman and Cragg, 2012). Our group has recently discovered several natural compounds from fungi and actinomycetes that inhibit cysteine synthase, an enzyme absent in humans and involved in de novo biosynthesis of L-cysteine (Mori et al., 2015). In this study, we identified 14 inhibitors of EhPanK; half of the inhibitors have sugar moieties, and some of them are aminoglycosides.
Teicoplanin showed moderate inhibitory activity with IC50 26.3 ± 2.6 μM and inhibition of E. histolytica trophozoite growth (IC50 15.1 ± 2.3 μM). Although Teicoplanin also demonstrated cytotoxicity to MRC-5 cells, preference towards E. histolytica cells was identified. Teicoplanin is a mixture of glycopeptide antibiotics used against Gram-positive bacteria including methicillin-resistant Staphylococcus aureus, with a mechanism of action inhibiting bacterial cell wall synthesis. Teicoplanin consists of the glycopeptide core, two carbohydrates (mannose and N-acetylglucosamine), and a side chain. The components of teicoplanin are classified by their side chain length and conformation; the major components are A2-1 through A2-5.
In conclusion, we have shown that pantothenate kinase and CoA biosynthesis are physiologically important for this parasite. We have also shown that potential EhPanK inhibitors were identified by screening the natural product library, suggesting that the enzyme can be a new target for the development of anti-amebic drugs.
Acknowledgements
We thank all members of Nozaki Lab, NIID, particularly, Kumiko Nakada-Tsukui, Yumiko Saito-Nakano, Takeshi Annoura, Herbert Santos, Yuki Hanadate, and Ratna Wahyuni. We thank Russell Miller for proofreading of the manuscript. We also thank RISET-Pro Kemenristekdikti, The Ministry of Research, Technology, and Higher Education, Indonesia. This work was supported in part by a grant for Science and Technology Research Partnership for Sustainable Development from Japan Agency for Medical Research and Development (AMED) and Japan International Cooperation Agency (JICA), a grant for Research on Emerging and Re-emerging Infectious Diseases from AMED, Grants-in-Aid for Challenging Research (Exploratory) (17K19416), for Scientific Research (15H04406) and for Scientific Research on Innovative Areas (23117001, 23117005, 23390099) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.02.004.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Fig. S1. Relative mRNA expression level of all enzymes in CoA biosynthesis. A) Relative levels in trophozoites of the E. histolytica HM1: IMSS cl6 (Penuliar et al., 2012, 2015) and G3 (Furukawa et al., 2013, 2012; Nakada-Tsukui et al., 2012) strain from array data. B) Relative levels in E. invadens encystation stage. Data was generated from the previous publication (De Cádiz et al., 2013).
Fig. S2. Phylogenetic tree of PanK from E. histolytica and other species. Optimal ML tree inferred by RAxML program with LG +Γ4 model was shown. Unambiguously aligned 145 positions from 81 sequences were used for the analysis.Branch lengths are proportional to estimated numbers of substitutions. Bootstrap proportion (BP) values (more than 50) of the rapid bootstrap analysis are shown on the internal branches. For each of the bacterial species, names a taxonomic group ID for Firmicutes (B31) in Table S1 is also indicated.
Fig. S3. Alignment of PanK protein sequences from E. histolytica and other organisms. The sequences were aligned using Muscle program (Edgar, 2004) in SeaView package version 4.6.1 (Gouy et al., 2010). The species names and the NCBI accession numbers are as follows: E. histolytica (XP_001913460); Entamoeba nuttali (XP_008855986); Homo sapiens PanK4 (NP_060686); Homo sapiens PanK3 (NP_078870); Arabidopsis thaliana (NP_001031768). Identical residues are denoted by asterisks (*). The glycine residue in the ATP binding sites are shown in the white letter on with a black background. Serine, arginine, and alanine residues highlighted with black letters on a grey background indicate residues implicated in pantothenate binding sites.
