Abstract
(1) Background: Aflatoxin contamination in food and grain poses serious problems both for economic development and public health protection, thus leading to a focus on an effective approach to control it; (2) Methods: Aflatoxin B1 (AFB1) degrading bacteria were isolated using a medium containing coumarin as the sole carbon source, and the biodegradation of AFB1 by the isolate was examined by high performance liquid chromatography, and liquid chromatography mass spectrometry; (3) Results: a bacterial strain exhibiting strong AFB1 degradation activity (91.5%) was isolated and identified as Bacillus velezensis DY3108. The AFB1 degrading activity was predominantly attributed to the cell-free supernatant of strain DY3108. Besides, it was heat-stable and resistant to proteinase K treatment but sensitive to sodium dodecyl sulfate treatment. The optimal temperature for the maximal degradation of AFB1 was 80 °C. Even more notable, the supernatant showed a high level of activity over a broad pH (4.0 to 11.0) and exhibited the highest degradation (94.70%) at pH 8.0. Cytotoxicity assays indicated that the degradation products displayed significantly (p < 0.05) lower cytotoxic effects than the parent AFB1; (4) Conclusions: B. velezensis DY3108 might be a promising candidate for exploitation in AFB1 detoxification and bioremediation in food and feed matrices.
Keywords: mycotoxin, aflatoxin B1, Bacillus velezensis, biodegradation, detoxification
1. Introduction
Aflatoxins are a group of toxic secondary metabolites primarily synthesized by filamentous fungal species including Aspergillus flavus and Aspergillus parasiticus in both field and storage conditions [1,2]. Aflatoxin B1, B2, G1 and G2 (AFB1, AFB2, AFG1 and AFG2) are four major aflatoxins, and AFB1 has been classified as a Group I naturally occurring carcinogen due to its hepatotoxic, carcinogenic, teratogenic, and immunosuppressive characteristics [3,4]. Furthermore, high aflatoxin B1 contamination in food can occur in the tropical region where fungal growth and proliferation are favored by high temperatures and humidity, and thus has attracted worldwide attention [5,6].
The prolonged contamination of agricultural and food products by aflatoxins has proposed an emergent demand to detoxify contaminated food and feed using different methods. Although several physical and chemical strategies have been proposed to degrade AFB1 [7,8,9], limitations such as not providing the desired efficacy, safety and nutrient retention along with cost requirements have made them less desirable [10,11]. However, as valuable alternatives to physicochemical methods, biological degradation of AFs are attracting substantial attention due to their additional benefits such as their minimal loss of product qualities, safety, efficiency, economic and eco-friendly nature [12,13].
There are two key directions in control aflatoxin contamination: preventing the growth of toxigenic A. flavus, namely prevention and if contamination occurs, then detoxify aflatoxin-contaminated commodities by removing the toxic compounds [14]. Over the past decades, several bacterial and fungal species have been exploited to manage aflatoxin contamination, including non-aflatoxigenic strains of A. flavus [15], saprophytic yeasts [16], Mycobacterium fluoranthenivorans [17], Rhodococcus erythropolis [18], Pseudomonas putida [2], Pontibacter sp. [5], Streptomyces sp. [19], and Rhodococcus pyridinivorans [20]. In addition, Bacillus spp. also appeared as valuable candidates for controlling filamentous fungal growth and inhibiting mycotoxin production [14]. Bacillus subtilis UTBSP1 can effectively restrict the growth of A. flavus growth and remove AFB1 in pistachio nut as previously described by Farzaneh et al. [21], while 67.2% AFB1 degradation by cell-free supernatant of Bacillus subtilis JSW-1 was also observed by Xia et al. [14]. Raksha et al. [22] reported that Bacillus licheniformis CFR1 can reduce AFB1 by 94.7% and eliminate the AFB1 induced mutagenicity, indicating that Bacillus spp. might be excellent candidates in the field of food safety. Despite these fungi and bacteria were able to degrade AFB 1 effectively, few strains have been applied commercially because the actual use of these microorganisms or their metabolites were affected by the long reaction times, narrow working temperatures, or relatively low degradative efficiency. Therefore, exploring microorganisms or their metabolites that degrade and detoxify AFB1 with excellent degradation efficacy, wide temperature ranges, or short degradation times would be highly beneficial [23].
In this study, we isolated a new Bacillus bacterium from the soil that exhibited high AFB1 degradation activity with broad pH tolerance and excellent thermostability. The optimal degradation conditions of AFB1 by the strain were determined, and the cytotoxic potential of the metabolites formed after degradation was also analyzed.
2. Results and Discussion
2.1. Isolation and Identification of AFB1-Degrading Bacteria
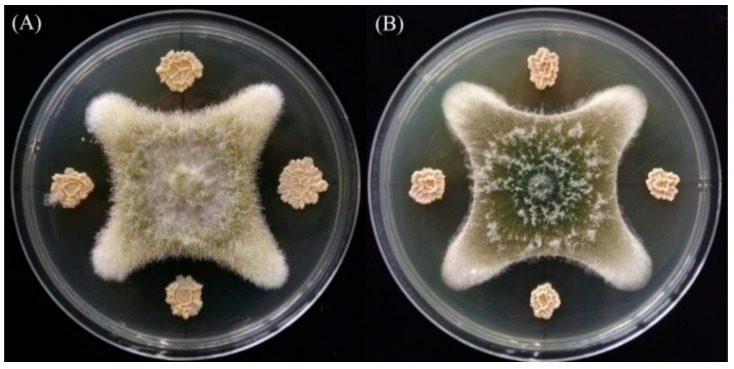
As the basic molecular structure of all aflatoxins, coumarin is considered to be a feasible, affordable, and effective tool to select AFB1-degrading microorganisms [24]. In this study, coumarin was used as the sole carbon source in the preliminary screen of bacteria with AFB1-degrading ability, and 13 bacteria were isolated from the soil samples. However, secondary screening results showed that strain DY3108 possessed the highest degradation rate of 91.5% after 96 h incubation at 30 °C. In addition, an in vivo antagonistic effect test showed that strain DY3108 could significantly reduce the mycelial growth of A. flavus and A. parasiticus (Figure 1). The greatest amount of degradation of AFB1 (66.43%) was recorded in the maize containing co-cultures of strain DY3108 and A. flavus strain 3.6305 (Table 1). Thus, this isolate was chosen for further study.
Figure 1.
In vitro inhibition of isolate DY3108 on Aspergillus flavus (A) and Aspergillus parasiticus (B) growth. Agar plug (5 mm diameter) from A. flavus or A. parasiticus was placed in the center of a potato dextrose agar (PDA) plate, and isolate DY3108 was cultured on the plate at four equidistance sites 3 cm apart from the center.
Table 1.
