Abstract
Association of felis catus papillomaviruses (FcaPVs) with feline squamous cell carcinoma (SCC) has been reported worldwide, while there is limited information about FcaPVs in Asia. In this study, 21 feline SCC biopsy samples from cats in Japan were analyzed by PCR with PV consensus primers and type-specific primers for FcaPV type 2 (FcaPV-2), FcaPV-3 and FcaPV-4 designed in this study. Sequence analysis revealed that one sample was FcaPV-3, and two were FcaPV-4. In both FcaPV-4 positive samples, 334th tryptophan in L1 ORF was deleted compared with the reference sequence. Moreover, immunohistochemistry showed that p16 protein was positive in both FcaPV-4 detected samples. This study would contribute to the molecular epidemiological and pathological understanding of FcaPV in Japan.
Keywords: FcaPV type 3, FcaPV type 4, Feline squamous cell carcinoma, Japan, type specific primer
In domestic cats, five types of papillomaviruses (PVs), designated as FcaPV (Felis catus papillomavirus), have been reported in association with cutaneous neoplastic lesions. Each genotype is classified by the sequence of L1 ORF, the major capsid protein of PV. FcaPV type 1 (FcaPV-1) DNA was detected from flat sessile cutaneous hyperkeratotic lesions [21], oral papillomas [12] and oral SCCs [17]. FcaPV-2 have been detected most frequently from feline cutaneous neoplastic diseases [10], including viral plaques [13], Bowenoid in situ carcinomas (BISCs) [8], and cutaneous SCCs [18]. The partial genome of FcaPV-3, previously described as FdPV-MY2, was detected in viral plaque [13], and also in SCCs [14]. Later on, the full genome of FcaPV-3 was identified from a multiple BISCs case of a domestic cat [15]. FcaPV-4 full genome DNA was identified from feline oral cavity with ulcerative gingivitis [4], and its partial genome DNA, named as FdPV-MY3 was reported from SCC cases in earlier study [14]. FcaPV-5 was recently identified from a feline viral plaque case [16]. These studies show that FcaPVs may associate with the cutaneous and mucosal neoplastic diseases in domestic cats, although the pathogenesis of FcaPVs are still uncertain.
SCC accounts for 15% of cutaneous neoplastic diseases in cats [11], and share the most in feline oral neoplastic diseases [20]. However, the definitive cause of feline SCC is not well understood. The exposure to ultraviolet (UV) is considered as one of the environmental risk factors in developing cutaneous SCCs [3] while a recent study revealed that FcaPV DNA was detected more in UV-protected SCCs than in UV-exposed cases [18], suggesting the involvement of PV in the pathogenesis of feline SCCs. FcaPVs have been detected from feline SCC lesions in Europe [5, 10], Oceania [18], and North America [19]. From those studies, FcaPV-2 have been detected the most in feline SCCs, and it is also suggested to infect the skin of cats asymptomatically [22]. In Asian countries including Japan, there is limited information about FcaPVs. Based on these backgrounds described above, we sought to detect FcaPV DNA from feline SCC lesions. Although PCR methods are commonly used, primers may affect the specificity and sensitivity for the amplification of PV DNA [18]. Therefore, in addition to the previously established consensus PV primers, the type specific primers designed in this study were used for detecting PVs from feline SCCs.
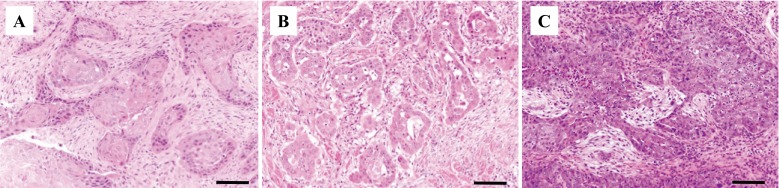
Twenty-one feline SCC biopsy samples, collected in Japan between 2013 and 2015, were fixed in 10% formalin and routinely embedded in paraffin and stained with hematoxylin and eosin for histopathological examination. Histopathological diagnoses of SCC were made on the consensus of two veterinary pathologists (certified by the Japanese College of Veterinary Pathologists) at the Department of Veterinary Pathology, the University of Tokyo. Clinical and histopathological findings of feline SCC cases are summarized in Table 1. The mean age at the diagnosis was 12.3 ± 2.3 (mean ± standard deviation) years with no sex predilection. In the present study, seven out of 21 cases were oral, nine were cutaneous, and the rest of the SCC case lesions were observed in other locations. Histopathological examination revealed the invasive growth of squamous epithelial tissue of the skin or the mucosa (Fig. 1). Abundant mitotic figures and severe nuclear atypia were observed. Nuclear inclusion bodies, indicative of papilloma virus infection, were not observed in any of the cases.
