Abstract
Background
Autosomal dominant tubulointerstitial kidney disease caused by mucin-1 gene (MUC1) mutations (ADTKD-MUC1) is characterized by progressive kidney failure. Genetic evaluation for ADTKD-MUC1 specifically tests for a cytosine duplication that creates a unique frameshift protein (MUC1fs). Our goal was to develop immunohistochemical methods to detect the MUC1fs created by the cytosine duplication and, possibly, by other similar frameshift mutations and to identify novel MUC1 mutations in individuals with positive immunohistochemical staining for the MUC1fs protein.
Methods
We performed MUC1fs immunostaining on urinary cell smears and various tissues from ADTKD-MUC1–positive and –negative controls as well as in individuals from 37 ADTKD families that were negative for mutations in known ADTKD genes. We used novel analytic methods to identify MUC1 frameshift mutations.
Results
After technique refinement, the sensitivity and specificity for MUC1fs immunostaining of urinary cell smears were 94.2% and 88.6%, respectively. Further genetic testing on 17 families with positive MUC1fs immunostaining revealed six families with five novel MUC1 frameshift mutations that all predict production of the identical MUC1fs protein.
Conclusions
We developed a noninvasive immunohistochemical method to detect MUC1fs that, after further validation, may be useful in the future for diagnostic testing. Production of the MUC1fs protein may be central to the pathogenesis of ADTKD-MUC1.
Keywords: Autosomal Dominant Tubulo-Interstitial Kidney Disease, MUC1, Inherited, kidney disease, Mucin-1 Kidney Disease, immunostaining, diagnosis
Autosomal dominant tubulointerstitial kidney disease (ADTKD) encompasses a broad group of inherited kidney diseases that are characterized by tubulointerstitial kidney disease and progressive kidney failure.1–5 Mutations in UMOD, REN, or MUC1 are the primary causes of ADTKD, with mutations in HNF1B6 and SEC61A14 as other causes of ADTKD.7
Autosomal dominant tubulointerstitial kidney disease caused by MUC1 mutations (ADTKD-MUC1) is caused by a cytosine duplication within a seven-cytosine tract in the variable number of tandem repeats (VNTR) region of the MUC1 gene. This duplication produces a frameshift during translation, resulting in a new protein, MUC1fs.3 The N-terminus and VNTR regions before the site of mutation will be translated normally, whereas the VNTR repeats after the frameshift have an 80% reduction in serine and threonine residues and the introduction of one cysteine and six basic amino acid residues per mutated VNTR unit.3 The new protein has a very high pI and is postulated to be toxic to renal tubular cells.8 Because of its new structure, unique antibodies can be developed that recognize the MUC1fs but not the wild-type MUC1.
The high guanosine/cytosine content and repetitive nature of the VNTR region means that mutational analysis of MUC1 has been extremely difficult. At this time, only the cytosine duplication can be identified by a Clinical Laboratory Improvement Amendments–approved genetic test as a cause of ADTKD-MUC1,9 and no truncating or missense MUC1 mutations have been found that result in ADTKD-MUC1. In addition, the MUC1 knockout mouse has not been found to have kidney disease.10 For this reason, it has been hypothesized that the specific MUC1fs produced by the cytosine duplication is critical to the pathogenesis of ADTKD-MUC1.3,11 It has been postulated that there may be other, distinct MUC1 mutations that produce the same MUC1fs and result in ADTKD-MUC1. A family with a deletion of 2 bp before the VNTR, resulting in the same MUC1fs protein, has also been reported.12
In this study we first performed immunostaining for the MUC1fs protein on epithelial tissues and urinary cell smears in individuals with ADTKD-MUC1 (genetically identified with the MUC1 cytosine duplication) and negative controls to determine if we could make a reliable diagnosis of ADTKD-MUC1 in this manner. We then tested epithelial tissue and urine from 37 ADTKD families with negative genetic testing for ADTKD genes and the MUC1 cytosine duplication and identified 17 families with positive urinary cells or tissue staining for the MUC1fs protein. Further genetic analyses on these families revealed five new distinct frameshift mutations in six families that predicted and resulted in the creation of the MUC1fs protein.
Methods
See Figure 1 for flow diagram.
Figure 1.
Work-flow diagram shows the distribution of samples used for validation of the methodology and for identification of families with ADTKD-MUC1.
Clinical Evaluation
The Wake Forest School of Medicine ADTKD registry comprises over 450 families referred to A.J.B. by physicians and/or family members since 1996. Families are screened by A.J.B., and the following data are collected: demographics, pedigree, medical history, laboratory values, imaging results, and biopsy reports.
UMOD/REN Sequencing and Standard MUC1 Genotyping
Whole blood or saliva was collected and DNA isolated by standard methodology. ADTKD-UMOD and ADTKD-REN genotyping was performed as previously described, using either a custom gene panel4 or candidate gene Sanger sequencing.5,13 ADTKD-MUC1 genotyping was performed by the Broad Institute (Cambridge, MA).9
Preparation of Urinary Cell Smears and Clinical Biopsy Material
Participants were asked to provide a 100 ml second morning spot urine sample, which was cooled and shipped overnight to Wake Forest School of Medicine with an ice pack. Evaluation of immediate and overnight delay of urine processing found that results were consistent when processing was delayed because of shipping (data not shown). Urine was centrifuged for 10 minutes at 1600×g. The supernatant was removed and the pellet was washed with 5 ml of wash buffer (2 mM EDTA, 0.1% BSA in 1× PBS). The sample was centrifuged again for 10 minutes at 1600×g. The supernatant was removed, and the pellet resuspended in 150 μl of wash buffer. Then, 30 μl of this suspension was pipetted onto a Polysine microscope adhesion slide (Thermo Fisher Scientific, Grand Island, NY) and smeared to create an even coating of the suspension over the slide surface. Slides were dried at ambient temperature for 30 minutes. The suspension was then fixed by placing the slides in ice-cold 100% methanol for 10 minutes at −20°C and drying the slides at ambient temperature. Slides could be stored at −20°C for up to 6 weeks and were shipped on dry ice to prevent particulate formation from water vapor in transit.
Multiple efforts were made to obtain residual paraffin slides not required for clinical diagnosis from as many prior clinical biopsy specimens in patients with ADTKD-MUC1 and ADTKD-UMOD as possible.
Immunostaining of Biopsy Specimens and Urinary Cell Smears
To detect MUC1fs, we prepared, obtained from collaborators, or purchased from various vendors, a series of 27 polyclonal and mAb raised against or interacting with various MUC1fs peptide fragments. These antibodies were tested under different conditions with positive and negative controls to determine which antibody functioned optimally for immunostaining of tissues and urine.
