Abstract
Background
The purpose of this study was to investigate the role of isocitrate dehydrogenase 1 (IDH1) expression on prognosis of patients with clear cell renal cell carcinoma (ccRCC) following nephrectomy.
Methods
We retrospectively enrolled 358 ccRCC patients undergoing nephrectomy in Renji Hospital. Clinicopathologic features, overall survival (OS) and recurrence-free survival (RFS) of ccRCC patents were all collected. IDH1 expression level was assessed by immunohistochemistry and its association with clinicopathologic features and outcomes were also evaluated. Kaplan-Meier method with the log-rank test was applied to compare survival curves. Multivariate cox regression models were applied to analyze the prognostic value of each factor on OS and RFS of ccRCC patients. Moreover, two nomograms with factors selected by multivariate analysis were constructed to evaluate the prognosis of ccRCC patients, and the calibration plots were built to assess the predictive accuracy of nomograms.
Results
Our data indicated that IDH1 expression level was down-regulated in ccRCC tissues, and it negatively correlated with tumor Fuhrman grade (p = 0.025). Low IDH1 expression was associated with worse OS and RFS for cccRCC patients (OS, p = 0.004; RFS, p = 0.03). In addition, IDH1 could significantly stratify patients’ OS and RFS in intermediate/high risk patients (UISS score ≥ 4) (p = 0.049 and p = 0.004, respectively). Furthermore, incorporating IDH1 with other prognostic factors could predict ccRCC patients’ OS and RFS (OS, c-index = 0.779; RFS, c-index = 0.798) and perform better than TNM and SSIGN system.
Conclusions
Low IDH1 expression level might be an adverse prognostic biomarker for clinical outcomes of ccRCC patients, and two nomograms with IDH1 are potential effective prognostic models for ccRCC.
Keywords: Clear-cell renal cell carcinoma, Isocitrate dehydrogenase 1, Prognosis, Overall survival, Recurrence-free survival
Background
Renal cell carcinoma (RCC) is the most common type of malignant tumor in kidney, which represents the sixth most common cancer in men and the tenth most common cancer in women [1]. Clear cell RCC (ccRCC) is the prominent histological subtype of RCC, which accounts for about 80% to 90% of all RCC patients [2]. However, despite nephrectomy may cure the majority of localized RCC, about one third of the patients undergo local recurrence or distant metastasis after nephrectomy [3]. Currently, several clinical or pathological outcome prediction systems have already been established to evaluate the outcomes of patients with RCC, such as the University of California Los Angeles integrated staging system (UISS), the Mayo clinic stage, size, grade, and necrosis score (SSIGN), and the TNM stage, Fuhrman grade, Eastern Cooperative Oncology Group performance status (ECOG PS) [4–6]. However, due to the heterogeneity of molecular phenotype, there is a long way to go to predict the clinical outcomes of ccRCC [7, 8]. Therefore, new prognosis prediction systems with high accuracy are imperative needed for patients with ccRCC.
Altered cellular metabolism in cancers was observed for many years [9]. Despite the presence of enough oxygen, cancer cells still show high levels of glycolysis, which means the altered cell metabolic regulation plays an important role in tumorigenesis [10]. Isocitrate dehydrogenases (IDHs) are comprised of three members: IDH1, IDH2 and IDH3 [11]. IDH1 mainly catalyzes the conversion of isocitrate to alpha-ketoglutarate (aKG), and provides sufficient NADPH and regulates the biosynthesis of cholesterol and fatty acid [12]. Recently, many studies showed that IDH1 is mutated in various human cancers, especially in low-grade glioma [13]. The most common mutation site of IDH1 is the R132H, which acquires the ability to catalyze the reduction of a-KG to 2-hydroxyglutarate (2-HG) [14]. Moreover, several studies indicated that the R132H mutation of IDH1 correlates with a favorable prognosis for patients with glioma and gastrointestinal cancer [15, 16]. Nevertheless, little research was conducted to investigate the relationship between wide-type IDH1 and tumors. Sun and colleagues reported that IDH1 was significantly higher in patients with non-small cell lung cancer than in healthy controls [17]. However, the exact role of IDH1 in ccRCC, especially patients with high risk, is remains unknown.
