Abstract
Mitochondria are a crucial target for the actions of neurotoxins, causing symptoms of Parkinson’s disease in various experimental animal models, and also neuroprotectors. There is evidence that mitochondrial dysfunction induced by the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) influences functioning of the ubiquitin-proteasomal system (UPS) responsible for selective proteolytic degradation of proteins from various intracellular compartments (including mitochondria) and neuroprotective effects of certain anti-Parkisonian agents (monoamine oxidase inhibitors) may be associated with their effects on the UPS. In this study, we have investigated the effect of the neurotoxin MPTP and neuroprotector isatin, and their combination on the profile of ubiquitinated brain mitochondrial proteins. The development of movement disorders induced by MPTP administration caused dramatic changes in the profile of ubiquitinated proteins associated with mitochondria. Pretreatment with the neuroprotector isatin decreased manifestations of MPTP-induced Parkinsonism, and had a significant impact on the profile of ubiquitinated mitochondrial proteins (including oxidative modified proteins). Administration of isatin alone to intact mice also influenced the profile of ubiquitinated mitochondrial proteins, and increased the proportion of oxidized proteins carrying the ubiquitination signature. These alterations in the ubiquitination of mitochondrial proteins observed within 2 h after administration of MPTP and isatin obviously reflect immediate short-term biological responses to these treatments.
Keywords: ubiquitin, proteasome, mouse brain mitochondrial proteins, proteomic profiling of ubiquitinated proteins, MPTP-induced Parkinsonism, neuroprotector isatin
1. Introduction
Mitochondria play an important role in molecular mechanisms of adaptive changes occurring in cells of various brain structures in response to altered physiological conditions or development of pathologies [1]. These intracellular organelles are a crucial target for various neurotoxins, causing symptoms of Parkinson’s disease [1,2,3]. In the context of experimental Parkinsonism induced by administration of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mitochondria are the principle organelles, where the key events in the development of this neurodegenerative disorder occur. Being a protoxin, MPTP undergoes bioactivation by monoamine oxidase B (MAO B), the enzyme of the outer mitochondrial membrane; the resultant neurotoxin MPP+ (1-methyl-4-phenylpyridinium) inhibits complex I of the mitochondrial respiratory chain, thus promoting the development of mitochondrial dysfunction and movement disorders typical of Parkinson’s disease [3,4,5]. Administration of MAO B inhibitors (e.g., deprenyl or isatin [6,7]) or substrates competing for the active site of this enzyme (e.g., phenylethylamine) [8] prevented not only metabolic activation of MPTP, but also deficiency of the neurotransmitter dopamine, and locomotor impairments typical of this disease.
Good evidence exists in the literature that neuroprotector mechanisms of certain anti-Parkisonian agents, inhibitors of monoamine oxidase (MAO), may be associated with their effects on the ubiquitin-proteasomal system (UPS) [9] responsible for selective proteolytic degradation of proteins from various intracellular compartments including mitochondria. Mitochondrial dysfunction induced by MPTP has a significant impact on the functioning of the ubiquitin-proteasome system (UPS) [10,11].
Ubiquitin, a 76-residue protein, is widely distributed in all eukaryotic cells, and targets proteins for subsequent degradation [12,13,14]. The ubiquitination process includes several sequential stages, which involve ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) [12,13,14]. The major function of ubiquitin consists of (poly)ubiquitination of proteins for their subsequent proteasomal degradation. In this context, the 19S proteasome subunits play a role in the ubiquitin receptor [15], which is responsible for the delivery of client proteins to the 20S proteasome, where subsequent proteolytic degradation occurs [16].
Recently, we have demonstrated that one of the 19S proteasome subunits, Rpn10, binds a wide range of mouse proteins associated with brain mitochondria [17]; and it demonstrated high affinity to both ubiquitinated and to non-ubiquitinated proteins [18]. The development of MPTP-induced movement impairments typical of experimental Parkinsonism in mice were accompanied by changes in the repertoire of Rpn10-binding proteins [17]. Pretreatment of mice with isatin significantly attenuated the manifestation of movement impairments, and decreased the repertoire of mitochondrial Rpn10-binding proteins [17]. However, the ubiquitination status of these proteins was not evaluated.
In this work, we have studied the profile of the mouse brain mitochondrial proteins carrying the ubiquitin signature and the possible contribution of cytosolic and mitochondrial ubiquitin conjugation machineries to the ubiquitination of mitochondrial proteins. We have also investigated the effect of the neurotoxin MPTP and the neuroprotector isatin on the profile of ubiquitinated proteins of the mouse brain mitochondrial fraction. The results of this study indicate that the development of MPTP-induced Parkinsonism had a significant impact on the profile of ubiquitinated brain mitochondrial proteins. Administration of the neuroprotector isatin either alone or before MPTP significantly altered the profile of ubiquitinated brain mitochondrial proteins.
2. Materials and Methods
2.1. Reagents
Sucrose, Triton X-100, triethylammonium bicarbonate, potassium phosphates, deoxycholic acid sodium salt, urea, 2-iodoacetoamide, MPTP, and isatin were purchased from Sigma (St. Louis, MO, USA); formic acid was from Merck (Darmstadt, Germany); acetonitrile was from Fisher Chemical (Leicestershire, UK); tris-(2-carboxyethyl) phosphine was obtained from Pierce-Thermo Scientific (Rockford, IL, USA). Trypsin (modified sequencing grade) was obtained from Promega (Madison, WI, USA). Deubiquitinase inhibitor degrasyn (WP-1130) was purchased from Selleck Chemicals (Houston, TX, USA).
2.2. Animals, MPTP, and Isatin Administration, and Behavioral Tests
Thirty two male C57BL/6 mice (20–25 g; n = 8 in each group), obtained from the Stolbovaya nursery (Moscow region), were used in this study. Experiments were performed one week after their arrival from the nursery. Animals were maintained at natural illumination and received a standard laboratory chow and water ad libitum. MPTP was injected intraperitoneally (i.p., 30 mg/kg). Isatin (100 mg/kg, i.p.) was injected 30 min before MPTP. Control mice were treated with intraperitoneal injection of saline (0.1 mL/kg). Behavioral changes induced by MPTP or isatin were analyzed 90 min after the last administration by means of the open field test [19]. The exploratory reaction of mice in the open field test was defined as a sum of horizontal activity (units) and vertical activity (units). All procedures were approved by local authorities for animal research.
2.3. Isolation of Mitochondrial Fraction and Sample Preparation for Mass Spectrometry
The animals were decapitated within 30 min after behavioral testing. All subsequent procedures were carried out at 4 °C. The brains, washed in ice-cold saline, were immediately dissected and homogenized in the isolation medium containing 0.32 M sucrose, 1 mM EDTA, 10 mM Tris-HCl buffer, pH 7.5, using an Ultra-Turrax T 10 homogenizer (IKA-Werke, Staufen Germany) at a low speed, to obtain 10% homogenate (w/v).
Isolation of the brain mitochondrial fraction was carried out as described previously [20]. Briefly, 10% homogenate was initially centrifuged at 1000× g for 10 min to remove sediment cell debris and nuclei. The resultant supernatant was further centrifuged at 10,000× g for 20 min to isolate the crude mitochondrial fraction, which was not subjected to additional purification procedures.
The mitochondrial pellets obtained at the previous stage were resuspended in 200 µL in the lyzing solution, containing 0.05 M potassium-phosphate buffer, pH 7.4, and 3% Triton X-100. After incubation for 60 min at 4 °C, the mitochondrial preparations were diluted three times with the same buffer but without Triton X-100 to obtain the final concentration of the detergent of 1%. Samples were centrifuged at 19,500× g for 20 min, and supernatants were used for subsequent proteomic analysis [21].
