Abstract
CSF1R-expressing tumor-associated macrophages (TAMs) induce a tumor-promoting microenvironment by regulating immunity. Evidence demonstrates that the expression and single nucleotide polymorphisms of CSF1R relate with survival and risk of lung cancer in never smokers. However, no previous studies have examined the association of CSF1R expression in TAMs with mortality or whether the prognostic association differs according to smoking status in lung adenocarcinoma. Quantitative phosphor-integrated dot staining was used to precisely assess CSF1R expression in TAMs. Using 195 consecutive cases of lung adenocarcinoma, we examined the association of CSF1R expression with mortality and whether the prognostic association differs according to smoking status. We observed high expression levels of CSF1R in TAMs in 65 of 195 (33%) cases of lung adenocarcinoma. High expression levels of CSF1R were associated with high lung cancer-specific mortality (log-rank p = 0.037; hazard ratio (HR) = 1.61, 95% confidence interval (CI) = 1.02−2.52, p = 0.043). This prognostic association differed according to smoking status (p for interaction = 0.049, between never-smoking and ever-smoking patients). The association between high expression levels of CSF1R and lung cancer-specific mortality was stronger in never-smoking patients (log-rank p = 0.0027; HR = 2.90, 95% CI = 1.41−6.11, p = 0.0041) than in ever-smoking patients (log-rank p = 0.73; HR = 1.11, 95% CI = 0.59−2.00, p = 0.73). The findings suggest that CSF1R-expressing TAMs may exert stronger tumor-promoting immunity in never-smoking patients with lung adenocarcinoma and serve as a therapeutic target in precision immunotherapies.
Keywords: CSF1R, tumor-associated macrophage (TAM), phosphor-integrated dot (PID), tobacco, non-small-cell lung cancer (NSCLC), immunohistochemistry, neoantigens, tumor mutational burden, immune checkpoint inhibitor, CD163
1. Introduction
Lung adenocarcinomas exhibit molecular features that differ according to the smoking history of the patient. Tobacco smoking creates DNA damage and induces neoantigens, which are important targets for antitumor immunity after administration of immune checkpoint inhibitors. Lung adenocarcinoma in smokers is characterized by a tobacco-induced mutational burden and pro-inflammatory tumor microenvironment, which may explain its responsiveness to treatment with immunotherapies [1,2,3]. In contrast, lung adenocarcinoma in never-smokers harbors a low mutational burden and immunosuppressive tumor microenvironment, which inactivates antitumor immunity and may lead to immunotherapy refractoriness [1,2,3]. Emerging evidence has demonstrated that the tumor microenvironment is differentially regulated by specific immune modulators that differ according to the smoking history of the patient [4,5,6,7].
CSF1R is a receptor tyrosine kinase that mediates tumorigenesis in tumor-immune microenvironments and is expressed on tumor-associated macrophages (TAMs) [8,9,10]; the high expression levels of CSF1R correlate with poor survival in patients with various malignancies [8,9,10,11]. Recently, CSF1R-targeted therapies have emerged as a promising new class of immune-modulatory drugs [8,9,10,12,13,14]. Evidence demonstrates that the expression and single nucleotide polymorphisms (SNPs) of CSF1R relate with survival and risk of lung cancer in never-smokers [15,16,17]; however, no studies have examined the association of CSF1R expression in TAMs with mortality or the prognostic interaction between CSF1R expression and smoking in lung adenocarcinoma. Elucidation of the prognostic association would inform future researches examining the role of CSF1R and the potential interplay of CSF1R expression in TAMs and smoking status.
Therefore, we examined the association between high expression levels of CSF1R in TAMs and patient mortality and assessed whether the prognostic association differs according to smoking status, using 195 consecutive cases of lung adenocarcinoma. Quantitative phosphor-integrated dot (PID) staining was used to precisely assess the expression levels of the CSF1R protein in TAMs [18,19].
2. Materials and Methods
2.1. Patients
On the basis of the availability of patient data on CSF1R expression status in TAMs, smoking status and survival, we enrolled 195 consecutive Japanese patients with lung adenocarcinoma who had undergone surgical resection between April 1995 and January 2002 at The Cancer Institute Hospital, Japanese Foundation for Cancer Research (JFCR), Tokyo, Japan [20,21,22]. Patients were observed until death or December 2016, whichever came first. Smoking histories were obtained from rigorous interviews of each patient by experienced thoracic surgeons. Pathological diagnoses were made by experienced pulmonary pathologists (Kentaro Inamura and Yuichi Ishikawa) basically according to the 2015 WHO classification of lung tumors [23]. All patients were pathologically staged according to the AJCC-TNM staging system, 7th edition [24]. The study protocol was approved by the International Review Board of JFCR on 27 October 2017 (ethic code: 2017-1085), and informed consent was obtained from all patients.
2.2. Immunostaining
Sequential triple immunostaining was carried out on previously constructed tissue microarrays [20,21,22] for CD68 [diaminobenzidine (DAB) staining], CD163 (HistoGreen staining), and CSF1R (PID staining). Four-micron-thick sections were deparaffinized and immersed in antigen retrieval solution (10 mM Tris buffer, pH 9) for 45 min at 95 °C. Endogenous peroxidase activity was blocked by treatment with 3% hydrogen peroxide for 15 min, followed by blockade of nonspecific reactions by immersion in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) for 15 min. The sections were incubated with anti-CD68 mouse monoclonal antibody (1:100; clone: PG-M1, Dako, Glostrup, Denmark) for 60 min at 25 °C, followed by treatment with the Universal Immunoperoxidase Polymer (Nichirei Biosciences, Tokyo, Japan) for 30 min at 25 °C. The sections were then visualized with DAB (Wako, Osaka, Japan) for 3 min at room temperature. After a second round of antigen retrieval and blockade of nonspecific reactions, the sections were incubated with anti-CD163 mouse monoclonal antibody (1:50; clone: 10D6, Abcam, Cambridge, UK) overnight at 4 °C, followed by treatment with the Universal Immunoperoxidase Polymer for 30 min at 25 °C. The sections were visualized with the addition of HistoGreen (AbCys, Paris, France) for 3 min at room temperature, followed by washing in PBS and distilled water. After blocking nonspecific reactions, the sections were incubated with anti-CSF1R rabbit monoclonal antibody (1:50; clone: SP211, Abcam) overnight at 4 °C, followed by treatment with biotinylated anti-rabbit monoclonal antibody (clone: LO-RG-1, Bio-Rad, Hercules, CA, USA) for 30 min at 25 °C. The sections were then visualized with PID-conjugated streptavidin (0.09 nM) for 120 min at 25 °C. After washing in PBS, the sections were fixed with 4% paraformaldehyde and immersed in hematoxylin for counterstaining. SK-BR3 cells (ATCC, Manassas, VA, USA), which express CSF1R, and human lymph nodes, which express CD68 and CD163, were used as positive controls. Sections processed with replacement of primary anti-CSF1R antibody with PBS containing 1% BSA were used as negative controls. To assess the heterogeneity of CSF1R expression in TAMs, we immunohistochemically stained 10 cases of lung adenocarcinoma for CSF1R, CD68 and CD163; we did not observe substantial intratumoral or peritumoral heterogeneity in terms of CSF1R expression in TAMs.
