Abstract
Background
Congenital anomalies of the kidney and urinary tract (CAKUT) are the most prevalent cause of kidney disease in the first three decades of life. Previous gene panel studies showed monogenic causation in up to 12% of patients with CAKUT.
Methods
We applied whole-exome sequencing to analyze the genotypes of individuals from 232 families with CAKUT, evaluating for mutations in single genes known to cause human CAKUT and genes known to cause CAKUT in mice. In consanguineous or multiplex families, we additionally performed a search for novel monogenic causes of CAKUT.
Results
In 29 families (13%), we detected a causative mutation in a known gene for isolated or syndromic CAKUT that sufficiently explained the patient’s CAKUT phenotype. In three families (1%), we detected a mutation in a gene reported to cause a phenocopy of CAKUT. In 15 of 155 families with isolated CAKUT, we detected deleterious mutations in syndromic CAKUT genes. Our additional search for novel monogenic causes of CAKUT in consanguineous and multiplex families revealed a potential single, novel monogenic CAKUT gene in 19 of 232 families (8%).
Conclusions
We identified monogenic mutations in a known human CAKUT gene or CAKUT phenocopy gene as the cause of disease in 14% of the CAKUT families in this study. Whole-exome sequencing provides an etiologic diagnosis in a high fraction of patients with CAKUT and will provide a new basis for the mechanistic understanding of CAKUT.
Keywords: Congenital Anomalies of the Kidney and Urinary Tract (CAKUT), Whole Exome Sequencing (WES), Vesico-ureteral Reflux (VUR), monogenic disease causation, renal developmental gene
Congenital anomalies of the kidney and urinary tract (CAKUT) constitute the most common cause of CKD in the first three decades of life.1,2 CAKUT can present as an isolated renal condition or as part of a clinical syndrome.3–7 Despite large differences in clinical manifestation, these conditions likely share a pathogenic origin in dysregulation of renal morphogenesis.8,9
We hypothesized that human CAKUT may be caused by mutations in distinct single monogenic genes. Previous supporting evidence for this hypothesis include (1) familial occurrence of CAKUT; (2) the presence of CAKUT as part of the phenotypic manifestation of known monogenic, multiorgan syndromes; (3) the presence of monogenic mouse models with CAKUT; (4) the congenital nature of CAKUT; and (5) the knowledge that specific master genes govern renal morphogenesis.4,10–12 To date, 40 monogenic causes for isolated CAKUT have been identified (Supplemental Table 1).5,6,12–37,39–41. We previously showed by gene panel sequencing that >10% of CAKUT were monogenic in origin,42 whereas another 2% of patients were explained by mutations in the Fraser complex of genes.33 With novel CAKUT gene discovery proceeding at an accelerating rate43,44 and considering that whole-exome sequencing (WES) is not limited to detection of a prespecified list of candidate genes, we hypothesize that, in >12% of patients with CAKUT, a monogenic cause can be detected by WES.
We and others have previously shown that a significant subset of patients with a clinical diagnosis of isolated CAKUT harbor mutations in known disease genes for syndromic forms of CAKUT (Supplemental Table 2).33,45,46 These patients did not exhibit syndromic CAKUT on clinical examination and are clinically indistinguishable from other patients with CAKUT.33,45,46 Two reasons were identified as possible explanations for this genotype-phenotype discrepancy. There may be mild extrarenal manifestations of the respective syndrome that are only unveiled after careful clinical re-evaluation after establishment of a molecular diagnosis.45,46 Alternatively, this broad phenotypic variability in the presence of mutations in syndromic disease genes can be due to an allelism of the underlying gene (Supplemental Figure 1).33 We and others have evaluated WES data from individuals with CAKUT; however the focus was often on specific subcategories of CAKUT.36,43,47–49 To date, only one publication has systemically evaluated WES in 62 CAKUT families.45
We attempted to quantify the prevalence of mutations in known CAKUT genes in a large cohort. On the basis of previous observations, we hypothesized that a significant proportion of individuals with a clinical diagnosis of isolated CAKUT will harbor disease-causing mutations in syndromic CAKUT genes as well as murine and novel “candidate” genes. We show that, in 14% of families with CAKUT, a likely pathogenic mutation in a known CAKUT gene or CAKUT phenocopy gene can be identified. Furthermore, WES facilitates the discovery of candidate variants for CAKUT as seen in 16% of families with CAKUT (Figure 1C-E).
Figure 1.
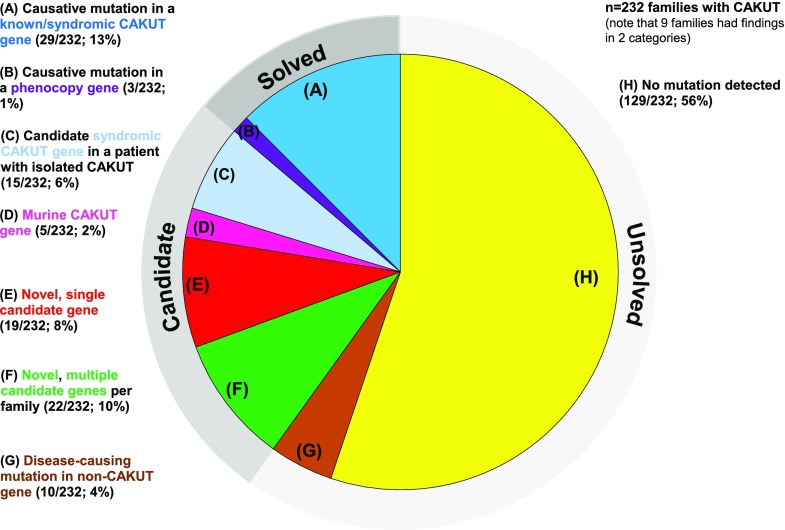
Number and percentage of 232 congenital anomalies of the kidney and urinary tract (CAKUT) families in which a causative mutation in a known monogenic CAKUT gene (14%) or a candidate gene(s) (16%) was detected by whole-exome sequencing. Blue color denotes that a mutation in a single causative gene was detected in a known isolated or syndromic CAKUT gene (dark blue), and purple color denotes that a mutation in a causative gene was known to phenocopy CAKUT (purple). Light blue denotes candidate mutations in a known syndromic CAKUT gene in families with isolated CAKUT (light blue). Pink was chosen if a candidate variant in a murine CAKUT gene was identified. Red was chosen if one potential novel CAKUT gene was detected in a family, or green was chosen if multiple novel candidate genes for CAKUT were detected in a family. (A) In 29 of 232 (13%) families with CAKUT (dark blue), a causative mutation was detected in one of 40 isolated CAKUT genes (Supplemental Table 1) or one of 179 known syndromic CAKUT genes (Supplemental Table 2). The individuals with mutations in a syndromic CAKUT gene exhibited the corresponding syndromic CAKUT phenotype. (B) In three of 232 (1%) families, a mutation was identified in a gene causing a kidney disease that may represent a phenocopy of CAKUT (purple; i.e., small kidneys of non-CAKUT origin). (C) In 15 of 232 (6%) families with predominantly isolated CAKUT, candidate mutations were detected in one of 179 syndromic CAKUT genes (light blue), indicating a “hypomorphic” effect of these mutations. (D) In five of 232 (2%) families, mutations in a known gene for murine CAKUT were identified (pink). (E) In 19 of 232 (8%) families, a single potential novel candidate gene for CAKUT was identified per family (red). (F) In 22 of 232 (9%) families, multiple potential novel candidate genes remained per family (green). (G) In ten of 232 (4%) families, we identified mutations in genes known to be causative of monogenic non-CAKUT diseases (brown). (H) In 129 of 232 (56%) families, no causative or candidate mutations were detected (yellow).
Methods
Human Subjects
The study was approved by the institutional review board of the University of Michigan and Boston Children’s Hospital as well as the institutional review boards of institutions where we have recruited families. From January 2010 to January 2017, patients with CAKUT were enrolled after obtaining informed consent. A total of 488 individuals (319 affected and 169 reportedly unaffected) from 232 different families were enrolled and had WES performed on DNA samples. All patients with CAKUT were referred to us by their pediatric nephrologist or urologist who made a clinical diagnosis of CAKUT on the basis of renal imaging studies. CAKUT was defined as demonstration of any abnormality of number, size, shape, or anatomic position of the kidneys or other parts of the urinary tract that included at least one of the following: renal agenesis, renal hypo-/dysplasia, multicystic dysplastic kidneys, hydronephrosis, ureteropelvic junction obstruction, hydroureter, vesicoureteral reflux, ectopic or horseshoe kidney, duplex collecting system, ureterovesical junction obstruction, epi-/hypospadias, posterior urethral valves, and cryptorchidism.