Fig. S4. Expression and purification of EhPanK in E. coli. A) Expression and purification of recombinant EhPanK. Protein samples at each step of purification were subjected to 12% SDS-PAGE under reducing conditions, and the gel was stained with Coomassie Brilliant Blue. B) Immunoblot analysis of purified recombinant EhPanK using anti His-Tag antibody.
Fig. S5. Optimum pH and temperature (A) and initial rate plots (B, C) of EhPanK. A) Enzyme specific activity of recombinant EhPanK was measured at the various pHs and temperatures indicated in the figure. B) The velocity of EhPanK reaction at various pantothenate concentrations in the presence of 25, 50, or 100 μM ATP. C). The velocity of EhPanK reaction at various ATP concentrations in the presence of 8, 64, or 128 μM pantothenate. Assays were performed as described in Materials and methods, with 50 ng of EhPanK recombinant enzyme, 15 mM HEPES, 20 mM NaCl, 10 mM MgCl2, 1 mM EGTA, 0.02% Tween-20, 0.1 mg/mL β−globulins. Assays for panes B and C were conducted at 37 °C at pH 6. Data are shown in mean ± SEM of three replicates.
Fig. S6. Cellular fractionation and immunoblot analysis of EhPanK. Trophozoites of Myc-EhPanK expressing transformant (Myc-PanK) were fractionated as described in Materials in Methods and subjected to immunoblot analysis using anti-Myc monoclonal antibody, anti-CPBF1, and anti-CS1 polyclonal antisera. CPBF1 and CS1 serve as a control of organelle and cytosolic proteins, respectively. The whole lysate of control strain transformed with empty vector was also subjected to analysis.
Fig. S7. Growth kinetics of E. histolytica transformants during 96 h incubation in BI-S-33 medium from two other independent experiments (A and B). Data are shown in mean ± SEM of three replicates.
Fig. S8. Dose-response data of inhibitors against EhPanK inhibitors. IC50 values were determined by dose-response-inhibition using GraphPad Prism (GraphPad Software Inc., San Diego, USA). Data are shown in mean of three replicates.
Detail information of pantothenate kinase from various organisms related to E. histolytica that used in phylogenetic analysis.
Purification of recombinant E. histolytica pantothenate kinase. Enzyme activity was measured as described in Materials and methods.
References
- Abiko Y. Metabolic Pathways. Academic Press; New York: 1975. Metabolism of coenzyme a; pp. 1–25. [Google Scholar]
- Ali V., Nozaki T. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin. Microbiol. Rev. 2007 doi: 10.1128/CMR.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthy D., Ambady A., Bhat J., Sheikh G., Ravishankar S., Subbulakshmi V., Mukherjee K., Sambandamurthy V., Sharma U. Essentiality and functional analysis of type I and type III pantothenate kinases of Mycobacterium tuberculosis. Microbiology. 2010;156:2691–2701. doi: 10.1099/mic.0.040717-0. [DOI] [PubMed] [Google Scholar]
- Begley T.P., Kinsland C., Strauss E. The biosynthesis of coenzyme A in bacteria. Vitam. Horm. 2001;61:157–171. doi: 10.1016/s0083-6729(01)61005-7. [DOI] [PubMed] [Google Scholar]
- Bracha R., Nuchamowitz Y., Anbar M., Mirelman D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006;2:431–441. doi: 10.1371/journal.ppat.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brand L.A., Strauss E. Characterization of a new pantothenate kinase isoform from Helicobacter pylori. J. Biol. Chem. 2005;280:20185–20188. doi: 10.1074/jbc.C500044200. [DOI] [PubMed] [Google Scholar]
- Bum S.H., Senisterra G., Rabeh W.M., Vedadi M., Leonardi R., Zhang Y.M., Rock C.O., Jackowski S., Park H.W. Crystal structures of human pantothenate kinases: insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J. Biol. Chem. 2007;282:27984–27993. doi: 10.1074/jbc.M701915200. [DOI] [PubMed] [Google Scholar]
