Abstract
In addition to energetically optimal structures, RNAs can fold into near energy suboptimal conformations that may be populated and play functional roles. The diversity of this structural ensemble can be estimated using a metric derived from the calculated RNA partition function: the ensemble diversity. In this report, 10 classes of functional RNAs were analyzed: the 5.8S and 5S rRNAs, ribozyme, RNase P, snoRNA, snRNA, SRP RNA, tmRNA, Vault RNA and Y RNA. Representative sequences from each class were mutagenized in two ways: firstly, all possible point mutations were generated and secondly, wild type sequences were randomized to generate multiple scrambled mutants. Compared to the mutants, the native RNA ensemble diversity was predicted to be lower. This finding held true when all available sequences (378,455 sequences) for each RNA class (archived in the RNAcentral database) were analyzed. This suggests that a compact structural ensemble is an evolved characteristic of functional RNAs.
Keywords: rRNA, Ribozyme, RNase P, snoRNA, snRNA, SRP RNA, tmRNA, Vault RNA, Y RNA
1. Introduction
A defining characteristic of functional RNA is the propensity of forming well-defined secondary and tertiary structure. RNA structure is essential for mediating interactions necessary for their function: e.g. in recognizing protein binding partners or other nucleic acids, performing catalysis, protecting RNA from degradation, etc. Previous work established that the predicted optimal folding energy of functional RNA is lower (more stable) than that of random sequence folding energy [1]; the order of a functional RNA sequence that has evolved over time, imbues it with unusual thermodynamic stability. When randomized, the native base pairing contacts in the evolved sequence are abolished, leading to less favorable, higher predicted energies. Thus, low (favorable) folding energy, in an evolved characteristic of functional RNAs. This property can be quantified by calculating a thermodynamic (ΔG) z-score, which compares the native predicted folding energy to random and normalizing by the standard deviation. Negative z-scores indicate the number of standard deviations more stable than random is a native RNA sequence [1]. The ΔG z-score is at the heart of some of the most effective noncoding (nc)RNA prediction algorithms [[2], [3], [4]], and has been successfully used in the analysis of human [5,6], viral [[7], [8], [9]] and other genomes [10,11].
In addition to the energetically optimal conformation, RNAs may fold into near-energy suboptimal conformations that may be populated and play functional roles [12,13]. Information about these suboptimal conformations can be derived from the calculated RNA secondary structure partition function [14]. Here, the thermodynamic states of the ensemble are the different RNA conformations. The complexity of the structural ensemble can be estimated by calculating the ensemble diversity (ED) metric. Here, the distance, measured as the number of base pairs different between Boltzmann weighted conformations, is averaged across the ensemble. Low ED indicates a single dominant conformation, while higher EDs suggest multiple diverse conformations or a lack of defined structure [15,16].
A program/webserver, RNA2DMut, was recently developed to analyze the effects of single nucleotide variations (SNVs) and other types of RNA mutations on the structure, folding energy and ED [17]. In the testing of this program, interesting features of several functional structured RNAs were uncovered: disruptive or stabilizing mutational “hot spots” (regions where SNVs could increase or decrease ED, respectively) could be found in functional RNAs, in some RNAs a single base substitution could abolish the native secondary structure, SNVs can stabilize biologically significant suboptimal folds and, importantly, compared to all possible SNVs the wild type (WT) sequence had lower ED. These finding provided the impetus for this current study, where the mutational analysis was extended to cover ten classes of structured non-coding (nc)RNAs.
2. Results
2.1. Compared to all possible SNVs, native ncRNA sequences have greater stability and lower ensemble diversity
All available sequences for ten major classes of ncRNA were acquired from RNAcentral: a comprehensive collection of ncRNA sequence data [18,19]. The classes of sequences analyzed are: the 5.8S ribosomal (r)RNA, 5S rRNA, hammerhead ribozyme, ribonuclease (RNase) P, small nucleolar (sno)RNA, small nuclear (sn)RNA, signal recognition particle (SRP) RNA, transfer-messenger (tm)RNA, Vault RNA and Y RNA. Representative sequences from each class were used in the structural analysis of all possible SNVs: 5.8S rRNA (Saccharomyces cerevisiae), 5S rRNA (Schizosaccharomyces pombe), hammerhead ribozyme (Schistosoma mansoni), RNase P (Bacillus subtilis), U3 snoRNA (Homo sapiens), U5 snRNA (H. sapiens), SRP RNA (Escherichia coli), tmRNA (Enterococcus durans), Vault RNA (Mus musculus), Y RNA (H. sapiens). The WT sequences were analyzed using the program RNA2DMut; experimental details are in the Methods section and sequences of WT and SNV mutants are in the RNA2DMut output (SI File 1), which also includes all predicted structures, folding energies and ED values.
A summary of results for each representative ncRNA appears in Table 1. Here, all WT sequences have lower (more thermodynamically stable) predicted folding energy (ΔG, in kcal/mol) than the average value of all possible SNVs. Likewise, in all cases the majority (>70%) of SNV mutant sequences had higher (less stable) energy (ΔGSNV) than the WT; this ranged from 70.15% (Y RNA) to 97.93% (ribozyme). Similarly, the ED of WT sequences is always lower (a more similar structural ensemble that may be more centered on a single dominant conformation) than the EDSNV average; this ranged from 84.83% for the ribozyme to 61.03% for the Vault RNA. To have a length-normalized metric to compare the distance of the WT ΔG and ED from the values calculated for all possible SNVs, the z-scoreSNV of each value was calculated: the difference of the WT value and average of SNVs is normalized by the standard deviation (the values given are the number of standard deviations more stable WT is vs. mutants).