The aim of this study was to reveal the clinical role of IDH1 in ccRCC patients. Moreover, two prognostic nomograms integrating IDH1 expression and clinical factors were established to predict the outcomes and may guide clinical decisions making for ccRCC patients.
Methods
Patients
A total of 358 ccRCC patients who underwent nephrectomy in the Department of Urology, Renji Hospital, during Jan 2005 and Dec 2008, were retrospectively included in our study. Patients’ clinicopathologic information, including gender, age, TNM stage, Fuhrman grade, tumor size, presence of tumor necrosis and SSIGN were collected. TNM stage was determined by one senior urologist according to the 2010 AJCC (the American Joint Committee on Cancer) TNM classification [18]. Fuhrman grade and tumor necrosis were determined according to the 2014 EAU guidelines and 2012 ISUP (International Society of Urological Pathology) consensus [19]. The SSIGN score were applied to stratify patient risks, just as previously reported [6]. OS and RFS were calculated as previously reported [20]. The last follow-up was at Apr. 30, 2016.
Tissue microarray (TMA) and immunohistochemistry
Tissue microarray (TMA) was constructed based on all the patients’ tumor samples, and were stained according to the standard method [21]. Primary antibody against human IDH1 (diluted:1:300; Abcam) was applied for immunohistochemistry (IHC) staining. Visualization reagent (Nikon eclipse Ti Microscope) was used to record the results. Tissue staining intensity and percentage were scored. Five areas of IDH1 positive stains were selected to estimate at low (× 40) or high (× 200) magnification. The intensity was scored as: 0 (negative), 1 (weak), 2 (moderate), and 3 (strong); the percentage of cells was scored into the following four categories: 1 (0–25%), 2 (26–50%), 3 (51–75%), or 4 (> 75%), and comprehensive score = staining percentage × intensity [22]. Finally, the expression level of IDH1 was defined into low expression and high expression according to the comprehensive score (cutoff value = 6) [23].
Statistical analysis
SPSS Statistics 22.0 and R software were used to performed statistics. Akaike information criterion (AIC) and Harrell’s concordance index (C-index) value were used to assess the predictive accuracy and sufficiency of different models [24]. Nomograms and calibration plots were generated by R software with “rms” package, and parameters included in nomograms were based on statistical significance in multivariate analysis. Two-sided p value < 0.05 was considered as statistically significant.
Results
Patients’ characteristics and association with IDH1 expression
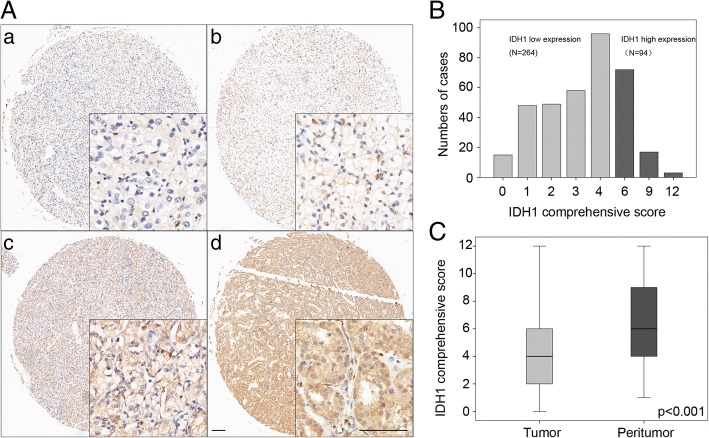
Patients’ clinical characteristics are shown in Table 1. To evaluate the expression level of IDH1 in ccRCC tumor tissues, IHC staining analysis was used in TMAs among all the ccRCC patients. IDH1 expression was predominantly found in the cytoplasm of tumor cells, with variable staining intensity in different specimens (Fig. 1A and B). Moreover, IDH1 was found to be down-regulated in ccRCC tumor tissues, compared with peritumor tissues (Fig. 1C, p < 0.001). After IHC evaluation, study cohort was divided into two groups: low IDH1 expression group (264 patients) and high IDH1 group (94 patients). The association between patients’ clinicopathological features and the IDH1 expression level are summarized in Table 1. IDH1 expression level in tumor tissues was negatively correlated with tumor Fuhrman grade (p = 0.025), whereas other patients’ clinicopathological features, including gender, age, TNM stage, T stage, N stage, M stage, tumor size, necrosis and SSIGN score, were not correlated with IDH1 expression (p > 0.05).