Table 1. Descriptions of the feline SCC samples, detection of FcaPV and the results of p16 immunohistochemical analysis.
| Sample ID | Breed | Age, in years | Sex | Anatomical site | PV genotype (GenBank accession number) |
p16 Immunohistochemistry |
|---|---|---|---|---|---|---|
| 13-060 | Mixed-breed | 13 | FXa) | Skin | NDd) | − |
| 13-0107 | Mixed-breed | 12 | FX | Oral (submandible) | ND | NAg) |
| 13-136 | Mixed-breed | 14 | FX | Skin | FcaPV-4 (LC333412)e) | + |
| 13-0153 | Mixed-breed | 12 | MXb) | Skin (eyelid, left) | FcaPV-3 (LC333418)f) | NA |
| 13-0297 | Mixed-breed | 12 | MX | Oral (lower gingia, right) | ND | − |
| 13-0317 | American shorthair | 15 | MX | Oral (tongue) | ND | − |
| 13-848 | Mixed-breed | 10 | FX | Auditory canal (left) | ND | − |
| 13-882 | Mixed-breed | 16 | FX | Oral (tongue) | ND | NA |
| 13-944 | Scottish fold | 10 | MX | Oral (tongue) | ND | − |
| 14-778 | Mixed-breed | 13 | Mc) | Skin (upper jaw, right) | ND | − |
| 14-1018 | Mixed-breed | 16 | FX | Auditory canal (left) | ND | − |
| 14-1110 | American shorthair | 10 | MX | Skin (forelimb, digit, right) | FcaPV-4 (LC333413) | + |
| 15-291 | Mixed-breed | 10 | FX | Oral (upper gingiva) | ND | − |
| 15-358 | Mixed-breed | 13 | FX | Skin (auditory canal, left) | ND | − |
| 15-498 | Mixed-breed | 9 | MX | Skin (external ear, left) | ND | − |
| 15-528 | Mixed-breed | 16 | MX | Skin (sublingual, right) | ND | − |
| 15-577 | Mixed-breed | 15 | FX | Oral (upper gingiva,right) | ND | − |
| 15-615 | Mixed-breed | 10 | MX | Esophagus | ND | − |
| 15-637 | Mixed-breed | 11 | FX | Skin (submandible) | ND | − |
| 5741-2013 | Mixed-breed | 12 | MX | Anus | ND | + |
| 6997-2015 | Mixed-breed | 9 | M | Oral (buccal mucosa) | ND | NA |
FcaPV genotype confirmed by sequence analysis are also shown for PV positive samples. a) Female (spayed); b) Male (castrated); c) Male; d) Not detected; e) Felis catus papillomavirus type 4; f) Felis catus papillomavirus type 3; g) Not available.
Fig. 1.
Representative histopathological findings of feline squamous cell carcinoma. Atypical squamous cells invade the fibrous tissue. (A) Case number 14-1110, FcaPV4-positive. (B) Case number 13-0153, FcaPV3-positive. (C) Case number 14-0778, FcaPV negative. Bars, 100 µm.