Tissue specimen immunodetection of MUC1fs was performed in formaldehyde-fixed human tissue using the methodology previously described.3 The choice of antibody was made on the basis of antibody availability over the time course of the study. For MUC1fs detection in kidney tissue, we used custom-prepared rabbit antibody PA4301 raised against the peptide SPRCHLGPGHQAGPGLHRPP (Open Biosystems, Huntsville, AL). These results were consistently confirmed with Fab fragment AbD22625 (selected from the Human Combinatorial Antibody Library (AbD Serotec, Puchheim, Germany) by screening with the peptide CHLGPGHQAGPGLHRPPSPR) and SISCAPA Peptide B antibody, which was raised against peptide CHLGPGEQAGPGLHR and provided by the Broad Institute. For MUC1fs detection in skin specimens we used the SISCAPA Peptide B antibody. For MUC1fs detection in all breast tissues we used Fab fragment AbD2265454 antibody (selected from the Human Combinatorial Antibody Library by screening with peptide GPGLHRPPSPRCHLGPGHQA). Confirmatory results were obtained with PA4301. For urinary cell testing, Fab fragment AbD2265454 was used.
The wild-type MUC1 protein was detected with monoclonal Mouse anti-Human Epithelial Membrane Antigen (EMA) antibody (Dako, Glostrup, Denmark), diluted at 1:400 in 5% BSA in PBS. Detection of bound primary antibody was achieved using either Dako EnVision+TM Peroxidase Rabbit Kit (Dako) or System-HRP labeled Polymer Anti-mouse (Dako), for rabbit or mouse antibodies, respectively, with 3,3′-diaminobenzidine as substrate. For frozen, fixed urinary cell smears, a 45-minute incubation in 5% FBS with 0.05% Tween 20 at room temperature was performed for protein blocking. After the protein blocking, slides were incubated with AbD22654 antibody diluted in 5% FBS with 0.05% Tween 20 overnight at 4°C. Incubation with the primary antibody was followed by four 2-minute washes with PBS with 0.05% Tween 20. Detection of bound primary antibody was achieved using a 60 minute incubation at 37°C with Mouse anti–V5-Tag AF488 antibody (AbD Serotec). After secondary antibody incubation, slides were washed four times for 2 minutes in PBS with 0.05% Tween 20 and mounted into ProLong Gold Antifade mountant with DAPI (Thermo Fisher Scientific). Slides were analyzed using confocal microscopy.
For parallel detection of MUC1fs and MUC1, protein-blocked slides were incubated with fluorescently labeled (Alexa 488) AbD22654 antibody (AbD Serotec, Puchheim, Germany) and monoclonal Mouse anti-Human EMA antibody, diluted 1:400 in 5% BSA in PBS. Detection of bound anti-EMA primary antibody was achieved using Donkey anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 555 (Thermo Fisher Scientific). Detection of uroplakin 1A (UPK1A) was achieved using polyclonal Rabbit anti-UPK1A (HPA049879; Sigma) and Donkey Anti-Rabbit AF555 (Thermo Fisher Scientific) as primary and secondary antibodies, respectively. Slides were mounted and analyzed by confocal microscopy.
Confocal Microscopy, Image Acquisition, and Analysis
XYZ images were sampled according to Nyquist criterion, using Leica SP8× laser scanning confocal microscopy, HC PL Apo objective (63×, N.A. 1.40), and 405, 488, and 555 laser lines. Images were restored using a classic maximum likelihood restoration algorithm in Huygens Professional Software (SVI, Hilversum, The Netherlands).
Formaldehyde-fixed tissue biopsy specimens and urinary cell smears were processed in a blinded manner by V.B. and M.Ž. Healthy controls, controls with CKD (defined as an eGFR<60 ml/min per 1.73 m2), and individuals with a genetic diagnosis of ADTKD-MUC1 or ADTKD-UMOD were included with individuals from families whose genetic testing was negative or not yet performed. Urinary smears were prepared at Wake Forest School of Medicine and mailed to the Charles University in four separate batches over time, as they were collected. The samples were analyzed at Charles University, and these results were then correlated at Wake Forest School of Medicine with clinical data. To explore the cause of false positive and false negative results, characteristics were compared between the proportion of true results (true positive and true negative) and false results (false positive and false negative) with a chi-squared or t test.
MUC1 VNTR Sequencing
Sanger Sequencing of Exon 1 of MUC1
Exon 1 of MUC1 was directly amplified from genomic DNA using primers gM1U-T7 (5′-AATACGACTCACTATAGTTGTCACCTGTCACCTGCTC-3′) and gM1L-RP (5′-GAAACAGCTATGACCATGGCATGACCAGAACCCGTAAC-3′). The resultant PCR products were purified and sequenced using the version 3.1 Dye Terminator cycle sequencing kit (Thermo Fisher Scientific) with electrophoresis on an ABI 3500XL Avant Genetic Analyzer (Thermo Fisher Scientific).
Illumina Sequencing of the VNTR Region of MUC1
The VNTR is very difficult to sequence because its repetitive nature and high guanosine and cytosine content. We developed a novel approach in which we looked specifically for frameshift mutations occurring within one of the VNTR repetitive units with the use of the Illumina HiSeq 2500 system. First, PCR amplification of the VNTR was performed, resulting in multiple repetitive DNA sequences of the VNTR. These sequences were then read with the Illumina system (see the following references for an explanation of the Illumina system).14,15 The VNTR region was directly amplified from genomic DNA using primers PS2F-T7 (5′-GGATCCTAATACGACTCACTATAGGAACAGACCACCATGGGAGAAAAGGAGACTTCGGCTACCCAG-3′) and PS3 (specified above) and long-range PCR (TaKaRa LA Taq DNA Polymerase with GC Buffers; Takara, Mountain View, CA). The resulting PCR products were purified using AmpureXP Beads fragmented to 400 bp with the Covaris E220 System and purified again with AmpureXP Beads. The sequencing library was prepared from the fragmented PCR product using the NEBNext UltraII DNA Library Prep Kit for Illumina and sequenced using the Illumina HiSeq 2500 system at the genomic facility in Motol University Hospital (Prague, Czech Republic). In this process, the individual nucleotide sequences derived from the PCR are sequenced in parallel, resulting in multiple, repeated sequence readings from the VNTR. The resulting sequence reads were analyzed directly, without mapping to the reference genome. Only samples that generated >10,000 reads were included in further analysis.