Table 1.
Association of IDH1 expression with clinicopathological characteristics in ccRCC patients
| Characteristics | Patients | IDH1 expression | |||
|---|---|---|---|---|---|
| n | % | Low | High | p-value | |
| All patients | 358 | 100 | 264 | 94 | |
| Gender | |||||
| Male | 254 | 70.9 | 189 | 65 | 0.654a |
| Female | 104 | 29.1 | 75 | 29 | |
| Age(years) | |||||
| ≤ 55 | 178 | 49.7 | 125 | 53 | 0.132a |
| > 55 | 180 | 50.3 | 139 | 41 | |
| TNM stage | |||||
| I | 286 | 79.9 | 208 | 78 | 0.384a |
| II + III + IV | 72 | 20.1 | 56 | 16 | |
| T stage | |||||
| T1 + T2 | 344 | 96.1 | 253 | 91 | 1.000b |
| T3 + T4 | 14 | 3.9 | 11 | 3 | |
| N stage | |||||
| N0 | 349 | 97.5 | 256 | 93 | 0.455b |
| N1 | 9 | 2.5 | 8 | 1 | |
| M stage | |||||
| M0 | 352 | 983 | 260 | 92 | 0.655b |
| M1 | 6 | 1.7 | 4 | 2 | |
| Fuhrman grade | |||||
| I + II | 297 | 83.0 | 212 | 85 | 0.025a* |
| III + IV | 61 | 17.0 | 52 | 9 | |
| Tumor size(cm) | |||||
| ≤ 5 | 186 | 52.0 | 134 | 52 | 0.447a |
| > 5 | 172 | 48.0 | 130 | 42 | |
| Necrosis | |||||
| Absent | 296 | 82.7 | 216 | 80 | 0.469a |
| Present | 62 | 17.3 | 48 | 14 | |
| SSIGN score | |||||
| < 4 | 266 | 74.3 | 193 | 73 | 0.386a |
| ≥ 4 | 92 | 25.7 | 71 | 21 | |
aChi-square test
bFisher’s exact test
*p < 0.05 indicates a significant association among the variables
Fig. 1.
A Representative immunohistochemical staining of IDH1 in ccRCC tissues. A (a). Negative staining of IDH1 in ccRCC, score 0. A (b). Weak staining of IDH1 in ccRCC, score 3. A (c). Moderate staining of IDH1 in ccRCC, score 6. A (d). Strong staining of IDH1 in ccRCC, score 12. Bar: 100 μm. B Frequency distribution of tumoral IDH1 immunohistochemistry integrated comprehensive score in 358 ccRCC samples. C Comprehensive score of IDH1 expression level in tumor and peritumor tissues. p value < 0.05 was regarded as statistically significant
Low IDH1 expression is associated with poor prognosis
Survival curves were performed to compare OS and RFS of ccRCC patients according to IDH1 expression. Interesting, patients with low IDH1 expression showed poorer OS (p = 0.004) and RFS (p = 0.03) than patients in high IDH1 expression group (Fig. 2a and d). Moreover, patients was classify into two groups: 0–3 (low-risk group), ≥4 (intermediate- and high-risk group) by SSIGN score. As is shown in Fig. 2c and f, IDH1 expression level showed as an adverse prognostic factor for both OS and RFS in intermediated- and high-risk groups (OS, p = 0.049; RFS, p = 0.004), while in the low-risk group it did not meet statistical significance.
Fig. 2.
Tumoral IDH1 expression stratified by SSIGN score and related Kaplan-Meier analyses of patient overall survival (OS) and recurrence free survival (RFS). a OS of all ccRCC patients according to IDH1 expression; (b-c) OS of patients in different SSIGN risk groups according to IDH1 expression; (d) RFS of all ccRCC patients according to IDH1 expression; (e-f) RFS of patients in different SSIGN risk groups according to IDH1 expression. p value < 0.05 was regarded as statistically significant
Further, multivariate analysis was used to assess whether IDH1 expression level is an independent prognostic factor for outcomes of ccRCC patients. As presented in Table 2, low IDH1 expression in tumor was an unfavorable independent predictor for OS and RFS of ccRCC patients (OS, HR, 0.500, 95% CI, 0.253–0.987, p = 0.046; RFS, HR, 0.463, 95% CI, 0.233–0.922, p = 0.028). In addition, TNM stage, N stage, Fuhrman grade, and tumor size were also considered as independent predictors for both OS and RFS.