Table 1.
Comparison of wild type folding metrics to SNV and randomized mutants.
| 5.8S | 5S | ribozyme | RNase P | snoRNA | snRNA | SRP | tmRNA | Vault | Y RNA | |
|---|---|---|---|---|---|---|---|---|---|---|
| WT ΔG | −51.10 | −38.40 | −27.10 | −136.20 | −23.90 | −33.20 | −57.00 | −84.70 | −33.30 | −31.30 |
| ΔGSNV average | −50.16 | −37.80 | −24.22 | −135.22 | −22.43 | −31.95 | −55.37 | −84.18 | −32.57 | −30.27 |
| ΔGrandom average | −38.35 | −35.70 | −10.21 | −114.34 | −12.72 | −22.90 | −41.78 | −70.76 | −26.50 | −14.96 |
| ΔGSNV > WT ΔG (%) | 74.51 | 70.27 | 97.93 | 76.08 | 84.65 | 78.51 | 79.44 | 70.15 | 72.21 | 79.69 |
| ΔGrandom > WT ΔG (%) | 100.00 | 82.93 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 95.12 | 100.00 |
| ΔG z-scoreSNV | −0.42 | −0.32 | −1.27 | −0.47 | −0.66 | −0.56 | −0.64 | −0.28 | −0.37 | −0.48 |
| ΔG z-scorerandom | −3.33 | −0.75 | −5.67 | −4.07 | −3.34 | −3.02 | −4.03 | −2.92 | −1.70 | −5.12 |
| WT ED | 16.94 | 9.63 | 0.65 | 40.88 | 1.78 | 10.99 | 5.23 | 47.81 | 15.65 | 8.36 |
| EDSNV average | 21.25 | 18.68 | 1.25 | 45.32 | 2.91 | 11.80 | 7.34 | 50.09 | 19.11 | 9.37 |
| EDrandom average | 33.38 | 27.38 | 6.77 | 99.88 | 15.55 | 25.31 | 29.59 | 80.24 | 24.75 | 19.94 |
| EDSNV > WT ED (%) | 72.57 | 71.89 | 84.83 | 73.34 | 74.27 | 72.21 | 78.87 | 62.11 | 61.03 | 71.38 |
| EDrandom > WT ED (%) | 92.68 | 97.56 | 95.12 | 97.56 | 97.56 | 92.68 | 97.56 | 97.56 | 78.05 | 92.68 |
| ED z-scoreSNV | −0.56 | −0.89 | −0.58 | −0.59 | −0.65 | −0.34 | −0.56 | −0.30 | −0.41 | −0.48 |
| ED z-scorerandom | −1.61 | −1.71 | −1.57 | −2.16 | −2.14 | −1.62 | −2.17 | −1.74 | −0.97 | −1.61 |
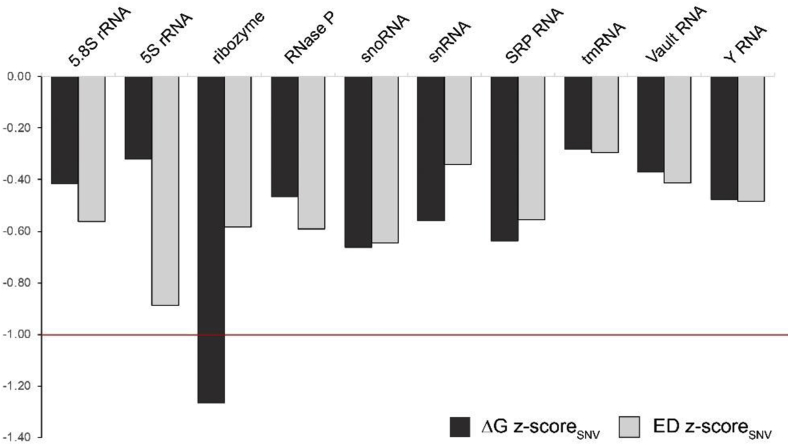
The ΔG and ED z-scoreSNV values are compared in Fig. 1. In all cases the z-scoreSNV is lower than zero, however, only in a single case, the S. mansoni hammerhead ribozyme, did the z-score (ΔG z-scoreSNV) go below −1. In most cases the ΔG and ED z-scoreSNV values were similar to each other (z-scores within ∼0.2 of each other). The exceptions were the S. mansoni ribozyme and S. pombe 5S rRNA: the ribozyme ΔG z-scoreSNV is over 2× lower than its ED z-scoreSNV (Table 1 and Fig. 1), whereas, the 5S rRNA ED z-scoreSNV is almost 3× lower than its ΔG z-scoreSNV.
Fig. 1.
Representative ncRNA SNV mutant z-scores. The z-scoreSNV values are calculated by taking the difference between the native folding free energy change (ΔG; dark bars) or ensemble diversity (ED; light bars) and the average value of all possible SNV mutants for each RNA, then normalizing by the standard deviation. A red line indicates z-scores one standard deviation lower than the mutant average.