In a pilot experiment, the lysates were incubated with the deubiquitinating enzyme inhibitor degrasyn (WP-1130) for the evaluation of a possible effect of endogenous deubiquitinases on ubiquitinated proteins.
2.4. Mass Spectrometry and Liquid Chromatography
High resolution mass spectrometry analysis was performed using an Orbitrap Fusion (Thermo Scientific, Rockford, IL, USA) with the installed ESI-NSI ion source. The instrument was operated in positive ionization mode with emitter voltage adjusted to 2.2 kV and drying gas temperature at 280 °C. Surveyed in a range of 400 m/z–1200 m/z precursor ions (maximum integration time was 80 ms) with charge states from z = 2+ to z = 6+ were isolated in the quadrupole mass analyzer within ±1.5 m/z, and triggered to fragmentation in the m/z range with a fixed lower mass (110 m/z) and a dynamic upper mass (depending on the charge state of the fragmented precursor) limited to 2100 m/z. Fragmentation and tandem scanning were performed in a MS3 synchronous precursor ions selection conditioned by mass difference between the fragment ions of either ΔM = 114.0429 u (corresponding to the ubiquitin tag GG) or ΔM = 383.2281 u (corresponding to the ubiquitin tag LRGG), both detected with an asymmetric mass tolerance of −3 ppm/+7 ppm. Only two (N = 2) ions were allowed for synchronous selection in the MS3 mode, provided that the mass difference between ions in pair was registered.
Liquid chromatography separation was accomplished on an Ultimate 3000 RSLCnano (Thermo Scientific, Rockford, IL, USA). Samples were loaded onto an enrichment Acclaim µ-Precolumn (0.5 mm × 3 mm, 5 µm) (Thermo Scientific, Rockford, IL, USA) at a flow rate of 15 µL/min for 3.5 minutes in 2% acetonitrile, supplied by 0.1% formic acid and 0.03% acetic acid. Analytical separation was carried out at a flow rate 0.3 µL/min using an Acclaim Pepmap® C18 (75 µm × 150 mm, 2 µm) (Thermo Scientific, USA) column in a gradient of mobile phase A (water with 0.1% formic acid and 0.03% acetic acid) and mobile phase B (acetonitrile with 0.1% formic acid and 0.03% acetic acid) in the following gradient: 2%–37% of mobile phase B for 45 min, followed by column washing in 90% of mobile phase B for 8 min, with equilibration of the column under initial gradient conditions (2% of mobile phase B) for 15 min before starting the next run.
Every mass spectrometry experiment was performed using pooled brain mitochondrial fractions isolated from two mice. For each group of animals, four independent experiments were carried out.
2.5. Protein Identification and GO Annotation
Raw data files were converted in MGF-files using MSConvert (Proteowizard, Palo Alto, CA, USA). Peak lists obtained from converted spectra were identified using X!Tandem Vengeance, version (2015.12.15.2). The search was conducted using SearchGUI, version 3.3.0 [22,23].
Protein identification was conducted against a concatenated target/decoy version of the Mus musculus (16,915 > 99.9%) complement of the UniProtKB fasta file (release February 2018). The decoy sequences were created by reversing the target sequences in SearchGUI. The identification settings were as follows: trypsin (specific), with a maximum of three missed cleavages within ±5.0 ppm tolerance at MS1 level, and ±0.03 Da tolerance for MS2 tolerances. The following variable modifications were set: carbamidomethylation of C (+57.021464 Da), oxidation of M (+15.994915 Da), ubiquitination of K as GG-tag (+114.042927 Da), and long ubiquitination tag of K (+383.228102 Da). Variable modifications refined after the search procedure. Peptides and proteins were inferred from the spectrum identification results using PeptideShaker version 1.16 (Compomics, Gent, Belgium). Peptide spectrum matches (PSMs), peptides, and proteins were validated at a 1.0% false discovery rate (FDR) estimated using the decoy hit distribution.
Proteins were classified by their cellular localization, molecular functions, and biological process involvement in terms of Gene Ontology (GO) annotations using STRAP software (version 1.5.0.0) [24].
Each protein listed in the Tables and the Supplementary Materials was identified at least in three independent experiments, each of which employed independent brain mitochondrial samples, as well as their chromatographic and proteomic processing.
3. Results
In accordance with results of our recent research [17,25], a single dose administration of MPTP (30 mg/kg, i.p.) caused a pronounced decrease (−76 ± 6%) in the locomotor activity of mice evaluated in this study in the open field test. Pretreatment with isatin (100 mg/kg, i.p.) significantly (p < 0.05) improved the locomotor activity, which however, was lower than in control animals (−44 ± 11%). Isatin administered to intact mice also decreased locomotor activity (−33 ± 8%). This effect may be attributed to known sedative activity as described in the literature (see for review, [26]).
Proteomic profiling of the brain mitochondrial fraction of the control mice resulted in reliable identification of 565 proteins (see Supplementary Materials). Among them, 301 proteins were annotated by GO as mitochondrial proteins. These included both intrinsic mitochondrial proteins, which localized in the inner mitochondrial membrane and matrix, and also proteins of the outer mitochondrial compartment, as well as proteins from extra-mitochondrial compartments associated with mitochondria. Good evidence exists that many extra-mitochondrial proteins interact with mitochondrial membranes, and some of them (e.g., histones [27,28]) are even inserted in the mitochondrial membranes. In this context, it is relevant to consider all of the identified proteins either as intrinsic mitochondrial proteins (i.e., proteins of the inner mitochondrial membrane and matrix) or as mitochondria associated proteins, including mitochondrial proteins of the outer mitochondrial membrane and of the intermembrane space. These mitochondria-associated proteins can be thus defined as proteins of the outer mitochondrial compartment. Such subdivision is important in the context of mitochondrial ubiquitination, because certain evidence exists that outer mitochondrial proteins and proteins of the outer surface of the inner mitochondrial membrane can be ubiquitinated by the cytosolic ubiquitin conjugation machinery [29,30]. Moreover, it appears that even UCP2 (mitochondrial uncoupling protein 2), which is located in the inner mitochondrial membrane [31], can be ubiquitinated by the cytosolic ubiquitin conjugating machinery [32].
