Abstract
The telomerase RNA in yeasts is large, usually >1000 nt, and contains functional elements that have been extensively studied experimentally in several disparate species. Nevertheless, they are very difficult to detect by homology-based methods and so far have escaped annotation in the majority of the genomes of Saccharomycotina. This is a consequence of sequences that evolve rapidly at nucleotide level, are subject to large variations in size, and are highly plastic with respect to their secondary structures. Here, we report on a survey that was aimed at closing this gap in RNA annotation. Despite considerable efforts and the combination of a variety of different methods, it was only partially successful. While 27 new telomerase RNAs were identified, we had to restrict our efforts to the subgroup Saccharomycetacea because even this narrow subgroup was diverse enough to require different search models for different phylogenetic subgroups. More distant branches of the Saccharomycotina remain without annotated telomerase RNA.
Keywords: non-coding RNA, telomerase RNA, secondary structure, synteny, homology search, yeast
1. Introduction
The linear chromosomes of eukaryotes require a specialized mechanism for completing duplication. Most commonly this is achieved by a special reverse transcriptase, telomerase, that carries a specific RNA that includes the template with telomeric sequence [1]. Most likely, this constitutes the ancestral state in eukaryotes. Despite its crucial function, telomerase has been lost several times in both animals (in particular insects) and possibly also in some plants [2]. In some cases, the ancestral telomere structure has been replaced by tandem arrays of DNA sequences that look much like heterochromatin and can be elongated by gene conversion. Specialized telomere-specific retrotransposons are at work in Drosophila [3].
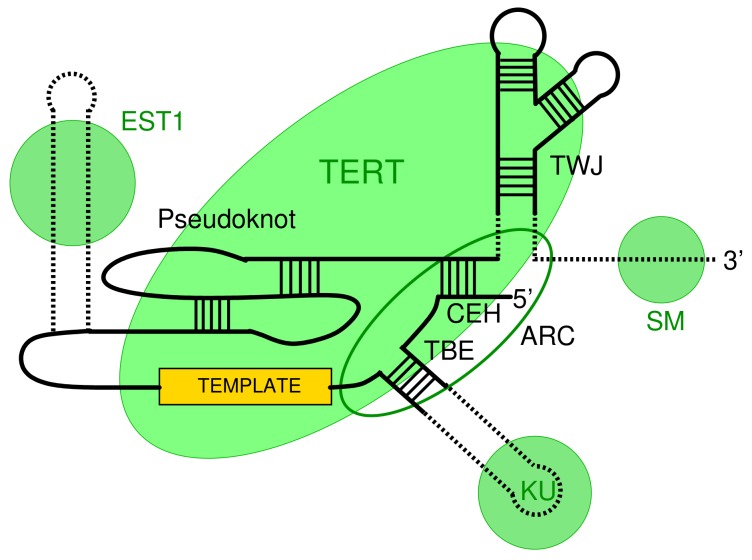
The telomerase (holo)enzyme consists of two main components, a specialized reverse transcriptase (TERT) and a RNA component (TER) that provides the template sequence. In addition, there are usually multiple clade-specific accessory protein components [4,5]. Four conserved regions in TER (Figure 1) are essential for telomerase activity: a core-enclosing helixig (CEH), template boundary element (TBE), the template sequence and a pseudoknot, are, in this order along the RNA, part of the the catalytic core [6]. The trans activating domain is involved in binding of TERT [7]. An Area of Required Connectivity (ARC) has been identified as the fifth essential element in Saccharomyces cerevisiae [6]. It is to some extent conserved in human telomerase RNA [8]. The ARC is not a localized sequence-structure feature but rather constitutes a combination of CEH, TBE, and some interactions between them. Hence, it is at least difficult to utilize the ARC for homology search. The three-way junction (TWJ) structure of this region region is widely conserved at least between animal and fungal telomerase RNAs (TER) [9]. It has been shown that, despite its conservation, the TWJ is not critical for TER function in S. cerevisiae [6,10,11]. The precisely defined template within TER is processively copied by TERT and regenerated, releasing a single-stranded DNA product [12].
Figure 1.
Schematic organization of telomerase RNA (TER). Contact regions for important binding sites are indicated by green circles (EST1, SM, and KU). The green ellipse denotes the contact region with the reverse transcriptase (TERT). Other major features are the template, the pseudoknot region, the template boundary element (TBE), the three-way junction (TWJ), and the Area of Required Connectivity (ARC). Adapted from [13].
Telomerase RNA is highly divergent. The TER in ciliates [14], human [15], and budding yeast [16,17,18] have a length of about 150 nt, 438 nt, and 1160 nt, respectively. A TER more than 2 kb in length has been reported for Candida glabrata [19], which, interestingly, seems to lack a TWJ. TERs in other kingdoms of eukaryotes have been discovered only quite recently in plants [20,21], excavates [22,23] and alveolates [24,25].
Despite their deeply conserved primary function and architectural similarities that seem to extend across eukaryotic kingdoms, TERs have turned out be very difficult to find by homology searches even within phylogenetically relatively narrow groups. Within the animal kingdom, even surveys of vertebrates turned out to be non-trivial [26]. Echinoderm TERs were found by deep sequencing of Strongylocentrotus purpuratus RNA pulled down with the TERT protein [27] after homology based searches remained unsuccessful. This opened the door to identifying TERs from other sea urchins, brittle stars, and a crinoid [28]. However, no TER from a protostome is known.
Within Fungi, the situation is similar: Thus far, TERs have been reported only for Ascomycota, while no candidates are known in Basidiomycota and any of the basal divisions. The TERs of Pezizomycotina and Taphrinomycotina share core features of vertebrate TERs. In particular, they have a fairly well-conserved secondary structure of the pseudoknot and the TWJ, and at least in these regions the sequence is sufficiently conserved for successful homology-based identification of TERs within these clades [29,30,31]. The TERs known for Saccharomycetes, the relatives of budding yeast, on the other hand, are sometimes remarkably large and present little similarity in sequence and secondary structure to vertebrate or ciliate TERs.
To-date, yeast TERs have been reported for three phylogenetically narrow subgroups (Saccharomyces spp. [16,17], Kluyveromyces spp. [9,32,33], and Candida spp. [34,35]), as well as some individual species such as Candida glabrata [19] and Hansenula polymorpha [36]. These sequences are already too diverse for reliable sequence alignments. It is not surprising, therefore, that simple sequence-based homology searches have not been successful in identifying TER in the majority of the saccharomycete genome sequences to-date. Even protein binding sites that are functionally important in budding yeast [37,38] are not widely conserved. For instance, Ku or Sm binding sites seem to be absent in the TERs of filamentous fungi [4,29].
