Abstract
The structure and functions of the extracellular matrix (ECM), its spatial distribution and pericellular association of ECM molecules remain poorly understood. Colocalization of ECM molecules with cell phenotypes through immunohistochemistry can provide crucial insights into their juxtacrine signaling role as well as their structural relevance to tissue architecture. As manual quantification of images introduces intra- and inter-user bias and is cumbersome for high-throughput approaches, we implemented an automated high-throughput method to quantify the spatial distribution and cellular association of one ECM molecule, thrombospondin 1 (TSP1) with two major cell phenotypes, neurons, and astrocytes. The distribution of TSP1 was homogeneous throughout the striatum and cortex along the anterior–posterior axis. TSP1 occupied 8.85% of the striatum and 7.40% in the cortex. TSP1 also associated with 94.58% and 88.45% of neurons in the striatum and cortex. The association with astrocytes was significantly lower at 47.55% and 28.09%. These findings highlight the key role that TSP1 plays in neuron physiology in a healthy brain, but also highlights key regional difference in astrocytes secreting ECM molecules. The semiautomated approach implemented here will improve the throughput and reliability of measuring the distribution and cellular colocalization of ECM molecules.
Keywords: astrocytes, automated analysis, CellProfiler, cortex, extracellular matrix, neurons, striatum, thrombospondin
Introduction
The extracellular matrix (ECM) is a functionally important, yet a commonly overlooked, complex structure of the brain and central nervous system (CNS), comprising nearly 20% of the neural tissue volume.1,2 Playing both structural and functional roles in the maintenance of the brain, ECM is comprised of many different molecules. The ECM forms the interstitial space in between the cellular elements, but also forms a sharp boundary, the so-called basement membrane, between the neural and vascular compartments.3,4 In addition to its structural role, ECM acts as a sink for growth factors that modulate cellular events and gene expression through juxtacrine, or contact dependent, cell signaling.5,6 The ECM plays an integral role in cellular events and signaling, yet a comprehensive quantification of the spatial and regional distribution of the ECM in the brain has not yet been done, although some characterizations have been reported.7–13 These analyses, however, are often restricted to a small area of interest or are semi-quantitative. A thorough characterization of the ECM will define both its regional and cellular distribution, including the area covered by these molecules and the percentage of major neural phenotypes that associate with these molecules. This information can then provide insight into how much these prominent ECM molecules associate with cells in the normal brain and can later serve as a basis for comparison in pathology. One such ECM molecule of interest is thrombospondin 1 (TSP1), as it has roles in CNS plasticity and synaptic repair after tissue damage.
TSPs are one family of ECM molecules and are comprised of five different TSPs (1 to 5) with TSP1 and TSP2, who share very similar structural domains, present in the adult CNS.7 Although TSP1 is produced and secreted by astrocytes, it has been shown to play roles in neuronal development, such as the aforementioned synaptic plasticity, encouraging neurite outgrowth, and cell migration.3,7,14–16 It had been found that, in early postnatal development, TSP1 is active in the brain and associates with most astrocytes.7 In the adult brain, however, TSP1 levels are much lower than in the developing brain and associations with astrocytes have been reported to be downregulated or completely absent.7,17 In line with its role in synaptic and behavioral recovery, TSP1 levels significantly increase and associate more with reactive astrocytes and microglia post-ischemia.7,15,18 Despite its role in neuronal cell events, the association between TSP1 and neurons has not been sufficiently investigated. These types of changes in associations with specific phenotypes during pathology are difficult to define without a more complete quantification of these associations in the normal brain. We, here, focus on TSP1 in the normal brain, as a quantification of its spatial distribution and cellular associations in the normal brain has not previously been quantified.