Table 2.
Multivariate cox regression analysis for overall survival and recurrence-free survival in ccRCC patientsᅟ
| Variables | Overall survival | Recurrence-free survival | ||||
|---|---|---|---|---|---|---|
| HR | (95% CI) | p* | HR | (95% CI) | p* | |
| IDH1 in cancer tissues | ||||||
| Low | 1 | 1 | ||||
| High | 0.500 | 0.253–0.987 | 0.046* | 0.463 | 0.233–0.922 | 0.028* |
| TNM stage | ||||||
| I | 1 | 1 | ||||
| II + III + IV | 3.866 | 1.185–12.610 | 0.025* | 6.453 | 1.645–25.309 | 0.007* |
| T stage | ||||||
| T1 + T2 | 1 | 1 | ||||
| T3 + T4 | 2.991 | 1.197–7.473 | 0.019* | 2.076 | 0.901–4.785 | 0.086 |
| N stage | ||||||
| N0 | 1 | 1 | ||||
| N1 | 3.884 | 1.418–10.640 | 0.008* | 3.319 | 1.253–8.792 | 0.016* |
| Fuhrman grade | ||||||
| I + II | 1 | 1 | ||||
| III + IV | 2.255 | 1.314–3.869 | 0.003* | 2.069 | 1.203–3.557 | 0.009* |
| Tumor size(cm) | ||||||
| ≤ 4 | 1 | 1 | ||||
| > 4 | 3.261 | 1.584–6.712 | 0.001* | 3.482 | 1.658–7.313 | 0.001* |
HR hazard ratio, 95% CI 95% confidence interval, *p < 0.05 was considered statistically significant
Comparison of the predictive abilities between IDH1 expression and other prognostic factors
To investigate the predictive ability of IDH1 expression in ccRCC, IDH1 was compared with several conventional ccRCC prognosis predictors, such as SSIGN outcome algorithm, TNM stage, T stage, N stage, Fuhrman grade and tumor sizes. As showed in Table 3, the C-indexes of IDH1 were 0.566 and 0.579 for OS and RFS respectively, which were higher than N stage (0.563and 0.55) and lower than SSIGN outcome algorithm, TNM stage, N stage, Fuhrman grade and tumor sizes. Moreover, the C-index of those models was increased when IDH1 expression factor was replenished both for OS and RFS, suggesting the expression level of IDH1 has a good predictive ability for ccRCC outcomes. Besides, the AIC value of all factors integrated model was lower than SSIGN outcome algorithm, which means the model integrated with all factors performed better than SSIGN to predict ccRCC prognosis.
Table 3.
