Abstract
Over the past decade, long noncoding RNAs (lncRNAs) have been identified as significant players in gene regulation. They are often differentially expressed and widely-associated with a majority of cancer types. The aberrant expression of these transcripts has been linked to tumorigenesis, metastasis, cancer stage progression and patient survival. Despite their apparent link to cancer, it has been challenging to gain a mechanistic understanding of how they contribute to cancer, partially due the difficulty in discriminating functional RNAs from other noncoding transcription events. However, there are several well-studied lncRNAs where specific mechanisms have been more clearly defined, leading to new discoveries into how these RNAs function. One major observation that has come to light is the context-dependence of lncRNA mechanisms, where they often have unique function in specific cell types and environment. Here, we review the molecular mechanisms of lncRNAs with a focus on cancer pathways, illustrating a few informative examples. Together, this type of detailed insight will lead to a greater understanding of the potential for the application of lncRNAs as targets of cancer therapies and diagnostics.
Keywords: lncRNA, Cancer, Chromatin, HOTAIR, TUG1, MEG3
1. Introduction to lncRNAs
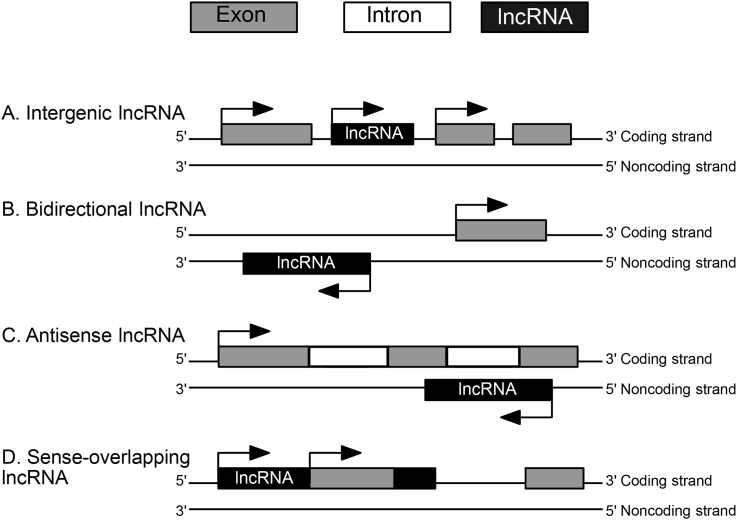
It is now well-recognized that over 75% of the human genome is transcribed, however, only about 2% of this transcription represents protein-coding potential [1]. Enhancements in high-throughput genomic technologies have led to the discovery of a myriad of previously unannotated non-protein coding transcription events in many previously-undescribed regions of the genome. One interesting class of noncoding RNAs to surface, are the numerous long noncoding RNAs (lncRNAs), stemming from tens of thousands (∼59,000) of RNA polymerase II transcription events [2]. These lncRNA transcripts are generally defined as greater then 200 nucleotides in length, typically ranging from 1000–10,000 nucleotides, with little to no protein-coding potential. LncRNAs resemble mRNAs as they are generally transcribed by RNA polymerase II, 5′ capped, 3′ polyadenylated, and often undergo splicing of multiple exons via canonical genomic splice motifs [[3], [4], [5]]. There are four main locations in which lncRNAs can originate that further aid in their classification. LncRNAs can be genomically located between two protein coding genes (intergenic lncRNA) (Fig. 1A), transcribed from a promoter of a protein-coding gene, yet in the opposite direction (bidirectional lncRNA) (Fig. 1B), originate from the antisense RNA strand of a protein coding gene (antisense lncRNA) (Fig. 1C), or overlap with one or more introns/exons of different protein-coding genes in the sense RNA strand (sense-overlapping lncRNAs) (Fig. 1D) [4,[6], [7], [8]]. Multiple transcripts originally classified as lncRNAs have been demonstrated to be transcribed within regions that have a clear function at the DNA level as enhancers, irrespective of whether the transcripts that are made within these regions have a function at the RNA level [[9], [10], [11]]. These observations make for a challenging task in dissecting whether these enhancer RNAs (eRNAs) could actually be enhancer-lncRNAs, meaning a functional RNA transcribed at an enhancer element. This review does not explicitly focus on enhancer-lncRNA mechanism, though it is possible that these RNAs share mechanisms with other lncRNA classes.
Fig. 1.
LncRNA classification based on genomic location. A) Intergenic lncRNAs are located between protein-coding genes. B) Bidirectional lncRNAs are transcribed from the same promoter as a protein-coding gene, but in the opposite direction. C) Antisense lncRNAs originate from the antisense RNA strand of a protein-coding gene. D) Sense-overlapping lncRNAs overlap with one or more introns and/or exons of a protein-coding gene in the sense RNA strand direction.
Despite minimal overall sequence conservation across species, many lncRNAs have evolutionarily conserved function, secondary structure, and regions of short sequence homology [[12], [13], [14], [15]]. Regulation of these transcripts is often controlled by well-studied transcription factors and epigenetic marks. LncRNA expression is often unique to specific cell types, tissues, developmental time frames, and disease [7,14,16,17]. Although only a small percentage of all lncRNAs identified have been studied in depth, they have emerged as key players in many diverse cellular contexts and biological processes. Thus far, lncRNAs have been implicated in X chromosome inactivation (Xi), genomic imprinting, nuclear compartmentalization, splicing, stem cell pluripotency, cell cycle progression, cellular reprogramming, apoptosis, and many diseases. They effect these processes through the regulation of gene expression, translational control, structural cellular integrity, protein localization and degradation [8,[17], [18], [19], [20], [21], [22], [23], [24]]. LncRNAs can associate with a wide range of interaction partners including RNA binding proteins (RBPs), transcription factors, chromatin-modifying complexes, nascent RNA transcripts, mature mRNA, microRNA, DNA, and chromatin.
2. Functional classifications of lncRNAs
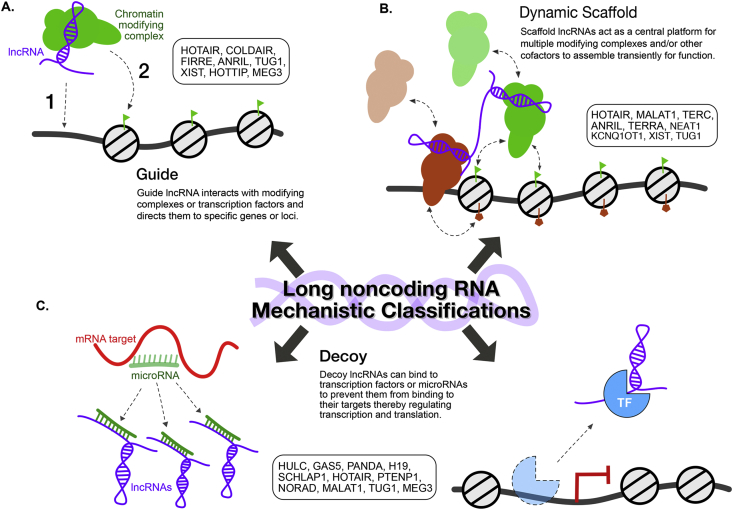
It has been challenging to clearly define and categorize lncRNA function due to the large numbers of identified transcripts, combined with the multitude of biological contexts in which they are involved. However, three general paradigms have emerged for broadly classifying lncRNA function: guides, dynamic scaffolds and molecular decoys [3,4,8,19]. Moreover, these classifications are not mutually exclusive, with the majority of well-studied lncRNAs acting under multiple modes, further illustrating the complexity at which these transcripts perform. An understanding of the reoccurring themes underlying these mechanisms may facilitate predictive models of lncRNA function and biological outcomes.