2.3. Measurement of CSF1R Expression in TAMs
TAMs were defined as intratumoral or peritumoral cells that stained positive for both CD68 (cytoplasmic DAB staining) and CD163 (membranous HistoGreen staining). An entire image of each case was acquired using an Aperio image analysis system (Leica Biosystems, New Castle, UK). In each case, bright- and dark-field images were taken in at least five fields [19,25] (with 196 × 147 µm in size) using a fluorescence microscope (BX63, Olympus, Tokyo, Japan) connected to a DP80 CCD camera (Olympus). In each field, the number of TAMs was counted, and the number of PID particles per TAM was measured using a software for analyzing PID (PID analyzer, Konica Minolta, Tokyo, Japan), as described previously [18]. Five fields were randomly selected, and the numbers of TAMs and PID particles per TAM in the five fields were averaged and used to score each case. The upper tertiles of the average numbers of TAMs and PID particles per TAM were defined as a high number of TAMs and a high expression level of CSF1R in each case, respectively. The lower and middle tertiles were defined as a low–moderate number of TAMs and a low–moderate expression level of CSF1R in each case, respectively.
2.4. Detection of EGFR and KRAS Mutations and ALK Fusions
Tumor specimens were snap-frozen in liquid nitrogen within 20 min of surgical removal and stored at −80 °C until use. DNA was extracted by standard proteinase K digestion and phenol–chloroform extraction. For analysis of EGFR mutations, we examined four exons (exons 18–21) that encode the tyrosine kinase domain of the EGFR gene. For exons 18 (G719X), 20 (S768I and T790M), and 21 (L858R and L861Q), the TaqManTM SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) was performed, according to the manufacturer’s instructions. Fragment analysis was conducted for the exon 19 deletion and the exon 20 insertion, as described previously [20,21]. To analyze KRAS mutations, we directly sequenced codons 12, 13 and 61, as described previously [20,21]. To detect ALK fusions, we performed ALK immunohistochemistry using an anti-ALK mouse monoclonal antibody (1:50; clone: 5A4, Leica Biosystems Newcastle Ltd., Newcastle, UK) and the Leica Bond III automated system (Leica Biosystems Melbourne Pty Ltd., Melbourne, Australia). The sections were incubated at pH 9 for 30 min at 100 °C. All fusions in the ALK-positive cases were confirmed by fluorescence in situ hybridization, as described previously [26].
2.5. Statistical Analysis
All statistical analyses were conducted using JMP 12 software (SAS Institute Inc., Cary, NC, USA). All two-sided p values less than 0.05 were considered statistically significant. To investigate the association of CSF1R expression status in TAMs (low–moderate vs. high) with clinicopathological and molecular features, we performed Chi-square or Fisher’s exact test appropriately.
For survival analyses, we used the Kaplan–Meier method and log-rank test. Survival time was defined as the duration from the date of surgery to death or the end of follow-up. In lung cancer-specific survival analysis, deaths as a result of other causes were censored. Cox proportional hazards regression models were used to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) for mortality, according to CSF1R expression status (low–moderate vs. high). In addition to CSF1R expression status, the multivariable model included age at surgery, gender, smoking status, pathological stage, tumor differentiation grade, EGFR status, KRAS status, ALK rearrangement and number of TAMs. A backward stepwise elimination with p equal to 0.05 as the threshold was performed to select variables for the final models. P values for interactions between CSF1R expression status and smoking status were assessed using the Wald test on the cross-product of the CSF1R expression status (low–moderate vs. high) and smoking status variables (never- vs. ever-smoker) in the Cox model.
3. Results
3.1. CSF1R Expression in TAMs
Of the 195 cases of lung adenocarcinoma, we observed 65 cases (33%) in which CSF1R expression in TAMs was high using PID immunohistochemistry (Figure 1). Table 1 summarizes the clinicopathological and molecular characteristics of cases of lung adenocarcinoma, according to CSF1R expression status (low–moderate vs. high). High expression levels of CSF1R were associated with a less-differentiated grade of adenocarcinoma (p = 0.012).
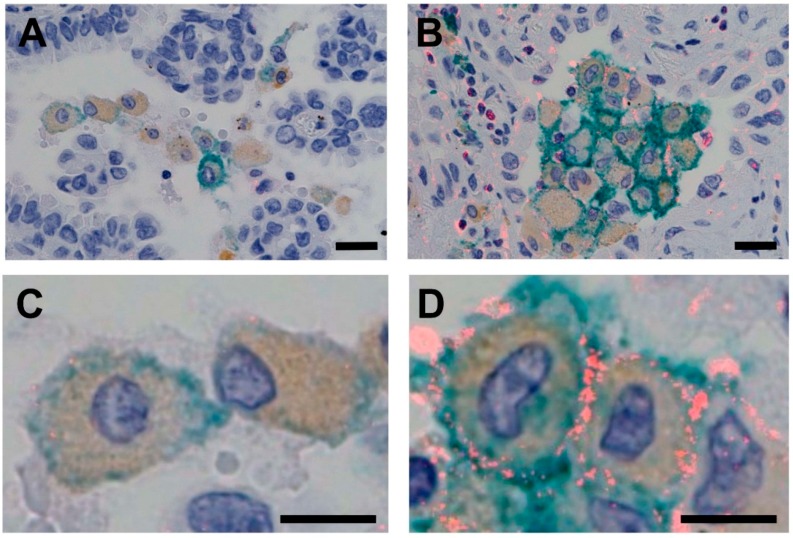
Figure 1.