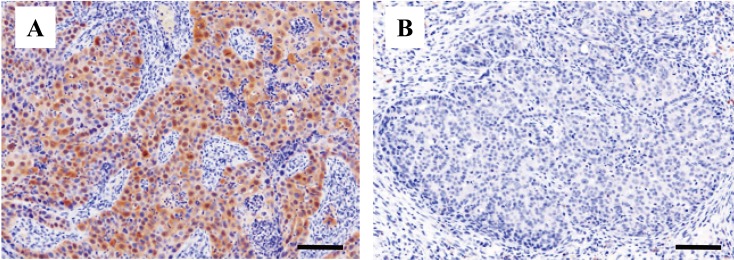
Immunohistochemistry (IHC) was conducted to detect the PV antigen and p16 protein, with mouse anti-PV (clone BPV-1/1H8 +CAMVIR, Abcam, Cambridge, U.K.) and mouse anti-CDKN2A/p16INK4a (Becton Dickinson Co., Franklin Lakes, NJ, U.S.A.) monoclonal antibodies, respectively. Horseradish perioxidase labeled detection kit (Envision + system, Dako Japan, Kyoto, Japan) was applied, and antigen-antibody complex was visualized by the chromogen treatment with 3,3′-diaminobenzidine (DAB). Sections were counterstained with Mayer’ hematoxylin. A section of cutaneous bovine papilloma, positive for bovine papillomavirus type-2, was used as the positive control for anti-PV antibody. For the negative control, the primary antibodies were substituted with tris-buffered saline. Positive staining of more than 10% of the neoplastic tissue was considered to be p16-positive. Immunohistochemical analysis for p16 showed diffuse positive staining in the neoplastic tissue of sample 13-136, 14-1110 (Fig. 2) and 5741-2013 (Table 1). For the immunohistochemical analysis for PV antigen, positive staining in the nucleus of epithelial cells in the section of bovine fibropapilloma was observed, while no nuclear-positive staining was detected in the feline SCC samples (data not shown). Some of the paraffin blocks, including FcaPV-3 positive sample (13-0153) were not available for IHC due to insufficient amount of paraffin blocks.
Fig. 2.
Immunohistochemical analysis against p16 protein in feline squamous cell carcinoma. (A) Intense positive p16 staining in the basal cell membrane (FcaPV4-positive, case number 14-1110). (B) Negative for p16 (FcaPV-negative, case number 14-0778). Bars, 100 μm.
DNA from 21 formalin-fixed paraffin-embedded tissue samples were extracted with Qiagen DNA FFPE Tissue kit (QIAGEN, Hilden, Germany) and the success in DNA extraction of all samples were confirmed by PCR using feline beta-actin primer pair, ACTB [7] (Table 2). To detect the PV genome, two consensus primer pairs, MY09/11 [9] and CP4/5 [23] (Table 2) were used. From two samples (Sample ID: 13-0153 and 14-1110), papillomaviral DNA was detected with the CP4/5 consensus primer pair, while no positive results were obtained with MY09/11 primer (data not shown). Accordind to the criteria of International Committee on Taxonomy of Viruses (ICTV), PVs are genotyped based on the sequence similarity of L1 ORF. Primer pair, CP4/5 is to detect the sequence of E1 ORF, so we have additionally designed type-specific primers for FcaPV-2, FcaPV-3 and FcaPV-4 L1 gene (Table 2) based on the reference sequence (GenBank accession number EU796884.1, NC_021472.1 and NC_022373.1 respectively) with GENETYX Ver.11 software (GENETYX Corp., Tokyo, Japan). Water in place of DNA template was added to the PCR mixture for the negative control. For the positive control of FcaPV-2 L1 primer, extracted DNA from a feline BISC case was applied. For FcaPV-3 and FcaPV-4 L1 primers, positive results for each FcaPV genotype was confirmed in this study (Table 1). PCR reactions were carried with the respective annealing temperature listed in Table 2, and each result was confirmed by electrophoresis. For the PCR positive samples, products were cloned and sequenced. Sequence analysis was performed using GENETYX Ver.11 software, and BLAST tool of the National Center for Biotechnology Information (NCBI) was used for the sequence similarity analysis. With FcaPV-3 type-specific primer, sample 13-0153 showed 99% sequence similarity with the reference FcaPV-3 (NC_021472.1). Sample 14-1110 also showed 99% sequence similarity with the reference FcaPV-4 (NC_022373.1) with FcaPV-4 type specific primer. Additionally, one sample (13-136) was detected with FcaPV-4 type-specific primer (Table 1), showing 95% sequence similarity with the reference FcaPV-4 (NC_022373.1). None of the samples were positive with FcaPV-2 L1 type specific primer (data not shown). The nucleotide sequences of the FcaPV-3 and FcaPV-4 identified in this study were deposited in GenBank (Table 1).