2.1. Guides
LncRNA guides are required for the proper localization/organization of factors at specific genomic loci for regulation of the genome. These transcripts bind to regulatory or enzymatically active proteins, such as transcription factors and chromatin modifiers, to direct them to precise locations in the genome at either cis (adjacent) or trans (distant) sites from their locus of transcription (Fig. 2A) [3,4,18]. One well-studied guide lncRNA is HOTAIR which functions in trans to direct the chromatin modifier Polycomb Repressive Complex 2 (PRC2) to the developmental HOXD locus and, when aberrantly overexpressed, to cancer-related genes, leading to gene repression [16,18,19,25]. The lncRNA MEG3 can recruit PRC2 to target genes via triple-helix formation with the underlying DNA [26]. An example of a lncRNA involved in chromosomal targeting via three-dimensional organization is Firre. Firre is transcribed from a genomic locus that escapes X chromosome inactivation (XCI). Firre can act in trans to form nuclear domains via interactions with hnRNP U to mediate co-localization of multiple chromosomal loci from chromosomes 2, 9, 15, and 17 [27]. Additionally, Firre acts in cis to help maintain XCI by positioning the inactive X chromosome near the nucleolus while also preserving H3K27me3 [28]. Thus, Firre is suggested to convey specificity in the organization of proper chromosomal domains within the nucleus through sequence specific interactions which may serve as a localization signal to initiate or maintain specific sub-compartments [27,29,30]. Specific targeting by guide lncRNAs are stimulated by RNA-DNA, RNA-RNA and RNA-Protein interactions.
Fig. 2.
General mechanisms for lncRNA classification. A) lncRNAs can act as guides to target chromatin-modifying complexes to specific genomic locations for the regulation of gene expression. B) lncRNAs can act as dynamic scaffolds for cofactors to transiently assemble together. C) lncRNAs can bind to microRNAs or transcription factors as decoys to sequester them away from their targets, affecting transcription and translation.
2.2. Dynamic scaffolds
LncRNAs acting as dynamic molecular scaffolds play a structural role by providing a central platform for the transient assembly of multiple enzymatic complexes and other regulatory co-factors. These often short-lived ribonucleoprotein (RNP) complexes can target specific genomic locations for regulation of gene expression. The combined catalytic activity of the various complexes within the RNP is often important for function (Fig. 2B) [3,20,31,32]. Telomerase RNA TERC is a classic example of an RNA scaffold that assembles the telomerase complex which maintains the ends of telomeres, combining reverse transcriptase activity with telomere targeting proteins in one RNP [33]. TERC served as a useful initial model to test whether newly-identified lncRNAs can form stable, homogeneous RNPs. However, little evidence exists for any recently-identified lncRNA to act as a stable molecular scaffold like TERC. Instead, lncRNAs may interact with proteins in more dynamic, low-affinity interactions, similar to mRNAs as they mature. This concept would explain some of the functional interactions that have been identified between lncRNAs and mRNA biogenesis factors such as heterogeneous nuclear ribonucleoproteins (hnRNPs). The dynamic scaffold model is also consistent with the diverse, sub-stoichiometric factors that have been identified through isolation of lncRNAs and proteomic analysis, such as for the Xist RNA [[34], [35], [36], [37]]. These dynamic interactions are also made with other non-canonical RNA-binding proteins such as chromatin modifying complexes. The lncRNAs TUG1, MALAT1 and ANRIL function as dynamic scaffolds linking chromatin modifying complexes PRC2 and PRC1 [[38], [39], [40]]. The imprinting-associated lncRNA Kcnq1ot1 scaffolds PRC2 and G9a to promote H3K27me3 and H3K9me3 for targeted genomic repression [41].
2.3. Decoys
The main function of decoy lncRNAs is to limit the availability of specific regulatory factors by acting as a molecular sink. This class of RNA modulates gene expression by sequestering RNA-binding proteins, transcription factors, microRNAs, catalytic proteins and subunits of larger modifying complexes (Fig. 2C) [3,20,21,42]. By titrating these factors away from interacting with their native target, decoys act by negatively regulating effector factors. Upon DNA damage, the lncRNA PANDA associates with the transcription factor NF-YA to prevent p53-mediated apoptosis. NF-YA activates several key genes for apoptosis and cell senescence, however PANDA binding to NF-YA titrates NF-YA away from target gene chromatin, thereby decreasing expression of apoptotic and senescence genes [[43], [44], [45]].
Additionally, several lncRNAs such as MEG3 and TUG1, have been shown to sequester various microRNA from protein and mRNA targets, resulting in altered protein translation and degradation [3,8,23,42,[46], [47], [48], [8]]. There is a much-debated hypothesis surrounding microRNA sponge function in lncRNAs, referred to as the competitive endogenous RNA (ceRNA) hypothesis. It proposes that specific transcripts can impair microRNA activity through sequestration, effectively de-repressing targets of that miRNA. This postulation has encountered skepticism, mainly due to the argument that physiological expression levels of an individual lncRNA would not be sufficient to suppress microRNA activity. However, subtle regulation by lowly expressed lncRNAs could be magnified through downstream processes, mainly through the upregulation of transcription factors that signal to multiple effector targets thus amplifying the outcome. Despite the controversy surrounding the ceRNA hypothesis, its widely acknowledged as a possible generic mechanism for regulating gene expression [[48], [49], [50]].