The obvious alternative is to increase the set of known TERs by finding homologs that are sufficiently similar to one of known yeast TERs, to allow the construction of multiple alignments of phylogenetically narrow subgroup. From these alignments, conserved elements can be extracted, which in turn form the basis for searches with tools such as fragrep [39] or infernal [40]. This strategy has been successful in previous searches for TER genes in both animals [26] and fungi [29], but thus far has not been successfully applied to Saccharomycetes. Here, sequences are highly divergent, so that plausible multiple sequence alignments and secondary structure models derived from them can be obtained only for quite narrow phylogenetic groups. The best studied case is Saccharomyces sensu stricto [17,41].
Until very recently, a phylogenetically local approach to homology search was also hampered by the lack of a trustworthy phylogeny of the Saccharomycotina. Recent updates in the International Code of Nomenclature for algae, fungi and plants [42,43] have substantially restructured the classification of fungi in general and of Saccharomycotina in particular. With large-scale efforts to sequence fungal genomes underway, first phylogenomic studies provide a trustworthy backbone of Saccharomycotina phylogeny [44], which we largely confirmed with an independent analysis.
2. Materials and Methods
2.1. Phylogenomics of Ascomycotes
Annotated protein sequences for 72 yeast species were downloaded from the National Center for Biotechnology Information (NCBI) refseq database (https://www.ncbi.nlm.nih.gov/refseq/). Initially, ProteinOrtho [45,46] was used to identify an initial set of 21,289 ortholog groups. Only 193 of these contained representatives of all 72 species. We therefore included all 1666 ortholog groups that covered at least 67 species. We used OMA (2.2.1, with default settings) [47,48] to decompose the ProteinOrtho groups further into clusters of 1-1 orthologs. This resulted in 6295 groups, of which 841 contained at least 67 species. This conservatively filtered dataset was then processed with Gblocks [49] (version 0.91b, with default parameters) to remove uninformative and potentially error-prone parts of the alignment, resulting in a dataset comprising 72 species and 248,581 characters. Phylogenetic trees were estimated with RAxML [50].
2.2. Ascomycote Telomerase RNAs
Telomerase RNA regions have been published for several Saccharomyces [16,17], Kluyveromyces [9,32,33], and Candida [19,34,35] species. Most of these published TER regions are collected in the telomerase database [51], which therefore provided a good starting point for our research. These sequences, however, are too diverse to construct multiple sequence alignments beyond the three genera individually. This effectively prohibits the automated discovery of novel TERs beyond close relatives with the help of either BLAST [52] (using sequence information alone) or infernal (relying on a combination of sequence and secondary structure information).
Therefore, we explored different strategies to overcome the limitations imposed by the extremely poor sequence conservation of saccaromycete telomerase RNAs. The basic idea is to use common features of the TERs to extract candidates from the genomes that can be analyzed and then inspected further using different techniques.
First, we attempted to learn TER-specific sequence patterns using MEME/GLAM2 [53], and also several machine learning techniques using k-mer distributions within sequence windows of the size of the known TERs. All attempts to learn from a training set covering the Saccharomycetaceae or all Saccharomycotina species failed.
There are several possible reasons. Machine learning methods crucially depend on training and test sets, both positive and negative. In our case, we have few positive samples, these have poorly defined features, and are very diverse as far as their sequences are concerned. It is unclear in this setting how a negative training set should be properly designed. The obvious choice of picking genomic sequence at random may be confounded by unintended strong signals, such as coding potential or repetitive sequence elements. It would appear that at the very least a more a careful construction of the positive and negative sets, and an appropriate normalization or scaling of the feature data will be required to make progress in this direction. Restricting the training phase to a more narrow phylogenetic range to reduce the inherent diversity of the training data, on the other hand, is infeasible due to the small number of known TER sequences.
The EDeN motif finder [54] was applied to 24 known TERs as positive set and 48 shuffled sequences as negative data. Only trivial sequence motifs such a poly-U stretch, presumably corresponding to part of the U-rich pseudoknot region, were found. Unsupervised clustering also remained unsuccessful.
2.3. Synteny-Based Homology Search
As an alternative strategy, we established a semi-automated workflow that aims at first extracting partially conserved RNA sequence-structure elements, which are then used to identify candidate loci. In response to the negative results of a direct pattern-based approach, we systematically used synteny to narrow down the search space in the initial phase. Starting from a whole genome alignment of phylogenetically related species, we used the positions of protein coding genes whose homologs are known to be adjacent in a closely related species to delimit the syntenic regions that are likely to contain a TER gene. These candidate regions were then analyzed in detail by means of pairwise or multiple sequence alignments. Whenever a global alignment of the entire candidate syntenic region did not yield a plausible alignment, we attempted to identify conserved motifs inside the syntenic region (usually the SM binding site and/or the template region, which is sometimes conserved between close relatives). Typically, these motifs were also sufficient to determine the correct reading direction of the TER candidate.
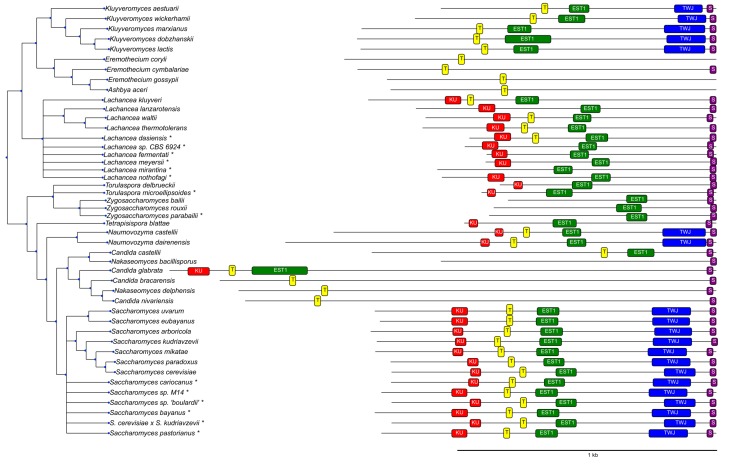
To identify known features in the candidate TER regions, we first constructed infernal [40] covariance models restricted to subgroups of Saccharomycetaceae covering only substructures, such as the Ku hairpin, Est1 binding site, and TWJ in the Saccharomyces and Kluyveromyces species. The alignments underlying the infernal models were constructed with the help of many software tools, including locARNA [55], MAFFT [56], mauve [57], MEME [53] and fragrep [39], as well as manual curation. Default settings were used for all tools. The consensus structure for the Ku binding motive corresponds to a recent crystal structure [58]. These models were then analyzed with CMCws [59] and used for precise localization of conserved TER elements in species that were: (a) taxonomically closely related, but not/only partially annotated in the literature (Saccharomyces uvarum, Saccharomyces sp. “boulardii”, Saccharomyces sp. M14, and Saccharomyces eubayanus); or (b) phylogenetically located in the subtree spanned by the Saccharomyces and Kluyveromyces species (Figure 2). Both the ViennaNGS [60] suite and custom Perl/Python scripts were used for handling and conversion of genomic annotation data.