Previous quantifications of TSP1 have focused on Western Blots or mRNA to assess general tissue levels, although no regional comparison have been performed. To quantify cellular associations and regional distributions, time-consuming immunohistochemistry and image analysis is required. However, large-scale quantification of histological images across multiple phenotypes and regions is cumbersome and inefficient when done through manual counting, including stereology. Manual counting can also introduce intra-user bias and if there is more than one counter, there is additionally the chance for inter-user bias.19,20 A versatile high-throughput approach is most appropriate for a comprehensive quantification and eliminates intra- and inter-user error, as the same results will be produced regardless of the time they are analyzed or by whom the analysis is run.21 Complete automation, however, can introduce other potential errors in correctly detecting and classifying cells, such as false negative and false positive detection.21 Some analyses may benefit from complete automation, where time and consistency are most important, while others may benefit from the introduction of manual checks done by the user where accuracy is most important. Eliminating human judgment in automated counting is, in one respect, beneficial when judgment varies within (i.e., same user counting the same image on 1 day vs. the next) or between users (i.e., user 1 counting the image vs. user 2). However, these types of manual checks are superior to complete automation when images are not of optimal quality or when there are image artifacts that an automated program is not able to account for.19,21–23 Thus, the automated method must be validated for accuracy against manual counts for a small sample subset, with the manual counts commonly being considered the gold standard method.19,20 Automation also allows more images to be processed, thus increasing the sample size and the confidence in the reliability and accuracy of the results.
Here, we focus on TSP1, as it plays significant roles in early synaptogenesis as well as in synaptic recovery after damage (such as stroke), and its distribution in the striatum and cortex.7,15,18 The striatum and cortex are two large adjacent and interconnected regions affected by pathologies, such as stroke, but also have distinct histological and cellular features. A high throughput, automated approach can be implemented to perform consistent image processing for both small and large batches of images.24 We here assess the feasibility and validity of an automated and quantitative approach that measures the spatial distribution of TSP1 and its association with neurons and astrocytes in normal rat brain striatum and cortex.
Materials and Methods
Animals and Tissue Preparation
All animal procedures complied with the Institutional Animal Care and Use Committee (IACUC), as well as National Institutes of Health (NIH) guidelines. Male Sprague-Dawley rats (n=5, 350–400 g, Taconic Labs) were perfused at 12 weeks old. Animals were euthanized by intraperitoneal injections of Pentobarbital Sodium (10 mg/100 g body weight, Fatal Plus, Vortech) and perfused transcardially with a peristaltic pump (MasterFlex, Cole Parmer) to flush out blood using ice cold Phosphate Buffered Saline (0.01 M PBS, P4417 Sigma) followed by tissue fixation using paraformaldehyde (4% in 0.01 M PBS, 15714 Electron Microscopy Sciences). Excised brains were postfixed in 4% paraformaldehyde overnight and then cryoprotected in 30% Sucrose (Sigma) along with the antimicrobial agent Sodium Azide (S2002 Sigma) at 4C. Fixed brains were embedded in frozen sectioning compound (Surgipath FSC 22, Leica) and sectioned at 50 µm thickness at 500 µm series intervals on a cryostat (Leica). Individual series were stored as floating sections in Tissue Cryopreservation Solution (TCS, 30% Ethylene Glycol, 25% Glycerol, 0.5% Sodium Azide in PBS) at −20C.
Immunohistochemistry
Sections were removed from storage in TCS and mounted onto glass slides. Sections were washed 3 × 5 min in PBS before overnight (16 hr, 4C) incubation with primary antibodies: NeuN for neurons, GFAP for astrocytes, and TSP1 for TSP1 (Table 1). These were diluted in PBS/0.5% Triton X-100 (Sigma) to the appropriate concentrations. Following overnight incubation, sections were washed 3× with PBS before application of corresponding AlexaFluor secondary antibodies in PBS (1:500, Molecular Probes) for 1 hr at room temperature (21C). Secondary antibodies were then removed and sections were washed 3× with PBS before counterstaining with the nuclear marker Hoechst (1 µg/mL in PBS, Sigma) for 5 min. Sections were washed a final 3× with PBS before being coverslipped with Vectashield (H1000, Vector Labs) mounting medium. All steps, except overnight incubation, were carried out on a rotator at minimum speed.