3. LncRNAs in cancer
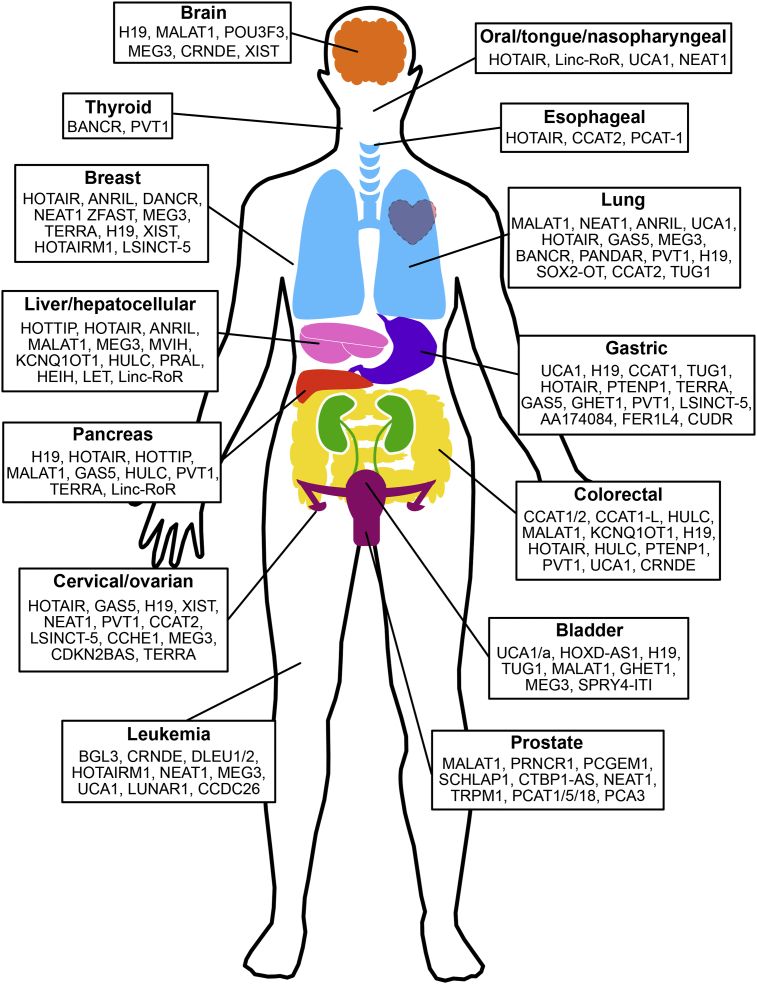
Cancer is largely caused by genetic alterations that result in aberrant gene expression. However, genome-wide association studies in cancer have revealed that more than 80% of cancer-associated SNPs occur in noncoding regions of the genome where, in some cases, cancer loci are transcribed into lncRNAs which play roles in tumorigenesis [46,[51], [52], [53], [54], [55]]. Numerous lncRNAs have been documented as aberrantly expressed in various cancers throughout the body, including the examples summarized in Fig. 3 [24,47]. As mentioned above, lncRNAs are a very heterogeneous group of transcripts that regulate gene expression by means of diverse mechanisms. Their expression has been correlated to distinct gene sets that influence cell cycle regulation, survival, mobility, immune response and pluripotency, among other functions, which contribute to the transformed phenotype of cancer cells. Moreover, they are found to be differentially expressed in tumors, where lncRNA dysregulation is associated with the cancer cell potential to initiate tumor growth, metastasis and decreased patient survival in a wide range of cancer types [46,47,[51], [52], [53], [54], [55]].
Fig. 3.
LncRNAs associated with various cancer types.
LncRNAs act through many different mechanisms to regulate cancer states. Studies so far suggest that many functional lncRNA transcripts contribute to epigenetic changes where lncRNAs have the potential to act as oncogenes and/or tumor suppressors. Oncogenic lncRNAs include ANRIL, BCAR4, CTBP-AS, H19, HEIH, HOTAIR, HOTTIP, HULC, KCNQ1OT1, LSINCT5, MALAT1, NEAT1, PCAT1/5/18, PCGEM1, PVT1, SCHLAP1, TUG1, UCA1, XIST. Tumor suppressor lncRNAs include BGL3, DILC, DLEU1/2, GAS5, MEG3, NBAT-1, PTENP1, TERRA, and others [24,47,54]. Moreover, lncRNAs have increasingly been shown to suppress microRNA levels by binding and sequestering them away from native targets, thereby upregulating miRNA target gene expression. Alterations in microRNAs are commonly observed in cancer, where activity is often found to be decreased [56], affecting various cellular processes, such as the cell cycle, proliferation, apoptosis and invasion [12,42,46,47]. Growing evidence suggests that lncRNAs play important roles in cancer development, however, what are the associated mechanisms that underlie lncRNA-mediated cancer phenotypes? To explore the potential of lncRNAs in cancer diagnosis and targeted therapy, it is important to understand each lncRNA in detail, by identifying the cellular functions, dissecting their molecular mechanisms and relating that back to their roles in disease. Below we explore some key cancer-related lncRNA mechanisms in detail.
4. HOTAIR mechanisms in cancer
One well-studied lncRNA regulator that is associated with multiple cancers is the Hox transcript antisense intergenic RNA (HOTAIR). This oncogenic lncRNA is involved in the progression of multiple human cancers including breast, gastric, pancreatic, liver, hepatocellular, colon, lung, colorectal and ovarian cancer. HOTAIR levels serve as predictive biomarkers in many cancers, where expression is highly correlated with patient prognosis. HOTAIR promotes different processes like tumor growth, metastasis, invasion and migration, epithelial to mesenchymal transition, and stemness via different pathways depending upon the cancer type. These cancer phenotypes predominantly occur through HOTAIR-mediated epigenetic changes, illustrating how lncRNA guide mechanisms can be hijacked in the context of cancer [46,[57], [58], [59], [60]].