Triple-stained images for PID, DAB and HistoGreen. CSF1R-expressing TAMs stained positive for CSF1R (red), CD68 (brown) and CD163 (green). (1) TAMs with low expression levels of CSF1R (A: low magnification, scale bar = 20 µm; C: high magnification, scale bar = 10 µm). (2) TAMs with high expression levels of CSF1R (B: low magnification, scale bar = 20 µm; D: high magnification, scale bar = 10 µm). DAB, diaminobenzidine; PID, phosphor-integrated dot; TAM, tumor-associated macrophage.
Table 1.
Clinicopathological and molecular characteristics of lung adenocarcinoma according to CSF1R expression status in tumor-associated macrophages.
| Variables |
N of Samples (%) |
CSF1R Expression | ||
|---|---|---|---|---|
| Low–Moderate 130 (67%) |
High 65 (33%) |
p Values | ||
| Age (years) | 0.40 | |||
| <60 | 73 (37%) | 46 (35%) | 27 (42%) | |
| ≥60 | 122 (63%) | 84 (65%) | 38 (58%) | |
| Gender | 0.92 | |||
| Male | 106 (54%) | 71 (55%) | 35 (54%) | |
| Female | 89 (47%) | 59 (45%) | 30 (46%) | |
| Smoking status | 0.54 | |||
| Never smoker | 84 (43%) | 54 (42%) | 30 (46%) | |
| Ever smoker | 111 (57%) | 76 (58%) | 35 (54%) | |
| Pathological stage | 0.68 | |||
| I | 112 (57%) | 76 (58%) | 36 (55%) | |
| II–IV | 83 (43%) | 54 (42%) | 29 (45%) | |
| Tumor differentiation | 0.012 | |||
| Well | 84 (43%) | 64 (50%) | 20 (31%) | |
| Moderate–poor | 110 (57%) | 65 (50%) | 45 (69%) | |
| EGFR status | 0.11 | |||
| Wild-type | 71 (54%) | 54 (59%) | 17 (44%) | |
| Mutant | 60 (46%) | 38 (41%) | 22 (56%) | |
| KRAS status | 0.22 | |||
| Wild-type | 114 (87%) | 78 (85%) | 36 (92%) | |
| Mutant | 17 (13%) | 14 (15%) | 3 (7.7%) | |
| ALK rearrangement | 0.43 | |||
| Negative | 187 (96%) | 124 (95%) | 64 (98%) | |
| Positive | 7 (3.6%) | 6 (4.6%) | 1 (1.5%) | |
| Number of TAMs | 0.83 | |||
| Low–moderate | 131 (67%) | 88 (68%) | 43 (66%) | |
| High | 64 (33%) | 42 (32%) | 22 (34%) | |
TAM, tumor-associated macrophage.
3.2. Association of CSF1R Expression in TAMs with Survival in Patients with Lung Adenocarcinoma
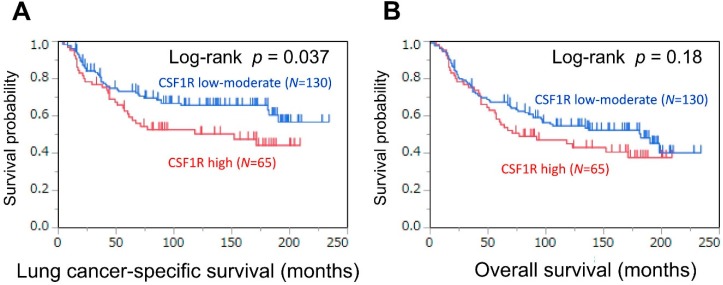
There were 101 deaths, including 77 lung cancer-specific deaths, during a median follow-up period of 134 months (interquartile range: 37–168 months) of 195 patients with lung adenocarcinoma. The 5 years lung cancer-specific survival and overall survival rates were 69% and 64%, respectively. We assessed the association between CSF1R expression in TAMs and survival (Figure 2 and Table 2). In a Kaplan–Meier analysis, high expression levels of CSF1R were associated with higher lung cancer-specific mortality (5 years survival: 61 months) than low–moderate expression levels of CSF1R (5 years survival: 73 months; log-rank p = 0.037) (Figure 2A). In a Cox regression analysis, high expression levels of CSF1R were associated with high lung cancer-specific mortality in both univariable (HR = 1.61, 95% CI = 1.02−2.52; p = 0.043) and multivariable analyses (HR = 1.32, 95% CI = 1.00−2.49, p = 0.048) (Table 2)
Figure 2.
Kaplan–Meier curves for lung cancer-specific (A) and overall (B) survival, according to CSF1R expression status of tumor-associated macrophages (low–moderate vs. high), in lung adenocarcinoma patients.
Table 2.