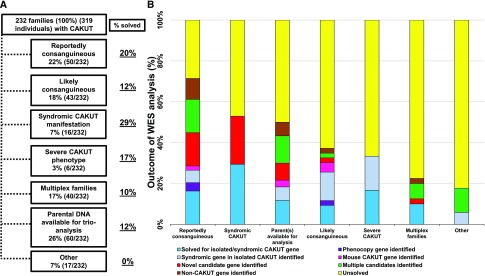
For evaluation using WES, families were divided into subgroups as follows: (1) reportedly consanguineous (50 of 232); (2) likely consanguineous (origin in a region with a high degree of remote consanguinity; 43 of 232); (3) syndromic manifestation of CAKUT (one or more extrarenal features; 16 of 232); (4) severe manifestation of CAKUT (i.e., unilateral renal agenesis or renal dysplasia; six of 232); (5) patients with multiplex cases of CAKUT (40 of 232); (6) parental DNA available for analysis (60 of 232); or (7) other (17 of 232) (Figure 2). Before being considered for WES, a selection of individuals with suggestive phenotypes were prescreened for mutations in the CAKUT genes EYA1, PAX2, HNF1B, GATA3, SIX1, and SIX5 using targeted sequencing approaches. In total, a causative mutation was identified in 70 families (78 individuals) after screening of 958 families (7.3%; 1111 affected individuals and 269 unaffected parents), and this information is not included in this study but is published elsewhere.21,33,42,50
Figure 2.
Inclusion criteria for 232 families to perform whole-exome sequencing (WES). (A) Individuals with congenital anomalies of the kidney and urinary tract (CAKUT) were prioritized for inclusion in WES on the basis of the following criteria: (1) reportedly consanguineous (50 of 232; 22%); (2) reportedly nonconsanguineous but origin in countries with increased rate of consanguinity and therefore, considered likely consanguineous (43 of 232; 18%); (3) syndromic CAKUT phenotype (16 of 232; 7%); (4) severe manifestation of CAKUT (renal agenesis or renal dysplasia; six of 232; 3%); (5) families with multiple affected family members (40 of 232; 17%); (6) DNA of additional family members available for a duo, trio, or quad analysis (60 of 232; 26%); and (7) other reasons to include in WES (e.g., family potentially related to a family to which the other criteria applied; 17 of 232; 7%). Outcome of WES analysis by “recruitment group.” (B) The seven recruitment groups for inclusion in CAKUT WES are sorted horizontally starting with the group with the lowest percentage of unsolved families and going to the group with the highest percentage unsolved. Each group is further subsorted into categories of identified genes. Categories are similar to those in Figure 1, and the colors used are the same as well: (1) solved for isolated or syndromic CAKUT gene (dark blue), (2) phenocopy gene (purple), (3) syndromic gene identified in patients with isolated CAKUT (light blue), (4) mouse CAKUT gene identified (pink), (5) single novel candidate gene identified (red), (6) multiple candidate genes identified (green), (7) non-CAKUT gene identified (brown), and (8) unsolved (yellow).
WES and Variant Calling
WES was performed as previously described.51 In brief, genomic DNA was isolated from blood lymphocyte or saliva samples and subjected to exome capture using Agilent SureSelect human exome capture arrays (Life Technologies) followed by next generation sequencing on the Illumina HighSeq sequencing platform. Sequence reads were mapped to the human reference genome assembly (NCBI build 37/hg19) using CLC Genomics Workbench (version 6.5.2) software (CLC Bio, Aarhus, Denmark). After alignment to the human reference genome, variants were filtered for most likely deleterious variants as previously described.52,53 Variants with minor allele frequencies >1% in the dbSNP (version 147) or the 1000 Genomes Project (1094 subjects of various ethnicities; May 2011 data release) databases were excluded, because they were unlikely to be deleterious. Synonymous and intronic variants that were not located within splice site regions were excluded. Kept variants, which included nonsynonymous variants and splice site variants, were then analyzed (Supplemental Figure 2).
Mutation Calling in Known Genetic Causes of Isolated Human CAKUT, Syndromic Human CAKUT, and Murine CAKUT Candidate Genes
We evaluated WES data for causative mutations in 40 monogenic genes for isolated CAKUT known at the time (Supplemental Table 1), 179 single-gene candidates for monogenic forms of known syndromic CAKUT (Supplemental Table 2), and 185 candidate genes for mutations in genes for murine CAKUT (Supplemental Table 3). Details on evaluation strategy are in Supplemental Figures 3 and 4. Remaining variants were ranked on the basis of their probable effect on the function of the encoded protein considering evolutionary conservation among orthologs across phylogeny using ENSEMBL Genome Browser and assembled using Clustal Omega as well as the web-based prediction programs PolyPhen-2, SIFT, and MutationTaster. Variant filtering on the basis of population frequency was performed using population databases (EVS server, ExAC, gnomAD, and 1000-genomes) to include only rare alleles (i.e., minor allele frequency <1%). Phenotype and functional aspects of each mutation/gene were discussed in a nephrogenetic panel with a minimum of five members for each of the 232 families before final candidate decisions were made (A.T.v.d.V, D.M.C, H.I., N.M., J.C., A.V., S.S., and F.H.) (Supplemental Figure 4). All variant calling was performed using our stringent a priori criteria (Supplemental Figure 4) along with the standards and guidelines set out by the American College of Medical Genetics.54
Remaining variants were confirmed in original patient DNA by Sanger sequencing. Whenever familial DNA (parents or siblings) was available, segregation analysis was performed. Although identification of copy number variants by WES is limited, WES data were analyzed using Conifer software to detect pathogenic copy number variants.
Targeted Search for Homozygously Mutated Novel Genetic Causes of CAKUT in Families with Significant Levels of Homozygosity
If no causative mutation was found in a monogenic cause of isolated, syndromic, or murine CAKUT and a family had significant levels of detected homozygosity (megabase pairs) after homozygosity mapping (≥60 megabase pairs (Mbp)), we proceeded to evaluate WES data for homozygous variants (Supplemental Figure 4). Homozygosity mapping data were generated from WES data using downstream processing of aligned BAM files using Picard and samtools.55 Single-nucleotide variant calling was performed using the Genome Analysis Toolkit (GATK),56 and the generated VCF file was subsequently used in the homozygosity mapper.57 All single heterozygous variants were excluded on the basis of an a priori recessive hypothesis. Remaining variants were ranked as described previously (Supplemental Figures 2 and 4).
Identification of Novel Genetic Causes of CAKUT by Familial Analysis (Duo, Trio, or Quad Analysis)
Data processing of FASTQs was performed by the Genomics Platform at the Broad Institute of Harvard and Massachusetts Institute of Technology (Broad Institute, Cambridge, MA). Single-nucleotide polymorphisms and insertions/deletions were jointly called across all samples using the GATK HaplotypeCaller. Default filters were applied to single-nucleotide polymorphisms and insertion/deletion calls using the GATK Variant Quality Score Recalibration approach. Lastly, the variants were annotated using Variant Effect Predictor.58 The variant call set was uploaded onto Seqr for analysis of the WES output.
Results
WES Identifies a Likely Pathogenic Monogenic Cause of CAKUT in 14% of Families with CAKUT
We performed WES in 232 families with CAKUT (319 affected individuals). Clinical characteristics for the 319 affected individuals are outlined in Supplemental Figure 5, Supplemental Table 5, and Table 1. In 14% (32 of 232) of CAKUT families, we identified mutations in 22 different monogenic genes known to cause isolated or syndromic CAKUT or phenocopies of CAKUT (32 different mutations in 22 genes) (Figure 1A and B, Table 2). Of the 32 different mutations identified in these 22 CAKUT genes, 16 of 32 (50%) are novel mutations not previously described in the literature. Specifically, we detected likely causative mutations in the following subgroup of CAKUT families.
Table 1.
Clinical characteristics of the 319 individuals (232 families) with congenital anomalies of the kidneys and urinary tract who were submitted for whole-exome sequencing analysis
| Patient Characteristics | Total Cohort | |
|---|---|---|
| n | Percentage | |
| Sex | ||
| Women | 129 | 40 |
| Men | 189 | 59 |
| Unknown | 1 | <1 |
| Total | 319 | 100 |
| Extrarenal manifestations | ||
| Yes | 79 | 25 |
| No | 240 | 75 |
| Total | 319 | 100 |
| Reported consanguinity | ||
| Yes | 59 | 18 |
| No | 260 | 82 |
| Total | 319 | 100 |
| Homozygosity on mapping ≥60 Mbp | ||
| Yes | 50 | 16 |
| No | 240 | 75 |
| Not enough SNPs to generate mapping | 29 | 9 |
| Total | 319 | 100 |
| CAKUT phenotype | ||
| Unilateral CAKUTa | 130 | 41 |
| Bilateral concordant CAKUTa | 111 | 35 |
| Bilateral discordant CAKUTa | 32 | 10 |
| Undefined CAKUT phenotype | 21 | 7 |
| Isolated PUV or epi-/hypospadias | 11 | 3 |
| PUV with an additional CAKUT phenotypes | 14 | 4 |
| Total | 319 | 100 |
SNP, single-nucleotide polymorphism; CAKUT, congenital anomalies of the kidney and urinary tract; PUV, posterior urethral value.