Comparison of the predictive accuracies of prognostic factorsᅟ
| Model | Overall Survival (N = 285) | Recurrence free survival (N = 285) | ||
|---|---|---|---|---|
| C-Index | AIC | C-Index | AIC | |
| IDH1 | 0.566 | 798.932 | 0.579 | 833.571 |
| TNM stage | 0.694 | 746.2725 | 0.724 | 779.0914 |
| TNM stage+IDH1 | 0.722 | 738.6321 | 0.749 | 769.0795 |
| T stage | 0.686 | 750.3168 | ||
| T stage+IDH1 | 0.716 | 738.1326 | ||
| N stage | 0.563 | 777.9406 | 0.55 | 819.1828 |
| N stage+IDH1 | 0.62 | 772.8811 | 0.615 | 813.6082 |
| Fuhrman grade | 0.663 | 781.1461 | 0.656 | 818.4013 |
| Fuhrman grade + IDH1 | 0.678 | 777.1598 | 0.68 | 813.3012 |
| Tumor size | 0.683 | 766.2329 | 0.703 | 793.0312 |
| Tumor size+IDH1 | 0.712 | 759.9979 | 0.738 | 785.7259 |
| Nomogram | 0.779 | 716.8505 | 0.798 | 749.2201 |
| SSIGN | 0.748 | 732.8423 | 0.77 | 764.8564 |
Prognostic nomogram of ccRCC patients
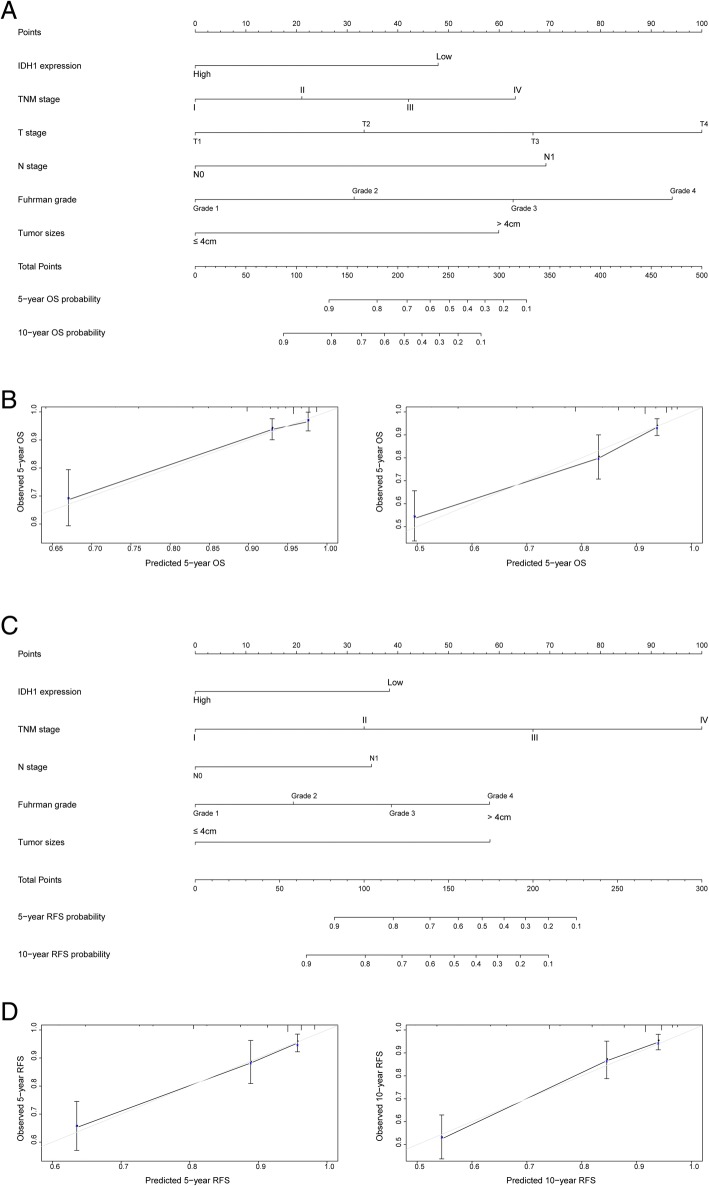
In order to use IDH1 as a prognostic factor, two nomograms were constructed to predict prognosis of ccRCC patients, via integrating the independent risk factors from multivariate analysis [25]. In addition, calibration plots of the nomograms for OS and RFS at 5 and 10 year revealed the good consistency between the actual and predicted survival of patients (Fig. 3). Our results suggest that these nomograms may be reliable prognostic models for ccRCC patients.
Fig. 3.
Nomograms and calibration plots for prognosis of OS and RFS in patients with ccRCC. a Nomogram for predicting the OS of ccRCC patients; (b) The calibration plots for overall survival at 5 and 10 years. c Nomogram for predicting the RFS of ccRCC patients; (d) The calibration plots for overall survival at 5 and 10 years
Discussion
IDH1, a NADP-dependent enzyme which involves in the control of oxidative cellular damage, was identified as a tumor suppressor since its inactivation plays a vital role in tumorigenesis [26, 27]. Although IDH1 have consistently been reported take part in the genesis of many cancers, the correlation between IDH1 expression and ccRCC outcomes remains unclear. In this study, we detected IDH1 was mainly expressed in the cytoplasm of ccRCC tumor cells, and low expression level of IDH1 in tumor correlated with an adverse outcomes of ccRCC, especially in patients with high SSIGN scores. Besides, IDH1 expression level was a risk factor of OS and RFS for ccRCC patients. Furthermore, by integrating with several factors from multivariate analysis, IDH1 expression in tumors could enhance the prognostic accuracy of these factors, including SSIGN outcome algorithm, TNM stage, N stage, Fuhrman grade and tumor sizes. Moreover, two prognostic nomograms models were constructed to predict OS and RFS of ccRCC patients, via integrating IDH1 with significant factors based on the multivariate analysis. Further c-index analysis indicated two prognostic nomograms had better prognostic capability compared with SSIGN prognostic model.