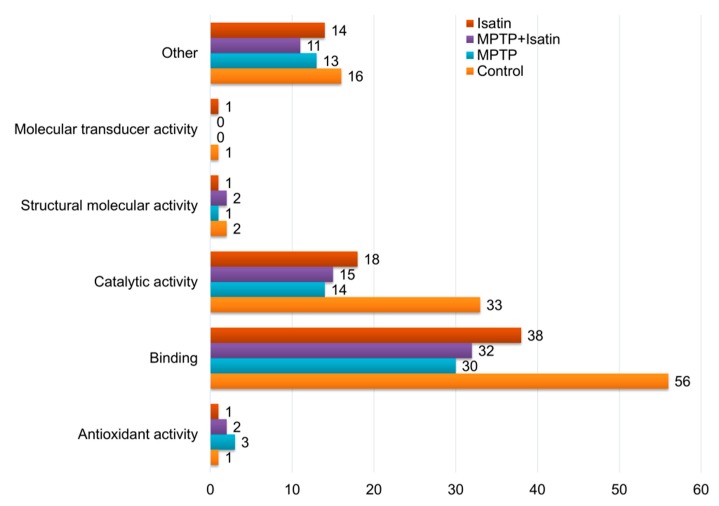
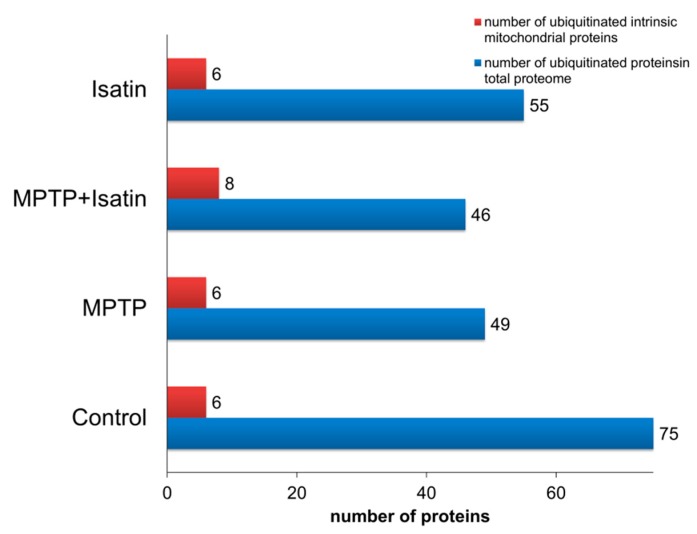
With this assumption, the brain mitochondrial fraction of control mice contained at least 75 individual ubiquitinated proteins; the number of the ubiquitinated sites in them varied from 1 to 4 (Figure 1; Supplementary Materials). In a pilot experiment, preincubation of brain mitochondrial lysates with the deubiquitinase inhibitor degrasyn (WP-1130; final concentration 10 µM) for 30 min at 37 °C had no influence of the profiles of the ubiquitinated proteins. This suggests that under our experimental conditions, endogenous deubiquitinases had insignificant impact on the content of ubiquitinated proteins in the brain mitochondrial fraction. Among these 75 proteins, only six proteins can be defined as proteins of the inner mitochondrial compartments. Their proportion in the studied groups varied from 8% (in control) to 17% (MPTP+isatin) (Figure 2). According to the GO annotation, they belonged to several functional groups (Table 1). In the context of molecular functions, ubiquitinated mitochondrial proteins of control animals were preferentially involved in catalytic and binding functions.
Figure 1.
Distribution of identified ubiquitinated proteins among GO groups. Numbers designate the number of identified ubiquitinated proteins in each experimental group of animals.
Figure 2.
Total number of ubiquitinated proteins identified in the mouse brain mitochondrial fraction.
Table 1.
Functional annotation of ubiquitinated proteins of mouse brain mitochondria in GO terms.
| Accession Number | UniProt ID | Recommended Protein Name | Sample Type | Extramitochondrial Compartment | Intramitochondrial Compartment | Molecular Function | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | MPTP | MPTP+isatin | Isatin | Antioxidant Activity | Binding | Catalytic Activity | Other | |||||
| CATD_MOUSE | P18242 | Cathepsin D | ● | ● | ● | |||||||
| APR_MOUSE | Q9JM54 | Phorbol-12-myristate-13-acetate-induced protein 1 | ● | ● | ||||||||
| TAP1_MOUSE | P21958 | Antigen peptide transporter 1 | ● | ● | ● | ● | ● | |||||
| AKT1_MOUSE | P31750 | RAC-alpha serine/threonine-protein kinase | ● | ● | ● | ● | ● | |||||
| KPYM_MOUSE | P52480 | Pyruvate kinase PKM | ● | ● | ● | ● | ||||||
| ACSS3_MOUSE | Q14DH7 | Acyl-CoA synthetase short-chain family member 3, mitochondrial | ● | ● | ● | ● | ||||||
| AL4A1_MOUSE | Q8CHT0 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | ● | ● | ● | ● | ||||||
| NALP5_MOUSE | Q9R1M5 | NACHT, LRR and PYD domains-containing protein 5 | ● | ● | ● | |||||||
| AT2A1_MOUSE | Q8R429 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | ● | ● | ● | ● | ● | |||||
| SUCA_MOUSE | Q9WUM5 | Succinate-CoA ligase (ADP/GDP-forming) subunit alpha, mitochondrial | ● | ● | ● | ● | ● | |||||
| OAT_MOUSE | P29758 | Ornithine aminotransferase, mitochondrial | ● | ● | ● | ● | ||||||
| MRP9_MOUSE | Q80WJ6 | Multidrug resistance-associated protein 9 | ● | ● | ● | ● | ||||||
| DLDH_MOUSE | O08749 | Dihydrolipoyl dehydrogenase, mitochondrial | ● | ● | ● | ● | ● | |||||
| CATB_MOUSE | P10605 | Cathepsin B | ● | ● | ● | ● | ||||||
| CLH1_MOUSE | Q68FD5 | Clathrin heavy chain 1 | ● | ● | ● | |||||||
| ADCYA_MOUSE | Q8C0T9 | Adenylate cyclase type 10 | ● | ● | ● | ● | ||||||
| FPPS_MOUSE | Q920E5 | Farnesyl pyrophosphate synthase | ● | ● | ● | ● | ||||||
| AATM_MOUSE | P05202 | Aspartate aminotransferase, mitochondrial | ● | ● | ● | ● | ||||||
| ERAL1_MOUSE | Q9CZU4 | GTPase Era, mitochondrial | ● | ● | ● | |||||||
| TO20L_MOUSE | Q9D4V6 | TOMM20-like protein 1 | ● | ● | ● | ● | ||||||
| PRDX1_MOUSE | P35700 | Peroxiredoxin-1 | ● | ● | ● | ● | ● | ● | ||||
| GSHR_MOUSE | P47791 | Glutathione reductase, mitochondrial | ● | ● | ● | ● | ● | |||||
Notes: color shows (extra) mitochondrial localization of the proteins.
MPTP administration to mice decreased the number of ubiquitinated proteins (n = 49; Figure 2). The list of ubiquitinated proteins identified in the brain mitochondrial fraction of MPTP-treated mice changed qualitatively, and contained only five proteins that were identified in intact animals. Poor correspondence between identified proteins from control and MPTP-treated mice suggested that the development of MPTP-induced toxicity had a significant impact on the ubiquitination of proteins associated with mitochondria (Figure 1, Table 1). Pretreatment of mice with isatin not only reduced manifestations of MPTP-induced neurotoxicity, but also influenced the profiles of all ubiquitinated proteins detected in the crude mitochondrial fraction of the mouse brain. The total number of ubiquitinated proteins in the crude mitochondrial fraction of the mouse brain and their molecular functions remained basically the same as in the crude mitochondrial fractions of the MPTP-treated mice (Table 1). However, despite similarity in molecular functions, ubiquitinated proteins in the fraction differed qualitatively as compared with both the control and the group of MPTP-treated mice (Table 1, Supplementary Materials).
Isatin administration to intact mice also caused significant changes in both the number of identified proteins in the fraction, as well as the profile of ubiquitinated proteins (Table 1, Supplementary Materials). The total number of mitochondrial proteins was lower than in both the control group and the group of MPTP-treated mice, and the same as in the group of mice treated with both MPTP and isatin. The profile of ubiquitinated proteins identified in the crude mitochondrial fraction of isatin-treated mice (n = 55; Figure 2) was also rather specific (Table 1, Supplementary Materials).
In the case of so-called intrinsic mitochondrial proteins, their ubiquitination profiles were highly specific and lacked any common components for all four groups of animals (Table 1).
It should be noted that the crude mitochondrial fraction of the mouse brain was isolated within 2 h after administration of MPTP and isatin to mice. Consequently, the changes in the ubiquitination state of mitochondrial proteins obviously reflected immediate short-term biological responses to these treatments that did not involve long-term adaptive mechanisms (mitochondrial biogenesis etc.).
Besides ubiquitination, we have also evaluated the oxidative status of mitochondrial proteins by the presence of oxidized methionine (see the Materials and Methods section). Although the number of mitochondrial proteins containing oxidized methionine residue(s) in the control group was somewhat higher, analysis of proteins carrying both the ubiquitin signature and oxidized methionine revealed very interesting tendencies. The percent of mitochondrial proteins containing both modifications increased in the following order: control (8% of total ubiquitinated proteins) > MPTP (12%) > MPTP + isatin (15%) > isatin (22%) (Supplementary Materials).