Table 2. The primer information used in this study.
| Primer pair | Alignment 5′→3′ | Target gene (location) | Amplicon length (bp) | Tm (°C) | Reference | |
|---|---|---|---|---|---|---|
| ACTB | Forward CAA CCG TGA GAA GAT GAC TCA GA | Feline beta actin | 410 | 54 | Kessler et al., 2009 | |
| Reverse CCC AGA GTC CAT GAC AAT AAC A | ||||||
| MY09/11 | Forward GCM CAG GGW CAT AAY AAT GG | PV L1 (Consensus) | approximately 450 | 55 | Manos et al., 1989 | |
| Reverse CGT CCM ARR GGA WAC TGA TC | ||||||
| CP4/5 | Forward ATG GTA CAR TGG GCA TWT GA | PV E1 (Consensus) | approximately 450 | 49 | Tieben et al., 1994 | |
| Reverse GAG GYT GCA ACC AAA AMT GRC T | ||||||
| FcaPV-2 L1 | Forward CGC AAG GAC AGA ATA ATG GTA TTT GCT | L1 | (7130–7156 nt) | 419 | 58 | This study |
| Reverse AAG ACG ATC CGA GAT GTC AAC AT | (7548–7525 nt) | |||||
| FcaPV-3 L1 | Forward TCT GGT AAT CAG TAT AGG GTG TTC AGA GT | L1 | (5868–5876 nt) | 473 | 59 | This study |
| Reverse ATT TCT AAA GGC ACC CCT GAT TTG TCT T | (6340–6312 nt) | |||||
| FcaPV-4 L1 | Forward CTT TGG TAA CCA GCG ATT CC | L1 | (6571–6590 nt) | 476 | 52 | This study |
| Reverse CAA TCT ATC CTT CAA GTC CAC TAC | (7046–7022 nt) | |||||
The name of the primer pairs, the target region of PV ORF, expected size of the PCR product, annealing temperature (Tm), and the reference information are shown respectively.
Sequence analysis revealed that two FcaPV-4 samples detected in this study showed a single deletion of tryptophan (W) in the position of 334th amino acid in the reference FcaPV-4 L1 sequence. To investigate whether the deletion of 334th tryptophan is limited to FcaPV-4, the amino acid sequences of the reference FcaPVs from genotype 1 to 5 (available on GenBank) were aligned. Interestingly, the 334th tryptophan was found only in the reference FcaPV-4 (NC_022373.1). In order to examine whether the deletion affects the structure of L1 protein, the translated amino acid sequence of FcaPV-4 samples, 13-136 and 14-110 was submitted to SWISS-MODEL (https://swissmodel.expasy.org/interactive). As results, no significant change of the structure of L1 was found. Further studies are required to figure out the significance of the 334th tryptophan.
In Table 1, the anatomical locations of the SCC lesion are summarized. Nine cases out of 21 were cutaneous, and eight cases were oral SCC. All three FcaPV-positive samples were detected from cutaneous SCCs (Table 1), which was consistent with the previous report that PV-associated SCCs in cats are common in cutaneous cases than oral SCCs [17].
In the present study, IHC for detecting p16 protein was conducted, as positive staining for p16 is one of the observations indicative of PV-infection in feline SCC [18]. Since PVs are recognized to asymptomatically infect the epidermis of cats [22], immunostaining to detect p16 within the lesions are preferable to support the involvement of PV to the lesion development. As expected, both of the FcaPV-4 positive samples, 13-136 and 14-1110 (Fig. 2) were immunopositive for p16 (Table 1). One sample, 5742-2013 was also positive for p16 while neither FcaPV-2, −3 and −4 was detected by PCR. This result suggests that non-PV factors or infection of other PV genotypes may be present. For the immunostaining of PV antigen, none of the feline SCC samples showed nuclear-positive staining. This result was not unexpected, because capsid antigen of PV is frequently disappeared during the malignant progression [1], and the antibodies used in this study (monoclonal BPV-1/1H8) react to a major capsid protein.
This study confirmed the detection of FcaPV-3 and FcaPV-4 by conventional PCR. As shown in Table 1, one additional sample, 13-136 was detected with the FcaPV-4 type-specific primer, but not by the consensus primer. Several mutations in sample 13-136 were observed within the sequence amplified with FcaPV-4 L1 type specific primer, assuming that additional mutations may be present in other genomic regions. As mutations in the primer attachment site can affect the primer specificity, this may be one of the reasons why PV-genome could not be detected with the consensus primers. Designing the PV-type specific primers are considered to be advantageous for detecting the target PV-genotype, while it may have difficulties in identifying the unknown PV genotypes [6]. In addition to FcaPV type-specific primers, designing the FcaPV consensus primers may be preferable for detecting the unknown FcaPV genotype.