Figure 2.
Features identified in TER sequences. (KU) ku binding hairpin, (T) template region, (EST1) Est1 binding site, (TWJ) three-way junction, and (SM1) SM1 binding site. Elements not shown are either not present in the corresponding species (e.g., the TWJ in Candida glabrata) or could not be located with reasonable certainty. Species marked by * are not part of the phylogenetic tree and were placed next to their closest related neighbor based on the similarity of their TER sequences.
We then extracted a sequence corresponding to the most closely related TER sequence as initial estimate of the full-length TER gene. We used MAFFT [56] to produce initial sequence-based alignments of candidate regions, which were then realigned with locARNA [55] to obtain RNA structural alignments. The latter was used with its free-end-gaps option, in particular in those cases where MAFFT was not sensitive enough to reliably estimate the TER boundaries. Conversely, MAFFT was able to identify and correctly align highly conserved subsequences, providing reliable anchors for the more divergent sequence regions. While locARNA is good at finding locally conserved structures in the whole alignment, we expected only parts of the TER sequences to be structurally conserved. Typically, multiple iterations of refinement of the TER boundaries were required to obtain the final TER candidate sequence.
Following this approach, we could localize TER regions for several members of the Saccharomycetacea clade. Subsequent alignment of candidate regions with known TERs allowed for exact localization of TERs.
2.4. Search for Candidates Using Telomere Template Sequences
The scope of the synteny-based approach is limited because fungal genomes are subject to frequent genome rearrangements at the time-scales of interest. We therefore attempted to identify candidate regions containing the template sequence for the telomere repeats (see [61] for a comprehensive review of the characteristics of different telomeric repeats). In genomes for which these sequences have not been reported, we searched chromosome ends for telomeric repeats. Unfortunately, most genome assemblies are not on chromosome level or do not include the telomere regions, hence we only succeeded to newly identify the template region of Ashbya aceri and Eremothecium cymbalariae this way. For the latter species, the pertinent information is available in [62], although the telomeric repeat is not explicitly reported. In addition, we used the published telomere sequences from the telomerase database [51].
We used the concatenation of two copies of telomeric repeat sequence as query for a BLAST [52] search against the whole genome (in case of longer, complex repeats) or against the syntenic region for shorter repeats. Other template regions were identified with by aligning them to known sequences and/or BLAST searches of known template regions in closely related species. A typical feature of the template region, which helped us to verify our hits, is the fact that it usually contains a few nucleotides repeated at both the beginning and the end of the template region [19].
2.5. BLAST Pipeline
BLAST [52] is by far the most commonly used tool for homology search. While it has been reported to have limited sensitivity for telomerase RNAs in previous studies [26,27,39], it has contributed significantly to the identification of the TER sequences in other ascomycete clades [29,31]. Here, we used a set of known TER regions as BLAST queries that comprises all Saccharomycetales TER regions that we found in literature, as well as all TERs newly identified in the contribution. As targets for BLASTn (with default parameters) we used the full genomes of species that are featured at the NCBI refseq database within the Saccharomycetales group (Taxonomy ID: 4892). The resulting BLAST hits were then filtered for E-values (), a minimum alignment length of 25 nt and a minimum identity of 60%. In addition, all hits on known telomeric regions were excluded. From the hits in genomes with known TERs, we computed the empirical false positive rate and found that the alignment length proved to be the most informative parameter. It has therefore been used to evaluate the reliability of hits, given their score.
The BLAST pipeline also contributed to the identification of the TER boundaries in some of the unannotated genomes. In cases where we initially chose the boundaries of our queries too generously and included neighboring coding regions or regulatory elements, the BLAST pipeline returned “false positive” hits. Thus, whenever multiple false positive hits in the beginning or the end of the query sequence occurred, we rechecked and, if necessary, improved the boundaries of the TER region.
3. Results
3.1. Phylogenomics of Saccharomycotina
The phylogenetic trees obtained of our phylogenomic analysis of the Saccharomycetales is essentially congruent with the one reported by Shen et al. [44] (see Figure A1 for more details). For consistency, we adopted the phylogenetic tree from the same publication as the basis for presenting our results.
3.2. Survey of Telomerase RNA Genes in Saccharomycotina
We initially screened 52 ascomycote genomes. Predominantly sequence-based methods (BLAST, but also MEME, GLAM2, and infernal) only contributed TERs from close relatives of baker’s yeast. The BLAST pipeline was applied to all 185 NCBI genomes Saccharomycetales, the subclade containing all known Saccharomycotina genomes. With the exception of the TER in Ogataea parapolymorpha, a very close relative of the known Ogataea polymorpha TER [36] all new sequences we found within the Saccharomycetaceae. We therefore restricted a more detailed analysis to this clade.
We found credible TER sequences in 46 of the 53 Saccharomycetaceae. Most of these TER sequences could be detected only after a short candidate region had been identified based on synteny. To our knowledge, at least 27 of these have not been reported previously.
3.3. Features of Telomerase RNA in Saccharomycetacea
To better understand the TER and its evolutionary constraints, at least within the Saccharomycetacea, we performed a detailed analysis of their structural features. Table 1 summarizes the results of the homology search and the functional features of the candidate TER genes. A graphical overview is given in Figure 2.
Table 1.
Overview of conserved TER substructures in Saccharomycetacea, as identified by the combined synteny/covariance model pipeline. The 3 end is defined as 10 nt downstream of the SM binding site. The 5 end is approximate. Citations refer to publication in which the sequence and/or the coordinates of respective features are reported explicitly. These annotations form the basis of Figure 2.