From comparison of ubiquitinated proteins associated with the mouse brain mitochondrial fraction (Supplementary Materials) with mouse brain mitochondrial proteins specifically bound to the proteasome ubiquitin receptor, the Rpn10 subunit [17], did not reveal any common proteins. This raises the possibility that ubiquitination of mitochondrial proteins is not directly linked to degradation in proteasomes.
4. Discussion
Ubiquitination plays an important role in targeting oxidized, misfolded, and damaged proteins from different intracellular compartments for subsequent proteasomal degradation [12,13,14,15]. This process is also important for protein modifications that are unrelated to proteasomal degradation but which are related to other processes, including the regulation of various cell functions [33,34]. Various (patho)physiological conditions, especially mitochondrial dysfunction, have a significant impact on UPS functioning [10,11].
In the context of MPTP-induced Parkinsonism, mitochondria are especially important “players” as the outer mitochondrial membrane enzyme, MAO B, catalyzes the conversion of MPTP in the active toxin, MPP+, which inhibits complex I of the respiratory chain, thus creating the conditions for the development of mitochondrial dysfunction [2,3,4,5]. In turn, the latter influences UPS [10,11] and promotes an increased accumulation of ubiquitin immunoreactivity in target cells [11] thus suggesting the formation of aggregates containing ubiquitinated proteins.
It should be noted that the analysis of the ubiquitination state of mitochondrial proteins is complicated by the existence of two mitochondrial compartments: (i) the outer compartment, which includes the outer mitochondrial membrane and the intermembrane space; (ii) the inner mitochondrial compartment, including the inner surface of the inner mitochondria membrane and the matrix. The outer mitochondrial compartment also contains numerous extramitochondrial proteins [31,32]. Such proteins can be ubiquitinated by the extramitochondrial ubiquitination machinery that are bound to mitochondrial membranes [35,36]. In the context of possible ubiquitination, these compartments are not identical. Proteins of the outer compartment (and even the inner mitochondrial membrane) can be ubiquitinated by the extramitochondrial ubiquitinating machinery [29,30,32]. Ubiquitination of proteins located in the inner compartment may obviously involve their own mitochondrial ubiquitin conjugation system, which still remains poorly investigated.
The results of our study indicate that ubiquitination of proteins located in the inner mitochondrial compartment covers just a few proteins (Supplementary Materials, Table 1), representing about 10% of the total pool of ubiquitinated proteins associated with brain mitochondria. Nevertheless, identification of such mitochondrial matrix proteins as Succinate-CoA ligase (EC 2.6.1.13), a tricarboxylic cycle enzyme that is responsible for Succinate-CoA conversion to succinate during substrate-coupled phosphorylation (with GTP formation), and other mitochondrial matrix enzymes listed in the table, provide convincing evidence for ubiquitination of intrinsic mitochondrial proteins by the intrinsic mitochondrial ubiquitin-conjugating machinery (Table 2).
Table 2.
Intrinsic brain mitochondrial proteins and their ubiquitination sites.
| UniProt ID | Protein Name | Gene Name | Sequence | Ubiquitination | Oxidized Residue | Confidence, % |
|---|---|---|---|---|---|---|
| O08749 | Dihydrolipoyl-dehydrogenase, mitochondrial | Dld | IPVNNRFQTKSTDR | K446 | 98.99 | |
| P29758 | Ornithine aminotransferase, mitochondrial | Oat | LFNYNKVLPMNTGVEAGETACK | K135 | M139 | 93.59 |
| Q14DH7 | Acyl-CoA synthetase short-chain family member 3, mitochondrial | Acss3 | TPPPGQAGK | K472 | 89.51 | |
| Q8CHT0 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | Aldh4a1 | NESVGYYVEPCIIESKDPQEPIMK | K437 | 95.86 | |
| Q9CZU4 * | GTPase Era, mitochondrial | Eral1 | LNPQVLQCLTKFSQVPSILVLN | K225 | 93.35 | |
| Q9WUM5 | Succinate-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | Suclg1 | KAKPVVSFIAGITAPPGR | K280 | 100.00 | |
| P05202 | Aspartate aminotransferase, mitochondrial | Got2 | GINVCLCQSYAKNMGLYGER | K279 | 93.83 | |
| P47791 | Glutathione reductase, mitochondrial | Gsr | RDAYVSRLNTIYQNNLTK | K141 | 92.89 | |
| P35700 | Peroxiredoxin-1 | Prdx1 | GSDTIKPDVNK | K185 | 99.48 |
Notes: *—long chain ubiquitination tag (LRGG).
Since ubiquitination profiles of intrinsic mitochondrial proteins are highly specific and involve only several individual proteins, it seems unlikely that their ubiquitination may be attributed to mitochondrial damage (and the involvement of the extramitochondrial ubiquitin conjugation machinery). This is consistent with previous results of in vitro studies: using biotinylated ubiquitin, we also detected the ubiquitination label in a few intramitochondrial brain proteins [20]. This proportion of mitochondrial ubiquitinated proteins corresponds to the results of bioinformatic analysis of human ubiquitinated proteins [37]: mitochondrial ubiquitinated proteins represent about 8% of the total pool of ubiquitinated proteins found in humans. The recent study performed using purified yeast mitochondria also revealed 36 ubiquitinated matrix proteins [38]. However, the mechanisms of their ubiquitination still remain poorly characterized. Thus, ubiquitination of intrinsic mitochondrial proteins of the brain can contribute to, but cannot determine the overall effect of the neurotoxin MPTP and the neuroprotector isatin on the overall ubiquitination profile of mitochondrial proteins.
A single dose administration of MPTP to mice significantly changed the profile of brain mitochondrial ubiquitinated protein. Since increased ubiquitin immunoreactivity is registered in the brain of experimental animals only after prolonged treatments with MPTP [11], the altered repertoire of endogenously ubiquitinated proteins observed within 2 h after a single dose of MPTP administration may be thus be considered as the earliest reaction of the ubiquitin conjugation machinery to the toxin. Interestingly, some of the ubiquitinated proteins associated with the brain mitochondrial fraction of MPTP-treated mice are involved in neurodegeneration (see Table 3).
Table 3.
Brain proteins ubiquitinated in mice treated with MPTP and their involvement in neurodegeneration.