Supplemental Figure 5 and Supplemental Table 5 have a breakdown of the CAKUT pathologies in individuals with unilateral or bilateral pathology.
Table 2.
Information on identified mutations in congenital anomalies of the kidney and urinary tract genes known to cause isolated or syndromic congenital anomalies of the kidney and urinary tract or mutations in genes known to phenocopy congenital anomalies of the kidney and urinary tract and the corresponding clinical phenotype
| Family Identificationa | Gene | Mode of Transmission | Nucleotide Change | Amino Acid Change | State | Evolutionary Conservationb | PP2 SIFT MT | CADD Score | EVSc | gnomADc | ACMGd | HGMDe | Phenotypes | Segregation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A617f | SALL1g | Dominant | c.703G>A | p.Ala 235Thr | Het | Danio rerio | 0.782 Del. D.C. | 18 | / | 0/3/277166 | Likely pathogenic | DM | Townes Brock syndrome | Variant inherited from father (affection status unknown) |
| BL VUR R duplex | ||||||||||||||
| A1041h | SRGAP1g | Dominant | c.1993C>A | p.Pro 665Thr | Het | Caenorhabditis elegans | 0.309 Del. D.C. | 24 | / | / | Pathogenic | DM | L horseshoe kidney | Variant inherited from mother (affection status unknown) |
| R MCDK | ||||||||||||||
| Cleft palate | ||||||||||||||
| Intellectual disability | ||||||||||||||
| A1147 | GATA3g | De novo dominant | c.708_709 insT | p.Ser 237Glnfs*67i | Het | — | / | NA | / | / | Pathogenic | Gene | BL VUR | De novo variant (paternity and maternity confirmed) |
| Septate uterus | ||||||||||||||
| Hearing loss | ||||||||||||||
| Progressive renal impairment | ||||||||||||||
| A1160j | Trisomy 20pg | De novo | — | / | / | L RA | De novo variant (paternity and maternity confirmed) | |||||||
| Mitral regurgitation | ||||||||||||||
| Hypotonia | ||||||||||||||
| Intellectual disability | ||||||||||||||
| A3346k | TRPS1g | Dominant | c.2795C>T | p.Ala 932Val | Het | Saccharomyces cerevisiae | 0.997 Del. D.C. | 21 | / | / | Likely pathogenic | DM | Trichorhinophalangeal syndrome | Variant inherited from affected father (trichorhinophalangeal syndrome) |
| BL RHD, VUR | ||||||||||||||
| A4450 | KMT2Dg | Dominant | c.6638G>A | p.Gly2213Asp | Het | Xenopus tropicalis | 0.186 Tol. D.C. | 4 | 0/1/4085 | 0/6/171078 | Likely pathogenic | DM | BL VUR | Variant inherited from father (mild facial dysmorphism) |
| Cleft palate | ||||||||||||||
| Facial dysmorphism | ||||||||||||||
| Protruding ears | ||||||||||||||
| Delayed development | ||||||||||||||
| A4478l | FAT4g | Compound het | c.9279A>C | p.Gln3093His | Het | D. rerio | Tol. D.C. | 16 | / | 0/10/244826 | Likely pathogenic | DM | L RA, R UVJO | Yes (unaffected parents het carriers, variant confirmed in affected siblings) |
| FAT4g | c.9313A>G | p.Ser3105Gly | Het | Xenopus tropicalis | Tol .D.C. | 4 | / | 0/5/245198 | Facial dysmorphism | |||||
| Psychomotor delay | ||||||||||||||
| Intellectual disability | ||||||||||||||
| Hypotonia | ||||||||||||||
| Sprengel deformity | ||||||||||||||
| Other skeletal deformities | ||||||||||||||
| BL cryptorchism | ||||||||||||||
| A4672f,m | HNF1Bg | Dominant | c.1024T>C | p.Ser 342Pro | Het | D. rerio | 0.767 Del. D.C. | 8 | / | 0/1/243686 | Likely pathogenic | DM | R RHD | Variant inherited from mother (affection status unknown) |
| Cystinuria | ||||||||||||||
| A4732h | SRGAP1g | Dominant | c.806G>A | p.Cys 269Tyr | Het | D. rerio | 0.840 Tol. D.C. | 25 | / | / | Pathogenic | DM | R MCDK | Variant inherited from affected mother (R duplicated kidney) |
| R ureterocele | ||||||||||||||
| A3403n | TRAP1g | Recessive | c.1406G>A | p.Arg 469His | Hom | S. cerevisiaeo | 0.997 Del. D.C. | 33 | 0/66/4234 | 11/1552/269946 | Pathogenic | DM | BL VUR | Yes (unaffected parents het carriers, variant segregates in two affected siblings) |
| A3880p | TBX18g | Dominant | c.1010delG | p.Gly 337Val fs*19i | Het | — | / | NA | / | / | Likely pathogenic | DM | UPJO | Yes (segregates in multiple affected family members) |
| HAGq | NRIP1g | Dominant | c.279del | p.Trp 93* (stop gain)i | Het | — | / | NA | / | / | Likely pathogenic | DM | RHD | Yes (segregates in multiple affected family members) |
| MCDK | ||||||||||||||
| Hydronephrosis | ||||||||||||||
| A1023r | FREM2g | Compound het | c.4031G>A | p.Arg 1344His | Het | D. rerio | 0.085 Del./ | 16 | 0/23/4277 | 12/102/277164 | Uncertain significance | DM | R RA | NA |
| FREM2g | c.7535G>A | p.Arg 2512His | Het | D. rerio | 0.929 Del./ | 18 | 0/10/4290 | 0/132/276840 | Uncertain significance | DM | Bladder calculi | |||
| A1220f | ROBO2g | Dominant | c.292G>T | p.Gly 98Trp | Het | C. elegans | 0.880 Del. D.C. | 19 | / | / | Uncertain significance | DM | R UPJO | NA |
| Renal stones | ||||||||||||||
| A1232r | FREM2g | Compound het | c.649C>T | p.Arg 217Cys | Het | X. tropicalis | 0.836 Del./ | 17 | / | 0/1/244270 | Uncertain significance | DM | PUV, R VUR, L UPJO | NA |
| FREM2g | c.4031G>A | p.Arg 1344His | Het | D. rerio | 0.085 Del./ | 16 | 0/23/4277 | 12/102/277164 | Uncertain significance | DM | Progressive renal impairment | |||
| B24s | ETV4g | Recessive | c.1244G>A | p.Arg 415His | Hom | Drosphilia melanogaster | 1.00 Del. D.C. | 36 | 0/1/4299 | 0/23/245730 | Likely pathogenic | Gene | R VUR | Variant het in unaffected mother, paternal DNA NA |
| B196 | CTU2g | Recessive | c.1399C>T | p.Arg 467Cys | Hom | D. melanogastert | 0.926 Del. D.C. | 21 | / | 0/1/245354 | Likely pathogenic | Gene | L hydronephrosis | Variant het in unaffected mother, paternal DNA NA |
| Facial dysmorphism | ||||||||||||||
| Microcephaly | ||||||||||||||
| Intellectual disability | ||||||||||||||
| Growth retardation | ||||||||||||||
| Pulmonary stenosis | ||||||||||||||
| Imperforate anus | ||||||||||||||
| Absent uterus | ||||||||||||||
| B268u | HPSE2g | Recessive | c.457C>T | p.Arg 153*i | Hom | — | / | 10 | / | 0/3/245274 | Pathogenic | DM | Urofacial syndrome | NA |
| L RHD, L UPJO | ||||||||||||||
| A3837 | TBX18g | Dominant | c.1802A>G | p.Gln 601Arg | Het | Ciona intestinalis | 0.932 Del. D.C. | 14 | / | 0/4/217866 | Uncertain significance | Gene | PUV, BL VUR | NA |
| A3900 | FRAS1g | Compound het | c.3998T>C | p.Val 1333Ala | Het | C. elegansv | 0.086 Tol. D.C. | 15 | / | 0/12/244984 | Uncertain significance | Gene | PUV | NA |
| FRAS1g | c.8131T>C | p.Tyr 2711His | Het | C. elegans | 0.928 Del. D.C. | 13 | / | 1/29/191356 | Uncertain significance | Gene | ||||
| B211 | Trisomy 18g | De novo dominant | — | / | / | R MCDK, L RHD | NA | |||||||
| Facial dysmorphism | ||||||||||||||
| Short palpebral fissures | ||||||||||||||
| Very small low-set ears | ||||||||||||||
| High arched palate | ||||||||||||||
| Congenital cardiopathy | ||||||||||||||
| Esophageal atresia and trachoesophageal fistula | ||||||||||||||
| High position of the anus | ||||||||||||||
| Generalized nail hypoplasia | ||||||||||||||
| Syndactyly of the feet | ||||||||||||||
| B630 | HNF1Bg | Dominant | 1.5-Mb deletion chromosome 17q12 | Pathogenic | DM | BL MCDK | NA | |||||||