2.2. The patterns of SNV-sensitive sites are distinct for ncRNAs
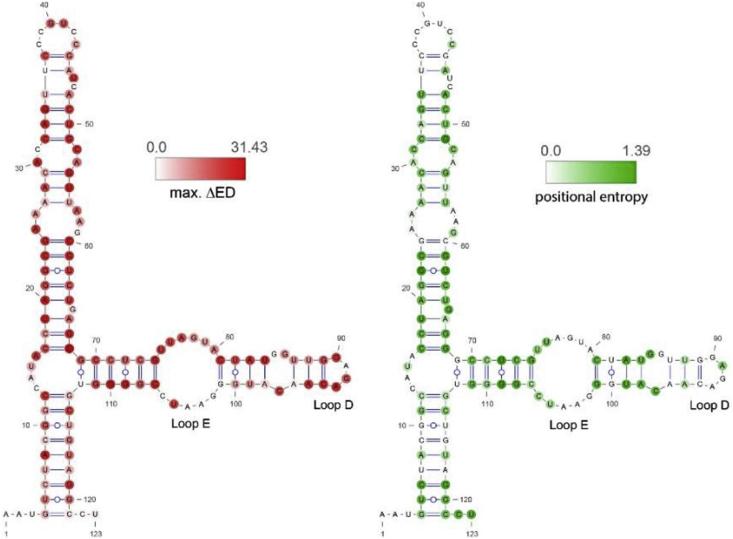
The ED maximizing and minimizing mutants for each position are generated as part of the RNA2DMut output. Results for each representative ncRNA appear in SI File 2. There are distinct patterns of sites that are sensitive to ED-changing mutations. Minimizing and maximizing SNVs tend to cluster together and range from “hot spots” that are extensive (e.g. for the Vault RNA) or highly localized (e.g. for the snRNA). There is a rough tendency of minimizing mutations to occur in loops and maximizing mutations to occur in helices. Both tendencies are best illustrated using the 5S rRNA as an example (SI File 2 and Fig. 2).
Fig. 2.
Schizosaccharomyces pombe 5S rRNA ensemble centroid models. (Left) shows the maximal change in calculated ensemble defect (mutant vs. WT) at each position, represented as a red heat map. (Right) shows the positional entropy calculated from an alignment of unique Ascomycota 5S rRNA sequences (N = 300).
As mentioned above, the 5S rRNA had the lowest ED z-scoreSNV of any evaluated ncRNA and most (71.89%) of mutants increased the ED. The remaining ED-minimizing mutations occurred primarily in loops, or at the ends of helices (SI File 2), where they added additional stabilizing base pairs to conformations predicted to be near-native conformations (SI File 1). ED-minimizing SNVs could also occur within helices, where they primarily converted GU wobble pairs into more thermodynamically stable Watson-Crick AU or GC pairs. In contrast, ED-maximizing mutations were widespread, occurring both in loops and helices, as well as of a much higher magnitude (Fig. 2; SI File 2). Other than the mutations that replaced wobble pairs with Watson-Crick ones, helical mutants almost always increased the ED by weakening pairs in the native structure (SI File 1). Interestingly, 5S rRNA loops also had many positions where SNVs could increase the ED (Fig. 2); in these cases, mutations stabilize base pairs in divergent alternative conformations (SI File 1).
2.3. Variability in natural 5S rRNA sequences
All Ascomycota 5S rRNA sequences archived in the 5S rRNA database [20] were aligned, and the nt positional entropy was mapped onto the predicted S. pombe ensemble centroid structure (Fig. 2, right). The positions with highest entropy (greatest variability across Ascomycota species) occurred primarily in the helical regions. Here the increased positional entropy occurs because of natural compensatory mutations (correlated double point mutations) that maintain base pairing (SI File 3). Loop regions of the 5S rRNA, in general, had low positional entropy. There are, however, exceptions where entropy in loops was high. For example, in loop D there was only a single residue (nt 91; Fig. 2, right) that had high positional entropy. Interestingly, this was the only residue in loop D that did not have SNVs with high ED (Fig. 2, left). Another noteworthy region is loop E, where (but for opposing nt 75/106 and 76/105) positional entropy was very low (Fig. 2). ED-Maximizing SNVs appeared along the 5′ side of this loop; however, the only residue on the 3′ side that has a high ED SNV is nt 105. It is interesting to see how the predicted ED maximizing mutations in this loop, and elsewhere in the 5S rRNA, that also overlap sites with high positional entropy behave.
Highly ED-maximizing mutants (with ED > 1σ the mutant average), that also occur in regions with high positional entropy are shown in SI File 4. In each case mutants increased the ED by stabilizing alternative long hairpin conformations. Ascomycota 5S rRNAs were analyzed to identify examples of sequences where the ED-maximizing (in S. pombe) variant nt occurs naturally (SI File 4). In all cases multiple mutations accumulate that disfavor this alternative conformation; this included two instances (P. fijiensis and C. sphaerospermum) where a natural base substitution forbids pairing to the variant site.
2.4. The stability and ensemble diversity of native ncRNAs are lower than random sequences
To see how more dramatic sequence disruptions (vs. SNVs) could affect the ΔG and ED, WT sequences were shuffled multiple times. The randomizations had a much higher disruptive effect on each metric and ncRNA than SNVs (Table 1 and SI File 5). Likewise, the percentage of mutants that had disruptive effects was higher using randomization for every ncRNA. The percentage of SNV mutants with less stable ΔG ranged from 70.15% (tmRNA) to 97.93% (ribozyme); while in almost all (7/10) ncRNAs, 100% of randomized sequences had less stable ΔG (the lowest percentage was for the 5S rRNA, where 82.93% of randomized sequences were less stable). Similar trends were observed comparing EDSNV vs. EDrandom values. In all cases, mutants were predicted to be (on average) disruptive—with random mutants being, in all instances, of greater magnitude than SNVs (Table 1). Similarly, in all cases the percentage of random mutants with higher ED than WT was greater than that of the SNV mutant populations.