Aflatoxin B1 inhibition in maize containing co-cultures of DY3108 and Aspergillus flavus.
| Conc. of AFB1 (μg/g) | Aflatoxin B1 Inhibition | |
|---|---|---|
| Maize grains + NB | ND 1 | / |
| Maize grains + DY108 | ND 1 | / |
| Maize grains + A.flavus | 4.23 ± 0.23 a | / |
| Maize grains + A.flavus + DY108 | 1.42 ± 0.21 b | 66.43% |
1 ND: not detected.
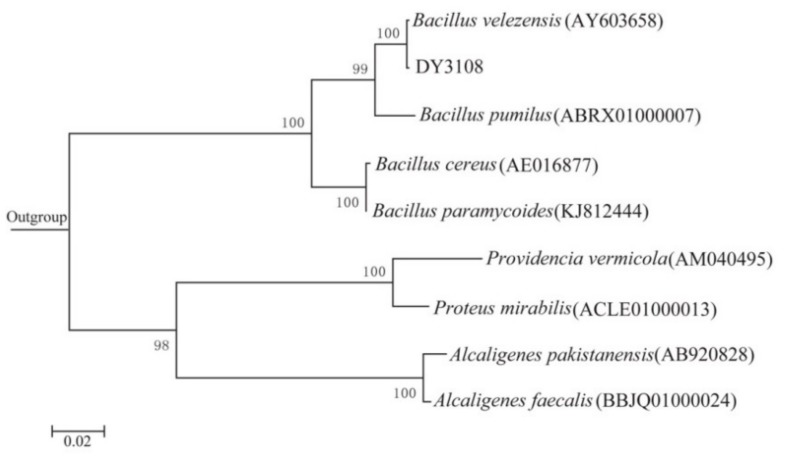
According to 16S rRNA gene sequence and phylogenetic evolution analysis, the closest relative of isolate DY3108 was Bacillus velezensis CR-502(T) with 99% similarity (Figure 2). In addition, strain DY3108 showed the typical characteristics of Bacillus sp. (Table S1). Therefore, the strain DY3108 identified as B. velezensis DY3108. B. velezensis CR-502(T) is a gram-positive, endospore-forming bacterium that can produce surfactant molecules [25]. In addition, several Bacillus species, such as Bacillus subtilis JSW-1 [14] and B. licheniformis CFR1 [22], have been reported to possess aflatoxins degrading capacity. However, this is the first study that showed that a B. velezensis strain could remove more than 91% of AFB1 from the liquid media.
Figure 2.
Phylogenetic tree built using Maximum likelihood method based on the 16S rRNA gene sequence of strain DY3108 and the sequences of representative strains from GenBank. The bar represents 0.02 substitutions per site. Sphingobacterium zeae (KU201960) was used as an outgroup.
2.2. AFB1 Degradation by B. Velezensis DY3108
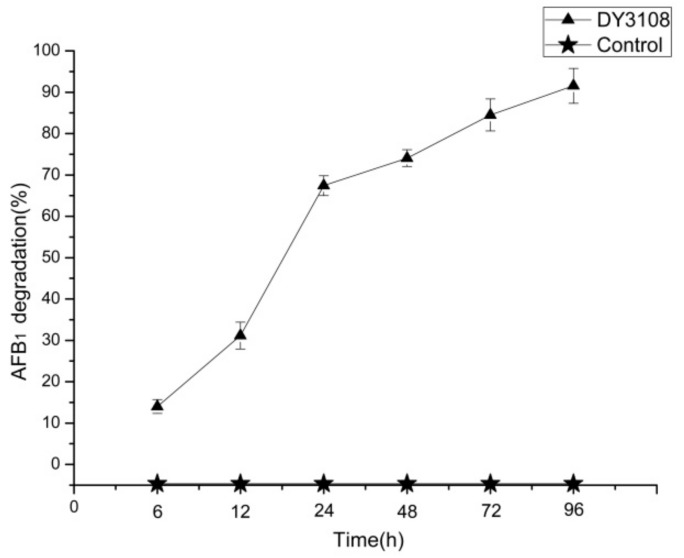
As observed by high-performance liquid chromatography (HPLC), B. velezensis DY3108 degraded AFB1 in a continuous process, and more than 90% of AFB1 was degraded in 96 h, while AFB1 concentration in the control broth was remaining stable from 0 to 96 h, with no significant difference (p < 0.05) observed (Figure 3).
Figure 3.
Kinetics of the Aflatoxin B1 (AFB1) degradation of by B. velezensis DY3108 over 96 h at 30 °C.
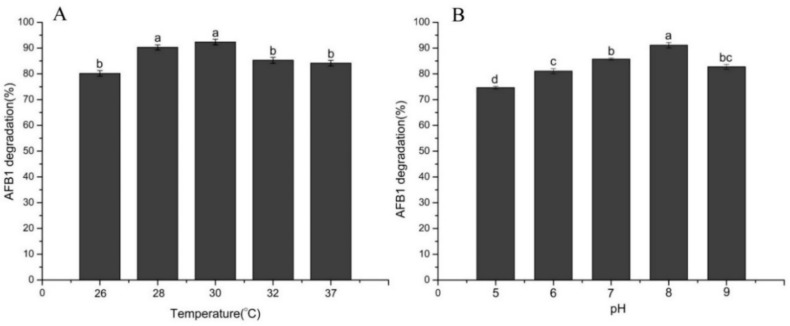
The effects of temperature and culture medium pH on the degradation of AFB1 are shown in Figure 4. In this study, AFB1 was degraded by B. velezensis DY3108 at all incubation temperatures (Figure 4A). This may occur because strain DY3108 could produce a range of extracellular enzymes or the compatibility of the enzymes to maintain catalytic activity over a wide range of temperatures. In addition, the degradation rate reached its maximum at 30 °C when the percentage of AFB1 degradation was 80.19%, 90.26%, 92.36%, 85.26% and 84.13% at 26 °C, 28 °C, 30 °C, 32 °C, and 37 °C, respectively. However, the degradation rate did not show a significant difference in the range of 28–30 °C (Figure 4A). This result agrees with that of Guan et al. [24], since they point out that the degradation rates of AFB1 demonstrated no statistically significant difference between 20 and 30 °C when Stenotrophomonas maltophilia 35-3 was used to detoxify AFB1. Maximum degradation of AFB1 was also observed at 30 °C and pH 6.0 for Rhodococcus erythropolis ATCC 4277 to degrade, while for Streptomyces strains, the optimal degradation conditions were 30 °C and pH 5.0 [26].
Figure 4.
Effect of the incubation temperature (A) and initial pH of the medium (B) on AFB1 degradation by B. velezensis DY3108 after 96 h incubation. Means with different letters are significantly different according to Tukey’s test (p < 0.05).
The effect of the initial pH of the culture on AFB1 degradation showed that the degradation efficiency of AFB1 was sensitive to initial pH of the medium. The degradation capability decreased in parallel with a decrease in pH (Figure 4B). B. velezensis DY3108 gained the highest AFB1 removal efficiencies (91.10%) at pH 8, while removal efficiency 74.64% was achieved at pH of 5. This was similar to the findings of Kong et al. [27], whose results showed that pH had a positive linear effect on the degradation of AFB1 by Rhodococcus erythropolis 4.1491, and increasing the pH properly is conducive to increasing degradation. Guan et al. [28] studied the initial pH of the medium on AFB1 biotransformation by Myxococcus fulvus ANSM068 and found that a pH value between 6.5 and 7.5 favored the reaction and that the optimal pH value was 7.5.