CSF1R expression in tumor-associated macrophages and patient mortality * in lung adenocarcinoma, stratified by smoking status.
| Patients and CSF1R Expression Status | Lung Cancer-Specific Mortality | Overall Mortality | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariable Analysis | Multivariable Analysis ** | Univariable Analysis | Multivariable Analysis ** | |||||
| HR (95% CI) |
p Values | HR (95% CI) |
p Values | HR (95% CI) |
p Values | HR (95% CI) |
p Values | |
| All patients | ||||||||
| CSF1R: low–moderate expression (N = 130) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 65) |
1.61 (1.02–2.52) |
0.043 | 1.32 (1.00–2.49) |
0.048 | 1.31 (0.87–1.95) |
0.19 | 1.09 (0.72–1.65) |
0.68 |
| Never-smoking patients | ||||||||
| CSF1R: low–moderate expression (N = 54) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 30) |
2.90 (1.41–6.11) |
0.0041 | 2.66 (1.28–5.66) |
0.0088 | 2.63 (1.37–5.09) |
0.0038 | 2.21 (1.05–4.77) |
0.037 |
| Ever-smoking patients | ||||||||
| CSF1R: low–moderate expression (N = 76) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 35) |
1.11 (0.59–2.00) |
0.73 | 1.21 (0.65–2.19) |
0.54 | 0.84 (0.48–1.43) |
0.53 | 0.84 (0.47–1.43) |
0.52 |
| p values for interaction *** | 0.049 | 0.12 | 0.0078 | 0.062 | ||||
* Cox proportional hazards regression models were used to calculate the HR and 95% CI. ** The multivariable model included age at surgery (<60 vs. ≥60 years), gender (male vs. female), smoking status (never- vs. ever-smoker), pathological stage (I vs. II−IV), tumor differentiation grade (well vs. moderate–poor), EGFR status (wild-type vs. mutant), KRAS status (wild-type vs. mutant), ALK rearrangement (negative vs. positive), and number of TAMs (low–moderate vs. high). A backward stepwise elimination with p equal to 0.05 as the threshold was performed to select variables for the final models. *** p values for interactions between CSF1R expression status and smoking status were assessed using the Wald test on the cross-product of the CSF1R expression status (low–moderate vs. high) and smoking status variables (never- vs. ever-smoker) in the Cox model. CI, confidence interval; HR, hazard ratio; TAM, tumor-associated macrophage.
3.3. Association of CSF1R Expression in TAMs with Survival, Stratified by Smoking Status
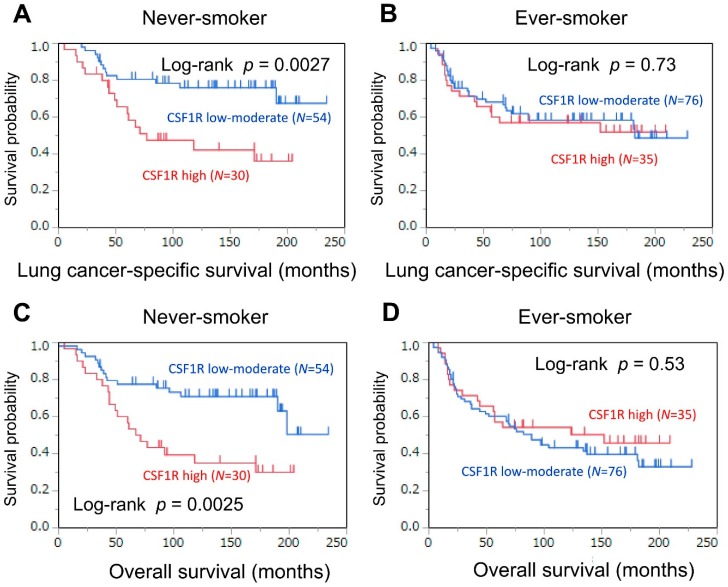
We examined whether the association of CSF1R expression in TAMs with mortality differed according to smoking status (Table 2 and Figure 3). High expression levels of CSF1R were associated with high lung cancer-specific mortality (log-rank p = 0.0027; Figure 3A) in never-smoking patients; however, high expression levels of CSF1R were not associated with lung cancer-specific mortality (log-rank p = 0.73; Figure 3B) in ever-smoking patients. In a Cox regression analysis (Table 2), high expression levels of CSF1R were associated with high lung cancer-specific mortality in both univariable (HR = 2.90, 95% CI = 1.41−6.11, p = 0.0041) and multivariable analyses (HR = 2.66, 95% CI = 1.28−5.66, p = 0.0088) in never-smoking patients; however, such associations were not observed in univariable (p = 0.73) or multivariable analyses (p = 0.54) in ever-smoking patients. In a multivariable Cox model, the P value for a prognostic interaction between CSF1R expression (low–moderate vs. high) and smoking status (never- vs. ever-smoker) was not significant (P for interaction = 0.12) after adjusting for the pathological stage of the adenocarcinoma. However, in a univariable Cox model, there was a significant prognostic interaction (p for interaction = 0.049) between CSF1R expression and smoking status (Table 2).
Figure 3.
Kaplan–Meier curves for lung cancer-specific (A) and overall (C) survival, according to CSF1R expression status in tumor-associated macrophages (low–moderate vs. high), in never-smoking patients. Kaplan–Meier curves for lung cancer-specific (B) and overall (D) survival, according to CSF1R expression status in tumor-associated macrophages (low–moderate vs. high), in ever-smoking patients.
3.4. Association of CSF1R Expression in TAMs with Survival in Female Patients, Stratified by Smoking Status
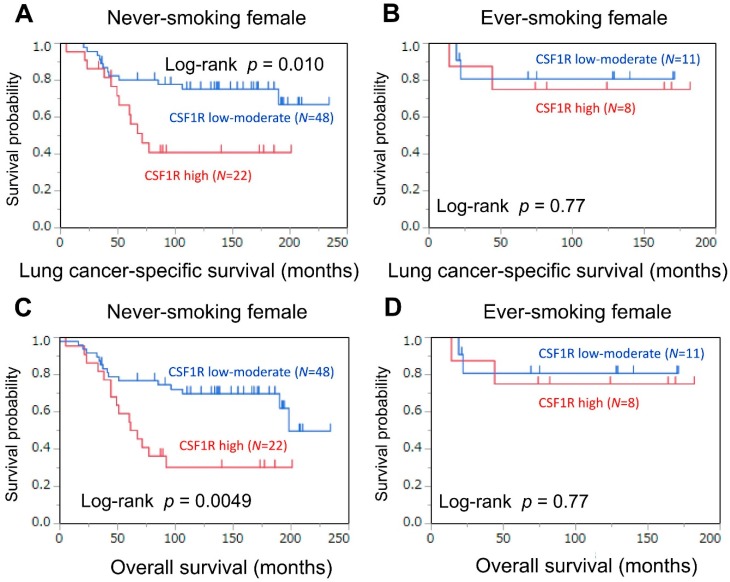
As an exploratory analysis, we assessed the association of CSF1R expression with mortality in female patients, stratified by smoking status (Table 3 and Figure 4), because CSF1R SNPs have been associated with both risk of lung cancer and survival in never-smoking females [15,16]. In never-smoking female patients, high expression levels of CSF1R were associated with high lung cancer-specific mortality (log-rank p = 0.010; univariable HR = 2.78, 95% CI = 1.22−6.32, p = 0.015). In contrast, such a prognostic association was not observed in ever-smoking female patients (log-rank p = 0.77). The p values for prognostic interactions between CSF1R expression and smoking status were not significant (p values for interaction >0.29), although the statistical power was limited in this subgroup analysis.