| UNIPROT Accession Number | Protein Name | Involvement in Neurodegeneration | Reference |
|---|---|---|---|
| Q6ZPY5 | Zinc finger protein 507 | Alterations in ZNFs are involved in the development of neurodegeneration | [39] |
| Q91YE6 | Importin-9 | Regeneration of injured neurons | [40] |
| P68368 | Tubulin alpha-4A chain | Alpha-tubulin levels decreased mainly in neurons containing neurofibrillary tau pathology | [41] |
| Q9QZ04 | MAGE-like protein 2 | MAGE proteins form complexes with E3 ubiquitin ligases | [42] |
| Q9R0G7 | Zinc finger E-box-binding homeobox 2 | It is involved in the regulation of microRNA in glioma stem cells | [43] |
| Q8BZ36 | RAD50-interacting protein 1 | It functions as a multitask protein, and is involved in genomic stability, ER homeostasis, and autophagy | [44] |
| Q8CE72 | Protein JBTS17 | Jbts17 mutant mice have cilia transition zone defects and related cerebellar anomalies | [45] |
| Q6P7F1 | MAGUK p55 subfamily member 4 | Plays a role in several CNS disorders | [46] |
| P43300 | Early growth response protein 3 | Gene encoding this protein is induced by alpha-synuclein | [47] |
| Q8C4A5 | Putative Polycomb group protein ASXL3 | Is upregulated in Alzheimer’s disease | [48] |
| Q9ESK9 | RB1-inducible coiled-coil protein 1 | Its insufficiency causes neuronal atrophy and is involved in the pathology of Alzheimer’s diseases | [49] |
| P47791 | Glutathione reductase, mitochondrial | Is implicated in glutathione reduction. GSH is important for pathogenesis of Parkinson’s disease | [50,51] |
| Q9CR16 | Peptidyl-prolyl cis-trans isomerase D | Binds to hyperphosphorylated Tau proteins in degenerating neurons | [52] |
| Q9EQK5 | Major vault protein | Is implicated in senescence-associated apoptosis resistance | [53] |
| Q3UUG6 | TBC1 domain family member 24 | Truncating mutation results in severe neurodegeneration | [54] |
| P23950 | mRNA decay activator protein ZFP36L1 | Involved in mRNA stability in the human brain | [55] |
| P97500 | Myelin transcription factor 1-like protein | Influences memory-related processes | [56] |
| Q9QZQ1 | Afadin | Maintenance of dendritic structure and excitatory tone | [57] |
| P81117 | Nucleobindin-2 | Altered levels found in neuropsychiatric disorders | [58] |
| P58006 | Sestrin-1 | A negative feedback regulator of TOR; its loss results in various TOR-dependent, age-related pathologies | [59] |
| Q9D2H8 | Fibronectin type III domain-containing protein 8 | Its expression is decreased in patients with PD with dementia | [60] |
| Q8C079 | Striatin-interacting protein 1 | Involved in the targeting, attachment, and cytoskeletal transport of autophagosomes, which are accumulated in neurodegenerative neurons | [61] |
| Q8BJQ2 | Ubiquitin carboxyl-terminal hydrolase 1 | Undergoes oxidative modification in both Alzheimer’s disease and Parkinson’s disease | [62] |
| Q91WJ8 | Far upstream element-binding protein 1 | Being a substrate for ubiquitination by Parkin, it plays an important role in development of Parkinson disease | [63] |
| P29758 | OAT, mitochondrial | Ornithine aminotransferase deficiency causes gyrate atrophy | [64] |
| P35700 | Peroxiredoxin-1 | Plays a protective role in counteracting Aβ injury by increasing cell viability preserving neurites, and decreasing cell death | [65] |
| Q8C4S8 | DENN domain-containing protein 2A | DENN proteins regulate autophagy | [66] |
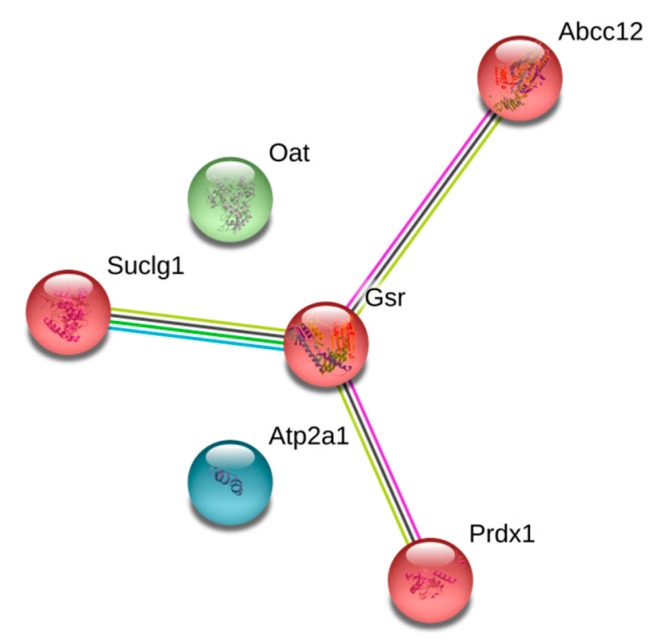
In the context of ubiquitination of intrinsic mitochondrial proteins, it should be noted that they were functionally linked only in the brain mitochondria of MPTP-treated mice (Figure 3).
Figure 3.
Functional links between mitochondrial ubiquitinated proteins of MPTP-treated mice. The links have been composed by the STRING database resource. Abbreviations used by the STRING resource designate the following proteins: Abcc12—multidrug resistance-associated protein 9; Atp2a1—sarcoplasmic/endoplasmic reticulum calcium ATPase 1; Gsr—mitochondrial glutathione reductase; Oat—mitochondrial ornithine aminotransferase; Prdx1—peroxiredoxin-1; Suclg1—mitochondrial succinate-CoA ligase (ADP/GDP-forming) subunit alpha. The links have been generated using the STRING high confidence score of 0.7. Other explanations are given in the text.
In these functional links, glutathione reductase (Gsr), catalyzing the reaction of NADPH-dependent glutathione reduction, was the core element (Figure 3). According to STRING database, Gsr has functional links with (Figure 3): (i) succinate CoA ligase, catalyzing substrate level phosphorylation reaction in the Krebs cycle, (ii) peroxiredoxin 1, the protein involved in the antioxidant defense system; (iii) multidrug resistance-associated protein 9, a novel member of the multidrug resistance-associated protein (MRP) family, contributing to decreased drug accumulation [67]. Peroxiredoxin 1 ubiquitination was detected in endothelial cells during ischemic insult; this targeting of peroxiredoxin 1 for degradation deteriorates ischemic brain damage [68]. Such links were not observed in the groups of animals treated with either MPTP or isatin, or only with isatin, due to different patterns of the mitochondrial ubiquitinated proteins. This suggests that the neuroprotector effect of isatin may be associated with blockade of functional links involving ubiquitinated proteins targeted for subsequent degradation.
It is known that the outer mitochondrial membranes and the proteins of the outer mitochondrial compartment are involved in numerous interactions between mitochondria and mitochondria associated membranes (MAM) [69]. Such interactions are crucial for various important cell functions, including mitochondrial morphology, apoptosis, autophagy, Ca2+ signaling, endoplasmic reticulum(ER)–mitochondria tethering, ER stress signaling [69]. In this context, the altered repertoire of ubiquitinated proteins determined in the brain mitochondrial fraction of MPTP-treated mice obviously reflects the impaired interactions between the mitochondria and the extra-mitochondrial compartments (MAM).
It is known that MPTP administration induces the development of oxidative stress, accompanied by oxidation of brain proteins [70]. Our study has shown that the repertoire of ubiquitinated proteins from the brain mitochondria of MPTP-treated mice includes oxidized proteins. Interestingly, their proportion was somewhat higher in the control, while the proportion of ubiquitinated and oxidized proteins increased in the following order: control > MPTP > MPTP + isatin > isatin. This suggests that the neuroprotector effect of isatin could be partially attributed to increased ubiquitination of oxidized mitochondrial proteins.
Isatin (indoledione-2,3) is an endogenous neuroprotector which is found in mammalian brain, peripheral tissues, and body fluids [26]. Besides the reversible inhibition of MAO B, which can account for the decreased bioactivation of MPTP, isatin interacts with numerous isatin-binding proteins located in various subcellular organelles, including mitochondria [25,26]. Previous proteomic profiling of brain isatin-binding proteins revealed several enzymes which were directly involved in UPS functioning [17]. However, isatin administration to mice significantly restricted the repertoire of brain mitochondrial proteins bound to the ubiquitin receptor, the 19S Rpn 10 subunit [17]. Moreover, proteomic profiles of ubiquitinated mitochondrial proteins identified in this study (Supplementary Materials) and mitochondrial proteins specifically bound to the proteasome ubiquitin receptor, the Rpn 10 subunit [17], lack any common elements. In addition, some isatin derivatives act as proteasome inhibitors [71,72]. Taken together, all these results suggest that besides increased ubiquitination of oxidized mitochondrial proteins, isatin reduces the possibility of their Rpn10-receptor mediated entry to, and subsequent degradation in proteasomes. Thus, it appears that the neuroprotector effect of isatin can be partially attributed to the switch of proteasomal degradation to selective autophagy, which includes a proteasome-independent route of ubiquitinated proteins [73,74].
