Abstract
Influenza (flu) D virus, a possible causative agent of bovine respiratory disease, is genetically classified into three clusters: D/OK-, D/660-, and D/Japan-lineages. To evaluate antigenic heterogeneity among these clusters, we compared antibody titers to each lineage virus using bovine sera collected over time following virus infection. Antibody titers to D/Japan-lineage virus rose rapidly in the acute phase of infection, and were 4 times higher than those to the other clustered viruses. In the later phase of infection, titers to D/Japan-lineage virus were equivalent to those to D/OK-lineage virus, and still higher than those to D/660-lineage virus. These results suggest the existence of common and lineage-specific antigenic epitopes in the hemagglutinin-esterase-fusion protein of flu D viruses.
Keywords: antigenicity, cattle, hemagglutinin-esterase-fusion protein, influenza D virus, serology
Influenza (flu) D virus was first isolated from pigs with respiratory illness in Oklahoma, U.S.A., in 2011 [7, 8]. Epidemiological analyses have indicated that cattle are the major reservoir of this virus [4, 7], and that flu D is potentially involved in the bovine respiratory disease complex [14, 18]. The high morbidity and mortality of this disease in feedlot cattle are due in part to co-infection with several viruses and bacteria. Flu D virus has also been detected in cattle and pigs with respiratory disease (and in some healthy cattle), small ruminants, camelids, and horses in several countries, which suggests that the virus is globally distributed in several animal species [1, 3, 6, 10, 11, 13, 15, 17, 19, 20].
In Japan, we detected flu D virus infection in a dairy cattle herd located in Ibaraki Prefecture in 2016 [15]. This was accomplished by assaying the seroconversion of virus-specific antibodies in paired serum samples collected before and after the onset of the symptoms. We also detected the viral genome in nasal swab samples using RT-PCR, thus identifying the first Japan flu D strain, D/bovine/Ibaraki/7768/2016 (D/Ibaraki). Phylogenetic analyses have revealed three clusters of the hemagglutinin-esterase-fusion (HEF) gene among flu D viruses: D/swine/Oklahoma/1334/2011 (D/OK)-, D/bovine/Oklahoma/660/2013 (D/660)-, and D/Ibaraki-lineages [15]. Interestingly, sera collected from D/Ibaraki-infected cows were found to react to the D/OK-lineage virus with higher hemagglutination-inhibition (HI) titers than to D/660-lineage virus, even though the HEF gene of D/Ibaraki is phylogenetically distant from that of the D/OK-lineage cluster. Although antigenic differences in the HEF protein have been identified between the D/OK- and D/660-lineage viruses using serology [2], comparisons have not yet been made between D/Ibaraki- and other lineage viruses.
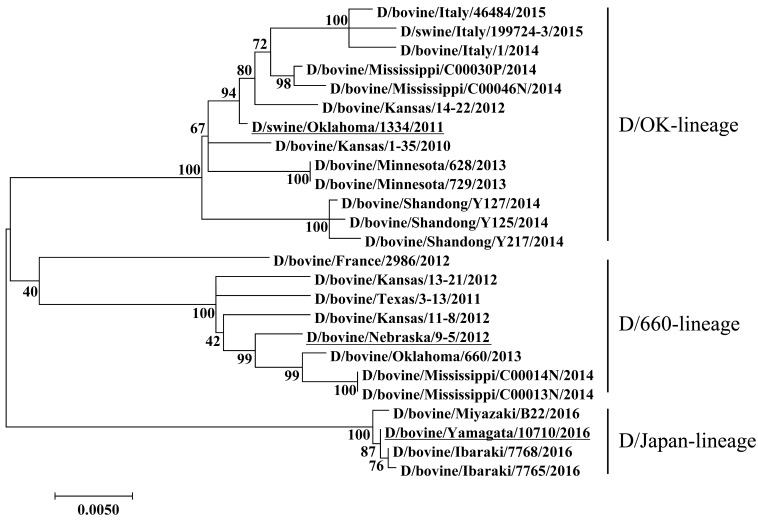
Recently, we successfully isolated a Japan strain (D/bovine/Yamagata/10710/2016; D/Yamagata) from a cow with respiratory illness in Yamagata Prefecture [16], which showed extremely high sequence homology to D/Ibaraki; identities of HEF were 99.8 and 100% at nucleotide and amino acid levels, respectively. To confirm that D/Yamagata is included in D/Ibaraki-lineage, we generated a phylogenetic tree based on the nucleotide sequence of the HEF gene from all reported Japan strains by maximum-likelihood analysis with ClustalW (MEGA7 software) [12]. This analysis confirmed that these strains form an independent cluster (termed as D/Japan-lineage) from the other two clusters (Fig. 1). These data strongly suggest homologous antigenicity of D/Japan-lineage virus HEFs. In this study, to evaluate antigenic heterogeneity among flu D viruses, we utilized three strains from different clusters as antigens. Serological testing was performed against serum samples collected from cattle at multiple time points following D/Ibaraki infection.