Table 3.
CSF1R expression in tumor-associated macrophages and female patient mortality * in lung adenocarcinoma, stratified by smoking status.
| Patients and CSF1R Expression Status | Lung Cancer-Specific Mortality | Overall Mortality | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariable Analysis | Multivariable Analysis ** | Univariable Analysis | Multivariable Analysis ** | |||||
| HR (95% CI) |
p Values | HR (95% CI) |
p Values | HR (95% CI) |
p Values | HR (95% CI) |
p Values | |
| Female patients | ||||||||
| CSF1R: low–moderate expression (N = 59) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 30) |
2.28 (1.07–4.88) |
0.033 | 4.16 (1.62–11.0) |
0.0034 | 2.23 (1.13–4.39) |
0.021 | 2.37 (1.19–4.73) |
0.015 |
| Never-smoking female patients | ||||||||
| CSF1R: low–moderate expression (N = 48) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 22) |
2.78 (1.22–6.32) |
0.015 | 3.31 (1.42–7.80) |
0.0060 | 2.69 (1.30–5.53) |
0.0081 | 3.18 (1.50–6.76) |
0.0028 |
| Ever-smoking female patients | ||||||||
| CSF1R: low–moderate expression (N = 11) |
1 (referent) |
1 (referent) |
1 (referent) |
1 (referent) |
||||
| CSF1R: high expression (N = 8) |
1.33 (0.16–11.1) |
0.77 | 0.92 (0.11–7.86) |
0.93 | 1.33 (0.16–11.1) |
0.77 | 0.92 (0.11–7.86) |
0.93 |
| p values for interaction *** | 0.49 | 0.29 | 0.51 | 0.31 | ||||
* Cox proportional hazards regression models were used to calculate the HR and 95% CI. ** The multivariable model included age at surgery (<60 vs. ≥60 years), gender (male vs. female), smoking status (never- vs. ever-smoker), pathological stage (I vs. II−IV), tumor differentiation grade (well vs. moderate–poor), EGFR status (wild-type vs. mutant), KRAS status (wild-type vs. mutant), ALK rearrangement (negative vs. positive), and number of TAMs (low–moderate vs. high). A backward stepwise elimination with P equal to 0.05 as the threshold was performed to select variables for the final models. *** p values for interactions between CSF1R expression status and smoking status were assessed using the Wald test on the cross-product of the CSF1R expression status (low–moderate vs. high) and smoking status variables (never- vs. ever-smoker) in the Cox model. CI, confidence interval; HR, hazard ratio; TAM, tumor-associated macrophage.
Figure 4.
Kaplan–Meier curves for lung cancer-specific (A) and overall (C) survival, according to CSF1R expression status in tumor-associated macrophages (low–moderate vs. high), in never-smoking female patients. Kaplan–Meier curves for lung cancer-specific (B) and overall (D) survival, according to CSF1R expression status in tumor-associated macrophages (low–moderate vs. high), in ever-smoking female patients.
4. Discussion
CSF1R-expressing TAMs induce a tumor-promoting microenvironment by regulating immunity. Evidence demonstrates that the expression and SNPs of CSF1R are associated with survival and risk of lung cancer in never smokers [15,16,17]; however, the association of CSF1R expression in TAMs with mortality and the prognostic interaction between CSF1R expression and smoking status have not been previously examined. Therefore, we conducted this study to examine the association of CSF1R expression in TAMs with mortality and determine whether this association differs according to smoking status in cases of lung adenocarcinoma using PID immunostaining. We found that high expression levels of CSF1R were associated with higher mortality in never-smoking patients compared with ever-smoking patients. Our results provide evidence for a potential interaction between CSF1R expression in TAMs and smoking status in the progression of lung adenocarcinoma. Our findings, if validated, would inform future researches examining the interplay of CSF1R expression in TAMs and smoking status.
Lung adenocarcinoma represents a group of clinicopathologically and molecularly heterogeneous diseases [23,27,28,29,30,31,32,33,34,35,36,37,38]. A history of smoking substantially affects the molecular features of these tumors. Lung adenocarcinomas in smokers, which are relatively susceptible to immune checkpoint inhibitors, exhibit a smoking-specific mutational signature, high mutational load, and pro-inflammatory tumor microenvironment [1,2,3]. In contrast, lung adenocarcinomas in never-smokers, which are often refractory to these immunotherapies, harbor less genomic complexity, lower mutational load, and immunosuppressive tumor microenvironment [1,2,3]. Emerging evidence has demonstrated that substantial differences exist in the lung adenocarcinoma microenvironment between smokers and non-smokers [4,5,6,7]. The tumor microenvironment appears to be differentially orchestrated by specific immune modulators in the context of smoking status [4,5,6,7].