| Species | Accession | Strand | TER Coordinates | Ku Binding Site | Template Region | Est1 Binding Site | TWJ | SM1 |
|---|---|---|---|---|---|---|---|---|
| K. aestuarii | AEAS01000245.1 | neg | 16,338–17,322 [32] | 16,940–16,966 | 16,794–16,862 [32] | 16,378–16,485 [9] | 16,350–16,359 | |
| K. wickerhamii | AEAV01000432.1 | pos | 250–1327 [32] | 662–693 | 765–858 [32] | 1183–1290 [9] | 1307–1316 | |
| K. marxianus | NC_036029.1 | pos | 506,443–507,711 [32] | 506,855–506,888 | 506,967–507,049 [32] | 507,518–50,7671 [9] | 507,691–507,700 | |
| K. dobzhanskii | CCBQ010000012.1 | pos | 461,805–463,090 [32] | 462,224–462,257 | 462,337–462,499 [32] | 462,905–463,051 [9] | 463,070–463,079 | |
| K. lactis | NC_006038.1 | pos | 611,456–612,727 [33] | absent [63] | 611,890–611,919 [64] | 612,006–612,090 [32] | 612,532–612,687 [9] | 612,708–612,716 [64] |
| E. coryli | AZAH01000001.1 | neg | 269,038–270,368 | 269,938–269,968 | ||||
| E. cymbalariae | NC_016454.1 | pos | 54,147–54,960 | 54,451–54,480 | ||||
| E. gossypii | NC_005782.2 | neg | 677,871–679,048 [65] | 678,276–678,305 [35] | ||||
| Ashbya aceri | CP006020.1 | neg | 693,543–694,708 | 693,942–693,973 | ||||
| L. kluyveri | CM000690.1 | pos | 348,600–349,844 | 348,876–348,930 | 348,957–348,982 [66] | 349,129–349,208 | 349,825–349,833 | |
| L. lanzarotensis | NW_019212880.1 | pos | 854,162–855,236 | 854,389–854,444 | 854,754–854,820 | 855,217–855,225 | ||
| L. waltii | AADM01000270.1 | neg | 134,961–136,000 | 135,698–135,756 [19] | 135,613–135,636 | 135,409–135,470 | 134,973–134,981 | |
| L. thermotolerans | NC_013079.1 | pos | 702,500–703,549 | 702,730–702,791 [19] | 702,853–702,876 | 703,022–703,083 | 703,530–703,538 | |
| L. dasiensis | LT598456.1 | pos | 682,034–682,916 | 682,124–682,181 | 682,261–682,283 | 682,900–682,905 | ||
| L. sp. CBS 6924 | LT598470.1 | neg | 441,802–442,700 | 442,582–442,638 | 442,229–442,292 | 441,811–441,820 | ||
| L. fermentati | LT598488.1 | neg | 306,329–307,150 | 307,076–307,129 | 306,786–306,850 | 306,339–306,348 | ||
| L. meyersii | LT598477.1 | pos | 575,851–576,676 | 575,886–575,941 | 576,233–576,294 | 576,657–576,666 | ||
| L. mirantina | LT598468.1 | pos | 690,800–691,797 | 691,218–691,282 | 691,777–691,786 | |||
| L. nothofagi | LT598449.1 | pos | 388,401–389,382 | 388,567–388,624 | 388,937–389,004 | 389,362–389,371 | ||
| T. delbrueckii | NC_016504.1 | pos | 709,007–709,780 | 709,057–709,086 | 709,267–709,336 | 709,761–709,770 | ||
| T. microellipsoides | FYBL01000005.1 | neg | 426,211–427,050 | 427,000–427,028 | 426,726–426,817 | 426,221–426,229 | ||
| Z. bailii | HG316456.1 | neg | 712,655–713,400 | 712,902–712,974 | 712,665–712,673 | |||
| Z. rouxii | NC_012990.1 | pos | 297,087–297,883 | 297,527–297,616 | 297,865–297,873 | |||
| Z. parabailii | CP019499.1 | pos | 455,564–455,975 [67] | 455,656–455,728 | 455,957–455,965 | |||
| T. blattae | NC_020193.1 | neg | 404,150–405,050 | 405,003–405,033 | 404,650–404,733 | 404,165–404,173 | ||
| N. castellii | NC_016499.1 | pos | 381,827–383,194 [64] | 382,404–382,432 [19] | 382,506–382,519 [64] | 382,647–382,710 [64] | 382,994–383,155 [64] | 383,176–383,184 [64] |
| N. dairenensis | NC_016479.1 | neg | 1,519,837–1,521,377 | 1,520,648–1,520,678 | 1,520,550–1,520,562 | 1,520,303–1,520,369 | 1,519,864–1,520,027 | 1,519,849–1,519,857 |
| C. castellii | CAPW01000002.1 | neg | 272,769–274,000 | 273,158–273,179 | 272,992–273,085 | 272,781–272,789 | ||
| N. bacillisporus | CAPX01000073.1 | pos | 1230–2215 | 2197–2204 | ||||
| C. glabrata | NC_006032.2 | neg | 419,194–421,150 [19] | 421,007–421,081 [19] | 420,914–420,932 [19] | 420,657–420,852 [19] | 419,206–419,214 [19] | |
| C. bracarensis | CAPU01000044.1 | pos | 2586–4361 | 2836–2854 | 4342–4350 | |||
| N. delphensis | CAPT01000167.1 | neg | 254,761–256,469 | 256,151–256,169 | 254,773–254,781 | |||
| C. nivariensis | CAPV01000033.1 | pos | 87,530–89,215 | 87,780–87,798 | 89,196–89,204 | |||
| S. uvarum | NOWY01000011.1 | pos | 45,720–46,940 | 45,996–46,050 | 46,193–46,203 | 46,301–46,377 | 46,703–46,848 | 46,921–46,929 |
| S. eubayanus | NC_030979.1 | pos | 476,134–47,7336 | 476,392–476,446 | 476,588–476,598 | 476,694–476,770 | 477,100–477,240 | 477,317–477,325 |
| S. arboricola | NC_026172.1 | pos | 287,410–288,645 | 287,705–287,739 | 287,888–287,898 | 288,019–288,096 | 288,417–288,558 | 288,626–288,634 |
| S. kudriavzevii | AY639012.1 | pos | 1–1215 [17] | 284–320 [17] | 424–434 | 585–662 | 981–1128 [9] | 1201–1209 |
| S. mikatae | AABZ01000048.1 | neg | 18,591–19,809 [17] | 19,497–19,532 [17] | 19,349–19,356 | 19,156–19,232 | 18,687–18,833 [9] | 18,603–18,611 |
| S. paradoxus | CP020294.1 | pos | 307,733–308,897 [17] | 308,010–308,045 [17] | 308,154–308,161 | 308,281–308,353 | 308,660–308,803 [9] | 308,878–308,886 |
| S. cerevisiae | NC_001134.8 | pos | 307,597–308,757 [17,18] | 307,880–307,914 [68] | 308,057–308,064 [64] | 308,185–308,256 [32] | 308,563–308,682 [9] | 308,737–308,746 [64] |
| S. pastorianus | AZCJ01000004.1 | neg | 478,773–479,970 [17] | 479,664–479,718 [17] | 479,512–479,520 | 479,340–479,417 | 478,866–479,012 [9] | 478,785–478,793 |
| S. cer. x S. kud. | AGVY01000004.1 | pos | 284,183–285,344 | 284,465–284,501 | 284,645–284,655 | 284,772–284,843 | 285,150–285,269 | 285,325–285,333 |
| S. bayanus | AACG02000058.1 | pos | 58,142–59,362 [17] | 58,418–58,472 [17] | 58,613–58,620 | 58,723–58,799 | 59,125–59,270 [9] | 59,343–59,351 |
| S. sp. ‘boulardii’ | CM003558.1 | pos | 287,536–288,696 | 287,818–287,854 | 287,998–288,008 | 288,124–288,195 | 288,502–288,621 | 288,677–288,685 |
| S. sp. M14 | MVPU01000005.1 | neg | 473,800–474,997 | 474,691–474,745 | 474,537–474,547 | 474,368–474,444 | 473,894–474,038 | 473,812–473,820 |
| S. cariocanus | AY639010.1 | pos | 1–1163 [17] | 278–313 [17] | 424–434 | 549–621 | 928–1072 [9] | 1147–1155 |
The exact genomic positions marking the 3 and 5 ends of the TER RNA are difficult to determine without additional experimental evidence. The 5 ends are therefore approximate. The 3 end of the mature TER is produced by splicing in most Ascomycota [31,69,70]. This mechanism, however, was lost at some point during the evolution of the Saccharomycotina. It has been reported in the Candida group and for Ogataea angusta (previously H. polymorpha), but it is missing in Saccharomyces and Kluyveromyces [31]; hence, we expect that the splicing-based 3-end processing was lost prior to the divergence of Saccharomycetacea. Indeed, no indication of a splice site was found for any of the TER sequences included in Table 1. We therefore used a position 10 nt downstream of the SM binding motif as approximation of the 3 end in Table 1.