2.3. AFB1 Degradation by Culture Supernatant of B. Velezensis DY3108
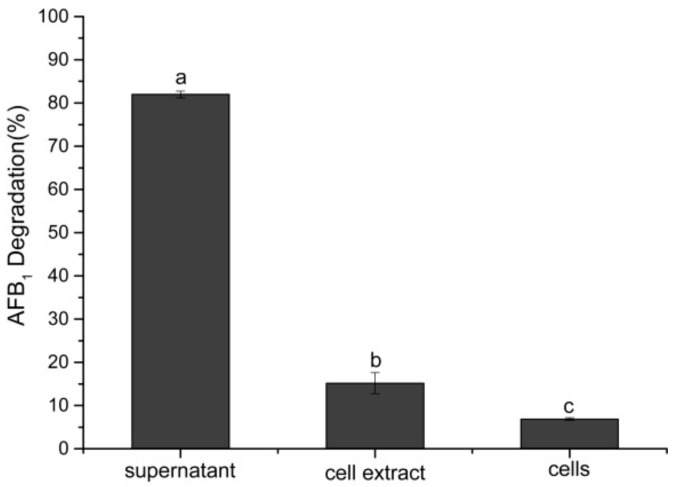
The culture supernatant of B. velezensis DY3108 appeared to be more effective in the degradation of AFB1 than viable cells and cell lysate (p < 0.05). After 72 h incubation, the culture supernatant could degrade 81.97% AFB1 compared to 6.87% and 15.17% by viable cells and cell lysate, respectively, indicating that B. velezensis DY3108 degrades AFB1 rather than absorbing it to the cell wall (Figure 5). Rao et al. [22] previously reported that approximately 94.73% of AFB1 was degraded when treated with the culture supernatant of B. licheniformis CFR1 for 72 h. Similarly, it was also found that the cell-free supernatant of Flavobacterium aurantiacum can degrade 74.5% of AFB1 after 24 h incubation [29], while AFB1 degradation primarily took place in the cell-free extracts of S. maltophilia 35-3 with 78.7% AFB1 degradation after 72 h incubation with culture supernatant [24]. According to Adebo et al. [5], the degradation of AFB1 by extracellular elements overcomes the disadvantages of applying a whole microorganism, which may impair the organoleptic and nutritional properties of the product.
Figure 5.
Biological degradation of AFB1 by the cell-free supernatant, cells, and cell extract of B. velezensis DY3108 at 30 °C after 72 h incubation. Values represent the mean ± SE (n = 3) and different letters indicate significant differences among them (p < 0.05).
2.4. Effect of Temperature, pH, Time, and Metal Ions on AFB1 Degradation
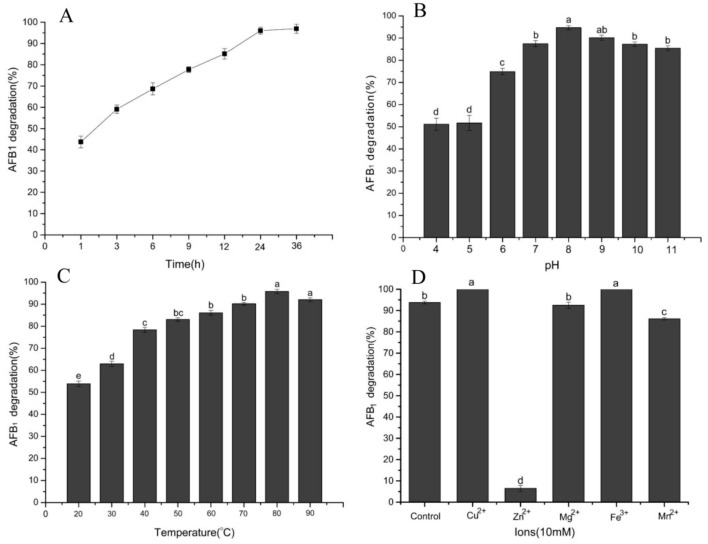
The influence of temperature, pH, time, and metal ions on the aflatoxin B1-degradation activity of cell-free extracts are shown in Figure 6. As shown in Figure 6A, it was a relatively rapid and sustained degradation process for the AFB1, since 68.66% of AFB1 was removed in the initial 6 h and 96.93% was removed after 48 h incubation. Rao et al. [22] indicated that the cell-free supernatant of B. licheniformis CFR1 could degrade 54% of AFB1 in the initial 12 h and remove 93.57% after incubation for 72 h. El-Deeb et al. [30] also indicated that the degradation of AFB1 by Bacillus sp. TUBF1 was primarily in the culture supernatant, and approximately 90% of AFB1 was removed within the first 12 h while it was reduced to undetectable levels after 24 h. The constant increase in degradation with time indicates that the removal of AFB1 by the cell-free supernatant of B. velezensis DY3108 is an enzymatically catalyzed reaction, and cell binding plays an insignificant role in AFB1 reduction.
Figure 6.
The influence of incubation conditions on the biodegradation of AFB1 after 24 h incubation with the cell-free supernatant of B. velezensis DY3108. (A) Influence of incubation time. (B) Influence of pH. (C) Influence of temperature. (D) Influence of metal ions. All of the degradation experiments were conducted at 80 °C except the temperature experiment. Values represent the mean ± SE (n = 3) and different letters indicate significant differences among them according to Tukey’s LSD test (p < 0.05).
The DY3108 culture supernatant worked well over a broad pH between 4.0 and 11.0 (Figure 6B). The amount of degradation increased with increasing of the pH. The highest degradation rate (94.70%) was obtained at pH 8.0, while the lowest removal efficiency was found at pH 4 (51.16%). The effect of pH on the degradation of AFB1 by the culture supernatant of S. maltophilia demonstrated a similar trend [24], since approximately 14% of AFB1 was degraded at pH 4, increased to 85% at pH 8 and decreased to 70% at pH 9. In addition, it is common knowledge that enzyme had maximal activity at their optimal pH, and the value beyond the optimal range may impair their activity. However, the DY3108 culture supernatant works well over a broad range of pH (pH 6–11, >70% AFB1 degradation), indicating that the enzyme involved in the degradation of AFB1 by the DY3108 is pH stable, which is advantageous for industrial applications in the area of food safety and quality.
Most of the strains that have been reported displayed AFB1 degradative ability in a narrow temperature range. Sangare et al. [12] found that Pseudomonas aeruginosa reduce >40% of AFB1 between 20–65 °C, while Zhang et al. [31] indicated that A. niger possessed approximately 25–45% degradation ratio at 20–50 °C. It was suggested that the cell free supernatant fluid of B. subtilis UTBSP1 could remove 60–80% AFB1 at 20–40 °C [32]. Interestingly, AFB1 degradation studies with different incubation temperatures showed that the DY3108 culture supernatant could significantly reduce AFB1 content over a broad temperature range between 20–90 °C and reached the maximal level of degradation at 80 °C (Figure 6C). Guan et al. [24] reported the maximum removal efficiency of AFB1 by S. maltophilia was at 37 °C, since the S. maltophilia strain was isolated from the feces of the warm-blooded organism Tapirus terrestris. Tan et al. [33] also reported that the optimal degradation of zearalenone by Pseudomonas otitidis TH-N1 occurred at 37 °C. However, Fusarium sp. WCQ3361 metabolites displayed excellent thermostability, since temperature change (0–90 °C) did not significantly affect the AFB1 degradation activity [23]. Similarly, the culture supernatant of DY3108 showed excellent thermostability, since the degradation rate increased with increasing temperature up to 80 °C. The reason may be that the active constituent of the culture supernatant could work well within a wide working temperature span or several active components which are presented in supernatant and could work under different temperatures. However, more importantly, the excellent thermostability indicates that DY3108 may be an excellent potential application for the degradation of AFB1.