CSF1R-expressing TAMs promote self-maintenance functions and tumorigenic processes, such as escape from immune surveillance [10,12]. Observational studies have demonstrated an association between high expression levels of CSF1R and poor survival in patients with various malignancies [8,9,10,11]. As a receptor tyrosine kinase, CSF1R is an attractive therapeutic target, considering the tumor-permissive and immunosuppressive characteristics of CSF1R-expressing TAMs. A variety of small molecules and monoclonal antibodies targeting CSF1R are in clinical development as monotherapies and combination therapies with chemotherapies or other immunotherapies. Given patient tolerance of CSF1R-targeted therapies, CSF1R inhibitors have emerged as a promising new class of immune-modulatory drugs [8,9,10,12,13,14]. In the current study, high expression levels of CSF1R were associated with high mortality in patients with lung adenocarcinoma; this prognostic association was stronger in never-smoking patients than in ever-smoking patients. These findings suggest that CSF1R-expressing TAMs may induce a tumor-promoting microenvironment, especially in never-smoking patients with lung adenocarcinoma. Never-smoking patients may thus be potential candidates for CSF1R-targeted therapies.
TAMs exhibit both anti-tumor and tumor-promoting functions, depending on their acquired immunophenotype (M1 or M2) [39,40,41,42]. M2-TAMs, which induce a tumor-promoting microenvironment, are characterized by co-expression of CD68 and CD163. High expression levels of CD163 in macrophages relate with high mortality in malignancies, including lung adenocarcinoma [39,40,41,42]. Furthermore, high expression levels of CD163 have been associated with high expression levels of CSF1, which is one of the ligands of CSF1R [41]. The association between expression levels of CD163 and CSF1R is intriguing, and thus, needs to be investigated.
Accumulating evidence indicates that tumor molecular alterations are associated with infiltration of specific immune-cell subtypes in tumor microenvironment [43,44,45]. This evidence supports the hypothesis that tumor cells may orchestrate their immune microenvironment [43]. In lung adenocarcinoma, EGFR wild-type tumors have been characterized by higher density of neutrophils and macrophages [43]. In the current study, expression levels of CSF1R in TAMs were not associated with driver genetic alterations. Further research is required to confirm our results.
The quantitative detection of PID nanoparticles allowed us to precisely measure the expression levels of CSF1R protein in TAMs. Although conventional immunohistochemistry with DAB is widely used, it has several limitations. The intensity of DAB staining depends on enzymatic activity and is substantially influenced by incubation time/temperature and signal enhancement [46]. Because of these factors, the sensitivity of immunohistochemistry with DAB is low [18]. Furthermore, co-localized proteins cannot be distinguished by the chromogenic method. In contrast, the PID method enables us to distinguish the PID-stained protein from the co-localized protein with chromogenic staining. Fluorescent immunohistochemistry has a relatively high quantitative sensitivity and produces a high signal-to-noise image under dark-field illumination; however, it exhibits poor photostability and is vulnerable to interference from tissue autofluorescence [18]. In contrast, PID staining, which was used in our study, produces an image with a high signal-to-noise ratio, even in the presence of tissue autofluorescence, owing to its brightness and photostability. Furthermore, the method used to process this type of image enables an automated calculation of the number of PIDs on the image, which corresponds to the expression levels of the target protein. Previous evidence has suggested that this method may dramatically improve the diagnostic capability of various targeted drug therapies [18,19]. The advantages of this novel technique have enhanced the credibility of our findings.
Limitations exist in this study. Its observational nature precludes the determination of a causal association between high expression levels of CSF1R in TAMs and mortality in never-smokers’ lung adenocarcinoma. The lack of a standardized evaluation method for CSF1R expression is another drawback. Nonetheless, we employed quantitative PID staining to precisely assess CSF1R expression in TAMs to minimize biases resulting from the subjective evaluation and susceptible intensities observed in DAB and fluorescent immunohistochemical staining [18,46]. Finally, this study may not be generalizable globally, because only Japanese patients at a single cancer hospital were enrolled. Therefore, our results must be validated in independent datasets.
5. Conclusion
In conclusion, the current study demonstrates that high expression levels of CSF1R in TAMs are associated with high mortality and that this prognostic association is stronger in never-smoking patients with lung adenocarcinoma than in ever-smoking patients. CSF1R-expressing TAMs may exert stronger tumor-promoting immunity in never-smoking patients than in ever-smoking patients; therefore, never-smokers with lung adenocarcinoma may be particularly responsive to CSF1R-targeted therapies. Given the growing popularity of immunotherapies [47,48,49], our findings, if validated, suggest the promising application of CSF1R-expressing TAMs as a biomarker or therapeutic target for the treatment of lung adenocarcinoma.
Acknowledgments
The authors thank Motoyoshi Iwakoshi, Miyuki Kogure and Tomoyo Kakita for their technical assistance and Yuki Takano and Ms. Chikako Yoshida for their secretarial expertise. This study was financially supported by Konica Minolta, Inc.
Abbreviations
The following abbreviations are used in this manuscript:
| BSA | bovine serum albumin |
| CI | confidence interval |
| DAB | diaminobenzidine |
| HR | hazard ratio |
| JFCR | Japanese Foundation for Cancer Research |
| PBS | phosphate-buffered saline |
| PID | phosphor-integrated dot |
| SNP | single nucleotide polymorphism |
| TAM | tumor-associated macrophage |
Author Contributions
K.I. conceived and designed the study. K.I., M.K., H.N., Y.N., H.S., K.T., E.F., S.O. and Y.I. contributed to the acquisition of clinical and tumor tissue data. K.I. performed data analyses. K.I., Y.S., M.K., H.N., H.S., K.T., E.F., H.K. and Y.I. contributed to the interpretation of the findings. K.I. drafted the manuscript. All authors contributed revisions and read and approved the final draft.
Funding
This research was funded by JSPS KAKENHI Grant Number JP16K08679 (K.I.).
Conflicts of Interest
K.I. received a research grant from Konica Minolta, Inc. H.S., K.T. and E.F. are employees of Konica Minolta, Inc. Y.I. received research grants from Daiichi Sankyo Co., Ltd., Chugai Pharmaceutical Co. Ltd., and Sony Corp. and is a consultant for Fujirebio Inc. All other authors declare no conflict of interest.