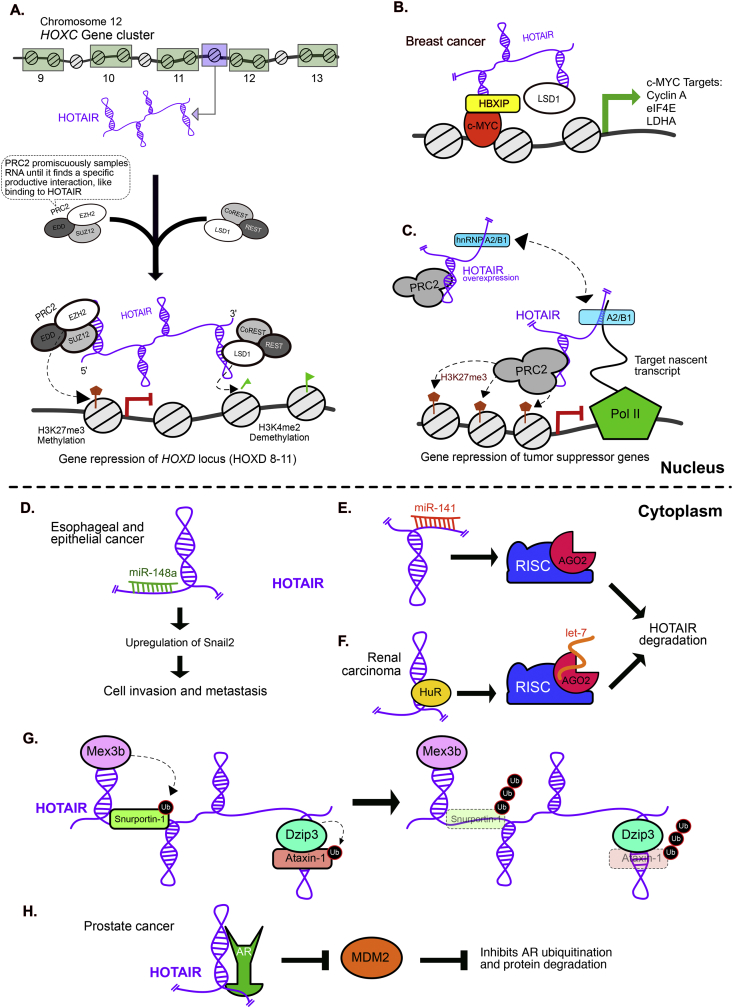
HOTAIR was first discovered while studying the transcriptional regulation of the HOX genes using tiling expression arrays in primary human fibroblasts. HOTAIR is an approximately 2200 nucleotide-long, spliced and polyadenylated transcript containing 6 exons. As its name indicates, it is transcribed from the antisense strand within an intergenic region of the HOXC gene cluster between HOXC11 and HOX12 on chromosome 12q13.13. HOX genes encode transcription factors that regulate segmental body patterning. Their precise temporal and spatial gene expression is under tight epigenetic control and in some cases regulated by neighboring HOX ncRNAs in cis on the same chromosome. In contrast to the cis-restricted mechanism, HOTAIR acts in trans to transcriptionally silence the HOXD cluster on chromosome 2, including HOXD8, HOXD9, HOXD10, and HOXD11 genes (Fig. 4A) [16,25,61,62].
Fig. 4.
Models of HOTAIR molecular mechanism in cancer. A) HOTAIR is transcribed from the HOXC locus on chromosome 12, scaffolds PRC2 and LSD1 chromatin-modifying complexes and targets them in trans to silence the HOXD locus on chromosome 2 through methylation of H3K27 and demethylation of H3K4 respectively. B) Model of c-Myc–mediated transcriptional activation in breast cancer, where the oncoprotein HBXIP acts as a co-activator by directly interacting with transcription factor c-Myc. HOTAIR scaffolds HBXIP and LSD1 to form a complex of c-Myc/HBXIP/HOTAIR/LSD1, which activates transcription of c-Myc target genes. C) Model for HOTAIR-PRC2 recruitment to specific genomic loci via RNA-RNA matchmaking, where hnRNP A2/B1 promotes HOTAIR binding to nascent transcripts of targets genes, resulting in PRC2-mediated transcriptional repression. D) In esophageal and epithelial cancers HOTAIR sequesters miR148-a, causing upregulation of Snail2 which results in increased cell invasion and metastasis. E) miR-141 can bind HOTAIR targeting it to AGO2, forming the Risc complex and subsequent HOTAIR degradation. F) In renal carcinoma, HOTAIR can bind to HuR, recruiting it to AGO2/let7, resulting in HOTAIR degradation. G) HOTAIR can bind E3 ubiquitin ligases Mex3b and Dzip3 and their target substrates Snurportin-1 and Ataxin-1 respectively, scaffolding them together and enhancing target substrate ubiquitination followed by protein degradation. H) In prostate cancer, HOTAIR can bind and stabilize the androgen receptor (AR), thereby blocking MDM2 association with AR and inhibiting ubiquitination and protein degradation of AR.
Mechanistic studies demonstrate that HOTAIR is a key regulator in gene silencing. The most well-studied HOTAIR mechanism involves interactions with two histone-modifying complexes, PRC2 and LSD1, responsible for changing methylation patterns on target loci. PRC2 is a multiprotein complex that catalyzes the silencing histone mark H3K27 tri-methylation through enzymatic activity of the EZH2 subunit along with other cofactors (SUZ12, EED, RBAP48/46) [16,18,19,25]. PRC2 binds HOTAIR on the 5′ end [19,32,63,64]. However, PRC2 also binds promiscuously to many other RNAs, indicating that PRC2 association alone is not responsible for specific targeting of HOTAIR function, suggesting the RNA itself may play a role in targeting [[65], [66], [67], [68], [69], [70]]. In contrast, histone H3K4 demethylase LSD1 binds to the 3′ end of HOTAIR. LSD1 also interacts with the repressor complex CoREST/REST to demethylate the active histone mark H3K4me2 (Fig. 4A) [19,25,71]. Both H3K27 methylation and H3K4 demethylation are important for gene silencing.
HOTAIR cancer phenotypes occur primarily due to HOTAIR overexpression [[57], [58], [59],[72], [73], [74]]. In epithelial cancer, HOTAIR overexpression can lead to genome-wide retargeting of PRC2 and H3K27 tri-methylation to hundreds of genes, including several metastasis suppressor genes including HOXD10, PGR, PCDH10, PCDHB5, and JAM2 [25,58,59,72,74,75]. In breast cancer cells, reprogramming of H3K27me3 profiles looked similar to embryonic fibroblasts resulting in an aggressive metastatic and invasive cancer outcome, where HOTAIR overexpression induced a 10-fold increase in metastasis to the lung from mouse tail xenografts [25]. Additionally, in breast cancer cells, HOTAIR can scaffold LSD1 to the oncogenic transcription factor c-Myc along with its co-activator HBXIP. This complex serves as a transcriptional activator of c-Myc target genes cyclin A, eIF4E, and LDHA, driving c-Myc-mediated oncogenesis (Fig. 4B) [71].
HOTAIR has been found to be highly structured, containing four independently folding modular domains, where in PRC2 binds domain 1 and LSD1 binds domain 4 [64]. Moreover, HOTAIR has been found to contain site-specific cytosine methylation within the vicinity of the LSD1 binding site. These types of post-transcriptional RNA modifications are known to modulate the structural organization and stability of tRNA and rRNA, suggesting these chemical modifications may also contribute to higher order folding and protein binding potential in lncRNAs [76]. This reinforces the idea that HOTAIR forms a modular platform by binding multiple proteins independently or cooperatively through it's different domains [64]. This platform may act as a dynamic scaffold bridging PRC2 and LSD1 complexes, while these factors sample other RNAs as well. A higher order dynamic structure may form transiently as it serves as a guide for these protein complexes to target locations for gene silencing [19]. But how is HOTAIR being specifically recruited to these loci? HOTAIR may be recruited to chromatin through direct RNA-RNA interactions with the nascent transcripts of target genes, mediated via the RBP hnRNP A2/B1. Many hnRNPs have been shown to aid in lncRNA function, though in many cases the mechanism of action is not clear [30,[77], [78], [79]]. HnRNP A2/B1 binds specifically to HOTAIR and also to HOTAIR target transcripts and was shown to stimulate RNA-RNA interactions between HOTAIR and one of its more abundant gene targets transcripts, JAM2. Moreover, knockdown of hnRNP A2/B1 in breast cancer cells overexpressing HOTAIR lead to a decrease of H3K27 tri-methylation at HOTAIR target genes [74]. More recently, genome-wide analysis in vivo has revealed extensive long-range RNA-RNA interactions across the transcriptome [[80], [81], [82]]. Recent evidence suggests that HOTAIR may also have the potential for RNA-DNA interactions via triplex formation [83]. Together these results support a model for how HOTAIR recruitment of chromatin modifiers to specific genomic locations may occur (Fig. 4C).