In mammal cells, three types of IDHs were discovered, including IDH1, IDH2, and IDH3. Although these three enzymes show the similar enzymatic reaction, they have different functions in different places. IDH1 performs its enzymatic activity mainly in the cytosol and the peroxisomes [28]. In recent years, mutations in IDH1 gene were reported in various tumors, which confer IDH1 the new enzymatic function of catalyzing α-KG to R-2-hydroxyglutarate (2-HG) [29]. The role of IDH1 gene mutation in esophageal squamous cell carcinoma [30], glioma [31] and acute myeloid leukemia (AML) have been successively reported [32]. The expression level of IDH1-R132H, which is the most common mutation type, correlates with poor outcomes in several cancers, such as gastrointestinal cancer and nonenhancing diffuse glioma [16, 33]. Besides, IDH1 wide-type expression was also explored in tumors. Overexpression of IDH1 was reported in non-small cell lung (NSCL) cancer, both in plasma and tumor tissues [17, 34]. However, our study revealed that low expression of IDH1 was associated with adverse ccRCC patients’ prognosis, which was inconsistent with that in NSCL cancer. Similarly, down-regulation of IDH1 was detected in kidney cancer [35], which means the loss of IDH1 expression might contribute ccRCC genesis.
The mechanism of low IDH1 participates in progression of cancers has not been well elaborated. IDH1 not only plays an important role on the biosynthesis of central metabolites in the tricarboxylic acid (TCA) cycle, but represents the major pathway for cellular NADPH generation in cells, a vital factor that regulates the amount of glutathione (GSH) and thioredoxin in cells [36]. GSH and thioredoxin are the main members of antioxidative systems, which could protect cells from oxidative damages by eliminating the reactive oxygen species (ROS) [37, 38]. Thus, loss the enzymatic function of IDH1 in tumor cells could impair detoxification procedure, which may result in DNA damages and genes mutations [39]. Besides, IDH could also regulate cellular apoptosis, and facilitate the development of a modifier of cancer chemotherapy [40].
Although the clinical significance of IDH1 in ccRCC was revealed, several limitations still exist in our study. Firstly, it is a retrospective cohort in a single center with limited patients, especially for the patients with advanced ccRCC. Therefore, patients from multicentric cohort are necessary to confirm our findings. Secondly, our study was based on the IHC staining and scored by two pathologists, and it would be more persuasive by measuring the mRNA or protein expression level of IDH1. Another problem is that, the time of patients follow up is not long enough to illustrate the clinical significance of IDH1, and longer follow up time is in need. Thus, more researches, especially mechanism studies, are needed to further understand the role of IDH1 in ccRCC progression.
Conclusions
In conclusion, we revealed low expression of IDH1 was significantly associated with poor ccRCC prognosis and could be used as a prognostic parameter to predict ccRCC patients’ outcomes. We also constructed two nomograms for ccRCC patients, which might be adapted into risk scoring systems and guide in clinical decisions for ccRCC patients.
Acknowledgements
We thank the Department of Pathology of Renji hospital for analyzing the immunohistochemistry results.
