Abstract
HIV-1 particle production occurs in a series of steps promoted by the viral Gag protein. Although it is well established that assembly and release take place at the plasma membrane, the nature of membrane assembly sites remains poorly understood. We show here that Gag specifically associates with cholesterol-enriched microdomains (“rafts”) at the plasma membrane. Kinetic studies demonstrate that raft association follows membrane binding, and the analysis of Gag mutants reveals that, whereas the N terminus of Gag mediates raft binding, this association is greatly enhanced by Gag–Gag interaction domains. We observe that depletion of cellular cholesterol markedly and specifically reduces HIV-1 particle production. Furthermore, treatment of virus-producing cells or virus particles with raft-disrupting agents significantly impairs virus infectivity. These results identify the association of Gag with plasma membrane rafts as an important step in HIV-1 replication. These findings may lead to novel strategies for suppressing HIV-1 replication in vivo.
The assembly of type C retroviruses and lentiviruses occurs at the plasma membrane (PM). Assembly involves multiple steps mediated by the viral Gag protein (reviewed in refs. 1 and 2). The Gag protein of HIV-1 is synthesized as a precursor polyprotein, Pr55Gag, which is composed of matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains, as well as the spacer peptides p2 and p1. Each of these domains is involved in specific steps in the virus assembly process (1, 2): the MA domain mediates targeting of Pr55Gag to the PM, Gag membrane binding, and incorporation of the viral envelope (Env) glycoproteins into virions; the C-terminal portion of CA, p2, and the N-terminal domain of NC promote Gag multimerization; and p6 stimulates the budding off of virus particles from the PM. Retroviral Gag proteins, including HIV-1 Gag, have been observed to be localized to small, discrete regions of the PM rather than being uniformly distributed at the cell surface (3–6). However, the nature of these regions, which may represent active centers of virus assembly and release, remains to be elucidated.
Several types of microdomains, each with a distinct lipid and protein composition, exist in the PM. Lipid rafts, which are highly enriched in sphingolipids and cholesterol, are one such microdomain (for recent reviews, see refs. 7 and 8). Lipid rafts can be isolated as detergent-resistant membrane (DRM) by Triton X-100 treatment at low temperature, followed by equilibrium flotation centrifugation (7, 8). By using this approach, several classes of proteins, including Src-family kinases and glycosylphosphatidylinositol (GPI)-anchored proteins, have been identified as being raft associated (7, 8). The use of cholesterol-depleting agents has provided evidence to support the hypothesis that rafts play a critical role in a number of biological processes such as signal transduction and protein trafficking (reviewed in refs. 7 and 8). In many cases, lipid rafts are thought to serve as concentrating platforms for molecules involved in these processes.
Over a number of years, virologists have observed that the lipid composition of envelope membranes from a variety of viruses, including several retroviruses, is distinct from that of the host PM from which they are derived (refs. 9–13; reviewed in 14). These observations led to the hypothesis that viruses may bud from specific PM microdomains (12, 15). Consistent with this model, a number of viral structural proteins, including those of Sendai (16), measles (17), and influenza viruses (18–21), reportedly associate with DRM. However, an impact of raft disruption on virus assembly/release remains to be demonstrated.
The HIV-1 lipid bilayer has long been known to be enriched, relative to the host cell PM, in sphingolipids and cholesterol (22). In addition, GPI-anchored proteins are incorporated into HIV-1 particles (23). Recently, it was reported that the non-raft protein CD45 is excluded from virus particles, and HIV-1 Gag and Env proteins were detected in DRM fractions (24). Thus, it is tempting to speculate that budding of HIV-1 occurs selectively from lipid rafts. However, several caveats apply to this hypothesis: the rhabdoviruses and alphaviruses, which are considered to be non-raft associated (15, 25), are also enriched in cholesterol (refs. 10 and 26; reviewed in ref. 14) and incorporate GPI-anchored proteins (27); HIV-1 can also incorporate DRM-excluded proteins [e.g., transferrin receptor (TfR) and VSV-G; refs. 23, 28, and 29]; exclusion of CD45 from HIV-1 virions is likely due to steric interference between its very long (707 aa) cytoplasmic domain and Gag, rather than its non-raft localization in the PM; and DRM association of viral proteins may simply reflect random targeting to both raft and non-raft domains at the PM. Most importantly, the physiological significance of rafts in virus assembly remains to be demonstrated.