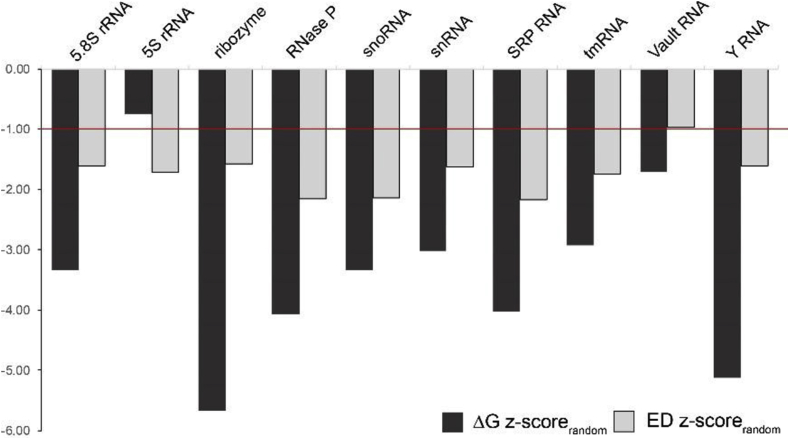
Z-scores were calculated comparing the WT metrics to randomized sequence averages. The ΔG and ED z-scorerandom values were universally lower than the z-scoreSNV values for each ncRNA (Table 1 and SI File 5). Almost every ncRNA had both z-score metrics that were lower than −1 (Fig. 3); the only exceptions were the 5S rRNA and Vault RNA, which had ΔG and ED z-scorerandom values, respectively, that were above −1. In all but one case (5S rRNA) the ΔG z-scores were lower than the ED z-scores, indicating that randomization has a greater disruptive effect on the folding energy than the ED. To determine if this is broadly true of ncRNAs, a larger dataset was analyzed.
Fig. 3.
Representative ncRNA randomized mutant z-scores. The z-scores are calculated by taking the difference between the native folding free energy change (ΔG; dark bars) or ensemble diversity (ED; light bars) and the average value of 40 nucleotide-randomized mutants for each RNA, then normalizing by the standard deviation. A red bar indicates z-scores one standard deviation lower than the mutant average.
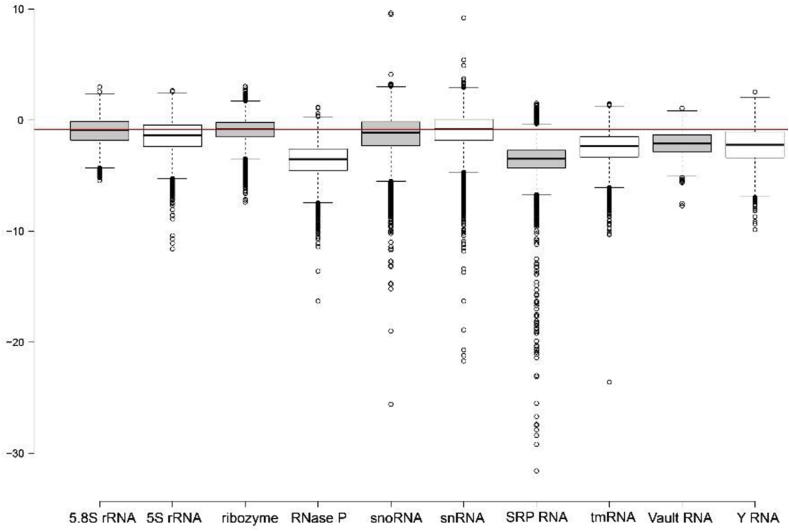
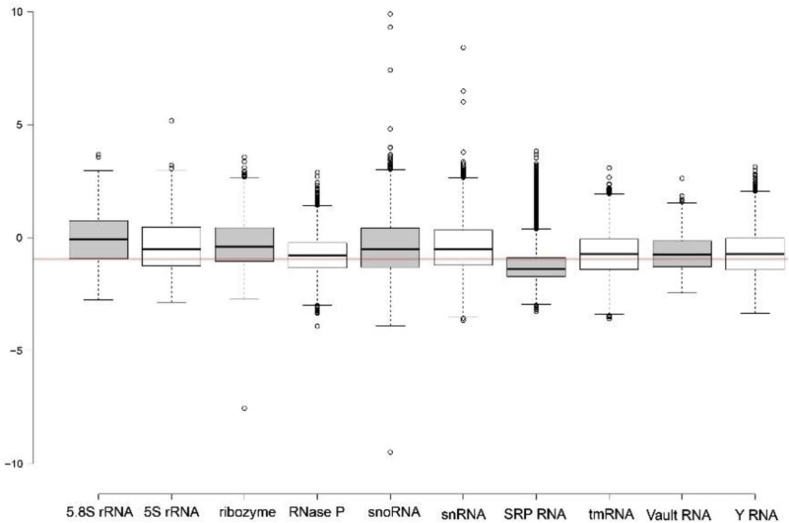
All sequences from each RNAcentral ncRNA class were randomized and evaluated to predict the ΔG and ED z-scorerandom values (complete results in SI File 6). The ΔG z-scorerandom distributions for each class are shown in the box plots on Fig. 4 (metrics in SI File 7). Similar to the results for representative ncRNA sequences, the distributions for all sequences in each class were shifted into the negative; in three cases (5.8S rRNA, ribozyme, and snRNA), however, the means were above −1. These results are consistent with previous analyses of ncRNAs, which were found to have ΔG z-scorerandom values shifted in the negative [1,2]. The means ranged from −0.79 (snRNA) to −3.51 (RNase P), with most (7/10) below −1 (SI File 7). An interesting feature of the ΔG z-scorerandom numbers, were the numerous outliers with very low z-scores. For example, there were SRP RNA sequences with z-scores over 30 standard deviations greater thermodynamic stability than random (Fig. 4)! These very structured RNAs had WT ΔGs that were much more stable than random due to the presence of very long hairpin with helixes with perfect, or near perfect, complementarity (SI File 6).