HOTAIR has also been suggested to function as a molecular decoy in tumors where it sequesters several miRNAs and RBPs. An example of this is in esophageal and epithelial cancer where HOTAIR acts as a ceRNA to negatively regulate miR-148a. Suppression of miR-148a promotes expression of Snail2, a key driver of EMT in cancer, to enhance cell invasion and metastasis (Fig. 4D) [84,85]. Furthermore, some miRNAs like miR-141 can regulate HOTAIR expression by targeting HOTAIR to the RISC complex for subsequent degradation by Ago2 induced cleavage (Fig. 4E) [85,86]. In renal carcinoma, HOTAIR can bind the RBP HuR (human antigen R), directing it to the let7 miRNA-Ago2 complex and leading to microRNA mediated suppression of HOTAIR through degradation (Fig. 4F) [73,87]. Therefore, miRNAs with sequence complementarity to HOTAIR or proteins that bind HOTAIR can repress each other's function adding another layer of complexity in regulation of HOTAIR activity and function.
Moreover, HOTAIR can function in the cytoplasm where it interacts with multiple E3 ubiquitin ligases suggesting a role in post translational protein degradation. Two such E3 ubiquitin ligases, Dzip3 and Mex3b, along with their respective targets Ataxin-1 and Snurportin-1 interact with HOTAIR. Dzip3 and its target Ataxin-1 were found to bind to HOTAIR between nucleotides 1028–1272, while Mex3b bound nucleotides 125–250. Mex3b's target, Snurportin-1 bound two regions within nucleotides 342–471 and 1142–1272 of HOTAIR. By scaffolding these complexes together, HOTAIR facilitates enhanced ubiquitination and target protein degradation of Snurportin-1 and Ataxin-1 (Fig. 4G) [87]. In contrast, HOTAIR in prostate cancer increases androgen receptor (AR) stability by binding its N-terminal domain to block interactions with the E3 ubiquitin ligase MDM2, thus preventing AR ubiquitination and protein degradation (Fig. 4H) [88].
5. TUG1 mechanisms in cancer
A recent player among the cancer-related lncRNAs is the taurine upregulated gene 1 (TUG1), which is expressed in a tissue-specific manner and exerts oncogenic and tumor suppressive functions in a number of human cancers contingent on the expression levels and gene targets involved [18]. Accumulating evidence has shown that TUG1 is a negative prognostic factor that is overexpressed in various solid tumors including osteosarcoma, bladder, esophagus, colorectal, gastric and liver cancer [89]. It has also been found to be downregulated in NSCLC, in addition to multiple myelomas, suggesting context-dependent roles in different cancer types [54,90,91]. TUG1 can promote cancer cell proliferation, migration and invasion, while also suppressing apoptosis. TUG1 mediates its biological functions in part by binding to PRC2 and PRC1 to repress gene expression in trans [38]. Moreover, TUG1 can also sequester microRNAs to influence cancer pathways [92]. Although the mechanisms of how TUG1 affects the tumorigenesis process remain to be fully elucidated, increasing studies suggest TUG1 offers potential as a diagnostic and prognostic biomarker, and as a therapeutic target in certain types of tumor.
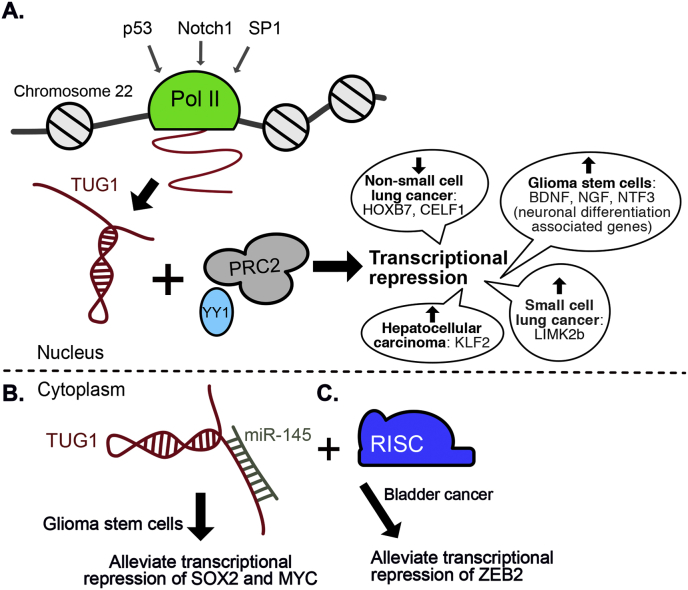
TUG1 is a 7.1 kb lncRNA that has multiple exons and is located on chromosome 22. TUG1 was first identified in a mouse screen for genes upregulated in developing retinal cells in response to taurine, and is required for the normal development of retina photoreceptors [93]. Previous studies have shown that TUG1 can be induced by Notch1 and SP1 in glioma stem cells (GSC) and hepatocellular carcinoma cells, respectively [89,90,94]. TUG1 functions as an epigenetic gene regulator by binding to PRC2 and directing H3K27me3 to specific targets (Fig. 5A). For instance, in glioma stem cells (GSC), TUG1 expression maintains GSC stemness through the repression of neuronal differentiation associated genes such as BDNF, NGF and NTF3 through the recruitment of PRC2 (via nucleotides 2316–2555) and transcription factor YY1 (via nucleotides 2746–2910) [95]. In hepatocellular carcinoma, upregulation of TUG1 causes PRC2 to be recruited to the promoter of tumor suppressor KLF2 (Kruppel like factor 2), thereby repressing its expression [96]. Additionally, TUG1 is overexpressed in small-cell lung cancer (SCLC) where expression was correlated with shorter patient survival time by regulating LIMK2b via PRC2. Downregulation of TUG1 expression in SCLC impaired cell proliferation and increased cell sensitivity to anticancer drugs, while TUG1 knockdown significantly promoted apoptosis and cell cycle arrest, in addition to inhibiting cell migration and invasion [97]. Conversely, downregulation of TUG1 in NSCLC led to an increase in oncogenes HOXB7 and CELF1, through the lack of PRC2 recruitment due to low TUG levels (Fig. 5A) [90,91].