In this report, we focus on several questions relating to the possible role of rafts in HIV-1 assembly: (i) Is Gag specifically targeted to rafts? (ii) If so, which domain(s) in Gag promote raft association? (iii) Is Gag–raft association necessary for efficient virus particle production and infectivity? By comparing total membrane binding and DRM association of Gag in kinetic analyses, we find that Gag is preferentially targeted to rafts after it binds membrane. Localization to rafts is primarily mediated by the N-terminal region of Gag, but the Gag–Gag interaction domain in NC greatly facilitates this association. Finally, disruption of Gag–raft binding by cholesterol depletion inhibits virus particle production and infectivity. Collectively, these results demonstrate that the association of Gag with rafts is an important step in HIV-1 replication.
Materials and Methods
Plasmids.
Molecular clones encoding Gag truncation mutants (30) or an inactive protease (PR; ref. 31) have been described previously. The NL4-3-based GagPol expression vector pCMVNLGagPolRRE was constructed by exchanging the BssHII-EcoRV fragment of pCMVGagPolRRE [a kind gift from D. Rekosh (32)] with the corresponding fragment from pNL4-3. pCMV5Fyn(16)eGFP (33), pT7VSVMWT (34), pNL4-3/HX 15A (35), pHCMV-G, and pCMVRev were kindly provided by M. Resh, Memorial Sloan-Kettering Cancer Center, P. Palese, Mount Sinai School of Medicine, J. Luban, Columbia University, J. Burns, University of California, San Diego, and S. Venkatesan, National Institutes of Health, respectively.
Gag Expression.
Gag was expressed either by transfecting cells with molecular clones or by infecting with high-titer vector virus stocks. The latter were obtained by cotransfecting 293T cells with pHCMV-G, pCMVNLGagPolRRE, and pNL4-3-derived molecular clones.
Antibodies and Reagents.
Antibodies were obtained from the following sources: rabbit polyclonal anti-caveolin (Cav) antibody (Transduction Laboratories, Lexington, KY); mouse anti-human TfR antibody (clone H68.4; Zymed); HIV Ig (cat. no. 3957) and AIDS patient sera (cat. nos. 1983 and 1984; the NIH AIDS Research and Reference Reagent Program); anti-human, anti-mouse, and anti-rabbit Ig antibodies conjugated with horseradish peroxidase (Amersham Pharmacia). Anti-VSV M monoclonal antibody (clone 23H12) was a kind gift from D. Lyles (36). Lipoprotein-deficient calf serum (LPDS) was either obtained from Sigma or prepared as described previously.† Methyl-β-cyclodextrin (MβCD) and mevalonic acid lactone were obtained from Sigma. Simvastatin, obtained from Calbiochem, was activated as described previously (38).
Assays for Membrane Binding and DRM Association.
Membrane binding and DRM association of Gag were analyzed by equilibrium flotation centrifugation as detailed previously (39). For DRM association, postnuclear supernatants were treated with Triton X-100 (final concentration 0.25%) for 20 min on ice before flotation centrifugation. In steady-state assays, fractions were analyzed by Western blotting as previously described (40); for the analysis of newly synthesized Gag proteins, cells were either pulse-labeled for 5 min and chased for 15 or 30 min, or labeled for 90 min with [35S]Met/[35S]Cys. Labeled cells were treated as above, and fractions were immunoprecipitated as described previously (41). For detailed procedures, see text in supporting information (Methods), which is published on the PNAS web site, www.pnas.org
Cholesterol Depletion and Virus Production Assays.
To deplete cellular cholesterol, HeLa cells were cultured for 2 days in DMEM containing 5% LPDS, then incubated in RPMI medium 1640 without methionine and cysteine (RPMI-Met−/Cys−) containing various concentrations of MβCD for 30 min at 37°C. Cells were labeled with [35S]Met/[35S]Cys for 90 min in RPMI-Met−/Cys− containing 2% LPDS. Cell lysates were immunoprecipitated with HIV Ig (41). Virus lysates were either directly loaded on SDS-polyacrylamide gels after boiling in sample buffer, or were immunoprecipitated (41). To inhibit cholesterol biosynthesis, HeLa cells were cultured for 2 days in DMEM containing 5% LPDS in the presence of 1, 2, or 5 μM simvastatin and 500 μM mevalonate. After starving in RPMI-Met−/Cys− containing simvastatin and 500 μM mevalonate for 30 min, cells were labeled in the same medium with [35S]Met/[35S]Cys for 90 min.
Results
Gag Is Associated with DRM.