From the previous studies in other geographical regions, FcaPV-2 has been reported to be found in feline SCC most frequently [10, 18, 19]. On the other hand, FcaPV-2 was not detected in this study. From the study on human papillomavirus (HPV), the distribution of high risk HPV type in cervical cancer differed depending on the geographical region [2]. Although the sample size of this study is small, the geographical difference may explain the discrepancies of dominated type of FcaPV found in SCCs.
To further understand the pathogenicity of FcaPVs and the aetiology of feline SCCs, concert effort is required including larger scale of PV molecular-epidemiological studies to determine the geographical FcaPV genotype distributions, pathological studies to determine the pathogenicity of FcaPVs for the SCC development, and clinical observation studies to investigate the malignance between FcaPV-positive and negative feline SCCs.
REFERENCES
- 1.Azzimonti B., Hertel L., Aluffi P., Pia F., Monga G., Zocchi M., Landolfo S., Gariglio M.1999. Demonstration of multiple HPV types in laryngeal premalignant lesions using polymerase chain reaction and immunohistochemistry. J. Med. Virol. 59: 110–116. doi: [DOI] [PubMed] [Google Scholar]
- 2.Crosbie E. J., Einstein M. H., Franceschi S., Kitchener H. C.2013. Human papillomavirus and cervical cancer. Lancet 382: 889–899. doi: 10.1016/S0140-6736(13)60022-7 [DOI] [PubMed] [Google Scholar]
- 3.Dorn C. R., Taylor D. O., Schneider R.1971. Sunlight exposure and risk of developing cutaneous and oral squamous cell carcinomas in white cats. J. Natl. Cancer Inst. 46: 1073–1078. [PubMed] [Google Scholar]
- 4.Dunowska M., Munday J. S., Laurie R. E., Hills S. F. K.2014. Genomic characterisation of Felis catus papillomavirus 4, a novel papillomavirus detected in the oral cavity of a domestic cat. Virus Genes 48: 111–119. doi: 10.1007/s11262-013-1002-3 [DOI] [PubMed] [Google Scholar]
- 5.Geisseler M., Lange C. E., Favrot C., Fischer N., Ackermann M., Tobler K.2016. Geno- and seroprevalence of Felis domesticus Papillomavirus type 2 (FdPV2) in dermatologically healthy cats. BMC Vet. Res. 2: 1–10. doi: 10.1186/s12917-016-0776-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haga T., Dong J., Zhu W., Burk R. D.2013. The many unknown aspects of bovine papillomavirus diversity, infection and pathogenesis. Vet. J. 197: 122–123. doi: 10.1016/j.tvjl.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 7.Kessler Y., Helfer-Hungerbuehler A. K., Cattori V., Meli M. L., Zellweger B., Ossent P., Riond B., Reusch C. E., Lutz H., Hofmann-Lehmann R.2009. Quantitative TaqMan real-time PCR assays for gene expression normalisation in feline tissues. BMC Mol. Biol. 10: 1–14. doi: 10.1186/1471-2199-10-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange C. E., Tobler K., Markau T., Alhaidari Z., Bornand V., Sto R.2009. Sequence and classification of FdPV2, a papillomavirus isolated from feline Bowenoid in situ carcinomas. Vet. Microbiol. 137: 60–65. [DOI] [PubMed] [Google Scholar]
- 9.Manos M. M., Ting Y., Wright D. K., Lewis A. J., Broker T. R., Wolinsky S. M.1989. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cell 7: 209–214. [Google Scholar]
- 10.Mazzei M., Forzan M., Carlucci V., Anfossi A. G., Alberti A., Albanese F., Binanti D., Millanta F., Baroncini L., Pirone A., Abramo F.2017. A study of multiple Felis catus in cat skin lesions in Italy by quantitative PCR. J. Feline Med. Surg. doi: 10.1177/1098612x17732255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller M. A., Nelson S. L., Turk J. R., Pace L. W., Brown T. P., Shaw D. P., Fischer J. R., Gosser H. S.1991. Cutaneous neoplasia in 340 cats. Vet. Pathol. 28: 389–395. doi: 10.1177/030098589102800506 [DOI] [PubMed] [Google Scholar]