Fig. 5.
LncRNA TUG1 models for molecular mechanism. A) TUG1 can be induced by p53, Notch1 and SP1. TUG1 can bind to PRC2 and YY1 to repress transcription. TUG1 is upregulated or downregulated in multiple cancer types where it affects target gene expression contributing to cancer phenotypes. B) In glioma stem cells, TUG1 suppresses miR-145 thereby alleviating the repression of SOX2 and MYC. C) In bladder cancer, TUG1 binding to miR-145 targets it to the Risc complex resulting in ZEB2 transcription.
Furthermore, TUG1 is known to act as a ceRNA by sponging microRNAs, in particular miR-145. In GSCs, TUG1 was found to prevent SOX2 and MYC mRNA degradation by sequestering miR-145 in the cytoplasm [89]. SOX2 and MYC are well-known stemness-associated transcription factors downstream of the Notch signaling pathway, as well as transcriptional activators of TUG1. This suggests TUG1/SOX2/MYC may form a reinforcing activation loop leading to mutual stabilization of their gene expression (Fig. 5B) [89,95]. In addition to targeting stemness-associated factors, miR-145 is also known to act as a tumor suppressor by decreasing proliferation, invasion, differentiation, angiogenesis in various cell types [98]. In bladder cancer, it was found that TUG1 and miR-145 were in the same RISC complex [99]. Interestingly, overexpression of miR-145 suppressed TUG1 levels whereas downregulation of miR-145 enhanced TUG1 levels, via a reporter assay. However, knockdown of TUG1 elevated miR-145 expression, signifying reciprocal suppression between TUG1 and miR-145. ZEB2, a key mediator of EMT in the development of cancer, was identified as a direct target of miR-145. ZEB2 can protect cancer cells from radiation-induced apoptosis, thus, suggesting a mechanism where TUG1 upregulation promotes ZEB2 expression by negatively regulating miR-145 expression, resulting in modulation of EMT, cell invasion and radio-resistance in bladder cancer (Fig. 5C] [99]. Recently TUG1 has also been shown to suppress other microRNAs in other cancers, including miR-138–5p in cervical cancer, miR-186 in colorectal cancer, miR-219 in oral squamous cell carcinoma and miR-132 in hepatocellular carcinoma [92,[100], [101], [102], [92]].
6. MEG3 mechanisms in cancer
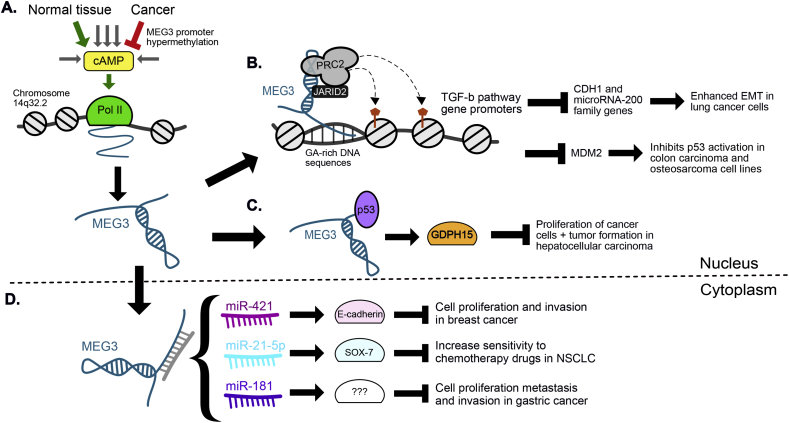
The maternally expressed gene 3 (MEG3) encodes an ∼1.6 kb long polyadenylated lncRNA, an imprinted gene belonging to the DLK1-MEG3 locus on chromosome 14q32.2 in humans [103]. There are currently sixteen different isoforms that have been reported via the alternative splicing of MEG3's 10 exons, with variant 1(NR_002766) as the predominant transcript [104]. MEG3 is highly expressed in normal human tissue where expression has been reported to be stimulated by cyclin AMP (cAMP). However, MEG3 expression is decreased or abolished in many cancers including, brain, lung, colon, liver, and leukemia [105]. Downregulation of MEG3 is thought to be caused, at least in part, by the hypermethylation of the MEG3 promoter region, thereby blocking the binding sites for cAMP-regulated transcription factors (Fig. 6A) [106,107]. MEG3 expression is under epigenetic control, and aberrant CpG methylation has been observed in several types of cancer [105,107]. MEG3 expression is involved in the activation of tumor suppressor p53 and modulation of TGF-β (transforming growth factor-b) pathway genes affecting cell invasion and immune regulation [26,44,105,108]. Overexpression of MEG3 has been shown to inhibit proliferation and promote apoptosis in various tumor types through interactions with different microRNAs [105,[109], [110], [111], [112]]. These studies indicate MEG3 can play a significant role as a tumor suppressor.
Fig. 6.
LncRNA MEG3 models of mechanism. A) MEG3 is expressed in normal tissue and significantly downregulated in many cancers, where hypermethylation of the MEG3 promoter inhibits normal activation by cAMP. B) MEG3 interacts with PRC2 and JARID2 to direct them to specific target promoters, via triplex formation with GA-rich DNA sequences, resulting in H3K27me3 and transcriptional repression. In lung cancer, MEG3-PRC2 represses TGF-β associated genes CDH1 and microRNA-200 family genes resulting in enhanced EMT. In colon and osteosarcoma carcinoma, MEG3 represses MDM2 which inhibits p53 activation. C) In hepatocellular cancer, MEG3 can bind to p53 to regulate p53 target gene expression, such as GDPH15. Upregulation of GDPH15 inhibits proliferation of cancer cells and tumor growth. D) Overexpression of MEG3 in multiple cancer types can suppress various microRNAs, thereby inhibiting the promotion of cancer phenotypes.
MEG3 interacts with PRC2 and its cofactor JARID2 to regulate genes in the TGF-β pathway, some of which were found to be direct targets of MEG3 via binding to promoter distal regulatory regions. These sites were found to be GA-rich sequences, able to guide MEG3 RNA to its target genes through RNA-DNA triplex formation, suggesting that this may be a common mechanism for target gene recognition by lncRNAs (Fig. 6B) [26]. In lung cancer cell lines, MEG3-dependent function of JARID2 and PRC2 was required for EMT-induced programs by regulating TGF-β EMT-related genes including E-cadherin, ZEB family and miR-200 family genes. Knockdown of MEG3 inhibited TGF-β-mediated changes in cell morphology and cell motility characteristic of EMT and counteracted TGF-β-dependent changes in the expression of EMT-related genes [113]. Mechanistic insight suggests MEG3 could associate with the regulatory regions of CDH1 and miR-200 family genes, possibly through RNA-DNA triplex formation [26], to induce the recruitment of PRC2 and subsequent H3K27me3 to these regions for transcriptional repression which is essential for TGF-β-induced EMT (Fig. 6B) [113].