| Hyperuricemia ADHD | ||||||||||||||
| Developmental delay | ||||||||||||||
| B1434 | CTU2g | Recessive | c.1399C>T | p.Arg 467Cys | Hom | D. melanogasterw | 0.926 Del. D.C. | 21 | / | 0/1/245354 | Likely pathogenic | Gene | R MCDK, L hydronephrosis | NA |
| Global developmental delay | ||||||||||||||
| Brain MRI; cave of septum pellucidium | ||||||||||||||
| B1435 | ACTG1g | De novo | c.464C>T | p.Ser 155Phe | Het | S. cerevisiae | 1.00 Del. D.C. | 18 | / | / | Pathogenic | DM | L partial duplex kidney, L hydroureter, L hydronephrosis, | NA |
| Facial dysmorphism, lissencephaly, Dany–Walker malformation, global developmental delay | ||||||||||||||
| Growth retardation | ||||||||||||||
| B1439 | SALL1g | Dominant | c.1666G>A | p.Gly 556Ser | Het | D. rerio | 0.999 Del. D.C. | 20 | / | 0/3/245992 | Uncertain significance | Gene | BL VUR, BL hydronephrosis | NA |
| B1316 | GREB1Lg | Dominant | c.4276G>A | p.Val 1426Ile | Het | D. rerioy | 0.079 Tol. D.C. | 14 | / | / | Likely pathogenic | Gene | BL RHD | NA |
| Facial dysmorphism | ||||||||||||||
| Short neck | ||||||||||||||
| Single transverse palmar crease | ||||||||||||||
| Brachydactyly | ||||||||||||||
| A1261x | GREB1Lg | Dominant | c.5068G>A | p.Val1690Met | Het | X. tropicalis | 0.681 Del. D.C. | 30 | / | / | Likely pathogenic | Gene | BL VUR | Yes (segregates in multiple affected family members) |
| L RA | ||||||||||||||
| Supernumerary nipple | ||||||||||||||
| F1436 | KAT6Bg | Dominant/de novo | c.4285G>A | p.Glu1429Lys | Het | D. rerio | 0.995 Tol. D.C. | 8 | / | / | Likely pathogenic | Gene | BL VUR, BL RHD | NA |
| Facial dysmorphism | ||||||||||||||
| Microcephaly | ||||||||||||||
| Developmental delay | ||||||||||||||
| Dysplastic ears | ||||||||||||||
| Muscle weakness (pectoralis and trapezius with limited mobility of shoulder) | ||||||||||||||
| B1650 | HPSE2g | Recessive | c.1099–2A>G | 100% ESS | Hom | — | / | NA | / | / | Pathogenic | Gene | Urofacial syndrome “inverted smile” | Yes (unaffected parents het carriers, variant segregates in two affected siblings) |
| BL VUR | ||||||||||||||
| Hinman syndrome | ||||||||||||||
| A3962 | NPHP1z | Recessive | c.1804delA | p.Ser 602Val fs*4i | Hom | — | / | NA | / | 0/1/245974 | Pathogenic | Gene | BL RHD | NA |
| A4235 | TMEM 231z | Recessive | c.119T>G | p.Leu40Arg | Hom | C. elegans | 0.951 Del. D.C. | 23 | / | / | Likely pathogenic | Gene | BL RHD, Facial dysmorphism, microcephaly, intellectual disability, polysyndactyly, heart anomalies, growth retardation. | Yes (segregates in two affected family members) |
| B1306 | NPHP4z | Compound het | c.3983C>T | p.Pro1328Leu | Het | C. elegans | 0.999 Del. / | 26 | 0/4/2047 | 0/62/268266 | Likely pathogenic | Gene | BL RHD | NA |
| NPHP4z | c.2021G>A | p.Arg674His | Het | C. elegans | 0.999 Del. / | 29 | 0/1/2089 | 0/8/277216 | Likely pathogenic | Gene | Growth retardation | |||
| Patent ductus arteriosus | ||||||||||||||
| Facial dysmorphism | ||||||||||||||
PP2, PolyPhen 2; SIFT, Sorting Intolerant from Tolerant; MT, Mutation Taster; CADD, Combined Annotation Dependent Depletion; EVS, Exome Variant Server; gnomAD, Genome Aggregation Database; ACMG, American College of Medical Genetics; Het, heterozygous; Del., deleterious; D.C., disease causing; /, data not available; DM, disease mutation; BL, bilateral; VUR, vesicoureteric reflux; R, right; L, left; MCDK, multicystic dysplastic kidney; —, not applicable; NA, not available; RA, renal agenesis; RHD, renal/hypodysplasia; Tol., tolerated; UVJO, ureterovesical junction obstruction; Hom, homozygous; UPJO, ureteropelvic junction obstruction; PUV, posterior urethral valve; ADHD, Attention Deficit Hyperactivity Disorder; Mb, megabase; MRI, magnetic resonance imaging; ESS, essential splice site.
For families in which the disease-causing variant has previously been reported in the literature, the corresponding reference is provided.
Evolutionary conservation was assessed across phylogeny over eight species: Mus musculus, Gallus gallus, Xenopus tropicalis, Danio rerio, Caenorhabditis elegans, Ciona intestinalis, Drosphilia melanogaster, and Saccharomyces cerevisiae. If conservation is interrupted in one species but otherwise preserved across phylogeny, additional information is provided.
Variant frequencies listed for homozygous/hemizygous (if applicable)/heterozygous/total alleles detected in the population.
ACMG American College of Human Genetics Standards and Guidelines Classification as pathogenic, likely pathogenic, or uncertain significance.66
HGMD, Human Gene Mutation Database; (https://portal.biobaseinternational.com/hgmd). If the exact variant has previously been reported and classified as a pathogenic mutation that is disease causing, the variant is denoted as DM. If the gene but not the exact variant has been reported for the corresponding phenotype, gene is indicated.
Ref. 42.
Mutations in the isolated or syndromic gene identified in families with the corresponding phenotype.
Ref. 21.
Frameshift, stop loss, stop gain, or nonsense variant.
Ref. 64.
Ref. 68.
Ref. 69.
Finding in more than two categories.
Ref. 36.
Interruption in conservation due to leucine present in C. intestinalis.
Ref. 23.
Ref. 43.
Ref. 33.
Ref. 32.
Interruption in conservation due to serine present in C. elegans.
Ref. 46.
Interruption in conservation due to arginine present in D. rerio.
Interruption in conservation due to serine present in C. elegans.
Ref. 44.
Interruption in conservation due to methionine present in M. musculus.
Mutations in phenocopy gene.
Detecting Mutations in Known Genes for Isolated or Syndromic CAKUT in 13% of Families with a Corresponding Phenotype
In 13% of CAKUT families (29 of 232), we detected a mutation in a gene that is known to cause isolated or syndromic CAKUT in patients exhibiting the corresponding isolated or syndromic CAKUT phenotype (Figure 1A, dark blue segment). In patients with isolated CAKUT, we detected mutations in 13 genes (five recessive [FRAS1, TRAP1, FREM2, ETV4, and HPSE2] and eight dominant [SALL1, SRGAP1, ROBO2, TBX18, HNF1B, NRIP1, GATA3, and GREB1L]) from the 40 genes that are known to cause isolated CAKUT when mutated (Supplemental Table 1). In addition, we detected six monogenic causes of syndromic CAKUT in patients with the corresponding syndromic CAKUT phenotype (three recessive [FAT4, CTU2, and TRPS1] and three dominant [ACTG1, KMT2D, and KAT6B]) as well as Trisomy 18 and Trisomy 20.
Detecting a Mutation in a Phenocopy Gene in 1% of Families with CAKUT
In three of the 232 families (1%), mutations in genes were identified that, if mutated, give rise to conditions that may phenocopy CAKUT (Figure 1B, purple segment). This pertained mostly to bilateral small kidneys that were thought to represent the CAKUT phenotype of renal hypo-/dysplasia but in fact, represented small cystic kidneys due to mutations in renal ciliopathy genes (NPHP1, NPHP4, and TMEM213). The molecular diagnosis after WES, therefore, differed from the previous clinical diagnosis in these three families.