Fig. 1.
Phylogenetic tree of the HEF segment of flu D viruses at the nucleotide level. Maximum-likelihood analysis in combination with 500 bootstrap replicates was used to derive tree based on nucleotide sequences of the HEF segment. Bootstrap values are shown above and to the left of the major nodes. Scale bars indicate the number of substitutions per site.Viruses used in this study are underlined.
To gain insight into antigenic heterogeneity among flu D viruses, we collected sera at several time intervals from cattle in the Ibaraki herd described above, in which the flu D outbreak had occurred [15]. The Committee of Animal Experiments at the Graduate School of Agricultural and Life Sciences, University of Tokyo, permitted our work (Permit Number: P12-652). The HI test was performed to detect anti-flu D virus antibody in the bovine sera, as per a protocol from the World Health Organization manual on animal influenza diagnosis and surveillance [24]. The samples were treated with receptor-destroying enzyme (RDE (II); Denka Seiken, Tokyo, Japan) at 37°C for 16 hr, followed by heat-inactivation at 56°C for 30 min. Serially diluted samples were then allowed to react to the flu D virus (4 hemagglutinin (HA) unit) for 30 min at room temperature, followed by incubation with a 0.6% suspension of turkey red blood cells (Nippon Bio-test Laboratories, Saitama, Japan) for 30 min at room temperature before reading the result. The HI titer of each sample was expressed as the reciprocal of the highest sample dilution that completely inhibited HA. The samples showing an HI titer ≥40 were considered as positive. The samples showing an HI titer ≥40 were considered as positive. An HI titer of 40 has been commonly used as the threshold for a seropositive result in flu D virus surveillance [4, 19]. This threshold authenticates reaction specificity in HI test [22].
We used three flu D strains, D/OK [8] (as D/OK-lineage virus), D/bovine/Nebraska/9-5/2013 [2] (D/NE; as D/660-lineage virus), and D/Yamagata [16] (as D/Japan-lineage virus) as antigens for the HI test. D/OK and D/NE were kindly provided by Dr. Benjamin Hause (Kansas State University). To prepare the viruses for the assay, they were inoculated in swine testicle cell culture (ATCC CRL-1746), which was grown at 37°C in Dulbecco’s modified Eagle’s medium (WAKO, Osaka, Japan) with 10% fetal calf serum and antibiotics, and then propagated in serum-free medium in the presence of TPCK-trypsin (1 µg/ml) (Worthington, Lakewood, NJ, U.S.A.).