Funding
This work was supported by Shanghai Committee of Science and Technology (13ZR1425100). All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AIC
Akaike information criterion
- AJCC
American joint committee on cancer
- ccRCC
Clear cell renal cell carcinoma
- CI
Confidence interval
- C-index
Concordance index
- ECOG PS
Eastern Cooperative Oncology Group performance status
- HR
Hazard ratio
- IDH1
Isocitrate dehydrogenase 1
- ISUP
International society of urological pathology
- NADPH
Nicotinamide adenine dinucleotide phosphate
- OS
Overall survival
- RFS
Recurrence-free survival
- SSIGN
Mayo clinic stage, size, grade and necrosis
- TCA
tricarboxylic acid
- TMA
Tissue microarray
- UISS
University of California Integrated Staging System
Authors’ contributions
PL for analysis and interpretation of data, statistical analysis and drafting of the manuscript; JW for acquisition of data, technical and material support; JZ for study concept and design, analysis and interpretation of data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This investigation was approved by the Ethics and Research Committees of Renji Hospital, Shanghai Jiao Tong University School of Medicine, and was conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines. All specimens were obtained from patients with written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pingcuo Laba, Email: 3237195990@qq.com.
Jianfeng Wang, Email: wjf_0509@sina.com.
Jin Zhang, Email: med-zhangjin@vip.sina.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Logan JE, Rampersaud EN, Sonn GA, Chamie K, Belldegrun AS, Pantuck AJ, et al. Systemic therapy for metastatic renal cell carcinoma: a review and update. Rev Urol. 2012;14:65–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 5.Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED, et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97:1663–1671. doi: 10.1002/cncr.11234. [DOI] [PubMed] [Google Scholar]
- 6.Ficarra V, Novara G, Galfano A, Brunelli M, Cavalleri S, Martignoni G, et al. The ‘Stage, size, grade and Necrosis’ score is more accurate than the University of California los Angeles Integrated Staging System for predicting cancer-specific survival in patients with clear cell renal cell carcinoma. BJU Int. 2009;103:165–170. doi: 10.1111/j.1464-410X.2008.07901.x. [DOI] [PubMed] [Google Scholar]
- 7.Harris WB. Biomarkers for evaluating racial disparities in clinical outcome in patients with renal cell carcinoma. Mol Asp Med. 2015;45:47–54. doi: 10.1016/j.mam.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Qu Y, Liu L, Wang J, Xi W, Xia Y, Bai Q, et al. Dot1l expression predicts adverse postoperative prognosis of patients with clear-cell renal cell carcinoma. Oncotarget. 2016;7:84775–84784. doi: 10.18632/oncotarget.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209:211–215. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernster L, Navazio F. The cytoplasmic distribution of isocitric dehydrogenases. Exp Cell Res. 1956;11:483–486. doi: 10.1016/0014-4827(56)90124-0. [DOI] [PubMed] [Google Scholar]
- 12.Shechter I, Dai P, Huo L, Guan G. IDH1 gene transcription is sterol regulated and activated by SREBP-1a and SREBP-2 in human hepatoma HepG2 cells: evidence that IDH1 may regulate lipogenesis in hepatic cells. J Lipid Res. 2003;44:2169–2180. doi: 10.1194/jlr.M300285-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penard-Lacronique V, Bernard OA. IDH1, histone methylation, and so forth. Cancer Cell. 2016;30:501. doi: 10.1016/j.ccell.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Kessler J, Guttler A, Wichmann H, Rot S, Kappler M, Bache M, et al. IDH1(R132H) mutation causes a less aggressive phenotype and radiosensitizes human malignant glioma cells independent of the oxygenation status. Radiother Oncol. 2015;116:381–387. doi: 10.1016/j.radonc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Huang J, Huang F, Jin Q, Zhu H, Wang X, et al. Decreased expression of IDH1-R132H correlates with poor survival in gastrointestinal cancer. Oncotarget. 2016;7:73638–50. [DOI] [PMC free article] [PubMed]
- 17.Sun N, Chen Z, Tan F, Zhang B, Yao R, Zhou C, et al. Isocitrate dehydrogenase 1 is a novel plasma biomarker for the diagnosis of non-small cell lung cancer. Clin Cancer Res. 2013;19:5136–5145. doi: 10.1158/1078-0432.CCR-13-0046. [DOI] [PubMed] [Google Scholar]