Identifying Hypomorphic Mutations in Known Genes for Syndromic CAKUT in 6% of Families with Isolated CAKUT
Because we previously found that null mutations in certain monogenic genes cause syndromic forms of CAKUT, whereas hypomorphic mutations in the same genes may cause isolated CAKUT (Supplemental Figure 1),33 we evaluated WES data for mutations in one of the 179 known causes of syndromic CAKUT in families with isolated CAKUT phenotypes. We detected deleterious mutations in 6% (15 of 232) of families in one of the following 12 dominant genes: AMER1, KAT6B, NOTCH2, KMT2D, EP300, NSDHL, TP63, OFD1, FGFR1, FGFR3, HOXA13, and FLNA (Figure 1C, light blue segment, Supplemental Table 6).
Identifying Novel Human CAKUT Genes Using Murine CAKUT Candidate Genes
Having detected likely causative mutations or candidate variants in 20% (47 of 232) of families (Figure 1, A–C), we proceeded to evaluate WES data of the remaining 185 of 232 unsolved families (80%) for mutations in potentially novel genetic causes of human CAKUT. By applying a search in the 185 known monogenic causes of murine CAKUT genes (Supplemental Table 3), we identified deleterious variants in 2% (five of 232) of families with CAKUT in three recessive genes (LAMA5, MEGF8, and TNS1) and one dominant gene (FOXC1) (Figure 1D, Supplemental Table 4, pink segment). Mutations in these genes have not yet been implicated in human CAKUT. However, given the corresponding phenotype in mouse models Supplemental Table 3, we consider them likely novel genes for human CAKUT.
Discovering 19 Potential Novel Unique Candidate Genes for CAKUT in 8% of Families
Having detected likely causative mutations or candidate variants in 22% of CAKUT families (Figure 1, A–D), we proceeded to evaluate 102 of 232 families (44%) that either were consanguineous or had a duo/trio/quad pedigree structure. We evaluated for either (1) novel recessive genes by evaluation of homozygous regions in consanguineous families (37 of 232; 16%) or (2) recessive and/or dominant mutations by duo, trio, or quad analysis depending on pedigree structure (65 of 232; 28%) (Supplemental Figures 3 and 4). After filtering of variants on the basis of a priori genetic criteria (Methods), we arrived at a single novel candidate gene in 8% of families (19 of 232) with CAKUT (Figure 1E, Supplemental Table 4, red segment). Search for additional CAKUT families with variants in these 19 novel candidate genes by GeneMatcher59 did not yield any additional families to date, a finding that is not uncommon in monogenic forms of CAKUT.38
Identifying Multiple Potential Novel Candidate Genes per Family in 10% of Families with CAKUT
In 10% of the families (22 of 232), multiple candidate CAKUT genes were identified after a priori filtering criteria (Methods, Figure 1F, Supplemental Table 4, green segment). No single gene per family could be prioritized on the basis of genetic criteria (e.g., nonsense versus missense variant) or protein information obtained from the literature.
Detecting Monogenic Causes for Non-CAKUT Diseases in Families with CAKUT
In 8% of CAKUT families (18 of 232), we detected mutations in 21 disease-causing genes known to be causative of non-CAKUT diseases (Supplemental Table 7). Twelve of these genes were identified in ten of 232 CAKUT families (4%); in these families, no CAKUT-causing mutation could be identified (Figure 1G). However, nine were identified in eight CAKUT families; in these families, we detected either a CAKUT-causing gene (Figure 1A and B) or a CAKUT candidate gene (Figure 1, C–F). These mutations are coded in brown in Supplemental Table 7 but highlighted to indicate their additional status as families with a CAKUT-causing mutation that was detected. These causative mutations in nonrenal disease genes were reported back to the referring physician according to the American College of Medical Genetics guidelines.60–62
Spectrum of Mutations in Known CAKUT Genes
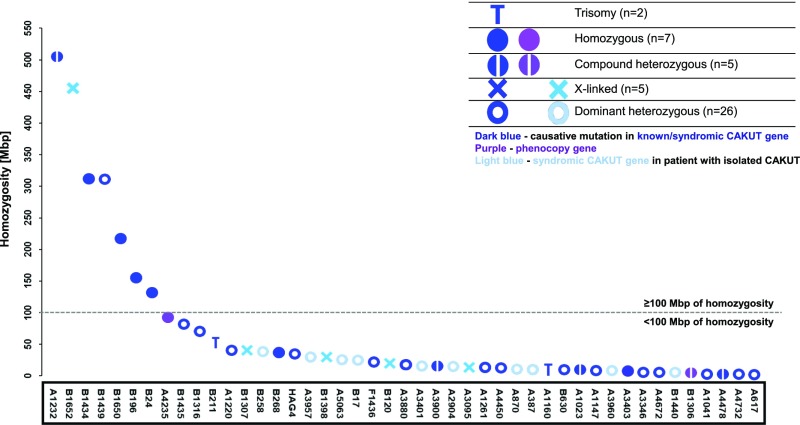
It is known that, in consanguineous pedigrees, the likelihood of detecting a homozygous causative mutation in a recessive gene rather than compound heterozygous mutations rises with the degree of relatedness or homozygosity across the genome.63 We, therefore, plotted homozygosity in descending order for families in which we identified a likely causative mutation or a candidate mutation in a CAKUT gene or a CAKUT phenocopy gene (Figure 3). In seven of 47 families that exhibited significant levels of homozygosity (≥100 Mbp), we identified four homozygous recessive mutations; in the 38 families that had homozygosity <100 Mbp, four had a compound heterozygous recessive mutation, whereas 25 had a dominant heterozygous mutation. Of note, in three families (B1316, B1439, and B1435) with homozygosity level of ≥60 Mbp, a single heterozygous disease-causing mutation in established isolated or syndromic CAKUT genes was detected (SALL1, ACTG1, and GREB1L) (Figure 3). Overall, the solve rate varies by pedigree structure, ranging from 10% in outbred multiplex families, 12% in pedigrees with a trio structure, 12% in families from regions where consanguinity is high, 17% in patients with severe CAKUT, and 20% in families that are consanguineous to 29% in families with syndromic CAKUT (Figure 2).
Figure 3.
Relationship between measured homozygosity and disease-causing mutations in congenital anomalies of the kidney and urinary tract (CAKUT). Homozygosity mapping was performed on the basis of single-nucleotide polymorphisms generated from whole-exome sequencing data. Data are shown for families in which a CAKUT-causing gene or a gene known to phenocopy CAKUT was identified (32 of 232) and families with isolated CAKUT in which a candidate gene in a known syndromic CAKUT gene was identified (15 of 232). A representative individual from each family was plotted from the highest to the lowest level of total homozygosity (megabase pairs) across the genome. In total, seven individuals had homozygosity ≥100 Mbp, whereas 38 individuals had homozygosity of <100 Mbp, which is denoted by the gray dashed line. In two families, homozygosity mapping could not be generated due to low coverage, and therefore, they are not included in this graph (A3887: TBX18 dominant heterozygous mutation and A2962: NPHP1 homozygous variant). Causative mutations in isolated/syndromic genes identified in CAKUT families with the corresponding phenotype are denoted by a dark blue color, phenocopy genes are denoted by a purple color, and candidate mutations in syndromic CAKUT genes identified in families with isolated CAKUT are denoted by a light blue color. Homozygous variants are denoted by filled circles, compound heterozygous variants are denoted by two half circles, dominant heterozygous variants are denoted by unfilled circles, X-linked variants are denoted by an “X,” and complex chromosomal rearrangements are denoted by a “T.” Note that, in the part of our cohort with homozygosity of ≥100 Mbp (the cluster left of the x axis), paradoxically, we identified causative mutations in heterozygous genes (e.g., B1439; SALL1). In patients with rare cases with extreme homozygosity, heterozygous disease-causing mutations can be identified. Such patients have previously been described in the literature.67
Syndromic CAKUT Genes Constitute Promising Candidate Genes for Isolated CAKUT Phenotypes
We previously described an allelic genotype-phenotype correlation, in which null mutations in known syndromic CAKUT genes (e.g., protein truncating) cause syndromic CAKUT phenotypes, whereas hypomorphic mutations in the same subset of syndromic CAKUT genes (e.g., missense) cause isolated CAKUT phenotypes.33 We, therefore, evaluated WES data for mutations in 40 genes that are known to cause isolated (i.e., nonsyndromic) CAKUT (Supplemental Table 1). Conversely, we also evaluated WES data for mutations in 179 genes that are known to cause syndromic CAKUT (Supplemental Table 2) in both patients with the corresponding phenotype and as a candidate gene hypothesis, additionally in families with an isolated CAKUT phenotype.