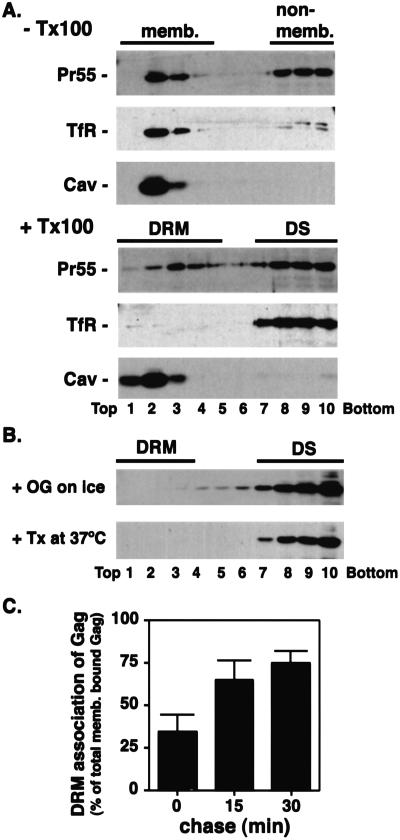
To determine whether HIV-1 Gag is associated with DRM, we homogenized Pr55Gag-expressing cells, divided the postnuclear supernatants into two aliquots, and treated them with or without 0.25% Triton X-100 on ice before equilibrium flotation centrifugation (Fig. 1A). Without detergent treatment, both the non-raft-associated protein TfR and the raft-associated protein Cav were recovered exclusively in the floated (membrane-containing) fractions. After detergent treatment, no TfR was detected in membrane fractions whereas almost all Cav remained associated with membrane. Greater than 90% of a Fyn-green fluorescent protein (GFP) chimeric protein (33), another raft marker, was membrane-bound, whereas ≈50% remained associated with DRM (Fig. 5A, which is published as supporting information on the PNAS web site). By using the same conditions, we observed that ≈50% of Pr55Gag was recovered in membrane fractions without detergent treatment (Fig. 1A). After cold Triton X-100 treatment, approximately half of the membrane-bound Pr55Gag remained associated with DRM (Fig. 1A). Similar data were obtained in the Jurkat T cell line (Fig. 5B). As is observed for other raft-localized proteins (42), the association of Pr55Gag with DRM was disrupted by treatment with octyl glucoside at 4°C or Triton X-100 at 37°C (Fig. 1B). Non-membrane-bound Gag complexes (43), still present after cold octyl glucoside or warm Triton X-100 treatments, were not able to float under these conditions (Fig. 5C), excluding the possibility that the Pr55Gag recovered in DRM fractions is derived from non-membrane-bound multimers. Non-myristylated mutant Pr55Gag, which shows a low level of membrane binding (39), was fully sensitive to Triton X-100 solubilization at 4°C (data not shown).
Figure 1.
HIV-1 Gag is associated with DRM. (A) Postnuclear supernatants derived from HeLa cells expressing HIV-1 Pr55Gag were treated with or without 0.25% Triton X-100 on ice and subjected to equilibrium flotation centrifugation. Pr55Gag, TfR, and Cav were detected by Western blotting. Membrane (memb.), non-membrane (non-memb.), DRM, and detergent-soluble (DS) fractions are shown. (B) Postnuclear supernatants derived from HeLa cells expressing HIV-1 Pr55Gag were treated with 30 mM octyl glucoside (OG) on ice or with 0.25% Triton X-100 at 37°C and analyzed as in A. (C) Pr55Gag-expressing cells were pulse-labeled for 5 min and chased as indicated. Postnuclear supernatants were treated and fractionated as in A. Fractions 1–5 and 6–10 were pooled, and Pr55Gag in these fractions was recovered by immunoprecipitation. The percentage of membrane-bound Gag that was DRM associated at each time point is indicated.
Association of Gag with DRM Occurs After Membrane Binding.
To further understand the process by which HIV-1 Gag interacts with DRM, we analyzed the kinetics of Pr55Gag membrane binding and DRM association. Gag-expressing HeLa cells were pulse-labeled with [35S]Met/[35S]Cys for 5 min and chased for 0, 15, and 30 min in unlabeled medium and subjected to flotation centrifugation with or without prior treatment in cold Triton X-100. Fractions (Frs.) containing floated (top five Frs.) and non-floated (bottom five Frs.) material were pooled, and Pr55Gag was immunoprecipitated. Approximately 30% of Pr55Gag was recovered in the membrane fraction immediately after the pulse; this amount increased to ≈40% during the chase period (data not shown). In contrast, the percentage of membrane-bound Gag that was DRM associated was low immediately after the 5-min pulse, then increased sharply during the 30-min chase period (Fig. 1C). In an experiment done with a shorter (2 min) pulse, around 20% of membrane-bound Gag was DRM associated after the pulse, whereas ≈70% was DRM associated after a 15-min chase (data not shown). These results indicate that DRM association of Gag occurs with a delay relative to membrane binding.
Gag Association with DRM Is Mediated by the N-Terminal Domain of Gag, but Is Enhanced by the C-Terminal “Interaction” Domain.