Potential effects of adding metal ions to the cell-free extracts of DY3108 on the AFB1 degradative ability were studied (Figure 6D). It showed that Cu2+ and Fe3+ enhanced removal efficiency, and Mg2+ displayed no significant effect, while Zn2+ strongly inhibited the degradative ability. These results suggest that Cu2+ and Fe3+ may play a role as activators or membrane stabilizers of enzyme to maintain the structural integrity and function of these proteins. The activation effect attained with Cu2+ is agreed with the study agrees with the results of Sangare et al. [12] who found that 10 mM of Cu2+ increased AFB1 degradation level by 29.6% using Pseudomonas aeruginosa N17-1, while Rao et al. [22] also indicated that Cu2+ and Mg2+ stimulated the degradation of AFB1 by cell-free extracts of B. licheniformis CFR1. However, it has been reported in Flavobacterium aurantiacum NRRL B-184 [29] that Mn2+ and Zn2+ can significantly decrease aflatoxin B1 degradation because Zn2+ may cause conformational changes in enzyme active sites, which leads to decreases in the affinity of AFB1. In addition, the effect of various metal ions on the AFB1 degradation activity of DY3108 further confirmed the enzyme’s involvement in AFB1 degradation.
2.5. Heat Treatment, SDS, and Proteinase K on AFB1 Degradation
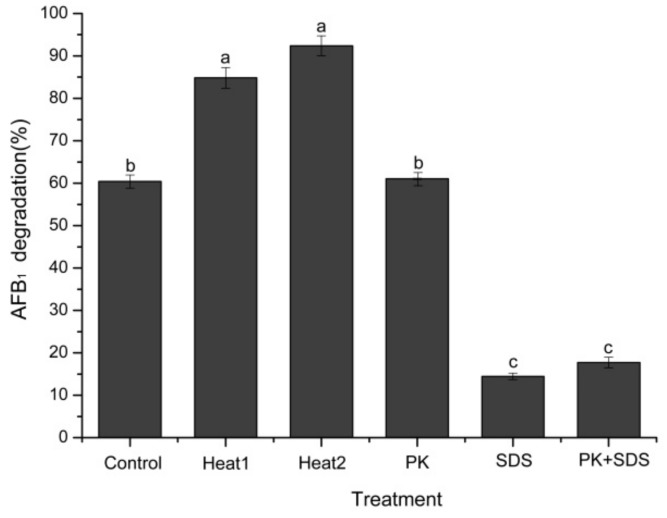
The AFB1 degradation ability of the culture supernatant significantly decreased (p < 0.05) after SDS and proteinase K plus SDS treatments, while the proteinase K treatment alone slightly affected the degradation activity (p > 0.05) (Figure 7). Similar results were also found in other aflatoxin B1 degrading bacteria, such as Mycobacterium fluoranthenivorans, P. aeruginosa, and R. erythropolis [11,13,24]. It showed that proteinase K-treated culture supernatant displayed low levels of AFB1 degradation activity, but when subjected to proteinase K plus SDS treatment, the AFB1 degradation activity was completely destroyed [18]. More importantly, when the supernatant was treated with heat (boiled or autoclaved for 30 min), the complete loss of AFB1 degradative activity was not detected, but a stimulated degradative ability was observed, which further proved that the enzymes involved in AFB1 degradation possessed excellent thermostability and high activity. The observed thermal activation of the enzymes was also found in other microorganisms. Sangare et al. [12] reported that the AFB1 degradation rate decreased significantly when the cell-free supernatant of P. aeruginosa N17-1 was treated with proteinase K plus SDS. However, when supernatant was subjected to heat treatment (boiling water bath for 10 min), AFB1 degradation activity was stimulated by thermal activation (increased by 3.49%). Kavitha et al. [34] isolated an antifungal protein with robust thermal stability from Bacillus polymyxa strain VLB16, which still maintained activity after sterilization (15 min at 121 °C). In addition, Wang et al. [23] found that the metabolites of Fusarium sp. WCQ3361 exhibited high thermal stability and 99.40% residual activity to degrade AFB1 was retained even after boiling for 10 min. Since mesophilic enzymes are often failed to endure the harsh reaction conditions required in industrial processes, it is extremely beneficial that thermostable enzymes may provide robust and efficient catalyst substitutes that can withstand the harsh reaction conditions required in industrial processes [35]. Therefore, the excellent thermostability of the enzyme’s involvement in AFB1 degradation by DY3108 offers a potentially valuable solution to remove aflatoxin from human diet and animal feed.
Figure 7.
Degradation of AFB1 by untreated, heat-treated, proteinase K-treated, SDS-treated, SDS plus proteinase K-treated cell-free supernatant of B. velezensis DY3108. heat1: boiled for 30 min; heat2: autoclaved for 30 min. Different letters indicate significant differences among them according to Tukey’s LSD test (p < 0.05).
2.6. Cytotoxicity Study
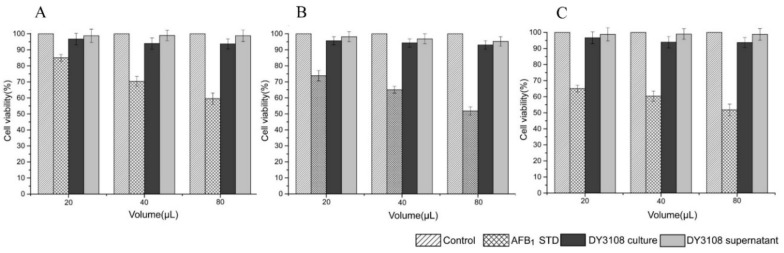
The cytotoxicity and mutagenicity of the degradation products of mycotoxins should not be neglected since certain degradable products may be toxic like their parent compounds [1]. Therefore, it is indispensable to clarify the toxicity of the AF biodegradation products by DY 3108 in comparison to the parent compound AFB1. In the present study, DMSO exposure alone at the test concentration did not significantly affect cell viability and the cytotoxicity of the biodegradation extract against human lymphocytes was evaluated via the MTT assay for 24, 48 and 72 h (Figure 8A–C). It is generally observed that the percentage of cell viability was related inversely to the AFB1 concentration. In addition, all of the bio-transformed extracts (culture and culture supernatant treatment) had more than 90% cell viability, comparable with those of the control sample. The corresponding diminution in the cytotoxic effect of AFB1 biodegradation extracts suggested that the AFB1-lactone ring had been cleaved and modified [36]. Similarly, high levels of cell viability of human lymphocytes were also reported by Adebo et al. [5] who found that human lymphocytes retained >90% viability when cells were exposed to AFB1 biodegradation extracts of Pontibacter sp. VGF1. Since the degradable or transformed products produced by DY3108 were less toxic than the parent compound AFB1, B. velezensis DY3108 has potential for use as a bio-detoxification agent for AFB1 in food commodities.