- Calder R.B., Williams R.S.B., Ramaswamy G., Rock C.O., Campbell E., Unkles S.E., Kinghorn J.R., Jackowski S. Cloning and characterization of a eukaryotic pantothenate kinase gene (PanK) from Aspergillus nidulans. J. Biol. Chem. 1999;274:2014–2020. doi: 10.1074/jbc.274.4.2014. [DOI] [PubMed] [Google Scholar]
- Chia M.-Y., Jeng C.-R., Hsiao S.-H., Lee A.-H., Chen C.-Y., Pang V.F. Entamoeba invadens myositis in a common water monitor lizard (Varanus salvator) Vet. Pathol. 2009;46:673–676. doi: 10.1354/vp.08-VP-0224-P-CR. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Mackey K. Modification of the TRI Reagent(TM) procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- De Cádiz A.E., Jeelani G., Nakada-Tsukui K., Caler E., Nozaki T. Transcriptome analysis of encystation in Entamoeba invadens. PLoS One. 2013;8:e74840. doi: 10.1371/journal.pone.0074840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond L.S., Harlow D.R., Cunnick C.C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Donaldson M., Heyneman D., Dempster R., Garcia L. Epizootic of fatal amebiasis among exhibited snakes: epidemiologic, pathologic, and chemotherapeutic considerations. Am. J. Vet. Res. 1975;36:807–817. [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.C., Trujillo C., Wang Z., Eoh H., Ehrt S., Schnappinger D., Boshoff H.I.M., Rhee K.Y., Barry C.E., Mizrahi V. Validation of CoaBC as a bactericidal target in the coenzyme A pathway of mycobacterium tuberculosis. ACS Infect. Dis. 2016;2:958–968. doi: 10.1021/acsinfecdis.6b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Cantellano M., Martínez-Palomo A. Pathogenesis of intestinal amebiasis: from molecules to disease. Clin. Microbiol. Rev. 2000 doi: 10.1128/cmr.13.2.318-331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A., Nakada-Tsukui K., Nozaki T. Cysteine protease-binding protein family 6 mediates the trafficking of amylases to phagosomes in the enteric protozoan Entamoeba histolytica. Infect. Immun. 2013;81:1820–1829. doi: 10.1128/IAI.00915-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A., Nakada-Tsukui K., Nozaki T. Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Griffin F.M. Failure of metronidazole to cure hepatic amebic abscess. N. Engl. J. Med. 1973;288 doi: 10.1056/NEJM197306282882610. 1397–1397. [DOI] [PubMed] [Google Scholar]
- Hart R.J., Cornillot E., Abraham A., Molina E., Nation C.S., Ben Mamoun C., Aly A.S.I. Genetic characterization of Plasmodium putative pantothenate kinase genes reveals their essential role in malaria parasite transmission to the mosquito. Sci. Rep. 2016;6:33518. doi: 10.1038/srep33518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtnagel K., Prokisch H., Meitinger T. An isoform of hPANK2, deficient in pantothenate kinase-associated neurodegeneration, localizes to mitochondria. Hum. Mol. Genet. 2003;12:321–327. doi: 10.1093/hmg/ddg026. [DOI] [PubMed] [Google Scholar]
- Huang L., Khusnutdinova A., Nocek B., Brown G., Xu X., Cui H., Petit P., Flick R., Zallot R., Balmant K., Ziemak M.J., Shanklin J., De Crécy-Lagard V., Fiehn O., Gregory J.F., Joachimiak A., Savchenko A., Yakunin A.F., Hanson A.D. A family of metal-dependent phosphatases implicated in metabolite damage-control. Nat. Chem. Biol. 2016;12:621–627. doi: 10.1038/nchembio.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain A., Jeelani G., Sato D., Nozaki T. Global analysis of gene expression in response to L-Cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genom. 2011;12:275. doi: 10.1186/1471-2164-12-275. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S. second ed. vol. 1. ASM Press; Washington DC: 1996. Biosynthesis of pantothenic acid and coenzyme A; pp. 687–694. (Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology). [Google Scholar]