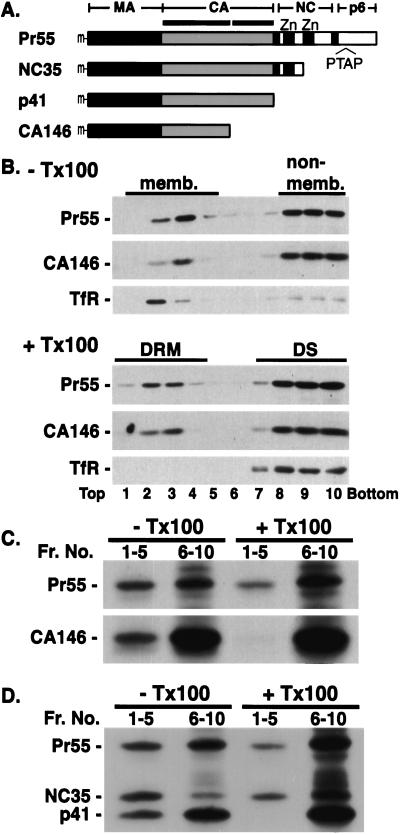
To identify the determinants of Pr55Gag association with DRM, we characterized a panel of C-terminally truncated Gag mutants (Fig. 2A; ref. 30) for their recovery in DRM. As we have shown previously (30), Pr55Gag and CA146 showed a comparable level of steady-state membrane binding (Fig. 2B, −Tx100). Approximately half of the membrane-bound CA146 and Pr55Gag was detected in DRM, suggesting that steady-state association of Gag with DRM is mediated by a region of Gag spanning MA and the N-terminal domain of CA. Data obtained by immunoprecipitation of fractions derived from cells metabolically labeled for 22 h also indicated that ≈50% of membrane-bound Pr55Gag and CA146 was recovered in DRM (data not shown).
Figure 2.
Determinants of Gag-DRM association. (A) Schematic representation of C-terminally truncated Gag mutants. Positions of N-terminal myristate (m), N-and C-terminal domains of CA (horizontal bars), the two zinc finger motifs in NC (Zn), and the p6 late domain (PTAP) are shown. (B) Cells singly expressing Pr55Gag and CA146 were pooled and analyzed as in Fig. 1A. (C and D) Cells singly expressing Pr55Gag and indicated mutants were metabolically labeled for 90 min, pooled and analyzed as in Fig. 1C.
We examined further membrane binding and DRM association of full-length and truncated Gag mutants early after their synthesis. Gag-expressing cells were labeled for 90 min with [35S] Met/[35S]Cys and homogenized, and postnuclear supernatants were subjected to flotation centrifugation with or without prior detergent treatment. Gag proteins in pooled fractions (Fig. 2C, Frs. 1–5 and Frs. 6–10) were immunoprecipitated. Newly synthesized CA146 was detected in membrane but not DRM fractions. In contrast, newly synthesized Pr55Gag was readily detectable in floated fractions with or without detergent treatment. To map the region of Gag required for the enhanced DRM association of full-length Pr55Gag, cells independently expressing Pr55Gag, NC35, or p41 were labeled as above and pooled before the preparation of postnuclear supernatant. Without Triton X-100 treatment, similar absolute amounts of full-length Pr55Gag, NC35, and p41 were recovered in the membrane-bound fractions (Fig. 2D, −Tx100, Frs. 1–5). After treatment with cold Triton X-100, full-length Pr55Gag and NC35 were recovered in DRM to a similar extent (Fig. 2D, +Tx100, Frs. 1–5). In contrast, no p41 was detected in DRM (Fig. 2D, +Tx100, Frs. 1–5). These results demonstrate that the sequence spanning p2 and the first 35 aa of NC enhances Gag DRM association.
Effects of Cholesterol Depletion on Virus Production and Infectivity.