Previous studies in colon and brain cancer first demonstrated MEG3's function as a tumor suppressor through activation of p53, leading to increased p53 protein levels and stimulation of p53-dependent transcription [108,114]. In hepatoma cells, MEG3 was shown to interact directly with p53 via its 3′ end (nucleotides 1067–1342) and the DBD (DNA binding domain) of p53. This interaction enhanced the stability of p53 and affected the regulation of a subset of p53 target genes including GDF15 (growth differentiation factor 15) [115]. GDF15 inhibits proliferation in several cancer cell types, as well as suppressing tumor formation (Fig. 6C). Additionally, MEG3 was shown to repress MDM2 levels in colon carcinoma and osteosarcoma cell lines, where MEG3 levels are significantly depleted [108]. MDM2, which is upregulated in a number of human cancers, contributes to tumorigenesis by inhibiting the activation of p53 (Fig. 6B) [107].
More recently, MEG3 has also emerged as having ceRNA function in many cancer types. MEG3 downregulation in breast cancer is associated with lymph node metastasis, invasion, and poor overall survival in patients and upregulation of MEG3 inhibits cell proliferation and invasion by negatively regulating E-cadherin expression via sequestration of miR-421 [116]. Overexpression of MEG3 in NSCLC resulted in an increased sensitivity to chemotherapy drugs through MEG3 suppression of miR-21–5p. miR-21–5p significantly disrupts the effects of MEG3 on drug resistance by modulating cell proliferation and apoptosis via SOX7, where MEG3 positively regulates SOX7 expression by suppressing miR-21–5p action [109]. Additionally, MEG3 is decreased in metastatic gastric cancer, where ectopic expression of MEG3 inhibited cell proliferation, migration, invasion and promoted cell apoptosis. MEG3 was found to affect gastric carcinoma phenotypes in a miR-181 site-dependent manner, occurring without changes in miR-181 levels, suggesting regulation by altering miR-181 targeting (Fig. 6D) [110]. Together these studies implicating MEG3 as a molecular decoy for cancer-associated microRNAs support the function of MEG3 as a tumor suppressor function and suggest the MEG3/microRNA regulatory axis may be a promising therapeutic target for multiple cancers.
7. Conclusion
LncRNAs are a diverse class of RNA molecules that have specialized function in a wide range of cellular processes. There are several molecular features that distinguish these transcripts from other genes, such as unique regulatory mechanisms, functional structured RNA domains and different forms of biogenesis. With the current advancements in high-throughput genomic technologies, the discovery of lncRNA species has increased dramatically, along with our appreciation of transcriptome complexity. However, the gap between identifying novel noncoding Pol II transcription and lncRNA confirmation and characterization has also increased, with only a fraction of well-annotated lncRNAs studied at the mechanistic level. Nevertheless, these well-described lncRNAs have provided significant insight into how these RNAs function. In cancer, aberrant expression, mutations and SNPs of lncRNAs are associated with cell transformation, tumorigenesis and metastasis. The majority of described lncRNAs are associated with different cancer types, where expression patterns are tumor specific and often predictive of patient prognosis. LncRNAs drive cancer phenotypes through the dysregulation of oncogenic and tumor suppressive gene networks via the variety of mechanisms discussed in this review. It is becoming more apparent that many cancer-related lncRNAs function through multiple recurring mechanisms to regulate specific targets in various tumor types. Thus, lncRNAs represent a relatively novel layer in cancer biology, providing a new paradigm for developing therapeutic strategies for cancer treatment.
Conflicts of interest
Declaration of interest: none.
Acknowledgements
We would like to thank Allison Porman for helpful comments on the manuscript. This work was supported by NIH grant R35GM119575 (A.M.J) and an RNA Scholar Award from the University of Colorado School of Medicine RNA Bioscience Initiative (M.M.B).
References
- 1.Djebali S. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer M.K. The landscape of long noncoding RNAs in the human transcriptome. Nature Publishing Group. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutenberg-Schoenberg M., Sexton A.N., Simon M.D. The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genom. Hum. Genet. 2016;17:69–94. doi: 10.1146/annurev-genom-090314-024939. [DOI] [PubMed] [Google Scholar]
- 6.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabili M.N. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa J.M. Revisiting lncRNAs: how do you know yours is not an eRNA? Mol. Cell. 2016;62:1–2. doi: 10.1016/j.molcel.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Paralkar V.R. Unlinking an lncRNA from its associated cis element. Mol. Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groff A.F. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 2016;16:2178–2186. doi: 10.1016/j.celrep.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsson P., Lipovich L., Grandér D., Morris K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn J.J. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016;30:191–207. doi: 10.1101/gad.272187.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hezroni H. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn J.L. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding RNAs. Nature Publishing Group. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 18.Khalil A.M. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai M.-C. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhan A., Mandal S.S. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–1956. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 23.Bhat S.A. Long non-coding RNAs: mechanism of action and functional utility. Non-coding RNA Research. 2016;1:43–50. doi: 10.1016/j.ncrna.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartonicek N., Maag J.L.V., Dinger M.E. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Canc. 2016:1–10. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta R.A. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondal T. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hacisuleyman E. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacisuleyman E., Shukla C.J., Weiner C.L., Rinn J.L. Function and evolution of local repeats in the Firre locus. Nat. Commun. 2016;7:11021. doi: 10.1038/ncomms11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa S., Hirano T. Gathering around Firre. Nat. Struct. Mol. Biol. 2014;21:207–208. doi: 10.1038/nsmb.2782. [DOI] [PubMed] [Google Scholar]
- 31.Schorderet P., Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002071. e1002071–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zappulla D.C., Cech T.R. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh C.A. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu C. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunwoo H., Wu J.Y., Lee J.T. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4216–E4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moindrot B., Brockdorff N. RNA binding proteins implicated in Xist-mediated chromosome silencing. Semin. Cell Dev. Biol. 2016;56:58–70. doi: 10.1016/j.semcdb.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 38.Yang L. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotake Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yap K.L. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandey R.R. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 42.Chen L.-L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016:1–12. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Hung T. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Publishing Group. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldassarre A., Masotti A. Long non-coding RNAs and p53 regulation. IJMS. 2012;13:16708–16717. doi: 10.3390/ijms131216708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng C. Over expression of long non-coding RNA PANDA promotes hepatocellular carcinoma by inhibiting senescence associated inflammatory factor IL8. Sci. Rep. 2017;7:4186. doi: 10.1038/s41598-017-04045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Canc. Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Canc. Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 49.Ebert M.S., Sharp P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 52.Rasool M. Non-coding RNAs in cancer diagnosis and therapy. Non-coding RNA Research. 2016;1:69–76. doi: 10.1016/j.ncrna.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khalil A.M. 2012. Emerging Roles for Long Non-coding RNAs in Cancer and Neurological Disorders; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang R., Xia L., Lu W., Zhang J., Zhu J.S. LncRNAs and cancer (review) Oncol Lett. 2016:1–7. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolha L., Ravnik-Glavač M., Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis. Markers. 2017;2017:1–14. doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 57.Biswas A., Desai K.V. The LncRNA HOTAIR-expression, regulation and function in cancer. Nucleus. 2017:1–10. [Google Scholar]