Targeted Genotyping of Identified Mutations
The 28dupA and 26_27 insG mutations were confirmed by a modified mass spectrometry-based assay9 at the Broad Institute. The 28dupA mutation was interrogated using the ADTKD-MUC1 7C probe (5′-CGGGCTCCACCGCCCCCCC-3′) and a nucleotide mix of dATP, ddCTP, and ddGTP. The 28dupA extension product is observed at 6571 D. The 26_27insG mutation was interrogated using the ADTKD-MUC1 6C probe (5′-CGGGCTCCACCGCCCCCCC-3′) with a nucleotide mix of dG and ddCTP in the probe extension reaction. The 26_27insG extension product is observed at 5944.85 D. For the 1_16dup and 23delinsAT mutation, the VNTR of MUC1 was amplified from genomic DNA using long-range PCR as described above. Obtained PCR products were cleaved by EciI and FokI, respectively, and resulting restriction fragments were analyzed using agarose gel electrophoresis.
Study Approval
This study was approved by the Institutional Review Boards of the First Faculty of Medicine, Charles University in Prague, and Wake Forest University Health Sciences Institutional Review Board (WFUHS IRB00000352). It adhered to the Declaration of Helsinki. Informed consent was obtained from all individuals. Authorization for release of biopsy materials by primary referral hospital/institute was also obtained.
Results
MUC1fs Immunostaining of Kidney Biopsy Specimens
Paraffin slides were obtained from prior kidney biopsies performed for clinical indications in 12 individuals with ADTKD-MUC1 and 11 with ADTKD-UMOD (Supplemental Table 1). Positivity was determined by the presence and intensity of MUC1fs intracytoplasmic granules found in the distal tubule and collecting duct cells (Figure 2, C and D), where wild-type MUC1 is constitutively expressed (Figure 2, A, B, E, and F). Eleven out of 12 samples from individuals with ADTKD-MUC1 were positive for the MUC1fs protein, with one patient having minimal/negative staining. Immunostaining was negative in control samples (Figure 2, G and H) in nine out of 11 samples, but positive in two individuals. The sensitivity for immunostaining of kidney biopsy specimens was 91.7% and specificity was 81.8%.
Figure 2.
Immunohistochemical staining of skin, sebaceous glands, and breast ducts shows positive MUC1fs staining in genetically affected individuals with ADTKD-MUC1 and negative MUC1fs staining in controls. (A) Survey of a kidney section showing positive staining for MUC1 in cortical distal tubules and collecting ducts in a patient with ADTKD-MUC1 and (B) a detailed view showing distal tubules positively stained for MUC1 with maximal staining intensity on the apical membranes of tubular cells. (C) Survey of a parallel kidney section from the patient with ADTKD-MUC1 stained with an antibody against MUC1fs with positivity in corresponding structures and (D) a detailed view showing finely granular intracellular MUC1fs staining pattern in distal tubules. (E) Survey of a control kidney section stained with an antibody against MUC1 and (F) a detailed view showing intracellular positivity in the distal tubule with an accent on the apical pole of the plasma membrane of tubular cells. (G) Survey of a parallel control kidney section stained with an antibody against MUC1fs and (H) a detailed view, both demonstrating negative staining in corresponding tubules. (I) Strong positivity for MUC1 in sebaceous glands in a skin biopsy specimen from a patient with ADTKD-MUC1 and (J) a detailed view. (K) Less intensive but distinct positivity of MUC1fs in sebaceous glands in a patient with ADTKD-MUC1 and (L) a detailed view of the patient’s MUC1fs-positive sebaceous glands. (M) Strong positivity of MUC1 in sebaceous glands in a control skin biopsy sample and (N) a detailed view. (O and P) Negative MUC1fs staining in sebaceous glands in a control skin biopsy sample demonstrated in a low power view (O) and (P) in detail. Epithelial cells in sweat glands (marked by arrows) displaying MUC1 positivity at the apical poles [demonstrated in a control skin biopsy sample in (M)] were MUC1fs-negative in a patient with ADTKD-MUC1 (K). (Q) Epithelial cells in breast ducts in a male patient with ADTKD-MUC1 display strong positivity of MUC1 at the apical poles and (R) distinct granular cytoplasmic positivity of MUC1fs. (S) Breast ducts in a male control display a similar staining pattern of MUC1 but (T) no immunostaining with the antibody detecting MUC1fs.
MUC1fs Immunostaining of Other MUC1-Expressing Epithelial Tissue Samples
Biopsy specimens from other MUC1-expressing epithelial tissue were analyzed in individuals with ADTKD-MUC1. This biopsy material was very limited, as only a few patients had undergone biopsy of nonrenal tissue. In two out of three skin biopsy samples from three individuals, MUC1fs was identified in the sebaceous glands (where wild-type MUC1 is constitutively expressed) (see Figure 2, K and L). The third sample lacked sebaceous glands. MUC1 and MUC1fs were identified in normal epithelial structures in two of three breast biopsy samples from three affected individuals (Figure 2, Q and R). In the third sample, MUC1fs was only identified in structures of carcinoma and less intensively in nontumor epithelium. There was also positive staining of the lung, colon, and fallopian tube (not shown).
MUC1fs immunostaining of nonrenal epithelial tissue was performed in three samples from two individuals with ADTKD-UMOD (negative controls). In a colon and skin biopsy specimen from one individual, the skin biopsy sample was positive and the colon biopsy sample was negative. A colon biopsy sample from another individual was negative.
Immunostaining of Urinary Cell Smears
Because of the limited quantity and quality of biopsy material, we decided on the alternative approach of staining urinary smears. Initially, we characterized expression of MUC1fs and MUC1 in urinary cells from ADTKD-MUC1 individuals in various CKD stages and controls (Figure 3). In affected individuals we observed strong diffuse to granular intracellular staining of MUC1fs in urothelial cells from all layers (Figure 3, A and B) that was absent in cells from healthy controls (Figure 3, C and D). In affected individuals and controls, MUC1 was detected in the form of cytoplasmic granules with distinct plasma membrane staining (Figure 3, E–H). Positive MUC1fs staining was detected in genetically affected individuals with normal kidney function (Figure 3, A and B), advanced CKD (Figure 3, K and L), and in an individual on dialysis (Figure 3, I and J). In advanced CKD in affected individuals, we further observed that cells that were strongly positive for both MUC1 and MUC1fs (Figure 3, I–L) and were much smaller in size (diameter approximately 15 µm) than cells from affected and unaffected individuals with normal kidney function (diameter 30–40 µm) (Figure 3, A–H).