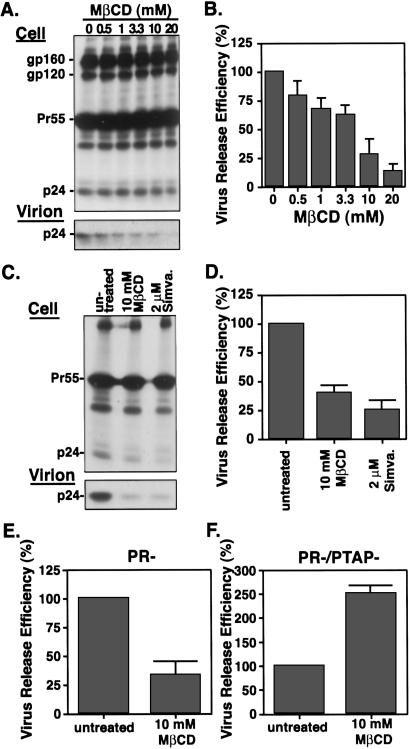
The experiments presented thus far indicate that HIV-1 Gag associates with DRM, measure the kinetics of DRM association, and map the determinants of this interaction. If the association of Gag with rafts plays a physiological role during virus assembly, we might predict that raft disruption would impair virus assembly and release. To address this issue, we used two classes of cholesterol-depleting drugs frequently used to disrupt rafts: MβCD, a cyclic sugar compound that acutely extracts cholesterol from the plasma membrane of treated cells, and the statins, which block cholesterol biosynthesis by inhibiting the key enzyme HMG-CoA reductase. Visualization of the cholesterol content in HeLa cells by staining with filipin, a fluorescent fungal metabolite that binds cholesterol (44), indicated that treatment with either 10 mM MβCD or 2 μM simvastatin caused a 3-fold reduction in filipin fluorescence intensity on the cell periphery (Fig. 6, which is published as supporting information on the PNAS web site). To examine the impact of this cholesterol depletion on virus production, HeLa cells transfected with the HIV-1 molecular clone pNL4-3 were treated with various concentrations of MβCD for 30 min at 37°C and were metabolically labeled with [35S]Met/[35S]Cys for 90 min. When cells were treated with 10–20 mM MβCD, the release of virion-associated p24 was reduced by 70–80% (Fig. 3 A and B). A significant reduction in virus release was also observed when cells were treated with simvastatin (Fig. 3 C and D) or lovastatin (data not shown). Virus-like particle production from HeLa cells expressing NL4-3/PR− was also significantly reduced after MβCD treatment (Fig. 3E). Jurkat cells expressing Gag showed a 3-fold reduction in virus release after 10 mM MβCD treatment for 15 min (Fig. 7, which is published as supporting information on the PNAS web site). In contrast, release of particle-associated VSV M from HeLa cells was not inhibited under identical conditions (Fig. 8, which is published as supporting information on the PNAS web site). Together, these data indicate that cholesterol depletion impairs HIV-1 particle production but not the release of a prototypical non-raft-associated virus (VSV).
Figure 3.
Effects of cholesterol depletion on virus production. (A and C) HeLa cells expressing NL4-3 (wt virus) were treated with the indicated concentrations of cholesterol-depleting drugs. MβCD treatments were performed for 30 min; cells were treated with simvastatin (simva.) for 2 days before metabolic labeling. Cells were washed and metabolically labeled for 90 min. Labeled viral proteins in cell and virion lysates were analyzed by SDS/PAGE followed by fluorography. Data were quantified by Phosphorimager (Fuji Film) analysis. Viral glycoproteins gp160 and gp120, Pr55Gag, and mature CA (p24) are shown. (B and D) Virus release efficiency represents the amount of virion-associated p24 as a fraction of total Gag (cell-associated Pr55Gag and p24 plus virion-associated p24). The relative virus release efficiencies in drug-treated vs. untreated cultures are indicated. (E) Quantification of virus release from HeLa cells expressing NL4-3/PR−. Virus release efficiency was calculated as the amount of virion-associated Pr55Gag as a fraction of total (cell plus virion) Gag. (F) Quantification of virus release from HeLa cells expressing NL4-3/PR−/PTAP−, calculated as in E.
We previously reported that cells expressing the NC35 Gag truncation mutant produce detectable levels of particle-associated Gag (30). Thus, we analyzed the effect of cholesterol depletion on NC35-mediated particle production. Interestingly, cholesterol depletion did not reduce but rather slightly increased the amount of NC35-derived particle-associated Gag released into the medium (data not shown). Because NC35 lacks the “late” domain in p6 implicated in virus budding (31, 45), we also tested virus production mediated by a p6 mutant (PR−/PTAP−; ref. 31), which contains substitutions in the p6 late domain motif Pro-Thr-Ala-Pro. Intriguingly, we observed that MβCD treatment increased (by ≈2.5-fold) the release of virion-associated PR−/PTAP− mutant Gag (Fig. 3F). Similarly, simvastatin treatment increased (by 2-fold) the release of the p6 mutant L1Term (ref. 31; data not shown). These results indicate that particle production mediated by p6 late domain mutants is enhanced by cholesterol depletion. In contrast, release of an assembly-deficient mutant (mutant 15A; ref. 35) that displays very low levels of virus production is reduced severalfold by cholesterol depletion (data not shown).