Figure 8.
Cell viability (percent of viable cells) of the DY3108 culture and the culture supernatant bio-transformed products (DY3108 supernatant) with different incubation times: (A) 24 h, (B) 48 h, and (C) 72 h.
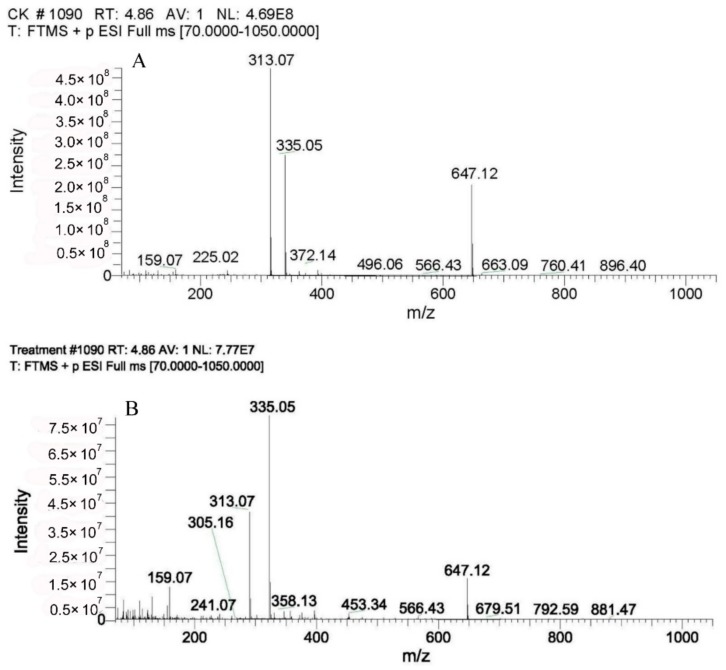
2.7. Analysis of AFB1 Degradation Products Using LC-MS
As shown in Figure 9A, molecular ions with m/z = 313, 335, and 647, which were specific for AFB1, presented in the full mass spectrum of AFB1 in nutrient broth (NB) medium (CK). From the MS analysis of the DY3108-treated samples it became evident that AFB1 was still present in the biodegradation extracts, but the incidence of its molecular peak declined sharply compared with that in the non-inoculated control (Figure 9B), which further affirmed the degradation of AFB1 by DY3108. However, in this study, it failed to identify any breakdown product of AFB1 by DY3108. Similar results have been reported by Farzaneh et al. [32], Rao et al. [22] and Sangare et al. [12]. It showed that Bacillus subtilis JSW-1 can degrade AFB1 effectively. However, no biodegradation products can be clearly identified by the liquid chromatography-mass spectrometry (LC-MS) analysis [14]. One reason for this is that AFB1 was most probably decomposed to certain components whose chemical characteristics were different from that of parent AFB1.
Figure 9.
Full mass spectra of AFB1: (A) AFB1 in nutrient broth (NB) culture medium (CK) (B) AFB1 in NB culture medium treated with B. velezensis DY3108.
3. Conclusions
In summary, a Bacillus strain that can degrade more than 90% AFB1 was isolated and identified as Bacillus velezensis DY3108. More importantly, to the best of our knowledge, this is the first study to demonstrate more than 90% degradation of AFB1 by a B. velezensis strain. It showed that the degradation capability was attributed to extracellular proteins or enzymes present in the culture supernatant. The degraded products differed chemically from AFB1 and showed significantly lowered cytotoxicity. In addition, these extracellular proteins or enzymes possess a broad reaction temperature range and pH tolerance, as well as excellent thermostability which will help them withstand the harsh conditions in food processing. Therefore, the enzymes or proteins in the supernatant are promising new agents for AF biodegradation from the human diet and animal feed. However, further research such as adequate purification and characterization of the enzymes or proteins in the supernatant are needed to exploit the probable use of the B. velezensis DY3108 in food and feed. Furthermore, it should be noted that the bacterial strains isolated from coumarin medium plates might have taken up coumarin from the original screen and that coumarin may have certain influence on aflatoxin degradation. Therefore, further research could focus on determination of coumarin content in bacterial strains (using HPLC, MS, etc.) to determine whether coumarin was taken up by bacterial strains and thus participated in degradation of aflatoxin. Meanwhile, more research is needed to identify coumarin-isolates interactions, and elucidate the mechanisms of coumarin uptake or degradation, to provide new insights into the aflatoxins bioremediation.
4. Materials and Methods
4.1. Chemicals and Medium
AFB1 standard reagent was purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and diluted into methanol (HPLC grade) to prepare AFB1 stock solution (at 10 ppm). All other analytical grade reagents and HPLC grade solvents were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Wild-type A. flavus strain 3.6305 and A. parasiticus strain 3.6155 were purchased from the China General Microbiological Culture Collection Center (CGMCC), and cultured on potato dextrose agar (PDA) medium at 28 °C.
4.2. Isolation of AFB1 Degrading Bacteria
4.2.1. Isolation of Microorganisms
Six soil samples were sampled from cropland of NongKang state farm, Bengbu, Anhui Province, China. The primary screening for microorganisms that able to remove or degrade AFB1 was carried out as described by Guan et al. [24] with small modifications.
Briefly, one gram of the soil sample was diluted in 9 mL sterile water and kept at room temperature under continuous shaking (200 rpm) for 6 h. And then, the samples were serially diluted (101–109) in sterilized distilled water. Aliquots (100 μL) of each dilution was spread on coumarin medium plates [24] and incubated at 30 °C for 4–5 days until visible colonies appeared on plates. The single colonies were picked from the plates and then streak on fresh plates. Repeat the above process 4–5 times and the pure isolates were finally preserved on LB agar and tested for AFB1-degradation activity.
4.2.2. Secondary Screening
AFB1-degradation activities of the chosen isolates were performed as follows: Briefly, each isolate was grown in nutrient broth (NB) (Oxoid Ltd., Hants, UK) at 30 °C for 20 h in a rotary shaker incubator (180 rpm). One hundred microliters AFB1 stock solution (10 ppm) was mixed with 1.9 mL culture broth to acquire the desired concentration (500 ppb), and then cultured in a rotary shaker incubator (180 rpm) at 30 °C for 4 days. AFB1 standard with sterile NB was used as—control. When the reaction is completed, the suspension was centrifuged at 8000× g for 20 min to recover the cell-free supernatant. For quantitation of residual AFB1, samples were extracted four times with chloroform, evaporated, and dissolved in 1 mL-methanol (HPLC grade), followed by filtering through 0.22 µm Nylon filter (Sangon Biotech Co., Ltd., Shanghai, China) and preserved at 4 °C until analysis.