As described in our previous report [15], sera were firstly collected from all cattle (n=25) in the Ibaraki herd on January 8, 2016 and tested for the HI antibody. Eight cattle were found to be antibody-positive with geometric mean titers of 67.2 to D/OK, 23.8 to D/NE, and 95.1 to D/Yamagata (Table 1; Group 1). Conversely, 17 cattle were antibody-negative to all three viruses (Group 2), indicating that Group 1 had a history of past flu D virus infection whereas Group 2 was immunologically naïve. Around January 15, a newly-introduced flu D virus (D/Ibaraki) infection occurred in this herd, and subsequently 4 cows in Group 2 exhibited respiratory illness. Sera were again collected from all cows on February 3, and naïve cows of Group 2 exhibited seroconversion of anti-flu D virus antibodies with geometric mean titers of 86.8 to D/OK, 76.8 to D/NE and 347.2 to D/Yamagata. The titers obtained to D/Yamagata, which were more than 4 times higher than the titers to the other foreign strains, confirmed heterologous antigenicity between D/Japan and the other clusters. The identical amino acid sequences of the D/Ibaraki and D/Yamagata HEFs correspond to the homologous antibody responses for these two strains. The serum collected on February 3, around two weeks after the primary infection of the D/Ibaraki, should contain virus-specific IgM antibody over IgG antibody, suggesting that the IgM antibody may react more strongly to the lineage-specific antigenic epitope than the IgG antibody. On the other hand, slight increases in HI titers were observed in the Group 1 cows on February 3, indicating that the animals contained antibodies induced by past infection with an undefined Japan strain. This provides additional evidence of homologous antigenicity among D/Japan-lineage viruses.
Table 1. Antibody kinetics against three different flu D lineage viruses in a herd exposed to D/Ibaraki.
| Cattle | Virus antigen | Geometric mean HI titer (± SD) | |||
|---|---|---|---|---|---|
| Serum sample | |||||
| 2016.1.8. | 2016.2.3. | 2016.7.4. | 2016.12.15. | ||
| Group 1 (N=8) | D/OK | 67.2 (± 0.7) | 95.1 (± 0.6) | 123.4 (± 0.5) | 95.1 (± 0.6) |
| D/NE | 23.8 (± 0.5) | 47.6 (± 0.5) | 56.6 (± 0.5) | 47.6 (± 0.5) | |
| D/Yamagata | 95.1 (± 0.7) | 160.0 (± 0.5) | 123.4 (± 0.5) | 146.7 (± 0.5) | |
| Group 2 (N=17) | D/OK | <40 (± 0.0) | 86.8 (± 0.9) | 57.7 (± 0.7) | 57.7 (± 0.8) |
| D/NE | <40 (± 0.0) | 76.8 (± 0.6) | <40 (± 0.5) | <40 (± 0.5) | |
| D/Yamagata | <40 (± 0.0) | 347.2 (± 0.9) | 60.1 (± 0.9) | 76.8 (± 0.8) | |
The animals were divided into two groups depending on antibody positivity of sera collected on January 8, 2016; Group 1 (N=8) includes antibody-positive cows, whereas Group 2 (N=17) includes antibody-negative cows. After flu D virus infection occurred around January 15 in this herd, sera were collected from all cows on the date shown, and tested for HI antibodies to three lineage viruses (D/OK, D/NE and D/Yamagata); geometric mean titers and standard deviations (SD) were shown.
To further compare antibody titers to each virus from the different clusters, sera were collected from all cows on follow-up sampling dates after the acute phase of virus infection and tested for HI antibodies (Table 1). In cows of Group 2, although lower titers were observed for all viruses compared to those in the acute phase, those to D/Yamagata decreased more relative to the other viruses on July 4, reaching titers equivalent to D/OK. Some samples that had tested positive to D/NE on February 3 became negative on July 4. HI titers to each virus did not change on December 15 compared to July 4. These results suggest a strong antibody response to the D/Japan-lineage-specific antigenic epitope of HEF in the acute phase, although antibody titers to this epitope were not maintained in the later phase of infection. No changes in titers to each lineage virus were observed for the period after February 3 in the cows of Group 1. This antibody kinetics indicated the presence of common and robust antigenic epitopes in HEF among flu D strains, authenticating a single serotype of flu D viruses.