- 58.Loewen G., Zhuo Y., Zhuang Y., Jayawickramarajah J., Shan B. lincRNA HOTAIR as a novel promoter of cancer progression. J. Canc. Res. Updates. 2014;3:134–140. doi: 10.6000/1929-2279.2014.03.03.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hajjari M., Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J., Zhang P., Wang L., Piao H.-L., Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim. Biophys. Sin. 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L. Targeted disruption of hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L., Murat P., Matak-Vinkovic D., Murrell A., Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Somarowthu S. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015:1–10. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davidovich C., Zheng L., Goodrich K.J., Cech T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kretz M., Meister G. RNA binding of PRC2: promiscuous or well ordered? Mol. Cell. 2014;55:157–158. doi: 10.1016/j.molcel.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Kaneko S., Son J., Bonasio R., Shen S.S., Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davidovich C., Cech T.R. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skalska L., Beltran-Nebot M., Ule J., Jenner R.G. Regulatory feedback from nascent RNA to chromatin and transcription. Nature Publishing Group. 2017;18:331–337. doi: 10.1038/nrm.2017.12. [DOI] [PubMed] [Google Scholar]
- 70.Wang X. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 2017;65:1056–1067. doi: 10.1016/j.molcel.2017.02.003. e5. [DOI] [PubMed] [Google Scholar]
- 71.Li Y. HBXIP and LSD1 scaffolded by lncRNA hotair mediate transcriptional activation by c-Myc. Canc. Res. 2016;76:293–304. doi: 10.1158/0008-5472.CAN-14-3607. [DOI] [PubMed] [Google Scholar]
- 72.Croce C.M. LINCing chromatin remodeling to metastasis. Nat. Biotechnol. 2010;28:931–932. doi: 10.1038/nbt0910-931. [DOI] [PubMed] [Google Scholar]
- 73.Loewen G., Jayawickramarajah J., Zhuo Y., Shan B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014;7:101–110. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meredith E.K., Balas M.M., Sindy K., Haislop K., Johnson A.M. An RNA matchmaker protein regulates the activity of the long noncoding RNA HOTAIR. RNA. 2016;22:995–1010. doi: 10.1261/rna.055830.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kogo R. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Canc. Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 76.Amort T. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013;10:1003–1008. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hasegawa Y. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 78.Carpenter S. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.P. K. Puvvula et al., Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun.. 5, 1–16 (1AD). [DOI] [PMC free article] [PubMed]
- 80.Engreitz J.M. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Z. RNA duplex map in living cells reveals higher- order transcriptome structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aw J.G.A. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 83.Kalwa M. The lncRNA HOTAIR impacts on mesenchymal stem cells viatriple helix formation. Nucleic Acids Res. 2016;44:10631–10643. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Z.-Y. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu F., Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed. Pharmacother. 2017;90:888–896. doi: 10.1016/j.biopha.2017.03.103. [DOI] [PubMed] [Google Scholar]
- 86.Pádua Alves C. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cell. 2013;31:2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 87.Yoon J.-H. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang A. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Z., Shen J., Chan M.T.V., Wu W.K.K. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–475. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang E.-B. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. 2014;5 doi: 10.1038/cddis.2014.201. e1243–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin P.-C. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Canc. 2016;16:583. doi: 10.1186/s12885-016-2569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J. The long noncoding RNA TUG1 acts as a competing endogenous RNA to regulate the Hedgehog pathway by targeting miR-132 in hepatocellular carcinoma. Oncotarget. 2017;8:65932–65945. doi: 10.18632/oncotarget.19582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Young T.L., Matsuda T., Cepko C.L. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 94.Huo X. 2017. Dysregulated Long Noncoding RNAs (LncRNAs) in Hepatocellular Carcinoma: Implications for Tumorigenesis, Disease Progression, and Liver Cancer Stem Cells; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Katsushima K. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang M.-D. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol. Canc. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Niu Y. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol. Canc. 2017:1–13. doi: 10.1186/s12943-016-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui S.-Y., Wang R., Chen L.-B. MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways. J. Cell Mol. Med. 2014;18:1913–1926. doi: 10.1111/jcmm.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tan J., Qiu K., Li M., Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589:3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 100.Zhu J., Shi H., Liu H., Wang X., Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8:65253–65264. doi: 10.18632/oncotarget.18224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li C., Gao Y., Li Y., Ding D. TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis. Biochem. Biophys. Res. Commun. 2017;491:552–557. doi: 10.1016/j.bbrc.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 102.Yan G., Wang X., Yang M., Lu L., Zhou Q. Long non-coding RNA TUG1 promotes progression of oral squamous cell carcinoma through upregulating FMNL2 by sponging miR-219. Am J Cancer Res. 2017;7:1899–1912. [PMC free article] [PubMed] [Google Scholar]
- 103.Miyoshi N. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Gene Cell. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 104.Zhang X. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao J., Zhang X., Zhou Y., Ansell P.J., Klibanski A. Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int. J. Biochem. Cell Biol. 2006;38:1808–1820. doi: 10.1016/j.biocel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 107.Benetatos L., Vartholomatos G., Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Canc. 2011;129:773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 108.Zhou Y. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 109.Wang P., Chen D., Ma H., Li Y. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. OncoTargets Ther. 2017;10:5137–5149. doi: 10.2147/OTT.S146423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng W. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J. Exp. Clin. Canc. Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Z. Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am J Transl Res. 2017;9:3326–3335. [PMC free article] [PubMed] [Google Scholar]
- 112.Su L., Han D., Wu J., Huo X. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour Biol. 2016;37:3925–3931. doi: 10.1007/s13277-015-4151-2. [DOI] [PubMed] [Google Scholar]
- 113.Terashima M., Tange S., Ishimura A., Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem. 2017;292:82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang P., Ren Z., Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 115.Zhu J. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139790. e0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang W. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 2017;91:312–319. doi: 10.1016/j.biopha.2017.04.085. [DOI] [PubMed] [Google Scholar]