5. Conclusions
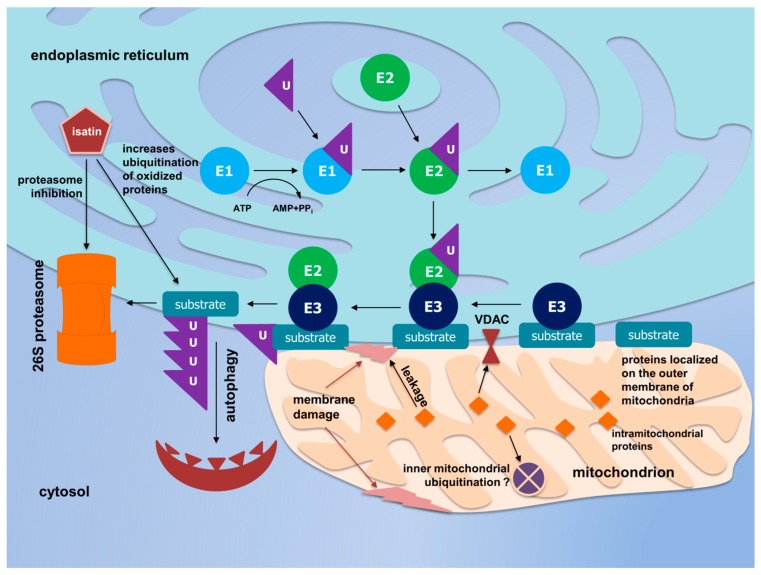
Administration of MPTP to mice caused locomotor impairments typical of Parkinson’s disease. This was accompanied by dramatic changes in the profile of mitochondrial ubiquitinated proteins. Pretreatment of mice with the neuroprotector isatin not only reduced manifestations of MPTP-induced neurotoxicity, but also influenced the profiles of all ubiquitinated proteins detected in the crude mitochondrial fraction of the mouse brain and increased the proportion of oxidized proteins carrying the ubiquitination signature. Since crude mitochondrial fraction of the mouse brain was isolated within 2 h after administration of MPTP and isatin, the changes in the ubiquitination state of mitochondrial proteins obviously reflect immediate short-term biological responses to these treatments that do not involve long-term adaptive mechanisms (mitochondrial biogenesis etc.). Figure 4 summarizes results of our studies.
Figure 4.
The scheme illustrating the proposed mechanism of neuroprotector action of isatin on metabolic routes of ubiquitinated proteins under conditions of MPTP-induced neurotoxicity. Isatin increases the ubiquitination of oxidized proteins associated with mitochondria, restricts binding of the proteins to the 26S proteasome ubiquitin receptor (Rpn10) and, thus, restricts the access of ubiquitinated proteins to the proteasome and shifts the flux of oxidized and ubiquitinated proteins to autophagy routes.
Results of our study indicate that ubiquitination of proteins located in the inner mitochondrial compartment covers just a few proteins, and major changes in the ubiquitination state occur in the outer mitochondrial compartment. Earlier we found that isatin also restricted the repertoire of mitochondrial proteins bound to the ubiquitin receptor, the 19S Rpn 10 subunit [17], directly participating in the proteasomal machinery. In addition, mitochondrial ubiquitinated proteins and mitochondrial Rpn10-binding proteins lack any common elements. This raises the possibility that ubiquitination of mitochondrial proteins is not directly linked to degradation in proteasomes. It is known in the literature that some isatin derivatives act as proteasome inhibitors [71,72]. In this context, the increased proportion of oxidized mitochondrial proteins carrying the ubiquitin signature may reflect their impaired proteasomal degradation. In this case the neuroprotector effect of isatin can be (at least partially) attributed to the switch of proteasomal degradation to selective autophagy, which includes a proteasome-independent route of ubiquitinated proteins [73,74].
Acknowledgments
The mass spectrometry identification of mouse brain mitochondrial proteins was performed using the “Human Proteome” Core Facility (Institute of Biomedical Chemistry, Moscow) supported by Ministry of Education and Science of the Russian Federation (a unique project identifier RFMEFI62117X0017).
Supplementary Materials
The Supplementary materials are available as a Microsoft Excel file, available online at http://www.mdpi.com/2073-4409/7/8/91/s1. It contains information about the whole proteomic profiles of crude mitochondrial fractions from all groups of animals, lists of ubiquitinated proteins, oxidized proteins, and ubiquitinated and oxidized proteins.
Author Contributions
O.B., A.K., V.Z., A.M. conceived and designed the experiments. I.K. and E.I. performed animal studies, O.B., A.K. performed sample preparation and mass spectrometry identification; A.M., A.K., O.B. made original draft preparation, wrote the paper. The study was supervised by V.Z. and A.M.
Funding
This work performed within the framework of the Program for Basic Research of State Academies of Sciences for 2013–2020 was partially supported by the Russian Foundation for Basic Research (project no. 16-04-00173).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Cheng A., Hou Y., Mattson M.P. Mitochondria and neuroplasticity. ASN Neuro. 2010;2:AN20100019. doi: 10.1042/AN20100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cookson M.R. The biochemistry of Parkinson’s disease. Annu. Rev. Biochem. 2005;74:9–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 3.Buneeva O.A., Medvedev A.E. Mitochondrial Disfunction in Parkinson’s Disease. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2011;5:313–336. doi: 10.18097/pbmc20115703246. [DOI] [PubMed] [Google Scholar]
- 4.Maret G., Testa B., Jenner P., el Tayar N., Carrupt P.A. The MPTP story: MAO activates tetrahydropyridine derivatives to toxins causing parkinsonism. Drug Metab. Rev. 1990;22:291–332. doi: 10.3109/03602539009041087. [DOI] [PubMed] [Google Scholar]
- 5.Park J.S., Davis R.L., Sue C.M. Mitochondrial Dysfunction in Parkinson’s Disease: New Mechanistic Insights and Therapeutic Perspectives. Curr. Neurol. Neurosci. Rep. 2018;18:21. doi: 10.1007/s11910-018-0829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y., Zhao Z.Q., Xie J.X. Effect of isatin on rotational behavior and DA levels in caudate putamen in Parkinsonian rats. Brain Res. 2001;917:127–132. doi: 10.1016/S0006-8993(01)02935-3. [DOI] [PubMed] [Google Scholar]
- 7.Hamaue N., Minami M., Terado M., Hirafuji M., Endo T., Machida M., Hiroshige T., Ogata A., Tashiro K., Saito H., et al. Comparative study of the effects of isatin, an endogenous MAO-inhibitor, and selegiline on bradykinesia and dopamine levels in a rat model of Parkinson’s disease induced by the Japanese Encephalitis virus. NeuroToxicology. 2004;25:205–213. doi: 10.1016/S0161-813X(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 8.Melamed E., Youdim M.B.H. Prevention of dopaminergic toxicity of MPTP in mice by phenylethylamine, a specific substrate of type B monoamine oxidase. Br. J. Pharmacol. 1985;86:529–531. doi: 10.1111/j.1476-5381.1985.tb08927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youdim M., Edmondson D., Tipton K. The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci. 2006;7:295–308. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 10.Hoglinger G.U., Carrard G., Michel P.P., Medja F., Lombes A., Ruberg M., Friguet B., Hirsch E.C. Dysfunction of mitochondrial complex I and the proteasome: Interactions between two biochemical deficits in a cellular model of Parkinson’s disease. J. Neurochem. 2003;86:1297–1307. doi: 10.1046/j.1471-4159.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- 11.Fornai F., Schluter O.M., Lenzi P., Gesi M., Ruffoli R., Ferrucci M., Lazzeri G., Busceti C.L., Pontarelli F., Battaglia G., et al. Parkinson-like syndrome induced by continuous MPTP infusion: Convergent roles of the ubiquitin proteasome system and α-synuclein. Proc. Natl. Acad. Sci. USA. 2005;102:3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 13.Hershko A., Ciechanover A., Varshavsky A. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz A.L., Ciechanover A. Targeting proteins for destruction by the ubiquitin system: Implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 15.Hamazaki J., Sasaki K., Kawahara H., Hisanaga S., Tanaka K., Murata S. Rpn10-Mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 2007;19:6629–6638. doi: 10.1128/MCB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeki Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017;161:113–124. doi: 10.1093/jb/mvw091. [DOI] [PubMed] [Google Scholar]