Fig. 4.
Distributions of ΔG z-scorerandom values for 10 classes of ncRNA. The z-score is calculated from the difference in the WT sequence vs. 40 randomized mutants. The red line indicates a value of −1. The shading of box plots is cosmetic. Distributions for pre-randomized control sequences are in SI File 8.
The distributions of ED values for each sequence is shown in Fig. 5. Congruent with results for representative ncRNAs (Fig. 3), all ncRNAs had ED z-scorerandom values shifted into the negative. Also consistent with results on representative ncRNAs, was the observation that the ED z-scores for each class had lower magnitude than ΔG z-scores: the mean values of each ED z-scorerandom distribution was higher than its corresponding ΔG z-scorerandom value (Fig. 4, Fig. 5; SI File 7). Only the SRP RNA had a mean ED z-scorerandom value less than −1 (−1.72). All other cases had higher mean values: e.g., the mean of the 5.8S rRNA class was only −0.07). Interestingly, in contrast to the ΔG z-scorerandom values, outliers were primarily positive; suggesting that sub-populations of sequences for each could be “tuned” (by evolution) to have more dynamic structural ensembles.
Fig. 5.
Distributions of ED z-scorerandom values for 10 classes of ncRNA. The z-score is calculated from the difference in the WT sequence vs. 40 randomized mutants. The red line indicates a value of −1. The shading of box plots is cosmetic. Distributions for pre-randomized control sequences are in SI File 9.
As a control, each WT RNA sequence was pre-randomized before calculation the ED and ΔG z-scorerandom values (compared to 40× additional randomizations of the pre-randomized input sequence). There is no bias in either metric for any class of ncRNAs (SI Files 8 and 9), indicating that the WT sequence order gives rise to the low ED and ΔG values. Most ncRNA classes had statistically significant differences between randomized and WT sequences for both the ΔG and ED metrics (Table 2). The 5.8S rRNA, tmRNA, and Vault RNA had p-values above the threshold of significance (0.05); The Y RNA class had a significant difference in the ED z-scorerandom distributions, but not the ΔG metric values.
Table 2.
P-values comparing the ΔG and ED z-scorerandom values of WT ncRNAs vs. pre-randomized controls.
| p-value ΔG | p-value ED | |
|---|---|---|
| 5.8S rRNA | 0.94 | 0.32 |
| 5S rRNA | 1.34E-03 | 6.76E-05 |
| ribozyme | 7.20E-14 | 1.56E-08 |
| RNase P | 1.78E-02 | 5.91E-03 |
| snoRNA | 1.59E-11 | 3.91E-11 |
| snRNA | 2.22E-11 | 2.11E-15 |
| SRP RNA | 1.87E-23 | 2.29E-12 |
| tmRNA | 0.67 | 0.44 |
| Vault RNA | 0.34 | 0.26 |
| Y RNA | 0.11 | 9.99E-03 |
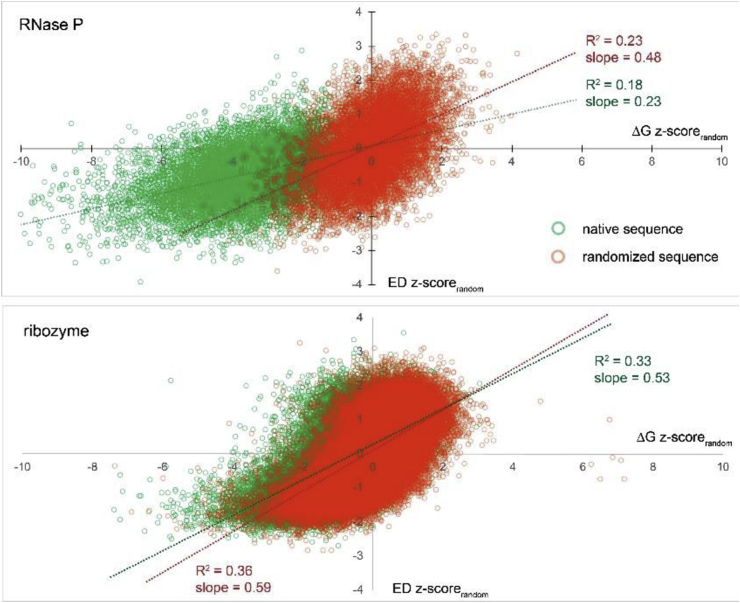
The WT and pre-randomized ED z-scorerandom values for each RNA sequence were plotted against corresponding ΔG z-scorerandom values; all results appear in SI File 10. The shape of the WT and pre-randomized distributions suggest that both z-scorerandom values have some degree of linearity (average R2 value of 0.32; SI File 10). Interestingly, the lowest correlations in the WT data were in ncRNA classes with the greatest negative shifts in ΔG and ED z-scorerandom values: e.g., RNase P (R2 = 0.18) and SRP RNA (R2 = 0.24). As observed in the box plots (Fig. 4, Fig. 5), the greatest shift in the data is toward negative ΔG z-scorerandom values; however, the ED z-scorerandom values can move the WT data away from the pre-randomized results. For example, the RNase P and the ribozyme ncRNA classes represent two extreme cases—with good and bad separation of the data, respectively (Fig. 6). Another interesting feature of these distributions is that the slope of the trendlines for the pre-randomized data is almost always higher than the WT data (SI File 10); this is most apparent in the well-separated data (e.g., RNase P; Fig. 6).