Figure 3.
Immunohistochemical staining of urinary smears from individuals with ADTKD-MUC1 reveals positive MUC1fs staining in all stages of kidney disease. Urinary pellets were smeared on glass slides, then fixed and stained with anti-MUC1fs and anti-MUC1 antibodies. Confocal images show DAPI (blue) and either MUC1fs or MUC1 (green). (A and B) Strong diffuse to granular intracellular staining of MUC1fs in superficial and intermediate urothelial cells in affected individuals. (C and D) Negative MUC1fs staining in urothelial cells from a healthy control. (E) Finely granular cytoplasmic staining in superficial urothelial cells and (F) distinct plasma membrane staining of MUC1 in intermediate urothelial cells in affected individuals that is similar to that in controls (G and H). Positive MUC1fs staining in urothelial cells was detected in (A and B) a genetically affected individual with normal kidney function, (I and J) in two individuals with advanced CKD on dialysis, (K) in a nondialyzed individual 1 month before transplantation, and (L) in another patient with advanced CKD on dialysis. Note that in individuals with advanced CKD, cells that stain strongly positive for both MUC1fs and MUC1 are much smaller in size (diameter ±15 µm) than controls (diameter 30–40 µm).
Illustrative immunofluorescence images of MUC1fs and MUC1 in urinary cells from individuals with ADTKD-MUC1, ADTKD-UMOD, and controls are shown in Figure 4. Urinary smears were positive for the MUC1fs protein in five patients with ADTKD-MUC1 who had undergone kidney transplantation 1.5, 4, 4, 7, and 8 years previously, suggesting that immunostaining for MUC1fs also occurs in cells that originate from the ureter, bladder, or urethra. Staining with the bladder cell marker UPK1A revealed that all UPK1A positive cells are positive for MUC1fs and that UPK1A-positive cells accounted for approximately 30% of cells that stained positive for MUC1fs (Supplemental Figures 1 and 2).
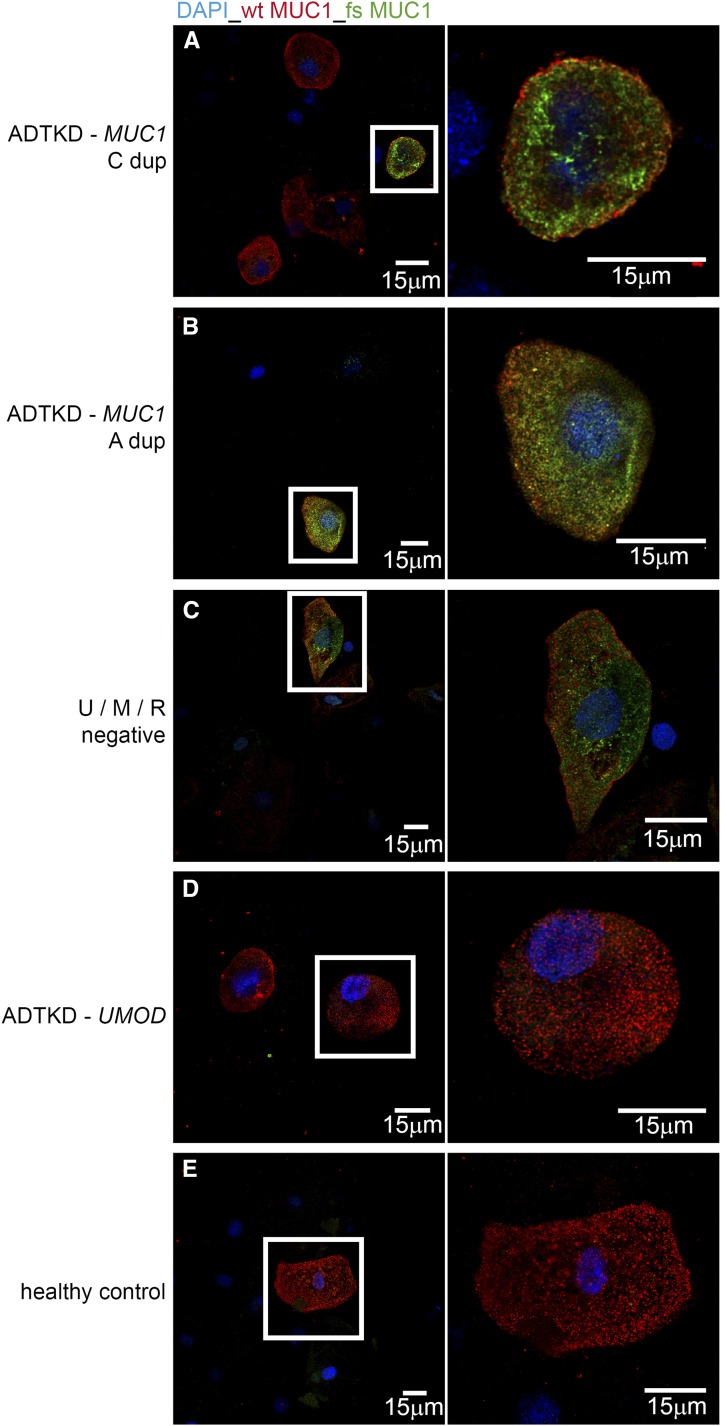
Figure 4.
Immunostaining for MUC1fs is positive in patients with different MUC1 mutations and is negative in a patient with ADTKD-UMOD. Urinary cell pellets were smeared on glass slides, then fixed and stained with anti-MUC1fs and anti-MUC1 antibodies. Merged confocal images show DAPI (blue), MUC1 (red), and MUC1fs (green). Panels illustrate the presence of MUC1fs in the cells of genotyped individuals with (A) the MUC1 27dupC mutation, (B) the MUC1 28dupA mutation, and (C) an unknown mutation. U/M/R negative denotes that the tested participant was negative for mutations in UMOD, REN, and the MUC1 mutations described here. Absence of MUC1fs is shown in an individual with (D) a UMOD mutation (ADTKD-UMOD) and (E) control. MUC1-positive cells outlined with a white rectangle are shown in detail in the corresponding image on the right.