- 17.Medvedev A.E., Buneeva O.A., Kopylov A.T., Tikhonova O.V., Medvedeva M.V., Nerobkova L.N., Kapitsa I.G., Zgoda V.G. The brain mitochondrial subproteome of Rpn10-binding proteins and its changes induced by the neurotoxin MPTP and the neuroprotector Isatin. Biochemistry (Moscow) 2017;82:330–333. doi: 10.1134/S0006297917030117. [DOI] [PubMed] [Google Scholar]
- 18.Buneeva O.A., Gnedenko O.V., Kopylov A.T., Medvedeva M.V., Zgoda V.G., Ivanov A.S., Medvedev A.E. Quantitative affinity interaction of ubiquitinated and non-ubiquitinated proteins with proteasome subunit Rpn10. Biochemistry (Moscow) 2017;82:1042–1047. doi: 10.1134/S0006297917090073. [DOI] [PubMed] [Google Scholar]
- 19.Prut L., Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: A review. Eur. J. Pharmacol. 2003;463:3–33. doi: 10.1016/S0014-2999(03)01272-X. [DOI] [PubMed] [Google Scholar]
- 20.Buneeva O., Medvedeva M., Kopylov A., Zgoda V., Medvsedev A. Use of biotinylated ubiquitin for analysis of rat brain mitochondrial proteome and interactome. Int. J. Mol. Sci. 2012;3:11593–11609. doi: 10.3390/ijms130911593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medvedev A., Buneeva O., Kopylov A., Gnedenko O., Ivanov A., Zgoda V., Makarov A.A. Amyloid-binding proteins: Affinity-based separation, proteomic identification, and optical biosensor validation. Methods Mol. Biol. 2015;1295:465–477. doi: 10.1007/978-1-4939-2550-6_33. [DOI] [PubMed] [Google Scholar]
- 22.Vaudel M., Burkhart J.M., Zahedi R.P., Oveland E., Berven F.S., Sickmann A., Martens L., Barsnes H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015;33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 23.Vaudel M., Barsnes H., Berven F.S., Sickmann A., Martens L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics. 2011;11:996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]
- 24.Bhatia V.N., Perlman D.H., Costello C.E., McComb M.E. Software tool for researching annotations of proteins: Open-source protein annotation software with data visualization. Anal. Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buneeva O.A., Kopylov A.T., Nerobkova L.N., Kapitsa I.G., Zgoda V.G., Medvedev A.E. The effect of neurotoxin MPTP administration to mice on the proteomic profile of brain isatin-binding proteins. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2018;12:22–26. doi: 10.1134/S1990750818010043. [DOI] [PubMed] [Google Scholar]
- 26.Medvedev A., Buneeva O., Gnedenko O., Ershov P., Ivanov A. Isatin, an endogenous non-peptide biofactor: A review of its molecular targets, mechanisms of actions and their biomedical implications. Biofactors. 2018;44:95–108. doi: 10.1002/biof.1408. [DOI] [PubMed] [Google Scholar]
- 27.Choi Y.S., Jeong J.H., Min H.K., Jung H.J., Hwang D., Lee S.W., Pak Y.K. Shot-gun proteomic analysis of mitochondrial D-loop DNA binding proteins: Identification of mitochondrial histones. Mol. BioSyst. 2011;7:1523–1536. doi: 10.1039/c0mb00277a. [DOI] [PubMed] [Google Scholar]
- 28.Cascone A., Bruelle C., Lindholm D., Bernardi P., Eriksson O. Destabilization of the outer and inner mitochondrial membranes by core and linker histones. PLoS ONE. 2012;7:e35357. doi: 10.1371/journal.pone.0035357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bragoszewski P., Gornicka A., Sztolsztener M.E., Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol. Cell. Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bragoszewski P., Turek M., Chacinska A. Control of mitochondrial biogenesis and function by the ubiquitin–proteasome system. Open Biol. 2017;7:170007. doi: 10.1098/rsob.170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand M.D., Esteves T.C. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Azzi V., Brandt M.D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 2009;123:578–585. doi: 10.1242/jcs.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vertegaal A.C. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buneeva O.A., Medvedev A.E. The role of atypical ubiquitination in cell regulation. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2017;11:16–31. doi: 10.1134/S1990750817010024. [DOI] [Google Scholar]
- 35.Marchi S., Patergnani S., Pinton P. The endoplasmic reticulum–mitochondria connection: One touch, multiple functions. Biochim. Biophys. Acta. 2014;1837:461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Plotegher N., Duchen M.R. Crosstalk between lysosomes and mitochondria in Parkinson’s disease. Front. Cell Dev. Biol. 2017;5:110. doi: 10.3389/fcell.2017.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehmann G., Udasin R.G., Ciechanover A. On the linkage between the ubiquitin-proteasome system and the mitochondria. Biochem. Biophys. Res. Commun. 2016;473:80–86. doi: 10.1016/j.bbrc.2016.03.055. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann G., Ziv T., Braten O., Admon A., Udasin R.G., Ciechanover A. Ubiquitination of specific mitochondrial matrix proteins. Biochem. Biophys. Res. Commun. 2016;475:13–18. doi: 10.1016/j.bbrc.2016.04.150. [DOI] [PubMed] [Google Scholar]
- 39.Cassandri M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M., Melino G., Raschellà G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanz S., Perlson E., Willis D., Zheng J.Q., Massarwa R., Huerta J.J. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/S0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 41.Perez M., Santa-Maria I., Gomez de Barreda E., Zhu X., Cuadros R., Cabrero J.R., Sanchez-Madrid F., Dawson H.N., Vitek M.P., Perry G., et al. Tau—An inhibitor of deacetylase HDAC6 function. J. Neurochem. 2009;109:1756–1766. doi: 10.1111/j.1471-4159.2009.06102.x. [DOI] [PubMed] [Google Scholar]
- 42.Doyle J.M., Gao J., Wang J., Yang M., Potts P.R. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu A., Yu Q., Peng Z., Huang Y., Diao S., Cheng J., Wang W., Hong M. Mir-200b inhibits CD133+ glioma cells by targeting the AKT pathway. Oncol. Lett. 2017;13:4701–4707. doi: 10.3892/ol.2017.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grigaravicius P., von Deimling A., Frappart P.-O. RINT1 functions as a multitasking protein at the crossroads between genomic stability, ER homeostasis, and autophagy. Autophagy. 2016;12:1413–1415. doi: 10.1080/15548627.2016.1191730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damerla R.R., Cui C., Gabriel G.C., Liu X., Craige B., Gibbs B.C., Francis R., Li Y., Chatterjee B., San Agustin J.T., et al. Novel Jbts17 mutant mouse model of Joubert syndrome with cilia transition zone defects and cerebellar and other ciliopathy related anomalies. Hum. Mol. Genet. 2015;24:3994–4005. doi: 10.1093/hmg/ddv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardoni F., Marcello E., Di Luca M. Postsynaptic density-membrane associated guanylate kinase proteins (PSD-MAGUKs) and their role in CNS disorders. Neuroscience. 2009;158:324–333. doi: 10.1016/j.neuroscience.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 47.Qin H., Buckley J.A., Li X., Liu Y., Fox T.H., III, Meares G.P., Yu H., Yan Z., Harms A.S., Li Y., et al. Inhibition of the JAK/STAT pathway protects against α-synuclein-induced neuroinflammation and dopaminergic neurodegeneration. J. Neurosci. 2016;26:5144–5159. doi: 10.1523/JNEUROSCI.4658-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto M., Bogdanovic N., Nakagawa H., Volkmann I., Aoki M., Winblad B., Sakai J., Tjernberg L.O. Analysis of microdissected neurons by 18O mass spectrometry reveals altered protein expression in Alzheimer’s disease. J. Cell. Mol. Med. 2012;16:1686–1700. doi: 10.1111/j.1582-4934.2011.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chano T., Okabe H., Hulette C.M. RB1CC1 insufficiency causes neuronal atrophy through mTOR signaling alteration and involved in the pathology of Alzheimer’s diseases. Brain Res. 2007;1168:97–105. doi: 10.1016/j.brainres.2007.06.075. [DOI] [PubMed] [Google Scholar]