Fig. 6.
Scatter plots showing the ΔG z-scorerandom vs. ED z-scorerandom values. (Top) and (Bottom) panels show data for the RNase P and ribozyme classes, respectively. Data for WT sequences appear as green circles and red circles show data from pre-randomized control sequences. Outliers occur outside of the plot area, but are omitted for space (all data are in SI File 10).
3. Discussion
The ensemble folding properties of RNA may be important in understanding ncRNA sequence evolution. This is particularly the case in loop regions. The effects of deleterious, ED increasing, mutations in helixes are compensated in a straightforward way: a compensatory change in its pairing partner to reform the base pair. Loop mutations can also disrupt ED (e.g., stabilizing alternative conformations); sequences can respond to these in complex ways (e.g., the examples in SI File 4). A better appreciation of how the RNA structure ensemble can affect sequence evolution, particularly in loops, may offer insights into the conservation patterns of ncRNA and facilitate RNA-based phylogenetic methods. This could also be helpful in ncRNA identification/discovery, for example, where the effects of loop mutations on the ED, could complement current methods that build covariance models for helical regions of RNA [21,22].
The results of the ED and ΔG z-scoreSNV calculations on representative ncRNA sequences (Fig. 1) indicate that the folding stability and ensemble diversity of WT sequences occupy a more thermodynamically stable region of “SNV space” with correspondingly lower amounts of conformational diversity. The majority of SNVs reduced stability and increased the structural diversity, suggesting that (in addition to stability) conformational equilibria (encoded within the sequence) is part of the RNA evolutionary landscape. In the 5S rRNA example, “deleterious” base changes (as predicted by RNA2DMut) could be compensated for in WT sequences. For example, in the S. pombe loop regions SNVs that stabilized base pairs in alternative conformations (increasing the ED) were offset in other Ascomycota by directly mutating the non-native pairing partner, or through the accumulation of other mutations compensatory to the native fold that simultaneously destabilized the alternative conformation (SI File 4). In the hammerhead ribozyme example, almost all SNVs disrupted the ΔG and ED metrics (97.93% and 84.83%, respectively); suggesting that this WT ribozyme sequence is in a particularly low evolutionary valley w/r to possible SNVs.
A number of predicted features of RNA folding (including the ΔG z-scorerandom metric) are useful in the discovery of functional ncRNA [15]. Likewise, additional genomic and sequence features of ncRNA can be used to improve prediction quality [23]. The results of this study suggest that the ED z-scorerandom metric could, in conjunction with other metrics, help to deduce functional ncRNA. Here it was shown that the ED of WT ncRNA sequences is lower than random, indicating that this is an evolved property of natural ncRNA sequences; the order of bases correlates with a more converged (less diverse) structural ensemble. This effect is of a lower magnitude than the ΔG z-scorerandom metric (Fig. 4, Fig. 5; SI Table 7); however, compared to pre-randomized control sequences, the ED z-scorerandom of the WT values are significantly lower (Table 2).
A potentially confounding factor is that the ΔG and ED z-scorerandom metrics show evidence of being correlated (SI File 10). The ED is linked to the ΔG in the calculation of the partition function, which means that ED and ΔG z-scorerandom values are linked (to a degree) by sequence content, as well as sequence order: thus the generally better linear fits of the pre-randomized control data for each ncRNA (vs. WT) in SI File 10. In the cases where the ED z-scorerandom values were most shifted to the negative (e.g. RNase P and SRP RNA; Fig. 5), the correlations between the two metrics were the weakest of any ncRNA class (Fig. 6 and SI File 10). The sequence order in these cases appears to be more important in the ED z-scorerandom values and their relationship to the ΔG z-scorerandom values. This suggests that these ncRNA classes may have been under greater pressure to maintain both a thermodynamically stable structure and a less diverse ensemble of potential suboptimal folds, which is encoded within their sequence order.
Detailed analyses of representative ncRNA sequences, as well as all available sequences for 10 classes of ncRNA found that the ensemble diversity of native sequences was lower than random: both with regards to randomly substituted bases and shuffled bases. This indicates that evolved ncRNA sequences are selected and ordered to be not only more thermodynamically stable than random, but also have a more compact structural ensemble. This feature can offer insight into ncRNA evolution as well as be a potentially useful feature in ncRNA prediction.