After developing this technique, we then evaluated in a blinded manner four sequential groups of urinary smears. Smear quality and the number of cells on each smear were graded before immunostaining. Optimally prepared smears contained at least 100 cells, of which approximately 25% stained positive for MUC1 in women and 35% in men. In affected individuals approximately 40% of MUC1-positive cells were positive for MUC1fs. Table 1 shows the characteristics of the sample population and the distribution of false positive/false negative results. There were 173 urinary cell smears tested from 69 genotyped individuals with ADTKD-MUC1, 45 individuals with ADTKD-UMOD, 35 healthy individuals, and 24 with other causes of CKD. The sensitivity was 94.2% and specificity was 88.6%. Comparisons between groups (see Table 1) showed no statistically significant differences that could explain false positive and false negative results. However, 14 out of 17 false positive/false negative results were from smears that were <60% confluent with <100 cells, with three out of 64 (4.6%) smears with >100 cells having false results versus 14 out of 109 (12.9%) of smears with <100 cells (P=0.08). Supplemental Figure 3 shows examples of false positive and false negative staining. Seventeen of nineteen individuals providing two samples had consistent results; in two, the first sample was a false negative and the second a true positive.
Table 1.
Characteristics of urinary samples
| Characteristics | n | True Positive | True Negative | False Positive | False Negative | Sensitivity, % | Specificity, % |
|---|---|---|---|---|---|---|---|
| Age | 173 | 45.7±14.3 | 39.3±15.4 | 49.7±16.3 | 36.5±17.3 | 94.2 | 88.6 |
| Sex | |||||||
| Men | 61 | 24 (39.3) | 30 (49.2) | 7 (11.5) | 0 | 100 | 81.8 |
| Women | 112 | 41 (36.6) | 61 (54.5) | 6 (5.4) | 4 (3.6) | 91.1 | 91.0 |
| Race | |||||||
| White | 135 | 46 (34.1) | 79 (58.5) | 9 (6.7) | 1 (0.7) | 97.9 | 98.8 |
| Black | 12 | 6 (50.0) | 4 (33.3) | 1 (8.3) | 1 (8.3) | 85.7 | 80.0 |
| Hispanic | 24 | 13 (54.2) | 6 (25.0) | 3 (12.5) | 2 (8.3) | 86.7 | 66.7 |
| Other | 1 | 0 | 1 (100) | 0 | 0 | 0 | 100 |
| eGFR, ml/min per 1.73 m2 | 173 | 50.3±25.7 | 70.9±42.7 | 66.7±33.0 | 66.6±31.6 | 94.2 | 88.6 |
| Batch 1 | 45 | 27 (60.0) | 11 (24.4) | 4 (8.9) | 3 (6.7) | 90.0 | 73.3 |
| Batch 2 | 55 | 15 (27.3) | 36 (65.5) | 3 (5.5) | 1 (1.8) | 93.8 | 92.3 |
| Batch 3 | 44 | 16 (36.4) | 27 (61.4) | 1 (2.3) | 0 | 100 | 96.4 |
| Batch 4 | 24 | 7 (29.1) | 17 (58.6) | 5 (17.2) | 0 | 100 | 77.3 |
| ADTKD-MUC1 with CKD | 52 | 50 (96.2) | 2 (3.9) | 96.2 | 0 | ||
| ADTKD-MUC1 without CKD | 17 | 15 (88.3) | 2 (11.8) | 88.2 | 0 | ||
| Controls with CKD | 24 | 22 (91.7) | 2 (8.3) | 0 | 91.7 | ||
| Controls without CKD | 35 | 28 (80.0) | 7 (20.0) | 0 | 80.0 | ||
| ADTKD-UMOD with CKD | 30 | 26 (86.7) | 4 (13.3) | 0 | 86.7 | ||
| ADTKD-UMOD without CKD | 15 | 15 (100) | 0 | 0 | 79.0 | ||
| Transplanted | 13 | 10 (76.9) | 3 (23.1) | 0 | 0 | 100 | 100 |
| Not transplanted | 160 | 55 (34.4) | 88 (55.0) | 13 (8.1) | 4 (2.5) | 93.2 | 87.1 |
| Low cell count (<100 cells per slide) | 109 | 43 (39.5) | 52 (47.7) | 11 (10.1) | 3 (2.8) | 93.5 | 82.5 |
| Optimal cell count (≥100 cells per slide) | 64 | 22 (34.4) | 39 (60.9) | 2 (3.1) | 1 (1.6) | 91.7 | 97.5 |
Continuous variables are represented as mean±SD. Discrete values are represented as n (%). There were no statistically significant differences between any of the groups. Sensitivity and specificity were calculated for each characteristic.
Immunostaining was also carried out in 60 individuals from 37 families with clinical features characteristic of ADTKD, but who were negative for mutations in ADTKD genes and the MUC1 cytosine duplication. Seventeen families were found to have biopsy samples or urinary cell smears that were positive for MUC1fs and underwent further genetic evaluation.
Identification of Novel MUC1 Mutations
To identify the addition or deletion of nucleotides within one of the VNTR units, the obtained DNA sequences were first aligned using the Basic Local Alignment Search Tool (BlastN)16 program. The most highly conserved ten nucleotide sequences (which we term sequence anchors) of the VNTR repeats were identified (Figure 5A). To maintain the original reading frame of MUC1, the distance between these repeating ten nucleotide sequences in neighboring VNTRs must be 3n nucleotides (three nucleotides for each amino acid produced). To find candidate frameshift mutations encoding the MUC1fs protein, we therefore searched for sequences with insertion of either 1, 4, 7, 10, 13, 16,…(3n+1) nucleotides or with deletion of 2, 5, 8, 11, 14,…(3n−1) nucleotides between the sequence anchors (Figure 5A).
Figure 5.