To determine whether raft disruption has an effect on virus infectivity, we tested the supernatant from simvastatin-treated cells in the single-cycle MAGI assay (46) and found a 5- to 7-fold reduction in its infectivity (Fig. 9, which is published as supporting information on the PNAS web site). This effect corresponds to a 2- to 3-fold reduction in relative infectivity when stocks are normalized for pelletable p24. Simultaneous treatment of virus-producing cells with simvastatin and MβCD almost completely abolished the production of infectious virus (data not shown).
Discussion
In this study, we examine whether Gag-raft association is a specific and physiologically relevant process in HIV-1 particle formation. We observe that ≈50% of membrane-bound Pr55Gag is associated with DRM at steady state (Fig. 1A), and that DRM association of Gag occurs more slowly than Gag membrane binding (Fig. 1C). Interestingly, the delay in DRM association vs. membrane binding has been observed for other raft-associated proteins, such as exogenously added CD59 (a GPI-anchored protein; ref. 47), and the Src-related kinase, Fyn (33). The slower kinetics of raft association rule out two major caveats relating to the specificity of DRM localization, i.e., that DRM association of Gag is an artifact of detergent treatment, or is due to random targeting of Gag to both DRM and non-DRM domains. If either of these possibilities applied, one would predict that the ratio between DRM-associated Gag and membrane-bound Gag would remain constant over time. However, this ratio increases steeply in a time-dependent manner (Fig. 1C). These results suggest that the observed recovery of Gag in DRM reflects a specific association of Gag with rafts.
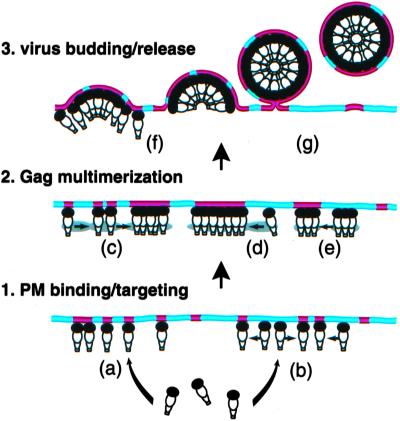
Under steady-state conditions, the truncated Gag mutant CA146 shows levels of DRM association similar to those observed with full-length Gag. In contrast, under short-term labeling conditions that allow the detection of newly synthesized protein, full-length and NC35 Gag are recovered in DRM fractions whereas CA146 and p41 are not (Fig. 2). Although DRM association of MA itself is difficult to evaluate because of its very low affinity for membrane (4, 39, 48), the analysis of an MA mutant (20LK) that displays increased membrane binding relative to wild type (40) indicates that MA is able to associate with DRM under steady-state conditions (unpublished data). Together, these results suggest that raft association of Pr55Gag is initiated by the MA domain of Gag and is subsequently stabilized by downstream sequences (in p2 and the N terminus of NC) that promote Gag–Gag interactions (30, 35). Oligomeric Gag may display a stronger affinity for DRM (Fig. 4e) because of an increased number of binding sites per complex, or an altered conformation induced by Gag–Gag interaction. Alternatively, rafts, which are thought to be small, unstable, and highly dynamic, may themselves be stabilized and coalesced to form DRM domains by Gag–Gag interaction (Fig. 4c). The observed punctate staining of HIV-1 Gag on the plasma membrane (4–6) may represent this larger, coalesced DRM. Larger rafts might, in turn, facilitate recruitment and concentration of Gag to preformed multimers (Fig. 4d). Delayed kinetics of Gag–DRM association (Fig. 1C) may reflect the kinetics of Gag oligomerization. It is worth noting that a number of raft-associated proteins have been observed to increase their interaction with DRM when cross-linked by antibodies or multivalent ligands (for examples, see refs. 49–51).
Figure 4.
Models for the role of rafts in HIV-1 assembly and release. Raft and non-raft domains are shown in red and blue, respectively. See text for details.
To address the physiological significance of Gag–raft association in virus particle assembly, we disrupted rafts by depleting cellular cholesterol. The levels of cholesterol depletion achievable with this approach markedly reduce HIV-1 particle production. A variety of points argue that drug treatment does not globally perturb the cell and impair virus production in a nonspecific manner. (i) Similar data were obtained with two distinct classes of compounds (MβCD and the statins) with different modes of action. The experimental conditions used in the present study, which closely parallel those used in previous reports, have been shown to be minimally cytotoxic (for examples, see refs. 44 and 52). (ii) In our system, Gag and Env protein synthesis, Env transport and processing, and cell surface Env expression are not affected by these reagents (Fig. 3 and unpublished data). (iii) Finally, and perhaps most importantly, particle production from cells expressing three Gag mutants (NC35, L1Term, and PR−/PTAP−) lacking the p6 late domain or expressing the VSV M protein is not reduced after cholesterol depletion (Fig. 3F, Fig. 8, and unpublished data). Together, these observations suggest that the reduction in wt virus production after cholesterol depletion is due not to gross effects on the cell, but rather to a specific disruption of a cholesterol-dependent process. At this time, we cannot exclude the possibility that a cellular process that depends on non-raft cholesterol, e.g., endocytosis (52, 53), may play an important role in virus production. Alphavirus exit is known to be reduced by cholesterol depletion even though alphaviruses do not appear to associate with rafts (15, 25, 54). In the case of HIV-1, however, considering the apparently specific association of Gag with DRM, it is likely that cholesterol depletion impairs virus particle production by disrupting rafts.