- 50.Gao H.M., Jiang J., Wilson B., Zhang W., Hong J.S., Liu B. Microglial activation-mediated delayed and progressive degeneration of rat nigral dopaminergic neurons: Relevance to Parkinson’s disease. J. Neurochem. 2002;81:1285–1297. doi: 10.1046/j.1471-4159.2002.00928.x. [DOI] [PubMed] [Google Scholar]
- 51.Martin H.L., Teismann P. Glutathione—A review on its role and significance in Parkinson’s disease. FASEB J. 2009;23:3263–3272. doi: 10.1096/fj.08-125443. [DOI] [PubMed] [Google Scholar]
- 52.Hamdane M., Smet C., Sambo A.V., Leroy A., Wieruszeski J.M., Delobel P., Maurage C.A., Ghestem A., Wintjens R., Bégard S., et al. Pin1: A therapeutic target in Alzheimer neurodegeneration. J. Mol. Neurosci. 2002;19:275–287. doi: 10.1385/JMN:19:3:275. [DOI] [PubMed] [Google Scholar]
- 53.Ryu S.J., Park S.C. Targeting major vault protein in senescence-associated apoptosis resistance. Expert Opin. Ther. Targets. 2009;13:479–484. doi: 10.1517/14728220902832705. [DOI] [PubMed] [Google Scholar]
- 54.Guven A., Tolun A. TBC1D24 truncating mutation resulting in severe neurodegeneration. J. Med. Genet. 2013;50:199–202. doi: 10.1136/jmedgenet-2012-101313. [DOI] [PubMed] [Google Scholar]
- 55.Alkallas R., Fish L., Goodarzi H., Najafabadi H.S. Inference of RNA decay rate from transcriptional profiling highlights the regulatory programs of Alzheimer’s disease. Nat. Commun. 2017;8:909. doi: 10.1038/s41467-017-00867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kepa A., Medina L.M., Erk S., Srivastava D.P., Fernandes A., Toro R., Lévi S., Ruggeri B., Fernandes C., Degenhardt F., et al. Associations of the intellectual disability gene MYT1L with helixloop-helix gene expression, hippocampus volume and hippocampus activation during memory retrieval. Neuropsychopharmacology. 2017;42:2516–2526. doi: 10.1038/npp.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava D.P., Copits B.A., Xie Z., Huda R., Jones K.A., Mukherji S., Cahill M.E., VanLeeuwen J.-E., Woolfrey K.M., Rafalovich I., et al. Afadin is required for maintenance of dendritic structure and excitatory tone. J. Biol. Chem. 2012;287:35964–35974. doi: 10.1074/jbc.M112.363358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Y., Li J., Wang H., Wang G. NUCB2/nesfatin-1: Expression and functions in the regulation of emotion and stress. Prog. Neuropsychopharmacol. Biol. Psychiatr. 2018;81:221–227. doi: 10.1016/j.pnpbp.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Lee J.H., Budanov A.V., Park E.J., Birse R., Kim T.E., Perkins G.A., Ocorr K., Ellisman M.H., Bodmer R., Bier E., et al. Sestrin as a feedback inhibitor of tor that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henderson-Smith A., Jason J., Corneveaux J.J., De Both M., Cuyugan L., Liang W.S., Huentelman M., Adler C., Driver-Dunckley E., Beach T.G., et al. Next-generation profiling to identify the molecular etiology of Parkinson dementia. Neurol. Genet. 2016;2:e75. doi: 10.1212/NXG.0000000000000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neisch A.L., Neufeld T.P., Thomas S., Hays T.S. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 2017;216:441–461. doi: 10.1083/jcb.201606082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bishop P., Dan Rocca D., Henley J.M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): Structure, distribution and roles in brain function and dysfunction. Biochem. J. 2016;473:2453–2462. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ko H.S., Kim S.W., Sriram S.R., Dawson V.L., Dawson T.M. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J. Biol. Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- 64.Ginguay A., Cynober L., Curis E., Nicolis I. Ornithine aminotransferase, an important glutamate-metabolizing enzyme at the crossroads of multiple metabolic pathways. Biology. 2017;6:18. doi: 10.3390/biology6010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park M.H., Jo M., Kim Y.R., Lee C.K., Hong J.T. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol. Ther. 2016;163:1–23. doi: 10.1016/j.pharmthera.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marat A.M., Dokainish H., McPherson P.S. DENN domain proteins: Regulators of Rab GTPases. J. Biol. Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dallas S., Miller D.S., Bendayan R. A novel member of the multidrug resistance-associated protein (MRP) family, contributing to decreased drug accumulation. Pharmacol. Rev. 2006;58:140–161. doi: 10.1124/pr.58.2.3. [DOI] [PubMed] [Google Scholar]
- 68.Tao R.-R., Wang H., Hong L.-J., Huang J.-Y., Lu Y.-M., Liao M.-H., Ye W.-F., Lu N.-N., Zhu D.-Y., Huang Q., et al. Nitrosative stress induces peroxiredoxin 1 ubiquitination during ischemic insult via E6AP activation in endothelial cells both in vitro and in vivo. Antioxid. Redox Signal. 2014;21:1–16. doi: 10.1089/ars.2013.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Vliet A.R., Verfaillie T., Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochim. Biophys. Acta. 2014;1843:2253–2262. doi: 10.1016/j.bbamcr.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 70.Yan M.H., Wang X., Zhu X. Mitochondrial defects and oxidative stress in Alzheimer disease and Parkinson disease. Free Radic. Biol. Med. 2013;62:90–101. doi: 10.1016/j.freeradbiomed.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirayama K., Aoki S., Nishikawa K., Matsumoto T., Wada K. Identification of novel chemical inhibitors for ubiquitin C-terminal hydrolase-L3 by virtual screening. Bioorg. Med. Chem. 2007;15:6810–6818. doi: 10.1016/j.bmc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 72.Zhang P., Bi C., Schmitt S.M., Li X., Fan Y., Zhang N., Dou Q.P. Metal-based 2,3-indolinedione derivatives as proteasome inhibitors and inducers of apoptosis in human cancer cells. Int. J. Mol. Med. 2014;34:870–879. doi: 10.3892/ijmm.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaid S., Brandts C.H., Serve H., Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ji C.H., Kwon Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells. 2017;40:441–449. doi: 10.14348/molcells.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.