MUC1 VNTR sequencing identifies novel mutations causing ADTKD-MUC1. (A) Sequence logo showing the most conserved regions of the VNTR repeats. Corresponding amino acid sequences of wild-type MUC1 (wt_AA) and MUC1fs (mut_AA) are shown below. To find novel frameshift mutations that change the open reading frame, different conserved 10-mers of the wild-type repeat were used as sequence anchors (underlined DNA sequence as an example). For each anchor pair, all sequences delimited by these two anchors that are changing an open reading frame (i.e., adding or deleting nucleotides) were selected from the FASTQ file. (B) Sequences of the canonical 60 nucleotide long wild-type VNTR repeat (wt) and candidate frameshift mutations identified in this study. (C) Random mutations are generated in DNA molecules during PCR amplification step. To find true germline mutations, the percentage of reads with a given sequence (putative frameshift mutation) from all reads was calculated for each of the analyzed samples (y-axis), and this needed to be higher than the average+2 SD of the nine wild-type control samples. Indicated are numbers of controls (wt), patients with individual MUC1 mutations (27dupC, 28dupA, 26_27insG, 1–16dup, 23delinsAT, 51dupC), and individuals with still unknown MUC1 mutation(s) who have urinary cell smears positive for MUC1fs and who tested negative for 27dupC by conventional genotyping assay (unknown). (D) 27dupC, confirmed by a mass spectrometry-based primer extension assay. The 27dupC extension product is observed at 5904 D (red asterisk). (E) 28dupA, confirmed by a mass spectrometry-based assay. The 28dupA extension product is observed at 6571 D (red asterisk). (F) 26_27insG, confirmed by a mass spectrometry-based assay. The 26_27insG extension product is observed at 5944.85 D (red arrow). (G) 1–16dup confirmed by restriction analysis. The mutation creates new restriction site for EciI enzyme. The electrophoretogram shows amplified VNTR regions of the affected patient (P1), two healthy relatives (H1, H2), and one unrelated control (NC) after (EciI) and before restriction by EciI (PCR). The patient’s (P1) mutated allele (5000 bp) was cut into two fragments of 3000 and 2000 bp. (H) 23delinsAT, confirmed by restriction analysis. The mutation creates new restriction site for FokI enzyme. The electrophoretogram is showing amplified VNTR regions of two affected patients (P1, P2) and one unrelated control (NC) after (FokI) and before restriction by FokI (PCR). The patients’ (P1, P2) mutated alleles (3000 bp) were cut into two fragments of 2000 and 1000 bp.
These frame-changing sequences were then counted in the DNA library created by Illumina sequencing, and their percentage from all obtained sequence reads was calculated for each particular sample. Because various mutations (that could also cause a frameshift) can be randomly introduced during the PCR amplification step, we considered as a real mutation only those that had the percentage of reads with a particular putative frameshift mutation higher than the mean plus 2 SDs of that in nine control samples. For example, a C duplication, 27dupC, is introduced in 0.36%±0.06% of reads randomly in wild-type DNA with PCR amplification. For a 27dupC duplication to be identified, we required at least 0.48% reads (mean+2 SD of the wild-type) to be present, as demonstrated by the 27dupC panel in Figure 5C. Using this approach, we were able to confirm the cytosine duplication (that we denote as 27dupC, see Figure 5B for numbering) in five individuals (Figure 5C) previously identified using the conventional primer extension assay (Figure 5D).
In six families we identified five novel frameshift mutations within the VNTR region of the MUC1 gene. These mutations included two families with an adenine duplication following a seven-cytosine tract (28dupA), one guanine insertion within the seven-cytosine tract (26_27insG), one 16 bp duplication (1_16dup), one deletion of a cytosine and parallel insertion of adenine and thymine in the seven-cytosine tract (23delinsAT), and one cytosine duplication in a four-cytosine tract (51dupC) (see Figure 5, B and C). All five mutations were predicted to encode the same MUC1fs protein that is found in patients with the 27dupC duplication. The existence and segregation of the 28dupA and 26_27insG were confirmed by a modified mass spectrometry-based assay9 at the Broad Institute (Figure 5, E and F). The 1_16dup and the 23delinsAT create within the VNTR region unique restriction sites within the VNTR for EciI and FokI, respectively, and PCR-RFLP assays using these two restriction enzymes were used for targeted genotyping (Figure 5, G and H). The 51dupC is not amenable to these alternative genotyping methods and therefore has not been confirmed yet.
The novel mutations segregated with clinical status in all six families, with all individuals with stage III or higher CKD found to have mutations (Figure 6, Table 2). Some younger individuals with the novel mutations did not have advanced CKD, as is often seen with ADTKD-MUC1 (see family trees in Figure 6).12,17
Figure 6.
Pedigrees of families with novel MUC1 mutations show characteristic autosomal dominant transmission. Clinically affected family members (defined as stage III-V CKD or requiring dialysis/kidney transplant) are shown with a black symbol, clinically unaffected family members are shown with a white symbol. Gray symbols indicate that clinical status is unknown. The plus sign (+) indicates that genotyping was performed and a mutation resulting in MUC1fs was identified in the individual. A dash (−) means the patient was genotyped and found not to have an MUC1 mutation.
Table 2.
Clinical characteristics of individuals in families with novel MUC1 mutations
| Individual | Mutation | Age, yr | eGFR, ESRD,a or Deceased | MUC1fs Positive | Other Clinical Findings |
|---|---|---|---|---|---|
| A-II-2 | 28dupA | 38 | ESRD | ||
| A-II-4 | 28dupA | 46 | ESRD | Gout at age 44 yr | |
| A-II-6 | No mutation | 66 | Not available | ||
| A-II-8 | No mutation | 66 | 90 | ||
| A-III-1 | 28dupA | 41 | ESRD | Kidney and breast biopsies | |
| A-III-2 | 28dupA | 45 | 49 | Urinary cell smear | |
| A-III-4 | 28dupA | 43 | 64 | Urinary cell smear | Enuresis past age 4 yr |
| A-III-6 | 28dupA | 37 | ESRD | ||
| A-III-8 | 28dupA | 45 | 53 | Urinary cell smear | |
| A-III-10 | No mutation | 40 | 90 | ||
| B-I-2 | Not tested | 45 | ESRD | ||
| B-II-2 | 28dupA | 19 | ESRD | ||
| B-III-1 | No mutation | 19 | 80 | Proteinuria | |
| C-I-1 | Not tested | 81 | Deceased | Uremia | |
| C-II-2 | Not tested | 59 | Deceased | CKD | |
| C-II-3 | Not tested | 50 | Deceased | ||
| C-III-1 | Not tested | 58 | 52 | High uric acid | |
| C-III-5 | No mutation | 61 | 102 | ||
| C-III-6 | 26_27insG | 32 | ESRD | ||
| C-IV-2 | No mutation | 40 | 123 | ||
| C-IV-3 | Not tested | 28 | ESRD | ||
| C-IV-4 | No mutation | 47 | Not available | ||
| C-V-1 | 26_27insG | 24 | 70 | Kidney biopsy | |
| D-I-1 | Not tested | 43 | ESRD | ||
| D-II-1 | Not tested | Not available | ESRD | High uric acid | |
| D-II-3 | Not tested | Not available | ESRD | High uric acid | |
| D-II-5 | Not tested | 40 | ESRD | Kidney biopsy | Gout |
| D-II-7 | 1_16dup | Clinically affected but no laboratory values can be obtained | |||
| D-III-4 | No mutation | 11 | 113 | ||
| E-I-2 | Not tested | Not available | ESRD | ||
| E-II-1 | Not tested | Clinically affected but no laboratory values can be obtained | |||
| E-II-4 | Not tested | 53 | ESRD | ||
| E-III-1 | Not tested | Clinically affected but no laboratory values can be obtained | |||
| E-III-4 | 23delinsAT | 57 | 24 | Kidney biopsy | |
| E-III-6 | 23delinsAT | 54 | ESRD | ||
| F-II-2 | Not tested | 89 | Deceased | CKD | |
| F-II-6 | Not tested | Not available | ESRD | ||
| F-II-8 | Not tested | Not available | ESRD | ||
| F-III-2 | Not tested | 52 | ESRD | ||
| F-III-8 | Not tested | 35 | ESRD | ||
| F-III-9 | Not tested | 31 | ESRD | ||
| F-III-15 | Not tested | Not available | ESRD | ||
| F-III-16 | Not tested | 28 | ESRD | ||
| F-IV-2 | 51dupC | 26 | 18 | Urinary cell smear |
The families identified with new mutations had clinical findings similar to those of other families with ADTKD-MUC1 because of a cytosine duplication. Supplemental Figure 4 shows the distribution of age of ESRD for families with the new mutations versus other families with the cytosine duplication.