Several of the features listed in Table 1 have been discussed in some detail in the literature. Not all of them were found in all the candidates reported here. This may, in some cases, be explained by sequences that are too divergent to be detected. In other cases, most likely the function is not preserved. Unfortunately, many studies report neither complete sequences nor coordinates, making it effectively impossible to accurately compare their results with the annotation reported here. References are included in Table 1 if sufficient information was included to locate the features unambiguously.
No Ku binding hairpin was recovered in Kluyveromyces or the Eremothecium species. This is not unexpected since there is experimental evidence that neither the Ku binding hairpin nor its function is present in Kluyveromyces lactis [63]. The putative Ku binding hairpin reported for C. glabrata in [19] lacks experimental support and contains long insertions that made it impossible to include it in our covariance model. Furthermore, this region of the TER sequence is very poorly conserved in the closest relatives of C. glabrata. While the TER of C. glabrata is among the longest known members of this gene family [19], its close relative Candida castellii features a TER that has been shortened drastically in its 3 half, with only ∼200 nt separating the EST1 and SM1 binding sites. Furthermore, the sequence GCUA, which is conserved in most known Ku binding sites, is not present within 600 nt upstream of the template region. The most likely explanation is that the TER of C. castellii (which, similar to C. glabrata, does not belong to the monophylogenetic genus Candida) (Appendix A) does not bind Ku. Of course, we cannot rule out without further experimental data that the motif has diverged beyond our ability to recognize it.
In a few species, we failed to identify the template region. In these cases (Lachancea, Zygosaccharomyces and Torulaspora species and Nakaseomyces bacillisporus), the telomeric repeat sequence is not known and seems to be very different from both the fungal consensus sequence TTAGGG [29] and the telomeric sequences found in closely related species.
The EST1 binding site forms a hairpin that is similar to the P3 domain of the RNase P and RNAse MRP RNAs [71]. We were not able to find an EST1 binding site in Eremothecium species, Lachancea dasiensis and in the C. glabrata group, even though it has been published for C. glabrata. While an EST1 binding site is present even in the more distantly related genus Candida [35], this motif is intrinsically too variable to be unambiguously recognizable in distant relatives. This pertains to both its sequence and the its base-pairing patterns.
Consistent with [19], we found no plausible secondary structure for the TWJ in C. glabrata, although the respective region of the sequence contains the highly conserved sequence AATA. It is worth noting in this context that the telomerase of the ciliate Tetrahymena has a stem-loop structure in place of the threeway junction [72]. TERs of the C. glabrata group thus may also have a functional trans-activation domain, albeit with an aberrant structure. Our TWJ covariance model, which was constructed from Kluyveromyces and Saccharomyces sequences only, also failed to detect a TWJ in Eremothecium and Lachancea. It remains an open question whether TERs of these species have a TWJ with a diverged structure that is just beyond our ability to detect, or whether trans-activation is achieved by different means. This is not implausible, given that the TWJ has been shown to be dispensable for TER function [6,10,11].
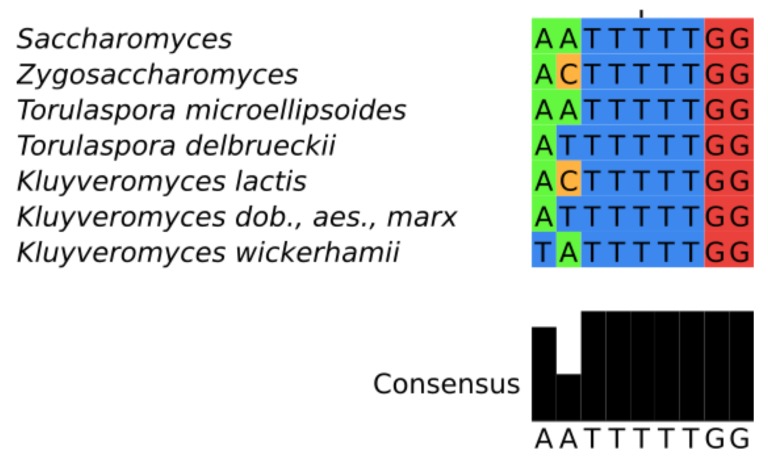
The sequence of the SM binding motif AATTTTTGG is perfectly conserved throughout much of the Saccharomycetaceae, with the notable exception of K. lactis [64] and additional small variations in other Kluyoveromyces species (Figure 3). We could not find this motif in species of the genus Eremothecium and the highly related species Ashba acerii.
Figure 3.
Alignment of the core SM-binding site motif: The common pattern of most Saccharomycetaceae (top); and species-specific variants (bottom).
4. Discussion
Although we succeeded in detecting 27 previously unknown TER sequences in Saccharomycetaceae, the main take-home message of this contribution is that homology search can be a terribly difficult problem. Although yeast TERs are quite long and fulfill a well-conserved function, their sequences are very poorly conserved. In this respect, yeast TER behaves similar to the the majority of long non-coding RNAs, which are also poorly conserved in sequence but often are evolutionary quite well conserved as functional entitied (see [73] for a recent review).