4.3. Analysis of Residual AFB1
The determination of AFB1 in samples was conducted by HPLC using LC-8A (Shimadzu, Kyoto, Japan) with a Varian C18 column (5 μm, 250 mm × 4.6 mm) and AOAC method as previously described by Xia et al. [14] with slight modifications. A water/acetonitrile/ methanol mix (1.0:1.5:1.5, v/v/v) was used as mobile phase, and the flow rate was 1 mL/min. AFB1 concentration was quantitatively determined using a fluorescence detector, and the detection wavelengths for excitation and emission were 365 nm and 418 nm, respectively. The limit of detection (LOD) was 5 ng/mL AFB1, and the limit of quantitation (LOQ) was 16 ng/mL AFB1 under the experimental conditions used.
In both cases, the AFB1 removal efficiency was calculated by following formula:
| (1 − AFB1 peak area in treatment/AFB1 peak area in control) ×100 | (1) |
4.4. Identification of the Isolate “DY3108”
Standard physical and biochemical tests of the DY3108 isolate were conducted using standard methods [37]. For the 16S rRNA gene sequence analysis, genomic DNA of strain DY3108 was isolated using an EasyPure Bacteria Genomic DNA Kit (TransGen Biotech Co., Ltd., Beijing, China). A 1471-bp 16S rRNA gene fragment was subsequently amplified with the 27F and 1492R primers [38] and sequenced as described by Huang et al. [39]. The sequence was then aligned against those found in the NCBI database, and on the EzBioCloud server [40] using the Basic Local Alignment and Search Tool (BLAST) algorithm. These sequences were then utilized to construct a phylogenetic tree using MEGA 7 with the maximum likelihood (ML) method [41]. The sequence has been deposited to the GenBank database under the accession number MG839279.
4.5. In Vitro Anti-Aflatoxigenic Effect
Biocontrol activity assays of DY3108 against AFB1 produced by A. flavus were conducted in 250 mL flasks as described by Gong et al. [42]. Briefly, 20 g of maize grains was added into 250 mL flasks and autoclaved at 121 °C for 30 min. After cooling to room temperature, maize grains in each flask were inoculated with 1 mL of 5 × 105 CFU/mL suspension AF conidia. The samples were divided equally into two parts: half was challenged with 10 mL DY3108 culture (grown on NB medium with 108 CFU/mL), and the other half was treated with 10 mL NB medium as a control. After 7 days incubation at 28 °C, samples in each treatment were collected and dried at 90 °C for 5 days. Then the dried samples were ground to a fine powder and subjected to aflatoxin analyses.
4.6. Effects of Culture Conditions of AFB1-Degrading Bacteria on Biodegradation
The effects of different culture conditions on AFB1-degradation activity of the DY3108 were conducted as follows: incubation temperature (26, 28, 30, 32, and 37 °C), culture medium pH (5, 6, 7, 8, and 9), and incubation time (6, 12, 24, 48, 72, and 96 h). The AFB1 concentration in the culture medium was 500 ppb. The reactions were manipulated in a rotary shaker incubator (180 rpm), and residual AFB1 was detected using HPLC as described previously.
4.7. AFB1 Degradation by Cells, Culture Supernatant, and Cell Lysate
The biodegradation of AFB1 by cells, cell-free supernatant, and cell lysate were conducted as described by Rao et al. [22] with slight modifications. Briefly, isolate DY3108 was grown in NB medium for 20 h at 30 °C. The cells were then harvested by centrifugation for 20 min at 8000× g. The supernatant was immediately filtered through 0.22 μm filter for further experiments, while the pellet was washed three times with sterile MilliQ water and dissolved in Phosphate buffer (0.1 M, pH 7.0). Then, the cell suspension was divided into two fractions: in one the cells were suspended in Phosphate buffer without any treatment (namely whole bacterial cell), and in the other, the cells were disintegrated using an ultrasonicator (Ningbo Scientz Biotechnlogy Co., Ltd., Ningbo, China), and centrifuged at 10,000× g for 40 min at 4 °C. After centrifugation, the obtained supernatant was filtered through 0.22 μm filter (namely cell lysate) and used in subsequent AFB1 degradation experiments. For AFB1 degradation, AFB1 (final concentration 500 ppb) were treated with the culture supernatant, whole bacterial cell, and cell lysate respectively, and then incubated at 30 °C for 72 h. Non-incubated cultures (NB + PBS) with AFB1 was used as control, and conducted as described previously.
4.8. Optimization of Conditions for Maximum Degradation of AFB1
The culture supernatant of strain DY3-108 was prepared as described previously. AFB1 (500 ppb) was incubated with the supernatant at 1, 3, 6, 9, 12, 24, and 36 h, and the analysis was carried out. AFB1 treated with uninoculated NB medium served as the control. In order to investigate the influence of temperature on the degradative efficiency, the supernatants combined with AFB1 were incubated at 20, 30, 37, 50, 60, 70, 80, and 90 °C for 24 h. The pH influence on degradation was conducted by adjusting the culture supernatant to pH 4–5 with citrate buffer, pH 6–8 with phosphate buffer, pH 9 with sodium carbonate buffer, and pH 10–11 with sodium carbonate/sodium bicarbonate buffer. Blank of uninoculated NB medium with a corresponding pH used as the control. The effect of various metal ions on the degradation was obtained by adding 10 mM of Mg2+ (MgCl2), Cu2+ (CuSO4), Mn2+ (MnCl2), Zn2+ (ZnSO4), or Fe3+ (FeCl3) to the supernatant and incubated with AFB1, while in the control the supernatant was replaced by NB. All of the AFB1 degradation experiments were conducted at 80 °C except for the temperature experiment.
4.9. Heat, SDS, and Proteinase K Treatment on AFB1 Degradation
In order to study the influence of heat treatment on degradation efficiency, the supernatant fractions were boiled for 30 min or placed in an autoclave for 30 min. Meanwhile, the effect of proteinase K and sodium dodecyl sulfate (SDS) treatments on the degradation ability was investigated by exposing them to 1 mg/mL proteinase K and 1% SDS, respectively, at 30 °C in the dark for 24 h as described by [21]. Blank of uninoculated NB medium and untreated supernatant culture were used as control. The extraction and the analysis of residual AFB1 were conducted as described previously.
4.10. Cytotoxicity Studies
Lymphocyte cells were purchased from Shanghai JRDUN Biotechnology Co., Ltd. (Shanghai, China) and cultured according to the method reported [5]. After the completion of culture, final cell concentrations were determined, and cells showing 100% vitality were used in subsequent trials.
The MTT assay was performed as described by Samuel et al. [2] and Adebo et al. [13] with minor modifications. Briefly, for preparing the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay, cell suspension (180 μL) which was stimulated with 10 mg/mL phytohemagglutinin-p (PHA-p) was pipetted into 96-well plate. Then the cells were respectively dealt with different volumes (20, 40 and 80 μL) of AFB1 standard solution (2.5 ppm, redissolved in DMSO with culture media), extracts of AFB1 degradation products (previously evaporated to dryness and redissolved in DMSO with culture media) as well as the DY3108 culture. Exactly 160 μL of culture media was then added to each well and cultured at 37 °C in a 5% CO2 humidified incubator for 24, 48 and 72 h respectively. After incubation, 30 μL of MTT solution (5 mg/mL in 0.14 M PBS) was added into each well and continued to be cultured for 3 h under the same conditions. Cells were exposed to the corresponding concentration of DMSO alone to assess the cytotoxic effects of DMSO on cell viability. Untreated cells were used as a negative control.