References
- 1.Herbst R.S., Soria J.C., Kowanetz M., Fine G.D., Hamid O., Gordon M.S., Sosman J.A., McDermott D.F., Powderly J.D., Gettinger S.N., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desrichard A., Kuo F., Chowell D., Lee K.-W., Riaz N., Wong R.J., Chan T.A., Morris L.G.T. Tobacco Smoking-Associated Alterations in the Immune Microenvironment of Squamous Cell Carcinomas. J. Natl. Cancer Inst. 2018 doi: 10.1093/jnci/djy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinoshita T., Kudo-Saito C., Muramatsu R., Fujita T., Saito M., Nagumo H., Sakurai T., Noji S., Takahata E., Yaguchi T., et al. Determination of poor prognostic immune features of tumour microenvironment in non-smoking patients with lung adenocarcinoma. Eur. J. Cancer. 2017;86:15–27. doi: 10.1016/j.ejca.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita T., Muramatsu R., Fujita T., Nagumo H., Sakurai T., Noji S., Takahata E., Yaguchi T., Tsukamoto N., Kudo-Saito C., et al. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann. Oncol. 2016;27:2117–2123. doi: 10.1093/annonc/mdw319. [DOI] [PubMed] [Google Scholar]
- 7.Haratani K., Hayashi H., Tanaka T., Kaneda H., Togashi Y., Sakai K., Hayashi K., Tomida S., Chiba Y., Yonesaka K., et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann. Oncol. 2017;28:1532–1539. doi: 10.1093/annonc/mdx183. [DOI] [PubMed] [Google Scholar]
- 8.Cannarile M.A., Weisser M., Jacob W., Jegg A.M., Ries C.H., Ruttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer. 2017;5:53. doi: 10.1186/s40425-017-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hume D.A., MacDonald K.P. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood. 2012;119:1810–1820. doi: 10.1182/blood-2011-09-379214. [DOI] [PubMed] [Google Scholar]
- 11.Koh Y.W., Park C., Yoon D.H., Suh C., Huh J. CSF-1R expression in tumor-associated macrophages is associated with worse prognosis in classical Hodgkin lymphoma. Am. J. Clin. Pathol. 2014;141:573–583. doi: 10.1309/AJCPR92TDDFARISU. [DOI] [PubMed] [Google Scholar]
- 12.Ries C.H., Cannarile M.A., Hoves S., Benz J., Wartha K., Runza V., Rey-Giraud F., Pradel L.P., Feuerhake F., Klaman I., et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulos K.P., Gluck L., Martin L.P., Olszanski A.J., Tolcher A.W., Ngarmchamnanrith G., Rasmussen E., Amore B.M., Nagorsen D., Hill J.S., et al. First-in-Human Study of AMG 820, a Monoclonal Anti-Colony-Stimulating Factor 1 Receptor Antibody, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2017;23:5703–5710. doi: 10.1158/1078-0432.CCR-16-3261. [DOI] [PubMed] [Google Scholar]
- 14.Candido J.B., Morton J.P., Bailey P., Campbell A.D., Karim S.A., Jamieson T., Lapienyte L., Gopinathan A., Clark W., McGhee E.J., et al. CSF1R(+) Macrophages Sustain Pancreatic Tumor Growth through T Cell Suppression and Maintenance of Key Gene Programs That Define the Squamous Subtype. Cell Rep. 2018;23:1448–1460. doi: 10.1016/j.celrep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H.G., Lee S.Y., Jeon H.S., Choi Y.Y., Kim S., Lee W.K., Lee H.C., Choi J.E., Bae E.Y., Yoo S.S., et al. A functional polymorphism in CSF1R gene is a novel susceptibility marker for lung cancer among never-smoking females. J. Thorac. Oncol. 2014;9:1647–1655. doi: 10.1097/JTO.0000000000000310. [DOI] [PubMed] [Google Scholar]
- 16.Yoo S.S., Kang H.G., Choi J.E., Do S.K., Lee W.K., Choi S.H., Lee S.Y., Lee S.Y., Lee J., Cha S.I., et al. Effects of polymorphisms identified in genome-wide association studies of never-smoking females on the prognosis of non-small cell lung cancer. Cancer Genet. 2017;212–213:8–12. doi: 10.1016/j.cancergen.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Szymanowska-Narloch A., Jassem E., Skrzypski M., Muley T., Meister M., Dienemann H., Taron M., Rosell R., Rzepko R., Jarzab M., et al. Molecular profiles of non-small cell lung cancers in cigarette smoking and never-smoking patients. Adv. Med. Sci. 2013;58:196–206. doi: 10.2478/ams-2013-0025. [DOI] [PubMed] [Google Scholar]
- 18.Gonda K., Watanabe M., Tada H., Miyashita M., Takahashi-Aoyama Y., Kamei T., Ishida T., Usami S., Hirakawa H., Kakugawa Y., et al. Quantitative diagnostic imaging of cancer tissues by using phosphor-integrated dots with ultra-high brightness. Sci. Rep. 2017;7:7509. doi: 10.1038/s41598-017-06534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaki S., Yanagimoto H., Tsuta K., Ryota H., Kon M. PD-L1 expression in pancreatic ductal adenocarcinoma is a poor prognostic factor in patients with high CD8(+) tumor-infiltrating lymphocytes: Highly sensitive detection using phosphor-integrated dot staining. Int. J. Clin. Oncol. 2017;22:726–733. doi: 10.1007/s10147-017-1112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inamura K., Yokouchi Y., Kobayashi M., Ninomiya H., Sakakibara R., Nishio M., Okumura S., Ishikawa Y. Relationship of tumor PD-L1 (CD274) expression with lower mortality in lung high-grade neuroendocrine tumor. Cancer Med. 2017;6:2347–2356. doi: 10.1002/cam4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inamura K., Yokouchi Y., Kobayashi M., Ninomiya H., Sakakibara R., Subat S., Nagano H., Nomura K., Okumura S., Shibutani T., et al. Association of tumor TROP2 expression with prognosis varies among lung cancer subtypes. Oncotarget. 2017;8:28725–28735. doi: 10.18632/oncotarget.15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inamura K., Yokouchi Y., Kobayashi M., Sakakibara R., Ninomiya H., Subat S., Nagano H., Nomura K., Okumura S., Shibutani T., et al. Tumor B7-H3 (CD276) expression and smoking history in relation to lung adenocarcinoma prognosis. Lung Cancer. 2017;103:44–51. doi: 10.1016/j.lungcan.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Travis W.D., Brambilla E., Burke A.P., Marx A., Nicholson A.G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th ed. IARC Press; Lyon, France: 2015. [DOI] [PubMed] [Google Scholar]