Because virus particle production takes place in a series of discrete steps (for review, see ref. 1), several models could explain the role of rafts in HIV-1 assembly and release (Fig. 4). (i) Rafts may serve as receptors for Gag at the PM. Gag may directly bind rafts, but acquire stable association over time (Fig. 4a). Alternatively, Gag may bind non-raft domains and move laterally to rafts (Fig. 4b). (ii) Rafts may function as oligomerization platforms for Gag, as mentioned above (Fig. 4 c–e). An analogous oligomerization platform model has been proposed for a raft-binding toxin (55). (iii) Rafts may modulate the late steps in virus release (Fig. 4 f and g). The composition of raft domains may influence bud formation or the membrane fusion step required for pinching off. Alternatively, host proteins that interact with late domains to modulate virion release (56) may themselves be raft associated. Our finding that the release of Gag mutants lacking the p6 late domain is not reduced but is actually increased by cholesterol depletion may suggest a relationship between rafts and late domain function. Experiments are needed to define further which step(s) in virus production are affected by raft disruption and to explore the role of rafts in late domain activity.
In summary, we show here that Gag association with rafts is likely a specific process that plays an important role in HIV-1 assembly and release. Because rafts are proposed to be concentrated at points of cell–cell contact in T cells (8, 57), targeting of HIV-1 assembly to rafts might facilitate cell-to-cell virus transmission. This proposed effect on virus spread would likely be greatest under conditions exhibiting a high degree of cell crowding [e.g., in lymphoid tissue; see supporting information (Discussion)]. In addition, we observe that virus derived from cholesterol-depleted cells displays impaired infectivity (Fig. 9), suggesting that raft targeting during assembly may enhance infectivity during the subsequent round of infection. The recent observation that raft disruption impairs the functional association of gp120 with CD4 and coreceptor during virus entry (37) highlights the importance of rafts throughout the HIV-1 replication cycle. Because effects observed in a single cycle of virus replication are amplified over multiple rounds of infection, the cumulative effect of cholesterol depletion on virus production plus subsequent infectivity would clearly have a major impact on virus replication in a spreading infection. Considering that simvastatin and related compounds are widely used clinically to treat high cholesterol, further analysis of Gag–raft interaction may provide insights into new strategies for controlling HIV-1 replication in vivo.
Supplementary Material
Acknowledgments
We thank D. Demirov and T. Murakami for helpful suggestions and critical review of the manuscript. We thank O. Schwartz for assistance with confocal microscopy and S. Bour for help in figure preparation. We acknowledge J. Burns, J. Luban, D. Lyles, P. Palese, D. Rekosh, M. Resh, and S. Venkatesan for reagents. The following reagents were obtained through the National Institutes of Health AIDS Research Reference and Reagent Program: AIDS patient (neutralizing) sera (from L. Vujcic), HIV Ig (from Nabi), and vTF7.3 (from B. Moss).
Abbreviations
- PM
plasma membrane
- MA
matrix
- CA
capsid
- NC
nucleocapsid
- DRM
detergent-resistant membrane
- TfR
transferrin receptor
- Cav
caveolin
- MβCD
methyl-β-cyclodextrin
- Env
envelope
- GPI
glycosylphosphatidylinositol
- LPDS
lipoprotein-deficient calf serum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Weinstein, D. B. (1979) Circulation 60, 204 (abstr.).