We detected likely causative mutations that were concordant (i.e., mutations in an isolated CAKUT gene in families with isolated CAKUT or mutations in syndromic CAKUT genes in families with syndromic CAKUT) in 13% of families (29 of 232) (Figure 1A). Interestingly, we also detected causative mutations that were discordant (i.e., mutations in syndromic CAKUT genes in families with isolated CAKUT) in 6% of families (15 of 232) (Figure 1C). Strikingly, 13 of these 15 mutations were hypomorphic mutations (i.e., 13 missense and two splice variants) (Supplemental Table 6, Table 3). The literature on 13 of these 15 mutations supports the genotype-phenotype correlation, in which hypomorphic mutations cause isolated CAKUT phenotypes (as shown here), whereas syndromic CAKUT was caused by null mutations in those same genes in 35%–97% of patients in the literature (Supplemental Table 6, column 7).
Table 3.
Information on identified candidate mutations in congenital anomalies of the kidney and urinary tract genes known to cause isolated or syndromic congenital anomalies of the kidney and urinary tract
| Family Identificationa | Gene | Mode of Transmission | Nucleotide Change | Amino Acid Change | State | Evolutionary Conservationb | PP2 SIFT MT | CADD Score | EVSc | gnomADc | ACMGd | HGMDe | Phenotypes | Segregation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A387 | KAT6Bf | Dominant | c.2152C>T | p.Arg 718Trp | Het | Saccharomyces cerevisiaeg | 0.983 Del. D.C. | 16 | / | / | Uncertain significance | Gene | L RA, R hydronephrosis; progressive renal impairment | Variant inherited from father (affection status unknown) |
| A870 | NOTCH2f | Dominant | c.3556T>A | p.Tyr 1186Asn | Het | Danio rerio | 0.854 Del. D.C. | 18 | 0/1/4299 | 0/12/277048 | Uncertain significance | Gene | BL VUR nevus pigmentosus R supranummery nipple | Variant inherited from mother (affection status unknown) |
| A3401 | NOTCH2f | Dominant | c.6767G>A | p.Arg 2256His | Het | Ciona intestinalish | 0.862 Del. D.C. | 15 | 0/1/4299 | 0/12/246058 | Uncertain significance | Gene | R RHD, L UVJO | Variant inherited from father (affection status unknown); segregates in two affected siblings |
| PUV | ||||||||||||||
| B17 | KMT2Df | Dominant | c.13190G>T | p.Gly 4397Val | Het | Drosphilia melanogaster | 0.960 Del. D.C. | 11 | / | / | Uncertain significance | Gene | L RA | NA |
| A3095 | NSDHLf | X-linked recessive | c. 842G>A | p.Arg 281His | Hemi | X. tropicalis | 0.899 Del. D.C. | 12 | / | 0/6/8/199924 | Uncertain significance | Gene | Prune belly syndrome | NA |
| L RA | ||||||||||||||
| A5063 | EP300f | Dominant | c.1781C>T | p.Thr594Met | Het | D. melanogasteri | 0.999 Del. D.C. | 32 | / | 0/17/246260 | Uncertain significance | Gene | BL MCDK | Variant inherited from father (affection status unknown); segregates in two affected siblings |
| B258 | TP63f | Dominant | c.799G>A | p.Val267Ile | Het | C. intestinalisj | 0.802 Tol. D.C. | 22 | / | 0/3/246070 | Uncertain significance | Gene | L UPJO | NA |
| A3957 | NOTCH2f | Dominant | c.6892C>T | p.Arg2298Trp | Het | D. rerio | 0.609 Del. D.C. | 11 | / | 0/1/245696 | Uncertain significance | Gene | BL hydronephrosis | NA |
| Anorectal malformation | ||||||||||||||
| A3960 | FGFR1f | Dominant | c.1426C>T | p.Arg476Trp | Het | D. rerio | 1.000 Del. D.C. | 6 | / | 0/4/277084 | Uncertain significance | Gene | L RA, R VUR | NA |
| Hypospadias | ||||||||||||||
| Bladder extrophy | ||||||||||||||
| B1307 | AMER1f | Dominant | c.185G>T | p.Gly62Val | Het | D. rerio | 0.813 Tol. D.C. | 19 | 0/1/1/2427 | 0/4/9/200427 | Uncertain significance | Gene | R MCDK | NA |
| B1398k | OFD1f | X-linked | c.936–2A>G | 100% ESS | Het | — | / | NA | 0/0/1/2427 | 0/8/23/198443 | Uncertain significance | Gene | BL VUR | Variant inherited from mother (affection status unknown) |
| R hydronephrosis | ||||||||||||||
| B1440 | HOXA13f | Dominant | c.25C>T | p.Pro9Ser | Het | D. rerio | 0.992 Tol. D.C. | 18 | / | 0/28/226114 | Uncertain significance | Gene | BL hydronephrosis | NA |
| BL RHD, PUV | ||||||||||||||
| Facial dysmorphism | ||||||||||||||
| A2904 | FGFR3f | Dominant | c.1663G>A | p.Val555Met | Het | D. melanogaster | 0.985 Del. D.C. | 19 | / | 0/48/274072 | Likely pathogenic | DM | R RHD, VUR | NA |
| B120 | OFD1f | Dominant | c.517+1G>A | 100% ESS | Het | / | NA | / | / | Pathogenic | Gene | R MCDK | NA | |
| L duplex | ||||||||||||||
| L VUR, ureterocoele | ||||||||||||||
| B1652k | FLNAf | X-linked recessive | c.6348C>G | p.His2116Gln | Hemi | Caenorhabditis elegans | 0.955 Del. D.C. | 12 | / | / | Uncertain significance | Gene | Prune belly syndrome | Variant inherited from mother (affection status unknown) |
| Neurogenic bladder | ||||||||||||||
| BL VUR |
PP2, PolyPhen 2; SIFT, Sorting Intolerant from Tolerant; MT, Mutation Taster; CADD, Combined Annotation Dependent Depletion; EVS, Exome Variant Server; gnomAD, Genome Aggregation Database; ACMG, American College of Medical Genetics; Het, heterozygous; Del., deleterious; D.C., disease causing; /, data not available; L, left; RA, renal agenesis; R, right; BL, bilateral; VUR, vesicoureteric reflux; RHD, renal/hypodysplasia; UVJO, ureterovesical junction obstruction; PUV, posterior urethral valve; NA, not available; Hemi, hemizygous; MCDK, multicystic dysplastic kidney; Tol., tolerated; UPJO, ureteropelvic junction obstruction; —, not applicable; ESS, essential splice site; DM, disease mutation.
Unique family identification number.
Evolutionary conservation was assessed across phylogeny over eight species: Mus musculus, Gallus gallus, Xenopus tropicalis, Danio rerio, Caenorhabditis elegans, Ciona intestinalis, Drosphilia melanogaster, and Saccharomyces cerevisiae. If conservation is interrupted in one species but otherwise preserved across phylogeny, additional information is outlined.
Variant frequencies listed for homozygous/hemi (if applicable)/het/total alleles detected in the population.
ACMG American College of Human Genetics Standards and Guidelines Classification as pathogenic, likely pathogenic, or uncertain significance.66
HGMD Human Gene Mutation Database; (https://portal.biobaseinternational.com/hgmd). If the exact variant has previously been reported and classified as a pathogenic mutation that is disease causing, the variant is denoted as DM. If the gene but not the exact variant has been reported for the corresponding phenotype, gene is indicated.
Mutations in syndromic genes identified in families with an isolated syndromic congenital anomaly of the kidney and urinary tract phenotype.
Interruption in conservation due to glutamine present in D. rerio and D. melanogaster.
Interruption in conservation due to leucine present in D. rerio.
Interruption in conservation due to glutamate present in C. elegans.
Interruption in conservation due to asparagine present in X. tropicalis.
Finding in more than two categories.
Discussion
WES Can Identify Likely Pathogenic Mutations in 14% of Families
We applied WES to a large cohort of 232 families with CAKUT. We showed that, in this patient cohort, WES detects a specific deleterious mutation in a known CAKUT or CAKUT phenocopy gene in 32 of 232 (14%) families with CAKUT. Mutations were identified in known genes for isolated or syndromic manifestations of CAKUT in 13% (29 of 232) of families exhibiting the corresponding phenotype (Figure 1A). In addition, we identified causative mutations in genes that may cause phenocopies of CAKUT in three of the 232 families (1%) (Figure 1B). Gene panel studies have shown that monogenic causation accounts for approximately 12% of patients with CAKUT.33,42 WES has the added advantage that detection of mutations is not limited to a prespecified list of candidate genes. We show here the utility of WES for the identification of monogenic, likely pathogenic mutations in 14% of families with CAKUT.