- Johnson P.J. Metronidazole and drug resistance. Parasitol. Today. 1993 doi: 10.1016/0169-4758(93)90143-4. [DOI] [PubMed] [Google Scholar]
- Koch C.J., Lord E.M., Shapiro I.M., Clyman R.I., Evans S.M. In Oxygen Transport to Tissue XIX. Springer; US: 1997. Imaging hypoxia and blood flow in normal tissues; pp. 585–593. [DOI] [PubMed] [Google Scholar]
- Kojimoto A, Uchida K., Horii Y., Okumura S., Yamaguch R., Tateyama S. Amebiasis in four ball pythons, Python reginus. J. Vet. Med. Sci. 2001;63:1365–1368. doi: 10.1292/jvms.63.1365. [DOI] [PubMed] [Google Scholar]
- Kotzbauer P.T., Truax A.C., Trojanowski J.Q., Lee V.M.-Y. Altered neuronal mitochondrial coenzyme A synthesis in neurodegeneration with brain iron accumulation caused by abnormal processing, stability, and catalytic activity of mutant pantothenate kinase 2. J. Neurosci. 2005;25:689–698. doi: 10.1523/JNEUROSCI.4265-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi R., Chohnan S., Zhang Y.M., Virga K.G., Lee R.E., Rock C.O., Jackowski S. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J. Biol. Chem. 2005;280:3314–3322. doi: 10.1074/jbc.M411608200. [DOI] [PubMed] [Google Scholar]
- Leonardi R., Zhang Y.M., Rock C.O., Jackowski S. Coenzyme a: back in action. Prog. Lipid Res. 2005 doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real- time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. doi:10.1006. [DOI] [PubMed] [Google Scholar]
- Mirelman D., Anbar M., Bracha R. Epigenetic transcriptional gene silencing in Entamoeba histolytica. IUBMB Life. 2008 doi: 10.1002/iub.96. [DOI] [PubMed] [Google Scholar]
- Mori M., Jeelani G., Masuda Y., Sakai K., Tsukui K., Waluyo D., Tarwadi, Watanabe Y., Nonaka K., Matsumoto A., Omura S., Nozaki T., Shiomi K. Identification of natural inhibitors of Entamoeba histolytica cysteine synthase from microbial secondary metabolites. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada-Tsukui K., Tsuboi K., Furukawa A., Yamada Y., Nozaki T. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell Microbiol. 2012;14:1299–1317. doi: 10.1111/j.1462-5822.2012.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D.J., Cragg G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012 doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T., Asai T., Sanchez L.B., Kobayashi S., Nakazawa M., Takeuchi T. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar regulation and possible function of the cysteine biosynthetic pathway in Entam. J. Biol. Chem. 1999;274:32445–32452. doi: 10.1074/jbc.274.45.32445. [DOI] [PubMed] [Google Scholar]
- Nozaki T., Bhattacharya A. 2015. Amebiasis: Biology and Pathogenesis of Entamoeba, Amebiasis: Biology and Pathogenesis of Entamoeba. [Google Scholar]
- Penuliar G.M., Furukawa A., Nakada-Tsukui K., Husain A., Sato D., Nozaki T. Transcriptional and functional analysis of trifluoromethionine resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 2012;67:375–386. doi: 10.1093/jac/dkr484. [DOI] [PubMed] [Google Scholar]
- Penuliar G.M., Nakada-Tsukui K., Nozaki T. Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman F.E., Pittmann J.C. Amebic liver abscess following metronidazole therapy for amebic colitis. Am. J. Trop. Med. Hyg. 1974;23:146–150. doi: 10.4269/ajtmh.1974.23.146. [DOI] [PubMed] [Google Scholar]
- Ralston K.S., Petri W. The ways of a killer: how does Entamoeba histolytica elicit host cell death? Essays Biochem. 2011;51:193–210. doi: 10.1042/bse0510193. [DOI] [PubMed] [Google Scholar]