- 12.Munday J. S., Fairley R. A., Mills H., Kiupel M., Vaatstra B. L.2015. Oral Papillomas Associated With Felis catus Papillomavirus Type 1 in 2 Domestic Cats. Vet. Pathol. 52: 1187–1190. doi: 10.1177/0300985814565133 [DOI] [PubMed] [Google Scholar]
- 13.Munday J. S., Peters-Kennedy J.2010. Consistent detection of Felis domesticus papillomavirus 2 DNA sequences within feline viral plaques. J. Vet. Diagn. Invest. 22: 946–949. doi: 10.1177/104063871002200615 [DOI] [PubMed] [Google Scholar]
- 14.Munday J. S., French A. F., Gibson I. R., Knight C. G.2013. The presence of p16 CDKN2A protein immunostaining within feline nasal planum squamous cell carcinomas is associated with an increased survival time and the presence of papillomaviral DNA. Vet. Pathol. 50: 269–273. doi: 10.1177/0300985812452582 [DOI] [PubMed] [Google Scholar]
- 15.Munday J. S., Dunowska M., Hills S. F., Laurie R. E.2013. Genomic characterization of Felis catus papillomavirus-3: a novel papillomavirus detected in a feline Bowenoid in situ carcinoma. Vet. Microbiol. 165: 319–325. doi: 10.1016/j.vetmic.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 16.Munday J. S., Dittmer K. E., Thomson N. A., Hills S. F., Laurie R. E.2017. Genomic characterisation of Felis catus papillomavirus type 5 with proposed classification within a new papillomavirus genus. Vet. Microbiol. 207: 50–55. doi: 10.1016/j.vetmic.2017.05.032 [DOI] [PubMed] [Google Scholar]
- 17.Munday J. S., French A. F.2015. Felis catus papillomavirus types 1 and 4 are rarely present in neoplastic and inflammatory oral lesions of cats. Res. Vet. Sci. 100: 220–222. doi: 10.1016/j.rvsc.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 18.Munday J. S., Gibson I., French A. F.2011. Papillomaviral DNA and increased p16CDKN2A protein are frequently present within feline cutaneous squamous cell carcinomas in ultraviolet-protected skin. Vet. Dermatol. 22: 360–366. doi: 10.1111/j.1365-3164.2011.00958.x [DOI] [PubMed] [Google Scholar]
- 19.O’Neill S. H., Newkirk K. M., Anis E. A., Brahmbhatt R., Frank L. A., Kania S. A.2011. Detection of human papillomavirus DNA in feline premalignant and invasive squamous cell carcinoma. Vet. Dermatol. 22: 68–74. doi: 10.1111/j.1365-3164.2010.00912.x [DOI] [PubMed] [Google Scholar]
- 20.Stebbins K. E., Morse C. C., Goldschmidt M. H.1989. Feline oral neoplasia: a ten-year survey. Vet. Pathol. 26: 121–128. doi: 10.1177/030098588902600204 [DOI] [PubMed] [Google Scholar]
- 21.Tachezy R., Duson G., Rector A., Jenson A. B., Sundberg J. P., Van Ranst M.2002. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology 301: 313–321. doi: 10.1006/viro.2002.1566 [DOI] [PubMed] [Google Scholar]
- 22.Thomson N. A., Dunowska M., Munday J. S.2015. The use of quantitative PCR to detect Felis catus papillomavirus type 2 DNA from a high proportion of queens and their kittens. Vet. Microbiol. 175: 211–217. doi: 10.1016/j.vetmic.2014.11.028 [DOI] [PubMed] [Google Scholar]
- 23.Tieben L. M., Berkhout R. J., Smits H. L., Bouwes Bavinck J. N., Vermeer B. J., Bruijn J. A., Van der Woude F. J., Ter Schegget J.1994. Detection of epidermodysplasia verruciformis-like human papillomavirus types in malignant and premalignant skin lesions of renal transplant recipients. Br. J. Dermatol. 131: 226–230. doi: 10.1111/j.1365-2133.1994.tb08496.x [DOI] [PubMed] [Google Scholar]