The remaining 11 families did not have one of the five newly identified mutations, and further genetic analysis of these families is underway.
Discussion
This investigation resulted in three significant advances in the study of ADTKD-MUC1. We developed and validated a new, noninvasive diagnostic method with immunohistochemical staining of urinary cell smears, we developed alternative genetic methods to identify frameshift mutations in the VNTR region of MUC1, and we identified in six families five new mutations that each encoded the same MUC1fs protein that is produced by the previously described and more common 27dupC mutation.
The majority of ADTKD disease-causing mutations in the MUC1 gene are believed to occur in the VNTR region. The high guanosine/cytosine content of this region prevents routine genetic approaches to identify a mutation. Given the inability to perform standard genetic sequencing, we decided to pursue an alternative approach, in which we tried to identify the MUC1fs protein by immunohistochemical methods in various tissues.
Although we were able to identify MUC1fs staining in 11 out of 12 kidney biopsy specimens, most patients do not undergo kidney biopsy and obtaining kidney samples can be associated with complications. Skin biopsy offers a more practical approach. However, MUC1 is only expressed in the sebaceous glands of the skin, and these glands are not consistently found in skin biopsy specimens. Thus, immunohistochemical staining of urinary cells provides the optimal diagnostic test. Urine is easily obtained, and epithelial cells present in urine express sufficient amounts of MUC1 for immunostaining. As in this case, multiple urine specimens can be obtained to optimize technique and validate results. We were able to develop a test that can easily be performed and has high sensitivity and specificity. The traditional urinalysis has been used to diagnose rare conditions such as adenine phosphoribosyltransferase deficiency18 and cystinuria,19 but we believe that this is the first time that immunohistochemical staining of the urine has been used systematically to diagnose an inherited kidney disease.
There were 17 false positive or false negative results out of 173 samples (9.8%). We were able to obtain repeat urine collections on two false negative samples that then returned as true positive. Fourteen of the 17 erroneous results occurred in samples that did not contain the optimal number of cells (approximately 100 per smear). We believe that poor smear quality (due to cell lysis, inadequate cells, or bacterial contamination) was the major cause of inaccurate results. Almost all of the samples obtained for this study were shipped to us overnight from many different regions of the United States. Future modifications will include obtaining larger urinary samples and the addition of antimicrobial agents to prevent contamination. We also need to consider the possibility that the frameshift mutation may occur late in the VNTR, resulting in very few of the repeat units having the frameshift sequence. Such a mutation would provide fewer sites for antigen recognition and reduce the sensitivity of immunodetection.
Urine testing remains a research test and should not be used at this time in the clinical diagnosis of individuals with this disorder. In families with a clinical history consistent with ADTKD, we believe that genetic testing for the 27dupC and 28dupA (as in this study) should be performed first, followed by immunohistochemical staining of urinary cell smears. Such testing should be performed in a laboratory that has experience and has validated the technique with positive and negative controls.
From 37 families with clinical features characteristic of ADTKD but negative for mutations in ADTKD genes and the MUC1 27dupC mutation, we found 17 families with MUC1fs protein consistently identified in urinary cells. Further study of these 17 families identified five more mutations that cause ADTKD-MUC1 and encode the same MUC1fs protein as encoded by the 27dupC mutation found in the majority of cases of ADTKD-MUC1. A family has been reported with a 2 bp deletion before the VNTR that also encoded the same MUC1fs protein.12 Thus, there are now 57 reported families with MUC1 mutations3,12,17,20–23 and 134 other families we have identified (A.J. Bleyer, S. Kmoch, unpublished data). Of these 191 families, 183 have the 27dupC mutation and seven have a different mutation. All 191 affected families produce the same frameshifted protein.
Identification of other genetic causes of ADTKD-MUC1 has been a high priority for investigators in this area. First, identification of other mutations will be helpful in the diagnosis of families who do not have the 27dupC mutation. Second, it is believed that the particular MUC1fs protein created by the cytosine duplication is central to the pathogenesis of this disorder. Identification of other mutations that result in the creation of the same MUC1fs protein will help confirm this hypothesis. In contrast, if other mutations are found in MUC1 that cause ADTKD but do not encode the MUC1fs protein, one might hypothesize that loss of function of the MUC1 protein is central to the disease.
There were several weaknesses to our study. First, although we identified new frameshift mutations resulting in the creation of the MUC1fs protein, we could not rule out that other mutations in MUC1 cause ADTKD-MUC1. This will only be accomplished when the entire MUC1 genomic region can be reliably sequenced and genetically analyzed for other mutations and their segregation within individual families. Another weakness is that the current Illumina method does not automatically identify mutations, but rather we must take multiple bioinformatic approaches to find each new mutation. At this time, we could not identify mutations in 11 out of 17 families. We continue to develop and refine new sequencing techniques using non-PCR-based approaches for MUC1 enrichment and long-read single molecule MUC1-VNTR sequencing using Nanopore and PacBio platforms to identify and position the causative mutation(s)20 and to apply novel bioinformatic tools for identifying mutations in these families.
We are interested in studying additional families and can provide genetic testing and the urinary screening described here, free of charge. Please contact ableyer@wakehealth.edu for evaluation or consultation regarding families with ADTKD.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank The National Center for Medical Genomics (grant LM2015091) for help in MUC1 sequencing.