Then, 50 μL of dimethylsulfoxide (DMSO) was pipetted into each well and an additional 2 h incubation was performed. After incubation, the absorbance was determined using a microplate reader that was set at a wavelength of 560 nm. Percentage of viable cell can be expressed as (AS/AC)/100, where AS and AC are the absorbance of the treated and untreated (control) samples.
4.11. Analysis of AFB1 Degradation Products by LC-MS
LC-MS/MS method was applied to analyze the AFB1 degradation products by using Thermo LXQ liquid chromatography coupled with ion trap mass spectrometry (Thermo Scientific, Waltham, MA, USA). Liquid chromatography was achieved using a Shim-pack VPODS C18 column (100 × 2.1 mm). The mobile phase used in this study comprised of 0.1% formic acid water and 0.1% formic acid acetonitrile, and the flow rate was 1 mL/min. MS was performed as follows for the positive ion mode: m/z range, 50–800; ion spray voltage 4.5 kV, sheath gas flow rate 36 L/min, auxiliary gas flow rate 10 L/min, and capillary temperature 300 °C.
4.12. Statistical Analysis
All analyses were conducted in triplicate, and the values represent the average of the measurements conducted from three independent assays and are expressed as the mean ± standard error of the mean (SEM). The data were further analyzed using an ANOVA at a 95% confidence level following Tukey’s test (SPSS 18.0, IBM, Somers, NY, USA).
Acknowledgments
This study was funded by the National Natural Science Foundation of China (31500531), the Science and Technology Service Program of the Chinese Academy of Sciences (KFJ-STS-ZDTP-002), the key program of 13th five-year plan, CASHIPS (No. kp-2017-21), the Grant of the President Foundation of the Hefei Institutes of Physical Science of the Chinese Academy of Sciences (YZJJ201619), the major special project of Anhui Province (16030701103) and the Natural Science Foundation of Anhui Province (1708085QC70).
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/10/8/330/s1, Table S1: Physiological and biochemical characteristic of the bacterial isolate DY3108.
Author Contributions
X.S.; S.H. and L.W. designed the experiments; X.S.; Q.Z.; Y.W. and M.L. performed the experiments; H.H.; Y.M.; X.C.; J.N. and W.Z. analyzed the data; X.S.; S.H. and L.W. wrote the paper. All authors have read and approved the final manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
Bacillus velezensis is firstly reported to degrade more than 90% of AFB1. The degradable products presented significantly lower cytotoxicity when compared with parent AFB1. Even more notable, proteins or enzymes in culture supernatant of DY3108 possess broad reaction temperature range (20–90 °C), strong pH tolerance (4–11), and excellent thermostability (bear sterilization), which provides a potential optimal solution to eliminate AFB1 contamination from the human diet and animal feed.
References
- 1.Adebo O.A., Njobeh P.B., Mavumengwana V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control. 2016;68:92–96. doi: 10.1016/j.foodcont.2016.03.021. [DOI] [Google Scholar]
- 2.Samuel M.S., Sivaramakrishna A., Mehta A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeter. Biodegr. 2014;86:202–209. doi: 10.1016/j.ibiod.2013.08.026. [DOI] [Google Scholar]
- 3.Gourama H., Bullerman L.B. Aspergillus flavus and Aspergillus parasiticus: Aflatoxigenic fungi of concern in foods and feeds: A review. J. Food Prot. 1995;58:1395–1404. doi: 10.4315/0362-028X-58.12.1395. [DOI] [PubMed] [Google Scholar]
- 4.Hussein H.S., Brasel J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology. 2001;167:101–134. doi: 10.1016/S0300-483X(01)00471-1. [DOI] [PubMed] [Google Scholar]
- 5.Adebo O.A., Njobeh P.B., Sidu S., Adebiyi J.A., Mavumengwana V. Aflatoxin B1 degradation by culture and lysate of a Pontibacter specie. Food Control. 2017;80:99–103. doi: 10.1016/j.foodcont.2017.04.042. [DOI] [Google Scholar]
- 6.Kim S., Lee H., Lee S., Lee J., Ha J., Choi Y., Yoon Y., Choi K.-H. Invited review: Microbe-mediated aflatoxin decontamination of dairy products and feeds. J. Dairy Sci. 2017;100:871–880. doi: 10.3168/jds.2016-11264. [DOI] [PubMed] [Google Scholar]
- 7.Méndez-Albores A., Arámbula-Villa G., Loarca-Piña M.G.F., Castaño-Tostado E., Moreno-Martínez E. Safety and efficacy evaluation of aqueous citric acid to degrade B-aflatoxins in maize. Food Chem. Toxicol. 2005;43:233–238. doi: 10.1016/j.fct.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Velazhahan R., Vijayanandraj S., Vijayasamundeeswari A., Paranidharan V., Samiyappan R., Iwamoto T., Friebe B., Muthukrishnan S. Detoxification of aflatoxins by seed extracts of the medicinal plant, Trachyspermum ammi (L.) Sprague ex Turrill—Structural analysis and biological toxicity of degradation product of aflatoxin G1. Food Control. 2010;21:719–725. doi: 10.1016/j.foodcont.2009.10.014. [DOI] [Google Scholar]
- 9.Wang B., Mahoney N.E., Pan Z., Khir R., Wu B., Ma H., Zhao L. Effectiveness of pulsed light treatment for degradation and detoxification of aflatoxin B1 and B2 in rough rice and rice bran. Food Control. 2016;59:461–467. doi: 10.1016/j.foodcont.2015.06.030. [DOI] [Google Scholar]
- 10.Adebo O.A., Njobeh P.B., Gbashi S., Nwinyi O.C., Mavumengwana V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. 2017;57:3208–3217. doi: 10.1080/10408398.2015.1106440. [DOI] [PubMed] [Google Scholar]
- 11.Teniola O.D., Addo P.A., Brost I.M., Färber P., Jany K.D., Alberts J.F., van Zyl W.H., Steyn P.S., Holzapfel W.H. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556T. Int. J. Food Microbiol. 2005;105:111–117. doi: 10.1016/j.ijfoodmicro.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Sangare L., Zhao Y., Folly Y., Chang J., Li J., Selvaraj J., Xing F., Zhou L., Wang Y., Liu Y. Aflatoxin B1 Degradation by a Pseudomonas Strain. Toxins. 2014;6:3028. doi: 10.3390/toxins6103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adebo O.A., Njobeh P.B., Sidu S., Tlou M.G., Mavumengwana V. Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains. Int. J. Food Microbiol. 2016;233:11–19. doi: 10.1016/j.ijfoodmicro.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Xia X., Zhang Y., Li M., Garba B., Zhang Q., Wang Y., Zhang H., Li P. Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control. 2017;75:92–98. doi: 10.1016/j.foodcont.2016.12.036. [DOI] [Google Scholar]