A previous study demonstrated heterologous antigenicity between D/OK- and D/660-lineage viruses using rabbit polyclonal antisera to each virus by HI test, and homologous antigenicity among the same lineage viruses [2]. Furthermore, the antigenic difference was determined to be caused by a difference in the amino acid at position 212 in the HEF; K for D/OK- and R for D/660-lineage viruses [2]. Notably, the D/Japan-lineage viruses uniformly possess S at this position [15]. Additionally, one putative N-glycosylation sequon was missing at positions 249–251 in the HEF of D/Japan-lineage strains. These molecular characteristics may contribute to the generation of D/Japan-lineage-specific antigenic epitopes in HEF, although further analyses using monoclonal antibodies are required.
Collectively, epidemiological and metagenomic data [21], as well as an experimental infection study [5], have indicated that the flu D virus is a causative agent of bovine respiratory disease. Several vaccines against bovine respiratory agents such as bovine herpesvirus 1, bovine respiratory syncytial virus, bovine parainfluenza virus 3, bovine viral diarrhea virus, and bovine adenovirus are currently available in Japan. However, the protective effects of these vaccines are unsatisfactory in some cases of respiratory illness, suggesting that the cases may be associated with flu D virus infection. Therefore, vaccine development for flu D should be considered [9, 23]. The antigenic heterogeneity among all flu D virus HEFs shown in this study emphasizes that, when developing flu D vaccine in Japan, the D/Japan-lineage virus should be used as a seed virus to yield maximal vaccine efficacy.
Acknowledgments
We thank Dr. Benjamin Hause (Veterinary Diagnostic Laboratory, Kansas State University, U.S.A.) for the D/OK and D/NE viruses. T. H. is supported, in part, by a Grant-in-Aid for Scientific Research (A) (grant number: 26252048) and by Livestock Promotion Funds from the Japan Racing Association. S. M. is supported by a Grant-in-Aid for the Encouragement of Young Scientists (A) (grant number: 17H05042).
REFERENCES
- 1.Chiapponi C., Faccini S., De Mattia A., Baioni L., Barbieri I., Rosignoli C., Nigrelli A., Foni E.2016. Detection of influenza D virus among swine and cattle, Italy. Emerg. Infect. Dis. 22: 352–354. doi: 10.3201/eid2202.151439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collin E. A., Sheng Z., Lang Y., Ma W., Hause B. M., Li F.2015. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 89: 1036–1042. doi: 10.1128/JVI.02718-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ducatez M. F., Pelletier C., Meyer G.2015. Influenza D virus in cattle, France, 2011-2014. Emerg. Infect. Dis. 21: 368–371. doi: 10.3201/eid2102.141449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson L., Eckard L., Epperson W. B., Long L. P., Smith D., Huston C., Genova S., Webby R., Wan X. F.2015. Influenza D virus infection in Mississippi beef cattle. Virology 486: 28–34. doi: 10.1016/j.virol.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson L., Olivier A. K., Genova S., Epperson W. B., Smith D. R., Schneider L., Barton K., McCuan K., Webby R. J., Wan X. F.2016. Pathogenesis of influenza D virus in cattle. J. Virol. 90: 5636–5642. doi: 10.1128/JVI.03122-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn O., Gallagher C., Mooney J., Irvine C., Ducatez M., Hause B., McGrath G., Ryan E.2018. Influenza D virus in cattle, Ireland. Emerg. Infect. Dis. 24: 389–391. doi: 10.3201/eid2402.170759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hause B. M., Collin E. A., Liu R., Huang B., Sheng Z., Lu W., Wang D., Nelson E. A., Li F.2014. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio 5: e00031–e14. doi: 10.1128/mBio.00031-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hause B. M., Ducatez M., Collin E. A., Ran Z., Liu R., Sheng Z., Armien A., Kaplan B., Chakravarty S., Hoppe A. D., Webby R. J., Simonson R. R., Li F.2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 9: e1003176. doi: 10.1371/journal.ppat.1003176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hause B. M., Huntimer L., Falkenberg S., Henningson J., Lechtenberg K., Halbur T.2017. An inactivated influenza D virus vaccine partially protects cattle from respiratory disease caused by homologous challenge. Vet. Microbiol. 