4. Materials and methods
4.1. Input data
All sequences from nine classes of ncRNAs (rRNA, hammerhead ribozyme, RNase P, snoRNA, snRNA, SRP RNA, tmRNA, Vault RNA and Y RNA) were downloaded from the RNAcentral database [18,19]. The sequences in the rRNA class were filtered and split into two sub-classes: 5.8S and 5S rRNA. These smaller rRNA species were analyzed, as they were in the size range of other classes. Additionally, the larger rRNA species were in a size range where singe-sequence in silico folding methods have reduced accuracy [12]. All sequences were filtered to remove polymorphisms, any base symbol other than A/G/C/U(T), using the Perl script “FilterPolymorphs.pl”. Next sequence length data for all sequences in each class was measured using the script “LengthAnalysis.pl” and all sequences within 1σ of the mean length were extracted with the script “LengthFilter.pl”. This was done to remove unusually long or short sequences and reduce possible length-associated artifacts in the structure analyses. All sequences and their accession numbers appear in SI File 1.
4.2. Data analysis
Representative sequences for each class were submitted to the RNA2DMut server Sequence Mutation tool - https://rna2dmut.bb.iastate.edu/. The Sequence Mutation tool generates all possible SNVs for each sequence and calculates their minimum free energy (ΔG) and a partition function, from which the ED metric is calculated. Additionally, the ensemble centroid structure (the structure with the shortest structure distance to all other conformations in the structural ensemble) is generated and output as image files (annotated where each base is colored with the maximizing and minimizing ED values). To calculate the ΔG and ED z-scoreSNV values, the RNA2DMut results (SI File 1, outfile1) were opened in Microsoft Excel and the WT ΔG and ED values were compared to the average values of SNV mutants according to the following equations:
Here, σ represents the standard deviation of the SNV mutant ΔG and ED values, respectively in each equation.
Each WT sequence was then submitted to the RNA2DMut Sequence Manipulation tool to generate 40× randomized mutant sequences, which were then evaluated using the Sequence Evaluation tool (generates the ΔG and ED values for each input sequence). All sequences and results appear in SI File 7. The results were opened in Microsoft Excel and the WT ΔG and ED values were compared to the average values of randomized mutants according to the following equations:
Here, σ represents the standard deviation of the randomized mutant ΔG and ED values, respectively in each equation.
For the large-scale analyses of ncRNAs, all sequences from each class were evaluated using the script “HTP_Z-Score.pl”, which takes a FASTA file as input and, for each input sequence, generates a user-defined (40× in this case) set of random mutants, then predicts their folding energy (ΔG) and ED (from the partition function). The ΔG and ED z-scorerandom values are calculated as in the equations above. The energy and partition function calculations make use of the program RNAfold [24].
All scripts used in this study are available on GitHub - https://github.com/walternmoss/RNA2DMut.
Author contributions
WM designed/conducted the study and wrote the manuscript.
Conflicts of interest
The author has no conflicts of interest to declare.
Acknowledgements
This work was supported by startup funds from the Iowa State University College of Agriculture and Life Sciences and the Roy J. Carver Charitable Trust, as well as grant 4R00GM112877-02 from the NIH/NIGMS. I would like to thank BBMB colleagues and members of the Moss Lab for helpful discussions and commentary.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ncrna.2018.04.005.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
SI File 1. Results from the RNA2DMut analysis of representative ncRNAs. Results for each ncRNA are named and get two worksheets. Outfile1 data are structured as follows: Column A contains names for Mutant_0 (the WT sequence) up to Mutant_X (where X is the highest number SNV mutant generated); column B contains the nt sequence; column C contains the MFE structure in dot-bracket notation; column D contains the MFE ΔG (in kcal/mol); column E contains the ensemble centroid structure in dot-bracket notation; and column F contains the ED. Outfile2 is formatted as follows: column A gives the nt position of the RNA; column B gives the WT base, column C gives the WT ED; columns D, G and J give the base substitution (SNV) of each mutant; columns E, H and K give the names of each mutant; and columns F, I, and L give the mutant ED values.
SI File 2. Secondary structure models of representative ncRNAs. The models shown are the predicted ensemble centroid secondary structure. For each ncRNA there are two models shown: the left-most model is annotated with the magnitude of the change in ED minimizing mutations (vs. WT), whereas the right-most model shows the magnitude of the change in ED maximizing mutations (vs. WT). The magnitude of minimizing and maximizing ED change is indicated by blue and red intensity, respectively (keys appear next to each model).
SI File 3. Sequence alignment of Ascomycota 5S rRNA sequences. The alignment was generated using MAFFT, then degapped to fit the WT S. pombe sequence. The top position of the alignment has the ensemble centroid bracket structure for S. pombe.
SI File 4. Secondary structure models of S. pombe 5S rRNA high ED mutants vs. WT Ascomycota sequences. Each pair of 2D structures contains the S. pombe SNV mutant to the left and the WT sequence that also bears the variant nt to the right (mutant and WT sequence names are below each structure model). The nt change to the S. pombe sequence is indicated with a red base and line pointing to the altered nt. The corresponding base in the WT sequence is also annotated in red. The base pairing type is annotated as follows: open circles are for GU “wobble” pairs, single solid lines are for AU pairs, double lines are for GC pairs and solid blue circles are for non-canonical (“inconsistent” pairs). The latter type, the non-canonical pairs, indicate places where the WT sequence potentially evolved to disfavor the alternative structure model stabilized by the SNV.
SI File 5. Results from the RNA2DMut Sequence Evaluation tool on randomized ncRNAs. Each representative ncRNA has a separate worksheet and data are organized as follows: Column A contains names for Mutant_0 (the WT sequence) up to Mutant_X (where X is the highest number of randomized mutants generated); column B contains the nt sequence; column C contains the MFE structure in dot-bracket notation; column D contains the MFE ΔG (in kcal/mol); column E contains the ensemble centroid structure in dot-bracket notation; and column F contains the ED.