Candidate Genes Can Be Derived from WES
In 6% (15 of 232) of CAKUT families with an isolated CAKUT phenotype, we were able to identify candidate mutations in a known syndromic CAKUT gene (Figure 1C). In five families (2%), we identified four novel murine candidate genes (Figure 1D). Additionally, in 19 families (8%), we identified single novel CAKUT candidates using targeted search for homozygously mutated genes in homozygous families or by trio evaluation in families in which parental DNA was available (Figure 1E). So far, mutation analyses have not yielded mutations in these genes in additional families with CAKUT. This rarity is not unexpected, because in many of recently identified CAKUT genes, very few families with mutations have been identified. Additional genetic and experimental evidence will help determine whether mutations in these newly identified genes are indeed disease causing in CAKUT. In 22 of the 232 families (9%), we were unable to identify a unique, potentially novel gene per family, but rather, after filtering of variants, we were left with multiple potential causative genes (Figure 1F).
Limitations of the Study
In total, 129 of 232 families (56%) remained without any findings (Figure 1H), the reason for which is likely multifold. First, it has been shown that up to 16.6% of individuals with CAKUT have a molecular diagnosis attributable to copy number variants, which can be difficult to detect using WES.64 Second, the coverage distribution across the exome is variable, which means that variants in some low-coverage areas may be missed.
Because our cohort was prescreened for CAKUT genes, this likely led to an overall underestimation of the true prevalence of monogenic causation within our CAKUT cohort.
In relation to causality, although we performed variant calling according to our stringent a priori criteria (which have been extensively published23,44,65) that adhere to standard classification as per the American College of Medical Genetics,66 functionality of each detected variant was not individually tested.
We show the success of WES in terms of obtaining a molecular diagnosis in families with CAKUT. The finding that, in 14% of families, a likely pathogenic gene can be identified is significant, further supporting the hypothesis that CAKUT is caused by mutations in monogenic genes.
Disclosures
D.M.C., A.T.v.d.V., H.I., N.M., M.N., J.C., A.V., D.y.H., J.S., D.A.B., J.M.S., D.S., R.S., J.K.W., A.D., A.J.M., W.T., T.J.S., T.H., E.W., S.A., A.A., C.A.H., H.H., T.M.K., F.K., C.M.K., R.D., and S.S., performed whole exome evaluation and mutation analysis. L.S., K.A., D.R.S., M.A.B., M.J.G.S., N.M.R., M.A.F., A.Z.T., G.H.D., R.B., N.S., N.A.S., J.A.K., S.E.D, H.M.F., D.M., M.A.S., H.S.A, L.A.E., A.K., P.S., S.S.C., R.S.L., S.B.B., W.L., H.M.R., V.T. and F.H. recruited patients and gathered detailed clinical information for the study. S.S., H.L.R., D.G.M.A., M.L., K.M.L., M.W.W., S.M.M., and R.P.L. performed whole exome sequencing and downstream data analysis. F.H. conceived of and directed the entire study and wrote the manuscript with D.M.C. and A.T.v.d.V.
Supplementary Material
Acknowledgments
We thank the physicians and the participating families for their contribution: S. Garcia-Minauz (Madrid, Spain), H. Fehrenbach (Memmingen, Germany), M.T.F. Wolf (Dallas, TX), J.P. Oliveirara (Porto, Portugal), R. Alfandary (Ramat Gan, Israel), N. Gruber (Ramat Gan, Israel), B. Hoppe (Cologne, Germany), J. Hofele (Munich, Germany), A. Riba (Magdeburg, Germany), R. Mallmann (Essen, Germany), B. Bartmann (Cologne, Germany), T. Neuhaus (Luzern, Switzerland), D. Mueller (Berlin, Germany), H. Jueppner (Boston), L. Patzer (Halle-Saale, Germany), S. Rains-Lynema (Michigan), A. Rissmann (Magdeburg, Germany), A. Schulte-Everding (Munster, Germany), U. John (Jena, Germany), A. Salerno (Boston), S. von Kleist (Freiburg, Germany), S. Hashmi (Karachi, Pakistan), Ali Gharavi (New York), and Elijah O. Kehinde (Astana, Kazakhstan).
A.T.v.d.V. is supported by postdoctoral research fellowship VE 969-7 from the German Research Foundation (DFG). D.M.C. is funded by Health Research Board, Ireland grant HPF-206-674; the International Pediatric Research Foundation Early Investigators’ Exchange Program; and the Amgen Irish Nephrology Society Specialist Registrar Bursary. N.M. is supported by funding from National Institutes of Health grant T32-DK007726-33 at Boston Children’s Hospital. M.N. is supported by a grant from the Japan Society for the Promotion of Science. A.V. is supported by a Manton Center for Orphan Diseases Research grant. A.J.M. is supported by Research Training in Pediatric Nephrology grant 2T32DK007726-31A1 and Harvard Stem Cell Institute Kidney Interlab Fellowship award F-KP-0003-17-00 at Harvard Medical School. J.K.W. is funded by the American Society of Nephrology and National Institutes of Health grant DK007726-31A1. W.T. is funded by the American Society of Nephrology and National Institutes of Health grant DK 007726-3A. T.J.-S. is supported by postdoctoral research fellowship Jo 1324/1-1 from the DFG. T.H. is supported by postdoctoral research fellowship HE 7456/1-1 from the DFG. E.W. is supported by German National Academy of Sciences Leopoldina grant LPDS-2015-07. T.M.K. is supported by a postdoctoral fellowship award from the KRESCENT Program, a national kidney research training partnership of the Kidney Foundation of Canada, the Canadian Society of Nephrology, and the Canadian Institutes of Health Research. Sequencing and data processing were performed by the Broad and Yale Centers for Mendelian Genomics funded by National Human Genome Research Institute grants UM1 HG008900 (to H.L.R. and D.G.M.) and U54 HG006504 (to R.P.L.). R.S.L. is supported by the National Institute of Health grant (DK096238). S.S., S.B.B. and F.H. are supported by the Begg Family Foundation. F.H. is the William E. Harmon Professor of Pediatrics at Harvard Medical School. This research was supported by National Institutes of Health grant DK076683 (to F.H.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017121265/-/DCSupplemental.
References
- 1.Chesnaye N, Bonthuis M, Schaefer F, Groothoff JW, Verrina E, Heaf JG, et al.: ESPN/ERA–EDTA registry : Demographics of paediatric renal replacement therapy in Europe: A report of the ESPN/ERA-EDTA registry. Pediatr Nephrol 29: 2403–2410, 2014 [DOI] [PubMed] [Google Scholar]
- 2.North American Pediatric Renal Transplant Cooperative Study: NAPRTCS 2008 Annual Report, Rockville, MD, The EMMES Corporation, 2008 [Google Scholar]
- 3.Vivante A, Hildebrandt F: Genetics of congenital anomalies of the kidneys and urinary tract. Pediatr Nephrol 29: 303, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F: Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol 29: 695–704, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont ME, Sullivan MJ, et al.: Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet 9: 358–364, 1995 [DOI] [PubMed] [Google Scholar]
- 6.Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, Sovik O: A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet 8: 2001–2008, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Soliman NA, Ali RI, Ghobrial EE, Habib EI, Ziada AM: Pattern of clinical presentation of congenital anomalies of the kidney and urinary tract among infants and children. Nephrology (Carlton) 20: 413–418, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa I, Kuwayama F, Pope JC 4th, Stephens FD, Miyazaki Y: Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int 61: 889–898, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Costantini F: Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip Rev Dev Biol 1: 693–713, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barratt TM, Avner ED, Harmon WE: Pediatric Nephrology, Baltimore, MD, Lippinkott Williams & Wilkins, 4th edition, 1999 [Google Scholar]
- 11.Davies JA: Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat (Basel) 156: 187–201, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Weber S, Thiele H, Mir S, Toliat MR, Sozeri B, Reutter H, et al.: Muscarinic acetylcholine receptor M3 mutation causes urinary bladder disease and a prune-belly-like syndrome. Am J Hum Genet 89: 668–674, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber S, Taylor JC, Winyard P, Baker KF, Sullivan-Brown J, Schild R, et al.: SIX2 and BMP4 mutations associate with anomalous kidney development. J Am Soc Nephrol 19: 891–903, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockschmidt A, Chung B, Weber S, Fischer DC, Kolatsi-Joannou M, Christ L, et al.: CHD1L: A new candidate gene for congenital anomalies of the kidneys and urinary tract (CAKUT). Nephrol Dial Transplant 27: 2355–2364, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Rivera E, Liu YP, Verbitsky M, Anderson BR, Capone VP, Otto EA, et al.: Genetic drivers of kidney defects in the DiGeorge syndrome. N Engl J Med 376: 742–754, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanna-Cherchi S, Sampogna RV, Papeta N, Burgess KE, Nees SN, Perry BJ, et al.: Mutations in DSTYK and dominant urinary tract malformations. N Engl J Med 369: 621–629, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, et al.: Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet 6: 2247–2255, 1997 [DOI] [PubMed] [Google Scholar]
- 18.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, et al.: Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet 11: 40–44, 1995 [DOI] [PubMed] [Google Scholar]