199: 47–53. doi: 10.1016/j.vetmic.2016.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horimoto T., Hiono T., Mekata H., Odagiri T., Lei Z., Kobayashi T., Norimine J., Inoshima Y., Hikono H., Murakami K., Sato R., Murakami H., Sakaguchi M., Ishii K., Ando T., Otomaru K., Ozawa M., Sakoda Y., Murakami S.2016. Nationwide distribution of bovine influenza D virus infection in Japan. PLoS One 11: e0163828. doi: 10.1371/journal.pone.0163828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang W. M., Wang S. C., Peng C., Yu J. M., Zhuang Q. Y., Hou G. Y., Liu S., Li J. P., Chen J. M.2014. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 49: 493–496. doi: 10.1007/s11262-014-1107-3 [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Stecher G., Tamura K.2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekata H., Yamamoto M., Hamabe S., Tanaka H., Omatsu T., Mizutani T., Hause B. M., Okabayashi T.2018. Molecular epidemiological survey and phylogenetic analysis of bovine influenza D virus in Japan. Transbound. Emerg. Dis. 65: e355–e360. doi: 10.1111/tbed.12765 [DOI] [PubMed] [Google Scholar]
- 14.Mitra N., Cernicchiaro N., Torres S., Li F., Hause B. M.2016. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 97: 1771–1784. doi: 10.1099/jgv.0.000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami S., Endoh M., Kobayashi T., Takenaka-Uema A., Chambers J. K., Uchida K., Nishihara M., Hause B., Horimoto T.2016. Influenza D virus infection in herd of cattle, Japan. Emerg. Infect. Dis. 22: 1517–1519. doi: 10.3201/eid2208.160362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatsu S., Murakami S., Shindo K., Horimoto T., Sagara H., Noda T., Kawaoka Y.2018. Influenza C and D viruses package eight organized ribonucleoprotein complexes. J. Virol. 92: e02084–e17. doi: 10.1128/JVI.02084-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedland H., Wollman J., Sreenivasan C., Quast M., Singrey A., Fawcett L., Christopher-Hennings J., Nelson E., Kaushik R. S., Wang D., Li F.2018. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 65: e148–e154. doi: 10.1111/zph.12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng T. F. F., Kondov N. O., Deng X., Van Eenennaam A., Neibergs H. L., Delwart E.2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 89: 5340–5349. doi: 10.1128/JVI.00064-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quast M., Sreenivasan C., Sexton G., Nedland H., Singrey A., Fawcett L., Miller G., Lauer D., Voss S., Pollock S., Cunha C. W., Christopher-Hennings J., Nelson E., Li F.2015. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 180: 281–285. doi: 10.1016/j.vetmic.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salem E., Cook E. A. J., Lbacha H. A., Oliva J., Awoume F., Aplogan G. L., Hymann E. C., Muloi D., Deem S. L., Alali S., Zouagui Z., Fèvre E. M., Meyer G., Ducatez M. F.2017. Serologic evidence for influenza C and D virus among ruminants and camelids, Africa, 1991–2015. Emerg. Infect. Dis. 23: 1556–1559. doi: 10.3201/eid2309.170342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su S., Fu X., Li G., Kerlin F., Veit M.2017. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 8: 1580–1591. doi: 10.1080/21505594.2017.1365216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trombetta C. M., Perini D., Mather S., Temperton N., Montomoli E.2014. Overview of serological techniques for influenza vaccine evaluation: past, present and future. Vaccines (Basel) 2: 707–734. doi: 10.3390/vaccines2040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan Y., Kang G., Sreenivasan C., Daharsh L., Zhang J., Fan W., Wang D., Moriyama H., Li F., Li Q.2018. A DNA vaccine expressing consensus hemagglutinin-esterase fusion protein protected guinea pigs from infection by two lineages of influenza D virus. J. Virol. 92: e00110-18. doi: 10.1128/JVI.00110-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization. Available: http://apps.who.int/iris/bitstream/10665/68026/1/WHO_CDS_CSR_NCS_2002.5.pdf [accessed on May 29, 2018].