A.J.B., P.C., P.L., D.M.C., A.W., B.B.B., T.S., S.R.-B., O.V., G.P., and C.D. referred families for this study. M.Ž., A.J.B., S.K., A.P., V.B., H. Hartmannová, K.H., V.S., A.V., P.V., J.Z., M.V., J.S., and H. Hůlková developed methodology and initial identification of novel MUC1 mutations. V.R. coordinated the collection of samples. S.K., A.J.B., S.L.A., and A.G. designed, developed and provided antibody. M.Ž., A.P., and V.B. performed immunohistochemical staining, microscopy, and interpretation of staining. K.K. and R.P. prepared urinary smears. K.K., V.R., and R.P. prepared blinded analyses. M.D., B.B., M.H., and A.G. developed assays for detection and verification of adenine and guanine insertions. K.K. analyzed results, performed data interpretation, and prepared figures and tables related to clinical interpretation. A.J.B., S.K., M.Ž., and K.K. drafted and revised the manuscript. All authors approved the final draft of the manuscript.
This work was supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases grant R21 DK106584, project NV17-29786A from the Ministry of Health of the Czech Republic, grant LQ1604 NPU II from the Ministry of Education of the Czech Republic, by institutional programs of Charles University in Prague (UNCE 204064, PROGRES-Q26/LF1, and SVV 260367/2017). This work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “MUC1 Makes Me Miserable,” on pages 2257–2258.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018020180/-/DCSupplemental.
References
- 1.Bleyer AJ, Woodard AS, Shihabi Z, Sandhu J, Zhu H, Satko SG, et al.: Clinical characterization of a family with a mutation in the uromodulin (Tamm-Horsfall glycoprotein) gene. Kidney Int 64: 36–42, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, et al.: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirby A, Gnirke A, Jaffe DB, Barešová V, Pochet N, Blumenstiel B, et al.: Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 45: 299–303, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolar NA, Golzio C, Živná M, Hayot G, Van Hemelrijk C, Schepers D, et al.: Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am J Hum Genet 99: 174–187, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zivná M, Hůlková H, Matignon M, Hodanová K, Vylet’al P, Kalbácová M, et al.: Dominant renin gene mutations associated with early-onset hyperuricemia, anemia, and chronic kidney failure. Am J Hum Genet 85: 204–213, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhave JC, Bech AP, Wetzels JF, Nijenhuis T: Hepatocyte nuclear factor 1β-associated kidney disease: More than renal cysts and diabetes. J Am Soc Nephrol 27: 345–353, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleyer AJ, Kidd K, Živná M, Kmoch S: Autosomal dominant tubulointerstitial kidney disease. Adv Chronic Kidney Dis 24: 86–93, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleyer AJ, Kmoch S: Autosomal dominant tubulointerstitial kidney disease: Of names and genes. Kidney Int 86: 459–461, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Blumenstiel B, DeFelice M, Birsoy O, Bleyer AJ, Kmoch S, Carter TA, et al.: Development and validation of a mass spectrometry-based assay for the molecular diagnosis of mucin-1 kidney disease. J Mol Diagn 18: 566–571, 2016 [DOI] [PubMed] [Google Scholar]
- 10.Spicer AP, Rowse GJ, Lidner TK, Gendler SJ: Delayed mammary tumor progression in Muc-1 null mice. J Biol Chem 270: 30093–30101, 1995 [DOI] [PubMed] [Google Scholar]
- 11.Staubach S, Wenzel A, Beck BB, Rinschen MM, Müller S, Hanisch FG: Autosomal tubulointerstitial kidney disease-MUC1 type: Differential proteomics suggests that mutated MUC1 (insC) affects vesicular transport in renal epithelial cells. Proteomics 18: e1700456, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto S, Kaimori JY, Yoshimura T, Namba T, Imai A, Kobayashi K, et al.: Analysis of an ADTKD family with a novel frameshift mutation in MUC1 reveals characteristic features of mutant MUC1 protein. Nephrol Dial Transplant 32: 2010–2017, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Vylet’al P, Kublová M, Kalbácová M, Hodanová K, Baresová V, Stibůrková B, et al.: Alterations of uromodulin biology: A common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int 70: 1155–1169, 2006 [DOI] [PubMed] [Google Scholar]
- 14.USD Bioinformatics : Next generation sequencing: Illumina sequencing, 2014. Available at: https://www.slideshare.net/USDBioinformatics/illumina-sequencing. Accessed June 25, 2018
- 15.Technology Spotlight: Illumina Sequencing Technology. Available at : https://www.illumina.com/documents/products/techspotlights/techspotlight_sequencing.pdf 2010. Accessed June 25, 2018
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410, 1990 [DOI] [PubMed] [Google Scholar]
- 17.Bleyer AJ, Kmoch S, Antignac C, Robins V, Kidd K, Kelsoe JR, et al.: Variable clinical presentation of an MUC1 mutation causing medullary cystic kidney disease type 1. Clin J Am Soc Nephrol 9: 527–535, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edvardsson V, Palsson R, Olafsson I, Hjaltadottir G, Laxdal T: Clinical features and genotype of adenine phosphoribosyltransferase deficiency in Iceland. Am J Kidney Dis 38: 473–480, 2001 [DOI] [PubMed] [Google Scholar]
- 19.Sumorok N, Goldfarb DS: Update on cystinuria. Curr Opin Nephrol Hypertens 22: 427–431, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenzel A, Altmueller J, Ekici AB, Popp B, Stueber K, Thiele H, et al.: Single molecule real time sequencing in ADTKD-MUC1 allows complete assembly of the VNTR and exact positioning of causative mutations. Sci Rep 8: 4170, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musetti C, Babu D, Fusco I, Mellone S, Zonta A, Quaglia M, et al.: Testing for the cytosine insertion in the VNTR of the MUC1 gene in a cohort of Italian patients with autosomal dominant tubulointerstitial kidney disease. J Nephrol 29: 451–455, 2016 [DOI] [PubMed] [Google Scholar]
- 22.Yu SM, Bleyer AJ, Anis K, Herlitz L, Živná M, Hůlková H, et al.: Autosomal dominant tubulointerstitial kidney disease due to MUC1 mutation. Am J Kidney Dis 71: 495–500, 2018 [DOI] [PubMed] [Google Scholar]
- 23.Si N, Zheng K, Ma J, Meng XL, Li XM, Zhang X: Genetic testing of the mucin 1 gene-variable number tandem repeat single cytosine insertion mutation in a chinese family with medullary cystic kidney disease. Chin Med J (Engl) 130: 2459–2464, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.