- 19.Kirby A, Gnirke A, Jaffe DB, Barešová V, Pochet N, Blumenstiel B, et al.: Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 45: 299–303, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skinner MA, Safford SD, Reeves JG, Jackson ME, Freemerman AJ: Renal aplasia in humans is associated with RET mutations. Am J Hum Genet 82: 344–351, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang DY, Kohl S, Fan X, Vivante A, Chan S, Dworschak GC, et al.: Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum Genet 134: 905–916, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimelli S, Caridi G, Beri S, McCracken K, Bocciardi R, Zordan P, et al.: Mutations in SOX17 are associated with congenital anomalies of the kidney and the urinary tract. Hum Mutat 31: 1352–1359, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivante A, Kleppa MJ, Schulz J, Kohl S, Sharma A, Chen J, et al.: Mutations in TBX18 cause dominant urinary tract malformations via transcriptional dysregulation of ureter development. Am J Hum Genet 97: 291–301, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gbadegesin RA, Brophy PD, Adeyemo A, Hall G, Gupta IR, Hains D, et al.: TNXB mutations can cause vesicoureteral reflux. J Am Soc Nephrol 24: 1313–1322, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, et al.: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins D, Bitner-Glindzicz M, Malcolm S, Hu CC, Allison J, Winyard PJ, et al.: De novo Uroplakin IIIa heterozygous mutations cause human renal adysplasia leading to severe kidney failure. J Am Soc Nephrol 16: 2141–2149, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Biason-Lauber A, Konrad D, Navratil F, Schoenle EJ: A WNT4 mutation associated with Müllerian-duct regression and virilization in a 46,XX woman. N Engl J Med 351: 792–798, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Mandel H, Shemer R, Borochowitz ZU, Okopnik M, Knopf C, Indelman M, et al.: SERKAL syndrome: An autosomal-recessive disorder caused by a loss-of-function mutation in WNT4. Am J Hum Genet 82: 39–47, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vivante A, Mark-Danieli M, Davidovits M, Harari-Steinberg O, Omer D, Gnatek Y, et al.: Renal hypodysplasia associates with a WNT4 variant that causes aberrant canonical WNT signaling. J Am Soc Nephrol 24: 550–558, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gribouval O, Gonzales M, Neuhaus T, Aziza J, Bieth E, Laurent N, et al.: Mutations in genes in the renin-angiotensin system are associated with autosomal recessive renal tubular dysgenesis. Nat Genet 37: 964–968, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, et al.: FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell 22: 1191–1207, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, van der Ven AT, Newman JA, Vivante A, Mann N, Aitkenhead H, et al.: ETV4 Mutation in a Patient with Congenital Anomalies of the Kidney and Urinary Tract. Int J Pediatr Child Health 4: 61–71, 2016. 22698282 [Google Scholar]
- 33.Kohl S, Hwang DY, Dworschak GC, Hilger AC, Saisawat P, Vivante A, et al.: Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 25: 1917–1922, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulum B, Özçakar ZB, Duman D, Cengiz FB, Kavaz A, Burgu B, et al.: HPSE2 mutations in urofacial syndrome, non-neurogenic neurogenic bladder and lower urinary tract dysfunction. Nephron 130: 54–58, 2015 [DOI] [PubMed] [Google Scholar]
- 35.Humbert C, Silbermann F, Morar B, Parisot M, Zarhrate M, Masson C, et al.: Integrin alpha 8 recessive mutations are responsible for bilateral renal agenesis in humans. Am J Hum Genet 94: 288–294, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saisawat P, Kohl S, Hilger AC, Hwang DY, Yung Gee H, Dworschak GC, et al.: Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int 85: 1310–1317, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardelin JP, Levilliers J, del Castillo I, Cohen-Salmon M, Legouis R, Blanchard S, et al.: X chromosome-linked Kallmann syndrome: Stop mutations validate the candidate gene. Proc Natl Acad Sci U S A 89: 8190–8194, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vivante A, Hildebrandt F: Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol 12: 133–146, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W: Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet 18: 81–83, 1998 [DOI] [PubMed] [Google Scholar]
- 40.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, et al.: SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A 101: 8090–8095, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoskins BE, Cramer CH, Silvius D, Zou D, Raymond RM, Orten DJ, et al.: Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am J Hum Genet 80: 800–804, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang DY, Dworschak GC, Kohl S, Saisawat P, Vivante A, Hilger AC, et al.: Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int 85: 1429–1433, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vivante A, Mann N, Yonath H, Weiss AC, Getwan M, Kaminski MM, et al.: A dominant mutation in nuclear receptor interacting protein 1 causes urinary tract malformations via dysregulation of retinoic acid signaling. J Am Soc Nephrol 28: 2364–2376, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanna-Cherchi S, Khan K, Westland R, Krithivasan P, Fievet L, Rasouly HM, et al.: Exome-wide association study identifies GREB1L mutations in congenital kidney malformations. Am J Hum Genet 101: 789–802, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bekheirnia MR, Bekheirnia N, Bainbridge MN, Gu S, Coban Akdemir ZH, Gambin T, et al.: Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genet Med 19: 412–420, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vivante A, Hwang DY, Kohl S, Chen J, Shril S, Schulz J, et al.: Exome sequencing discerns syndromes in patients from consanguineous families with congenital anomalies of the kidneys and urinary tract. J Am Soc Nephrol 28: 69–75, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brophy PD, Rasmussen M, Parida M, Bonde G, Darbro BW, Hong X, et al.: A gene implicated in activation of retinoic acid receptor targets is a novel renal agenesis gene in humans. Genetics 207: 215–228, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Tomasi L, David P, Humbert C, Silbermann F, Arrondel C, Tores F, et al. : Mutations in GREB1L cause bilateral kidney agenesis in humans and mice. Am J Hum Genet 5: 803–814, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosfeld A, Kreuzer M, Daniel C, Brand F, Schäfer AK, Chadt A, et al.: Whole-exome sequencing identifies mutations of TBC1D1 encoding a Rab-GTPase-activating protein in patients with congenital anomalies of the kidneys. Hum Genet 135: 69, 2016 [DOI] [PubMed] [Google Scholar]
- 50.Saisawat P, Tasic V, Vega-Warner V, Kehinde EO, Günther B, Airik R, et al.: Identification of two novel CAKUT-causing genes by massively parallel exon resequencing of candidate genes in patients with unilateral renal agenesis. Kidney Int 81: 196–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, et al.: Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 48: 457–465, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, et al.: Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85: 880–887, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, et al.: SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ACMG Board of Directors : ACMG policy statement: Updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 17: 68–69, 2015 [DOI] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.: 1000 Genome Project Data Processing Subgroup : The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al.: From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 1–33, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seelow D, Schuelke M, Hildebrandt F, Nurnberg P: HomozygosityMapper–an interactive approach to homozygosity mapping. Nucleic Acids Res 37: W593–599, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al.: Exome Aggregation Consortium : Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobreira N, Schiettecatte F, Valle D, Hamosh A: GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat 36: 928–930, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al.: American College of Medical Genetics and Genomics : ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15: 565–574, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al.: CORRIGENDUM: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 19: 606, 2017 [DOI] [PubMed] [Google Scholar]
- 62.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al.: Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med 19: 249–255, 2017 [DOI] [PubMed] [Google Scholar]
- 63.Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, et al.: A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 5: e1000353, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanna-Cherchi S, Kiryluk K, Burgess KE, Bodria M, Sampson MG, Hadley D, et al.: Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 91: 987–997, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braun DA, Rao J, Mollet G, Schapiro D, Daugeron M-C, Tan W, et al.: Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat Genet 49: 1529–1538, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al.: ACMG Laboratory Quality Assurance Committee : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beckmann JS: The Réunion paradox and the digenic model. Am J Hum Genet 59: 1400–1402, 1996 [PMC free article] [PubMed] [Google Scholar]
- 68.Tasic V, Gucev Z, Ristoska-Bojkovska N, Janchevska A, Ludecke HJ: Tricho-rhino-phalangeal syndrome in a 13-year-old girl with chronic renal failure and severe growth retardation. Renal Failure 36: 619–622, 2014 [DOI] [PubMed] [Google Scholar]
- 69.van der Ven AT, Shril S, Ityel H, Vivante A, Chen J, Hwang DY, et al.: Whole-exome sequencing reveals FAT4 mutations in a clinically unrecognizable patient with syndromic CAKUT: A case report. Mol Syndromology 8: 272–277, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.