- 24.Edge S.B., Byrd D.R., Compton C.C., Fritz A.G., Greene F.L., Trotti A. AJCC Cancer Staging Manual. 7th ed. Springer; New York, NY, USA: 2010. [Google Scholar]
- 25.Jung K.Y., Cho S.W., Kim Y.A., Kim D., Oh B.C., Park D.J., Park Y.J. Cancers with Higher Density of Tumor-Associated Macrophages Were Associated with Poor Survival Rates. J. Pathol. Transl. Med. 2015;49:318–324. doi: 10.4132/jptm.2015.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., Asaka R., Hamanaka W., Ninomiya H., Uehara H., et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 27.The Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z., Fillmore C.M., Hammerman P.S., Kim C.F., Wong K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varn F.S., Tafe L.J., Amos C.I., Cheng C. Computational immune profiling in lung adenocarcinoma reveals reproducible prognostic associations with implications for immunotherapy. Oncoimmunology. 2018;7:e1431084. doi: 10.1080/2162402X.2018.1431084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 31.Vargas A.J., Harris C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer. 2016;16:525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devarakonda S., Morgensztern D., Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:e342–e351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 33.Barlesi F., Mazieres J., Merlio J.P., Debieuvre D., Mosser J., Lena H., Ouafik L., Besse B., Rouquette I., Westeel V., et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 34.Campbell B.B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R., Davidson S., Edwards M., Elvin J.A., Hodel K.P., et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell. 2017;171:1042–1056. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamay T.N., Zamay G.S., Kolovskaya O.S., Zukov R.A., Petrova M.M., Gargaun A., Berezovski M.V., Kichkailo A.S. Current and Prospective Protein Biomarkers of Lung Cancer. Cancers (Basel) 2017;9:155. doi: 10.3390/cancers9110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legras A., Pecuchet N., Imbeaud S., Pallier K., Didelot A., Roussel H., Gibault L., Fabre E., Le Pimpec-Barthes F., Laurent-Puig P., et al. Epithelial-to-Mesenchymal Transition and MicroRNAs in Lung Cancer. Cancers (Basel) 2017;9:101. doi: 10.3390/cancers9080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofman V., Lassalle S., Bence C., Long-Mira E., Nahon-Esteve S., Heeke S., Lespinet-Fabre V., Butori C., Ilie M., Hofman P. Any Place for Immunohistochemistry within the Predictive Biomarkers of Treatment in Lung Cancer Patients? Cancers (Basel) 2018;10:70. doi: 10.3390/cancers10030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heusinkveld M., van der Burg S.H. Identification and manipulation of tumor associated macrophages in human cancers. J. Transl. Med. 2011;9:216. doi: 10.1186/1479-5876-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Condeelis J., Pollard J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Baghdadi M., Endo H., Takano A., Ishikawa K., Kameda Y., Wada H., Miyagi Y., Yokose T., Ito H., Nakayama H., et al. High co-expression of IL-34 and M-CSF correlates with tumor progression and poor survival in lung cancers. Sci. Rep. 2018;8:418. doi: 10.1038/s41598-017-18796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medrek C., Ponten F., Jirstrom K., Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mansuet-Lupo A., Alifano M., Pecuchet N., Biton J., Becht E., Goc J., Germain C., Ouakrim H., Regnard J.F., Cremer I., et al. Intratumoral Immune Cell Densities Are Associated with Lung Adenocarcinoma Gene Alterations. Am. J. Respir. Crit. Care Med. 2016;194:1403–1412. doi: 10.1164/rccm.201510-2031OC. [DOI] [PubMed] [Google Scholar]
- 44.Zdanov S., Mandapathil M., Abu Eid R., Adamson-Fadeyi S., Wilson W., Qian J., Carnie A., Tarasova N., Mkrtichyan M., Berzofsky J.A., et al. Mutant KRAS Conversion of Conventional T Cells into Regulatory T Cells. Cancer Immunol. Res. 2016;4:354–365. doi: 10.1158/2326-6066.CIR-15-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W.Q., Liu L., Xu H.X., Wu C.T., Xiang J.F., Xu J., Liu C., Long J., Ni Q.X., Yu X.J. Infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br. J. Surg. 2016;103:1189–1199. doi: 10.1002/bjs.10187. [DOI] [PubMed] [Google Scholar]
- 46.Thunnissen E., Allen T.C., Adam J., Aisner D.L., Beasley M.B., Borczuk A.C., Cagle P.T., Capelozzi V.L., Cooper W., Hariri L.P., et al. Immunohistochemistry of Pulmonary Biomarkers: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2018;142:408–419. doi: 10.5858/arpa.2017-0106-SA. [DOI] [PubMed] [Google Scholar]
- 47.Forde P.M., Chaft J.E., Smith K.N., Anagnostou V., Cottrell T.R., Hellmann M.D., Zahurak M., Yang S.C., Jones D.R., Broderick S., et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N. Engl. J. Med. 2018;378:1976–1986. doi: 10.1056/NEJMoa1716078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon S.R., Maute R.L., Dulken B.W., Hutter G., George B.M., McCracken M.N., Gupta R., Tsai J.M., Sinha R., Corey D., et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., Choksi S., Liu Z.G. Butylated hydroxyanisole blocks the occurrence of tumor associated macrophages in tobacco smoke carcinogen-induced lung tumorigenesis. Cancers (Basel) 2013;5:1643–1654. doi: 10.3390/cancers5041643. [DOI] [PMC free article] [PubMed] [Google Scholar]