- 18.Edge SB, Compton CC. The American joint committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 19.Delahunt B, Srigley JR, Egevad L, Montironi R. International Society of Urological Pathology grading and other prognostic factors for renal neoplasia. Eur Urol. 2014;66:795–798. doi: 10.1016/j.eururo.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Margonis GA, Gani F, Buettner S, Amini N, Sasaki K, Andreatos N, et al. Rates and patterns of recurrence after curative intent resection for gallbladder cancer: a multi-institution analysis from the US Extra-hepatic Biliary Malignancy Consortium. HPB (Oxford) 2016;18:872–878. doi: 10.1016/j.hpb.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matos LL, Trufelli DC, de Matos MG, da Silva Pinhal MA. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010;5:9–20. doi: 10.4137/BMI.S2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Xu Y, Zhu L, Zou Y, Kong W, Dong B, et al. High expression of Stearoyl-CoA Desaturase 1 predicts poor prognosis in patients with clear-cell renal cell carcinoma. PLoS One. 2016;11:e0166231. doi: 10.1371/journal.pone.0166231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hang J, Hu H, Huang J, Han T, Zhuo M, Zhou Y, et al. Sp1 and COX2 expression is positively correlated with a poor prognosis in pancreatic ductal adenocarcinoma. Oncotarget. 2016;7:28207–28217. doi: 10.18632/oncotarget.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng L, Xu L, Yang Y, Zhou L, Chang Y, Shi T, et al. High expression of FUT3 is linked to poor prognosis in clear cell renal cell carcinoma. Oncotarget. 2017;8:61036–61047. doi: 10.18632/oncotarget.17717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y, Liu L, Xia Y, Wang J, Xi W, Bai Q, et al. Low CCL17 expression associates with unfavorable postoperative prognosis of patients with clear cell renal cell carcinoma. BMC Cancer. 2017;17:117. doi: 10.1186/s12885-017-3106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohba S, Hirose Y. Biological Significance of Mutant Isocitrate Dehydrogenase 1 and 2 in Gliomagenesis. Neurol Med Chir (Tokyo) 2016;56:170–179. doi: 10.2176/nmc.ra.2015-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Ling ZQ. Role of isocitrate dehydrogenase 1/2 (IDH 1/2) gene mutations in human tumors. Histol Histopathol. 2015;30:1155–1160. doi: 10.14670/HH-11-643. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Li Q, Wang C, Xu W, Han L, Liu Y, et al. Prognostic and diagnostic potential of isocitrate dehydrogenase 1 in esophageal squamous cell carcinoma. Oncotarget. 2016;7:86148–86160. doi: 10.18632/oncotarget.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waitkus MS, Diplas BH, Yan H. Isocitrate dehydrogenase mutations in gliomas. Neuro-Oncology. 2016;18:16–26. doi: 10.1093/neuonc/nov136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassereddine S, Lap CJ, Haroun F, Tabbara I. The role of mutant IDH1 and IDH2 inhibitors in the treatment of acute myeloid leukemia. Ann Hematol. 2017;96:1983–1991. doi: 10.1007/s00277-017-3161-0. [DOI] [PubMed] [Google Scholar]
- 33.Olar A, Raghunathan A, Albarracin CT, Aldape KD, Cahill DP, 3rd, Powell SZ, et al. Absence of IDH1-R132H mutation predicts rapid progression of nonenhancing diffuse glioma in older adults. Ann Diagn Pathol. 2012;16:161–170. doi: 10.1016/j.anndiagpath.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Tan F, Jiang Y, Sun N, Chen Z, Lv Y, Shao K, et al. Identification of isocitrate dehydrogenase 1 as a potential diagnostic and prognostic biomarker for non-small cell lung cancer by proteomic analysis. Mol Cell Proteomics. 2012;11:M111.008821. doi: 10.1074/mcp.M111.008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26:103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogdanovic E. IDH1, lipid metabolism and cancer: shedding new light on old ideas. Biochim Biophys Acta. 2015;1850:1781–1785. doi: 10.1016/j.bbagen.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Lu J, Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 38.Lu SC. Regulation of glutathione synthesis. Mol Asp Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 40.Lee SM, Park SY, Shin SW, Kil IS, Yang ES, Park JW. Silencing of cytosolic NADP(+)-dependent isocitrate dehydrogenase by small interfering RNA enhances the sensitivity of HeLa cells toward staurosporine. Free Radic Res. 2009;43:165–173. doi: 10.1080/10715760802653661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.