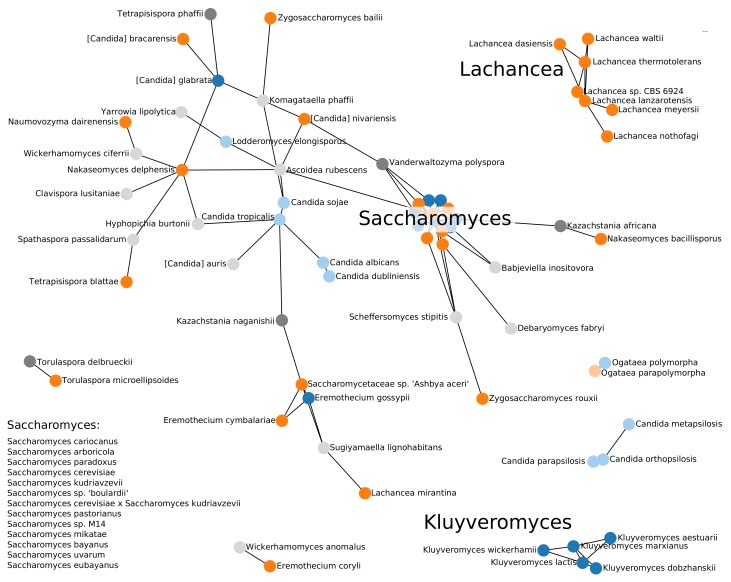
The “BLAST graph” in Figure 4 highlights the practical problem. Sequence comparison methods identify homology only in closely related species. A comparison of Figure 4 and a corresponding graph based on the previously published TER sequences only (see Supplementary Materials) shows that the larger set of queries identifies many additional connections and thus improves the situation at least within the Saccharomycetacea. Even within the clade, however, we have been unable to confirm the candidate hits in Kasachstania. The tree in Figure A1 indicates longer branch lengths leading to Kasachstania; it appears that the accelerated evolution of these genomes is already sufficient to hide the TER genes from our homology search methods.
Figure 4.
Summary of the BLAST-based survey of TER genes. Blue nodes show TERs described in the literature, orange nodes represent TERs that we identified, and grey nodes are additional candidates for which we could not validate characteristic features. TERs outside the Saccharomycetaceae group are presented in light colors. The length of the edges are weighted by the inverse of the length of the BLAST hit. Note that distances in drawing between nodes not connected by an edge are not indicative of their evolutionary distance.
While the direct sequence-based search against complete genomes was not very successful, we observed that the synteny-based approach worked remarkably well. This is not entirely unexpected, since the restriction to the interval between a pair of coding genes effectively reduces the size of the target from several million nucleotides to a few thousand. Unfortunately, the applicability of synteny-based methods is limited to relatively narrow phylogenetic scales. On longer time-scales, genome rearrangements are likely to disrupt syntenic conservation. A systematic exploitation of synteny similar to the work described here for Saccharomycetacea would most likely be successful in a survey for TER in the Candida group. In fact, synteny has been employed to find some of the known TERs in this clade [17,35].
The study presented here was largely conducted using publicly available tools complemented by some custom scripting. It also highlights the need for customized tools to conduct difficult homology searches. In particular, specific alignment tools and viewers to efficiently evaluate the synteny-based candidates relative to known template sequences and alignments of the better conserved regions would facilitate the manual curation efforts, which we found to be indispensable.
Finally, it remains on open question whether direct machine learning methods can be adapted as homology search tools, and, if so, whether such a strategy can be more effective than sequence comparison methods. It is likely that such efforts failed so far because of the difficulties inherent in the construction of a suitable negative training set that is not confounded by frequent genomic features such as coding sequence. Furthermore, the small number of positive samples was presumably insufficient to capture the full variability of TER sequences.
Complementarily, a phylogenetically dense sample of TERs that are sufficiently similar to support global sequence alignments might help to better understand the rapid divergence of TER sequences. This might be helpful not only to identify informative features for machine learning applications, but also to design modified sequence comparison algorithms that better reflect the peculiarities of rapidly evolving long non-coding RNAs. In this contribution, we have provided such a set of TERs for the Saccharomycetaceae.
Acknowledgments
The research reported here is the outcome of a PhD level seminar taught by P.F.S. at Univ. Vienna in the fall term 2017/2018.
Supplementary Materials
Machine readable Supplemental Information, in particular accession numbers, TER sequences, alignments of conserved features, and covariance models are available at http://www.bioinf.uni-leipzig.de/Publications/SUPPLEMENTS/18-048/.
Appendix A. Phylogenomics of the Saccharomycetales
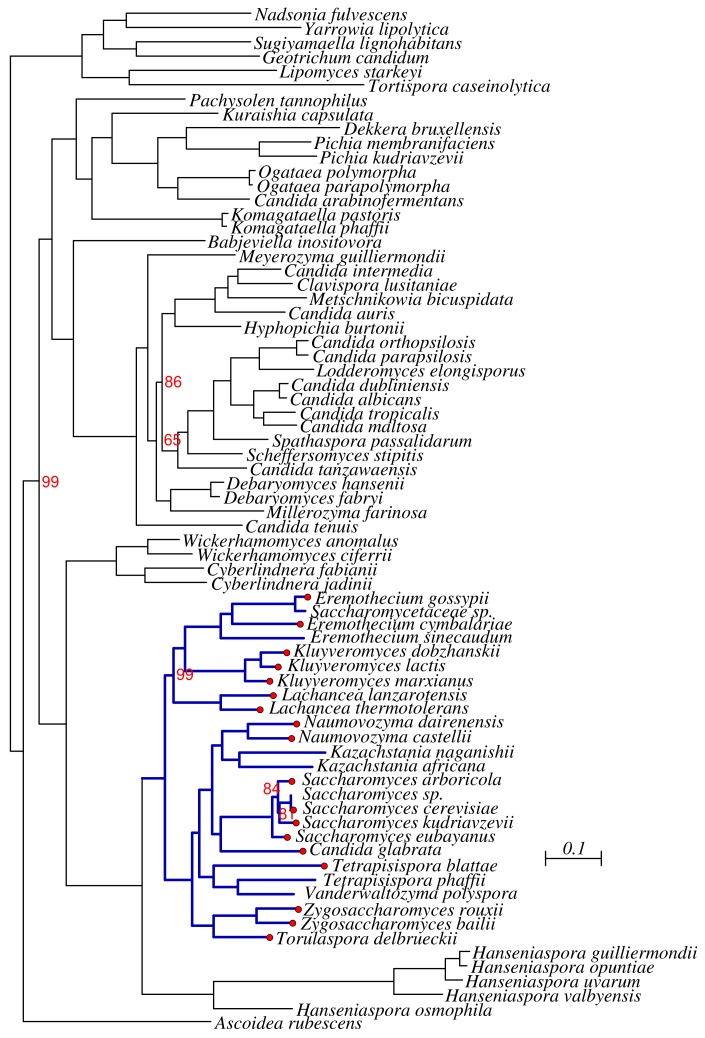
The maximum likelihood tree obtained from 841 orthologous groups of proteins present in at least 67 of the 72 species is shown in Figure A1. The phylogeny is nearly identical to the tree reported in [44]. In particular, it provides strong support for monophyletic Saccharomycetacea (comprising in particular the genera Saccharomyces and Kluyveromyces), and the Candida group. Noteworthy, “Candida glabrata” is nested within the Saccharomycetacea as a close relative of Saccharomyces rather than appearing as member of the Candida clade.