- Reddy B.K.K., Landge S., Ravishankar S., Patil V., Shinde V., Tantry S., Kale M., Raichurkar A., Menasinakai S., Mudugal N.V., Ambady A., Ghosh A., Tunduguru R., Kaur P., Singh R., Kumar N., Bharath S., Sundaram A., Bhat J., Sambandamurthy V.K., Björkelid C., Jones T.A., Das K., Bandodkar B., Malolanarasimhan K., Mukherjee K., Ramachandran V. Assessment of Mycobacterium tuberculosis pantothenate kinase vulnerability through target knockdown and mechanistically diverse inhibitors. Antimicrob. Agents Chemother. 2014;58:3312–3326. doi: 10.1128/AAC.00140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C.O., Karim M.A., Zhang Y.M., Jackowski S. The murine pantothenate kinase (Pank1) gene encodes two differentially regulated pantothenate kinase isozymes. Gene. 2002;291:35–43. doi: 10.1016/s0378-1119(02)00564-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Cold Spring Harb. Lab. Press. Cold Spring Harb; NY: 2001. Molecular Cloning: a Laboratory Manual; p. 999. [Google Scholar]
- Song W.J., Jackowski S. Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli. [Erratum to document cited in CA119(7):64394q] J. Bacteriol. 1993;175:2792. doi: 10.1128/jb.174.20.6411-6417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan B., Baratashvili M., van der Zwaag M., Kanon B., Colombelli C., Lambrechts R.A., Schaap O., Nollen E.A., Podgoršek A., Kosec G., Petković H., Hayflick S., Tiranti V., Reijngoud D.-J., Grzeschik N.A., Sibon O.C.M. Extracellular 4′-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat. Chem. Biol. 2015;11:784–792. doi: 10.1038/nchembio.1906. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Philip N., Hughes K.R., Georgiou K., MacRae J.I., Barrett M.P., Creek D.J., McConville M.J., Waters A.P. Stage-specific changes in Plasmodium metabolism required for differentiation and adaptation to different host and vector environments. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanley S.L. Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- Strauss E. Comprehensive Natural Products II Chemistry and Biology. 2010. Coenzyme a biosynthesis and enzymology; pp. 351–410. [Google Scholar]
- Takagi M., Tamaki H., Miyamoto Y., Leonardi R., Hanada S., Jackowski S., Chohnan S. Pantothenate kinase from the thermoacidophilic archaeon Picrophilus torridus. J. Bacteriol. 2010;192:233–241. doi: 10.1128/JB.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallari D.S., Jackowski S., Rock C.O. Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 1987;262:2468–2471. [PubMed] [Google Scholar]
- Yang K., Eyobo Y., Brand L.A., Martynowski D., Tomchick D., Strauss E., Zhang H. Crystal structure of a type III pantothenate kinase: insight into the mechanism of an essential coenzyme A biosynthetic enzyme universally distributed in bacteria. J. Bacteriol. 2006;188:5532–5540. doi: 10.1128/JB.00469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Alramini H., Tran V., Singh U. Nucleus-localized antisense small RNAs with 5’-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J. Biol. Chem. 2011;286:44467–44479. doi: 10.1074/jbc.M111.278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.M., Rock C.O., Jackowski S. Feedback regulation of murine pantothenate kinase 3 by coenzyme A and coenzyme A thioesters. J. Biol. Chem. 2005:32594–32601. doi: 10.1074/jbc.M506275200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detail information of pantothenate kinase from various organisms related to E. histolytica that used in phylogenetic analysis.
Purification of recombinant E. histolytica pantothenate kinase. Enzyme activity was measured as described in Materials and methods.