SI File 6. Structure data for ten classes of ncRNAs from the RNAcentral database. Each ncRNA class has a separate worksheet, which is organized as follows: column A has the sequence accession and name; column B has the ncRNA sequence, column C has the MFE predicted structure in dot-bracket notation; column D has the MFE ΔG (in kcal/mol); column E has the ΔG z-scorerandom value; column F has the fraction of mutants with ΔG < the WT input sequence; column G has the ensemble centroid structure model in bracket notation; column H has the WT ED value; column I has the ED z-scorerandom value; column J has the fraction of mutants with ED < the WT input sequence; columns K–P correspond to D–J, only the input sequence analyzed was the “pre-randomized” WT control sequence; columns Q–T give the number of A, G, C, and U residues, respectively, in the WT and mutant sequences.
SI File 7. ED and ΔG z-scorerandom metrics for ncRNA classes. These data are associated with the box plots in Figures 4 and 5.
SI File 8. Distributions of ΔG z-scorerandom values for 10 classes of ncRNA. The z-score is calculated from the difference in the pre-randomized control sequence vs. 40 additional randomized mutants.
SI File 9. Distributions of ED z-scorerandom values for 10 classes of ncRNA. The z-score is calculated from the difference in the pre-randomized control sequence vs. 40 additional randomized mutants.
SI File 10. Scatter plots comparing the ΔG vs. ED z-scorerandom values for 10 classes of ncRNA. Each ncRNA has a separate worksheet that is organized as follows: column A has the ncRNA accession number and name; columns B and C have the WT sequence ΔG and ED z-scorerandom values, respectively; while columns E and F has those for pre-randomized control sequences. Scatter plots for both kinds of data appear to the right where WT data is in green and pre-randomized control data is in red. Linear trendlines for each distribution are included and the associated R2 values appear next to the line.
References
- 1.Clote P., Ferre F., Kranakis E., Krizanc D. Structural rna has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruber A.R., Findeiss S., Washietl S., Hofacker I.L., Stadler P.F. Rnaz 2.0: improved noncoding RNA detection. Pac Symp Biocomput. 2010:69–79. [PubMed] [Google Scholar]
- 4.Gorodkin J., Hofacker I.L., Torarinsson E., Yao Z., Havgaard J.H., Ruzzo W.L. De novo prediction of structured RNAs from genomic sequences. Trends Biotechnol. 2010;28:9–19. doi: 10.1016/j.tibtech.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washietl S., Hofacker I.L., Lukasser M., Huttenhofer A., Stadler P.F. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 6.Andrews R.J., Baber L., Moss W.N. RNAStructuromeDB: a genome-wide database for RNA structural inference. Sci. Rep. 2017;7:17269. doi: 10.1038/s41598-017-17510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss W.N., Priore S.F., Turner D.H. Identification of potential conserved RNA secondary structure throughout influenza a coding regions. RNA. 2011;17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moss W.N., Steitz J.A. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence rna. BMC Genom. 2013;14:543. doi: 10.1186/1471-2164-14-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofacker I.L., Stadler P.F., Stocsits R.R. Conserved RNA secondary structures in viral genomes: a survey. Bioinformatics. 2004;20:1495–1499. doi: 10.1093/bioinformatics/bth108. [DOI] [PubMed] [Google Scholar]
- 10.Raghavan R., Groisman E.A., Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swiercz J.P., Hindra, Bobek J., Bobek J., Haiser H.J., Di Berardo C., Tjaden B., Elliot M.A. Small non-coding RNAs in streptomyces coelicolor. Nucleic Acids Res. 2008;36:7240–7251. doi: 10.1093/nar/gkn898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews D.H., Moss W.N., Turner D.H. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a003665. a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wuchty S., Fontana W., Hofacker I.L., Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 15.Freyhult E., Gardner P.P., Moulton V. A comparison of RNA folding measures. BMC Bioinf. 2005;6:241. doi: 10.1186/1471-2105-6-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulton V., Zuker M., Steel M., Pointon R., Penny D. Metrics on RNA secondary structures. J. Comput. Biol. 2000;7:277–292. doi: 10.1089/10665270050081522. [DOI] [PubMed] [Google Scholar]
- 17.Moss W.N. RNA2DMut: a web tool for the design and analysis of RNA structure mutations. RNA. 2018 Mar;24(3):273–286. doi: 10.1261/rna.063933.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman A., Agrawal S., Birney E., Bruford E.A., Bujnicki J.M., Cochrane G., Cole J.R., Dinger M.E., Enright A.J., Gardner P.P. RNAcentral: a vision for an international database of RNA sequences. RNA. 2011;17:1941–1946. doi: 10.1261/rna.2750811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The R.C. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017;45:D128–D134. doi: 10.1093/nar/gkw1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szymanski M., Zielezinski A., Barciszewski J., Erdmann V.A., Karlowski W.M. 5SRNAdb: an information resource for 5s ribosomal RNAs. Nucleic Acids Res. 2016;44:D180–D183. doi: 10.1093/nar/gkv1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawrocki E.P., Kolbe D.L., Eddy S.R. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu L., Di C., Kai M., Yang Y.C., Li Y., Qiu Y., Hu X., Yip K.Y., Zhang M.Q., Lu Z.J. A common set of distinct features that characterize noncoding RNAs across multiple species. Nucleic Acids Res. 2015;43:104–114. doi: 10.1093/nar/gku1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. Viennarna package 2.0. Algorithm Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.