- 15.Chang P.K., Hua S.S. Nonaflatoxigenic Aspergillus flavus TX9-8 competitively prevents aflatoxin accumulation by A. flavus isolates of large and small sclerotial morphotypes. Int. J. Food Microbiol. 2007;114:275–279. doi: 10.1016/j.ijfoodmicro.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Afsah-Hejri L. Saprophytic yeasts: Effective biocontrol agents against Aspergillus flavus. Int. Food Res. J. 2013;20:3403–3409. [Google Scholar]
- 17.Hormisch D., Hormisch D., Brost I., Kohring G.W., Giffhorn F., Kroppenstedt R.M., Stackebradt E., Färber P., Holzapfel W.H. Mycobacterium fluoranthenivorans sp. nov., a Fluoranthene and Aflatoxin B1 Degrading Bacterium from Contaminated Soil of a Former Coal Gas Plant. Syst. Appl. Microbiol. 2004;27:653–660. doi: 10.1078/0723202042369866. [DOI] [PubMed] [Google Scholar]
- 18.Alberts J.F., Engelbrecht Y., Steyn P.S., Holzapfel W.H., van Zyl W.H. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006;109:121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Harkai P., Szabó I., Cserháti M., Krifaton C., Risa A., Radó J., Balázs A., Berta K., Kriszt B. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeter. Biodegr. 2016;108:48–56. doi: 10.1016/j.ibiod.2015.12.007. [DOI] [Google Scholar]
- 20.Prettl Z., Dési E., Lepossa A., Kriszt B., Kukolya J., Nagy E. Biological degradation of aflatoxin B1 by a Rhodococcus pyridinivorans strain in by-product of bioethanol. Anim. Feed Sci. Technol. 2017;224:104–114. doi: 10.1016/j.anifeedsci.2016.12.011. [DOI] [Google Scholar]
- 21.Farzaneh M., Shi Z.Q., Ahmadzadeh M., Hu L.B., Ghassempour A. Inhibition of the Aspergillus flavus Growth and Aflatoxin B1 Contamination on Pistachio Nut by Fengycin and Surfactin-Producing Bacillus subtilis UTBSP1. Plant Pathol. J. 2016;32:6. doi: 10.5423/PPJ.OA.11.2015.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raksha R.K., Vipin A.V., Hariprasad P., Anu Appaiah K.A., Venkateswaran G. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control. 2017;71:234–241. doi: 10.1016/j.foodcont.2016.06.040. [DOI] [Google Scholar]
- 23.Wang C., Li Z., Wang H., Qiu H., Zhang M., Li S., Luo X., Song Y., Zhou H., Ma W., et al. Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agric. 2017;97:1342–1348. doi: 10.1002/jsfa.7872. [DOI] [PubMed] [Google Scholar]
- 24.Guan S., Ji C., Zhou T., Li J., Ma Q., Niu T. Aflatoxin B1 Degradation by Stenotrophomonas maltophilia and Other Microbes Selected Using Coumarin Medium. Int. J. Mol. Sci. 2008;9:1489. doi: 10.3390/ijms9081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-García C., Béjar V., Martínez-Checa F., Llamas I., Quesada E. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005;55:191–195. doi: 10.1099/ijs.0.63310-0. [DOI] [PubMed] [Google Scholar]
- 26.Eshelli M., Harvey L., Edrada-Ebel R., McNeil B. Metabolomics of the Bio-Degradation Process of Aflatoxin B1 by Actinomycetes at an Initial pH of 6.0. Toxins. 2015;7:439. doi: 10.3390/toxins7020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Q., Zhai C., Guan B., Li C., Shan S., Yu J. Mathematic Modeling for Optimum Conditions on Aflatoxin B1 Degradation by the Aerobic Bacterium Rhodococcus erythropolis. Toxins. 2012;4:1181. doi: 10.3390/toxins4111181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan S., Zhao L., Ma Q., Zhou T., Wang N., Hu X., Ji C. In Vitro Efficacy of Myxococcus fulvus ANSM068 to Biotransform Aflatoxin B1. Int. J. Mol. Sci. 2010;11:4063. doi: 10.3390/ijms11104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’souza D.H., Brackett R.E. The Role of Trace Metal Ions in Aflatoxin B1 Degradation by Flavobacterium aurantiacum. J. Food Prot. 1998;61:1666–1669. doi: 10.4315/0362-028X-61.12.1666. [DOI] [PubMed] [Google Scholar]
- 30.El-Deeb B., Altalhi A., Khiralla G., Hassan S., Gherbawy Y. Isolation and Characterization of Endophytic Bacilli Bacterium from Maize Grains Able to Detoxify Aflatoxin B1. Food Biotechnol. 2013;27:199–212. doi: 10.1080/08905436.2013.811083. [DOI] [Google Scholar]
- 31.Zhang W., Xue B., Li M., Mu Y., Chen Z., Li J., Shan A. Screening a Strain of Aspergillus niger and Optimization of Fermentation Conditions for Degradation of Aflatoxin B1. Toxins. 2014;6:3157. doi: 10.3390/toxins6113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farzaneh M., Shi Z.Q., Ghassempour A., Sedaghat N., Ahmadzadeh M., Mirabolfathy M., Javan-Nikkhah M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control. 2012;23:100–106. doi: 10.1016/j.foodcont.2011.06.018. [DOI] [Google Scholar]
- 33.Tan H., Zhang Z., Hu Y., Wu L., Liao F., He J., Luo B., He Y., Zuo Z., Ren Z., et al. Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading Zearalenone. Food Control. 2015;47:285–290. doi: 10.1016/j.foodcont.2014.07.013. [DOI] [Google Scholar]
- 34.Kavitha S., Senthilkumar S., Gnanamanickam S., Inayathullah M., Jayakumar R. Isolation and partial characterization of antifungal protein from Bacillus polymyxa strain VLB16. Process Biochem. 2005;40:3236–3243. doi: 10.1016/j.procbio.2005.03.060. [DOI] [Google Scholar]
- 35.Turner P., Mamo G., Karlsson E.N. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Fact. 2007;6:9. doi: 10.1186/1475-2859-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motomura M., Toyomasu T., Mizuno K., Shinozawa T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003;158:237–242. doi: 10.1078/0944-5013-00199. [DOI] [PubMed] [Google Scholar]
- 37.Holt J.G. Bergey’s Manual of Determinative Bacteriology. 9th ed. Baltimore Williams and Wilkins; Philadelphia, PA, USA: 1994. [Google Scholar]
- 38.Heuer H., Krsek M., Baker P., Smalla K., Wellington E.M. Analysis of actinomycete communities by specific aplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microb. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S., Sheng P., Zhang H. Isolation and Identification of Cellulolytic Bacteria from the Gut of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae) Int. J. Mol. Sci. 2012;13:2563. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon S.H., Ha SM., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong A.D., Li H.P., Shen L., Zhang J.B., Wu A.B., He W.J., Yuan Q.S., He J.D., Liao Y.C. The Shewanella algae strain YM8 produces volatiles with strong inhibition activity against Aspergillus pathogens and aflatoxins. Front. Microbiol. 2015;6:1091. doi: 10.3389/fmicb.2015.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.