Figure A1.
Phylogeny of the Saccharomycetales. Bootstrap support is 100% unless otherwise indicated. The Saccharomycetacea are indicted in dark blue. A red dot at tip of the tree indicates a TER sequences listed in Table 1.
Author Contributions
P.F.S. and M.T.W. conceived the study. M.W., B.C.T., R.O., and M.T.W. analyzed the TER sequences and structures. A.H. conducted the phylogenomic analysis. J.V.d.A.O. and M.E.M.T.W. contributed machine learning approaches. All authors contributed to writing the paper and approved of the submitted version.
Funding
This research was funded in part by the Doktoratskolleg RNA Biology at Univ. Vienna, the Austrian Science Fund (grant numbers SFB 4305-B09, I 2874-N28, I-1303 B21), Sinergia (grant number CRSII3_154471/1), and the German Federal Ministery for Education and Research (grant number BMBF 031A538A, de.NBI/RBC).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 2.Mason J.M., Reddy H.M., Frydrychova R.C. Telomere maintenance in organisms without telomerase. In: Seligmann H., editor. DNA Replication—Current Advances. InTech; Rijeka, Croatia: 2011. Chapter 15. [Google Scholar]
- 3.Pardue M., Rashkova S., Casacuberta E., DeBaryshe P.G., George J.A., Traverse K. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Podlevsky J.D., Chen J. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016;13:720–732. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musgrove C., Jansson L.I., Stone M.D. New perspectives on telomerase RNA structure and function. Wiley Interdiscip. Rev. RNA. 2018;9:e1456. doi: 10.1002/wrna.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mefford M.A., Rafiq Q., Zappulla D.C. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. EMBO J. 2013;32:2980–2993. doi: 10.1038/emboj.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J., Greider C.W. An emerging consensus for telomerase RNA structure. Proc. Natl. Acad. Sci. USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mefford M.A., Zappulla D.C. Physical connectivity mapping by circular permutation of human telomerase RNA reveals new regions critical for activity and processivity. Mol. Cell. Biol. 2015 doi: 10.1128/MCB.00794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown Y., Abraham M., Pearl S., Kabaha M.M., Elboher E., Tsfati Y. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livengood A.J., Zaug A.J., Cech T.R. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zappulla D.C., Goodrich K., Cech T.R. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 2005;12:1072. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 12.Wu R.A., Upton H.E., Vogan J.M., Collins K. Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 2017;86:439–460. doi: 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb C.J., Zakian V.A. Telomerase RNA is more than a DNA template. RNA Biol. 2016;13:683–689. doi: 10.1080/15476286.2016.1191725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greider C.W., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 15.Feng J., Funk W.D., Wang S.S., Weinrich S.L., Avilion A.A., Chiu C.P., Adams R.R., Chang E., Allsopp R.C., Yu J. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 16.Singer M.S., Gottschling D.E. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 17.Dandjinou A.T., Lévesque N., Larose S., Lucier J.F., Abou Elela S., Wellinger R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 18.Bosoy D., Peng Y.P., Mian I.S., Lue N.F. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 19.Kachouri-Lafond R., Dujon B., Gilson E., Westhof E., Fairhead C., Teixeira M.T. Large telomerase RNA, telomere length heterogeneity and escape from senescence in Candida glabrata. FEBS Lett. 2009;583:3605–3610. doi: 10.1016/j.febslet.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 20.Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D.E. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. USA. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Beilstein M.A., Brinegar A.E., Shippen D.E. Evolution of the Arabidopsis telomerase RNA. Front. Genet. 2012;3:188. doi: 10.3389/fgene.2012.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S.K., Kolet L., Doniger T., Biswas V.K., Unger R., Tzfati Y., Michaeli S. The Trypanosoma brucei telomerase RNA (TER) homologue binds core proteins of the C/D snoRNA family. FEBS Lett. 2013;587:1399–1404. doi: 10.1016/j.febslet.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Sandhu R., Sanford S., Basu S., Park M., Pandya U.M., Li B., Chakrabarti K. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013;23:537–551. doi: 10.1038/cr.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakrabarti K., Pearson M., Grate L., Sterne-Weiler T., Deans J., Donohue J.P., Ares M., Jr. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–1939. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Religa A.A., Ramesar J., Janse C.J., Scherf A., Waters A.P. P. berghei telomerase subunit TERT is essential for parasite survival. PLoS ONE. 2014;9:e108930. doi: 10.1371/journal.pone.0108930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie M., Mosig A., Qi X., Li Y., Stadler P.F., Chen J.J.L. Size variation and structural conservation of vertebrate telomerase RNA. J. Biol. Chem. 2008;283:2049–2059. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Marz M., Qi X., Podlevsky J.D., Hoffmann S., Stadler P.F., Chen J.J.L. Identification of Purple Sea urchin telomerase RNA using a next-generation sequencing based approach. RNA. 2013;19:852–860. doi: 10.1261/rna.039131.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podlevsky J.D., Li Y., Chen J.J. Structure and function of echinoderm telomerase RNA. RNA. 2016;22:204–215. doi: 10.1261/rna.053280.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi X., Li Y., Honda S., Hoffmann S., Marz M., Mosig A., Podlevskya J.D., Stadler P.F., Selker E.U., Chen J.J.L. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuprys P.V., Davis S.M., Hauer T.M., Meltser M., Tzfati Y., Kirk K.E. Identification of telomerase RNAs from filamentous fungi reveals conservation with vertebrates and yeasts. PLoS ONE. 2013;8:e58661. doi: 10.1371/journal.pone.0058661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi X., Rand D.P., Podlevsky J.D., Li Y., Mosig A., Stadler P.F., Chen J.J. Prevalent and distinct spliceosomal 3′-end processing mechanisms for fungal telomerase RNA. Nat. Commun. 2015;6:6105. doi: 10.1038/ncomms7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto A.G., Livengood A.J., Tzfati Y., Blackburn E.H., Cech T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McEachern M.J., Blackburn E.H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 34.Hsu M., McEachern M.J., Dandjinou A.T., Tzfati Y., Orr E., Blackburn E.H., Lue N.F. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. USA. 2007;104:11682–11687. doi: 10.1073/pnas.0700327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunisova S., Elboher E., Nosek J., Gorkovoy V., Brown Y., Jean-François L., Laterreur N., Wellinger R.J., Tzfati Y., Tomaska L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–559. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smekalova E.M., Malyavko A.N., Zvereva M.I., Mardanov A.V., Ravin N.V., Skryabin K.G., Westhof E., Dontsova O.A. Specific features of telomerase RNA from Hansenula polymorpha. RNA. 2013;19:1563–1574. doi: 10.1261/rna.038612.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stellwagen A.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zappulla D.C., Goodrich K.J., Arthur J.R., Gurski L.A., Denham E.M., Stellwagen A.E., Cech T.R. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA. 2011;17:298–311. doi: 10.1261/rna.2483611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosig A., Chen J.L., Stadler P.F. Homology Search with Fragmented Nucleic Acid Sequence Patterns. In: Giancarlo R., Hannenhalli S., editors. Lecture Notes in Computer Science, Proceedings of the International Workshop on Algorithms in Bioinformatics (WABI 2007), Philadelphia, PA, USA, 8–9 September 2007. Volume 4645. Springer; Berlin/Heidelberg, Germany: 2007. pp. 335–345. [Google Scholar]
- 40.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zappulla D.C., Cech T.R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawksworth D.L. A new dawn for the naming of fungi: Impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA Fungus. 2011;2:155–162. doi: 10.5598/imafungus.2011.02.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNeill J., Barrie F.R., Buck W.R., Demoulin V., Greuter W., Hawksworth D.L., Herendeen P.S., Knapp S., Marhold K., Prado J., et al., editors. Regnum Vegetabile. Volume 154 Koeltz Scientific Books; Oberreifenberg, Germany: 2012. International Code of Nomenclature for algae, fungi and plants. [Google Scholar]
- 44.Shen X.X., Zhou X., Kominek J., Kurtzman C.P., Hittinger C.T., Rokas A. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 2016;6:3927–3939. doi: 10.1534/g3.116.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: Detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechner M., Hernandez-Rosales M., Doerr D., Wieseke N., Thévenin A., Stoye J., Hartmann R.K., Prohaska S.J., Stadler P.F. Orthology detection combining clustering and synteny for very large datasets. PLoS ONE. 2014;9:e105015. doi: 10.1371/journal.pone.0105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth A.C.J., Gonnet G.H., Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinform. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altenhoff A.M., Gil M., Gonnet G.H., Dessimoz C. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS ONE. 2013;8:e53786. doi: 10.1371/journal.pone.0053786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podlevsky J.D., Bley C.J., Omana R.V., Qi X., Chen J.J.L. The telomerase database. Nucleic Acids Res. 2007;36:D339–D343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa F.C., De Grave K. Fast neighborhood subgraph pairwise distance Kernel. In: Fürnkranz J., Joachims T., editors. Proceedings of the 27th International Conference on International Conference on Machine Learning (ICML’10); Haifa, Israel. 21–24 June 2010; Madison, WI, USA: Omnipress; 2010. pp. 255–262. [Google Scholar]
- 55.Will S., Reiche K., Hofacker I.L., Stadler P.F., Backofen R. Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comp. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darling A.E., Mau B., Perna N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H., Xue J., Churikov D., Hass E.P., Shi S., Lemon L.D., Luciano P., Bertuch A.A., Zappulla D.C., Géli V., et al. Structural insights into yeast telomerase recruitment to telomeres. Cell. 2018;172:331–343. doi: 10.1016/j.cell.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eggenhofer F., Hofacker I.L., Höner zu Siederdissen C. CMCompare webserver: Comparing RNA families via covariance models. Nucleic Acids Res. 2012;41:W499–W503. doi: 10.1093/nar/gkt329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolfinger M.T., Fallmann J., Eggenhofer F., Amman F. ViennaNGS: A toolbox for building efficient next-generation sequencing analysis pipelines. F1000Research. 2015;4:50. doi: 10.12688/f1000research.6157.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teixeira M.T., Gilson E. Telomere maintenance, function and evolution: The yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- 62.Wendland J., Walther A. Genome evolution in the Eremothecium clade of the Saccharomyces complex revealed by comparative genomics. Genes Genomes Genet. 2011;1:539–548. doi: 10.1534/g3.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabaha M.M., Zhitomirsky B., Schwartz I., Tzfati Y. The 5′ arm of Kluyveromyces lactis telomerase RNA is critical for telomerase function. Mol. Cell. Biol. 2008;28:1875–1882. doi: 10.1128/MCB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Telomerase Database-Secondary Structures. [(accessed on 30 April 2018)]; Available online: http://telomerase.asu.edu/structures.html#secondary.
- 65.Dietrich F.S. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 66.Genome Resources for Yeast Chromosomes Database-TLC1 (Telomerase RNA Template) [(accessed on 30 April 2018)]; Available online: http://gryc.inra.fr/index.php?page=locus&seqid=SAKL0D04356r.
- 67.Ortiz-Merino R.A., Kuanyshev N., Braun-Galleani S., Byrne K.P., Porro D., Branduardi P., Wolfe K.H. Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. PLoS Biol. 2017;15:e2002128. doi: 10.1371/journal.pbio.2002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peterson S.E., Stellwagen A.E., Diede S.J., Singer M.S., Haimberger Z.W., Johnson C.O., Tzoneva M., Gottschling D.E. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001;27:64. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- 69.Box J.A., Bunch J.T., Tang W., Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 70.Kannan R., Helston R.M., Dannebaum R.O., Baumann P. Diverse mechanisms for spliceosome-mediated 3′ end processing of telomerase RNA. Nat. Commun. 2015;6:6104. doi: 10.1038/ncomms7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemieux B., Laterreur N., Perederina A., Noël J.F., Dubois M.L., Krasilnikov A.S., Wellinger R.J. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–1181. doi: 10.1016/j.cell.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh M., Wang Z., Koo B.K., Patel A., Cascio D., Collins K., Feigon J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nitsche A., Stadler P.F. Evolutionary clues in lncRNAs. Wiley Interdiscip. Rev. RNA. 2017;8:e1376. doi: 10.1002/wrna.1376. [DOI] [PubMed] [Google Scholar]