References
- 1.Freed E O. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 2.Swanstrom R, Wills J W. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 3.Jones T A, Blaug G, Hansen M, Barklis E. J Virol. 1990;64:2265–2279. doi: 10.1128/jvi.64.5.2265-2279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandefur S, Varthakavi V, Spearman P. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono A, Orenstein J M, Freed E O. J Virol. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermida-Matsumoto L, Resh M D. J Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown D A, London E. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 8.Simons K, Toomre D. Nat Rev. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 9.McSharry J J, Wagner R R. J Virol. 1971;7:59–70. doi: 10.1128/jvi.7.1.59-70.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renkonen O, Kaarainen L, Simons K, Gahmberg C G. Virology. 1971;46:318–326. doi: 10.1016/0042-6822(71)90033-x. [DOI] [PubMed] [Google Scholar]
- 11.Quigley J P, Rifkin D B, Reich E. Virology. 1972;50:550–557. doi: 10.1016/0042-6822(72)90406-0. [DOI] [PubMed] [Google Scholar]
- 12.Pessin J E, Glaser M. J Biol Chem. 1980;255:9044–9050. [PubMed] [Google Scholar]
- 13.Slosberg B N, Montelaro R C. Biochim Biophys Acta. 1982;689:393–402. doi: 10.1016/0005-2736(82)90274-7. [DOI] [PubMed] [Google Scholar]
- 14.Aloia R C, Curtain C C, Jensen F C. In: Advances in Membrane Fluidity. Aloia R C, Curtain C C, editors. Vol. 6. New York: Wiley-Liss; 1992. pp. 283–304. [Google Scholar]
- 15.Scheiffele P, Rietveld A, Wilk T, Simons K. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 16.Sanderson C M, Avalos R, Kundu A, Nayak D P. Virology. 1995;209:701–707. doi: 10.1006/viro.1995.1308. [DOI] [PubMed] [Google Scholar]
- 17.Vincent S, Gerlier D, Manie S N. J Virol. 2000;74:9911–9915. doi: 10.1128/jvi.74.21.9911-9915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skibbens J E, Roth M G, Matlin K S. J Cell Biol. 1989;108:821–832. doi: 10.1083/jcb.108.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundu A, Avalos R T, Sanderson C M, Nayak D P. J Virol. 1996;70:6508–6515. doi: 10.1128/jvi.70.9.6508-6515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Pekosz A, Lamb R A. J Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali A, Avalos R T, Ponimaskin E, Nayak D P. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aloia R C, Tian H, Jensen F C. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott D E. Rev Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D H, Hildreth J E. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y E, Kielian M. J Virol. 2000;74:7708–7719. doi: 10.1128/jvi.74.17.7708-7719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patzer E J, Moore N F, Barenholz Y, Shaw J M, Wagner R R. J Biol Chem. 1978;253:4544–4550. [PubMed] [Google Scholar]
- 27.Calafat J, Janssen H, Demant P, Hilgers J, Zavada J. J Gen Virol. 1983;64:1241–1253. doi: 10.1099/0022-1317-64-6-1241. [DOI] [PubMed] [Google Scholar]
- 28.Akkina R K, Walton R M, Chen M L, Li Q X, Planelles V, Chen I S. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 30.Ono A, Demirov D, Freed E O. J Virol. 2000;74:5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjold M L, Rekosh D. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van't Hof W, Resh M D. J Cell Biol. 1997;136:1023–1035. doi: 10.1083/jcb.136.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harty R N, Paragas J, Sudol M, Palese P. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cimarelli A, Luban J. J Virol. 2000;74:6734–6740. doi: 10.1128/jvi.74.15.6734-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefrancois L, Lyles D S. Virology. 1982;121:157–167. [PubMed] [Google Scholar]
- 37.Manes S, del Real G, Lacalle R A, Lucas P, Gomez-Mouton C, Sanchez-Palomino S, Delgado R, Alcami J, Mira E, Martinez A C. EMBO Rep. 2000;1:190–196. doi: 10.1093/embo-reports/kvd025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keyomarsi K, Sandoval L, Band V, Pardee A B. Cancer Res. 1991;51:3602–3609. [PubMed] [Google Scholar]
- 39.Ono A, Freed E O. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiernan R E, Ono A, Englund G, Freed E O. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown D A, Rose J K. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y M, Yu X F. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 44.Keller P, Simons K. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimpton J, Emerman M. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg C W, Cinek T, Hallett M B, Horejsi V, Morgan B P. J Cell Biol. 1995;131:669–677. doi: 10.1083/jcb.131.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, Resh M D. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Field K A, Holowka D, Baird B. J Biol Chem. 1997;272:4276–4280. doi: 10.1074/jbc.272.7.4276. [DOI] [PubMed] [Google Scholar]
- 50.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janes P W, Ley S C, Magee A I. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodal S K, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subtil A, Gaidarov I, Kobylarz K, Lampson M A, Keen J H, McGraw T E. Proc Natl Acad Sci USA. 1999;96:6775–6780. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marquardt M T, Phalen T, Kielian M. J Cell Biol. 1993;123:57–65. doi: 10.1083/jcb.123.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abrami L, van der Goot F G. J Cell Biol. 1999;147:175–184. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogt V M. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. Science. 1999;283:680–682. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.