Table 1.
Antibodies Used for Immunohistochemistry.
| Antibody | Host | Company | Catalog No. | Concentration |
|---|---|---|---|---|
| NeuN | Rabbit | Abcam | ab177487 | 1:500 |
| GFAP | Chicken | Abcam | ab4674 | 1:3000 |
| TSP1 | Mouse | Abcam | ab1823 | 1:100 |
Abbreviations: GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Validation of immunohistochemistry was performed using negative controls, where the primary antibodies were omitted, on rat tissue to determine potential unspecific staining of secondary antibodies. No unspecific staining of the pre-adsorbed secondary antibodies was evident (Supplementary Fig. 1A). To avoid mouse-on-mouse staining, a preconjugated primary antibody of the same clone (A6.1; 1:300; sc-599887 AF594, Santa Cruz) was used to stain mouse liver tissue in wild-type and TSP1 knockout mice to validate the specificity of the antibody for TSP1. Autofluorescence in mouse livers was quenched using Sudan Black (1.5% in 70% industrially methylated spirit for 20 min),25 before application of the preconjugated TSP1 antibody and 4’,6-diamidino-2-phenylindole (DAPI) costaining. No binding of the TSP1 antibody was evident in TSP1 knockout tissue (Supplementary Fig. 1B). This clone from different providers has been widely used and cross-validated to study TSP1,26–28 including brain.29
Image Acquisition
Images were acquired using an AxioImager M2 microscope (Zeiss) in conjunction with Stereo Investigator software (MBF) interfaced with a 120 W metal halide arc lamp (012-63000, X-Cite) light source. Whole hemisphere images were acquired using automated image stitching in the VirtualTissue module and a 10× objective (420341-9911, numerical aperture [NA] = 0.3; Fig. 1A). To afford a comparison between manual and computational analyses, 15 fields of view (FOVs) were acquired per region (striatum and cortex) using a 20× objective (420650-9901, NA = 0.8) with fixed acquisition parameters between images and animals (Table 2). FOVs were spread out evenly throughout the regions along the anterior-posterior axis (four sections per animal at 1.0, 0.5, 0.0, and −0.5 mm from Bregma; Fig. 1A). To afford a regional visualization and occupancy of TSP1, images were taken in both the striatum and cortex (Fig. 1B). High magnification z-stacks to demonstrate the extracellular nature of TSP1 were acquired in the striatum and cortex with a 40× oil immersion objective (420460-9900, NA = 1.3).
Figure 1.
Regional visualization of TSP distribution and FOVs. (A) FOVs were taken on stitched whole brain images along the anterior to posterior axis with representative sections at indicated Bregma values shown. (B) Example FOV for both the striatum and the cortex, taken with a 20× objective, showing the qualitatively even distribution of TSP. Scale bar is 200 μm. Abbreviations: FOV, field of view; GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Table 2.
Emission/Exposure Settings for Image Acquisition.
| Objective | Channel (nm) | Antibody | Exposure (ms) |
|---|---|---|---|
| 10× | DAPI (350) | Hoechst | 64 |
| GFP (488) | NeuN | 226 | |
| dsRED (555) | TSP1 | 150 | |
| Cy5 (660) | GFAP | 2,657 | |
| 20× | DAPI (350) | Hoechst | 8 (striatum and cortex) |
| GFP (488) | NeuN | 27 (striatum and cortex) | |
| dsRED (555) | TSP1 | 60 (striatum and cortex) | |
| Cy5 (660) | GFAP | 496 (striatum), 200 (cortex) |
For all settings, gain was set to 0. Abbreviations: DAPI, 4’,6-diamidino-2-phenylindole; dsRED, Discosoma sp. red fluorescent protein; GFAP, glial fibrially acid protein; GFP, green fluorescent protein; TSP1, thrombospondin 1.
Image Processing
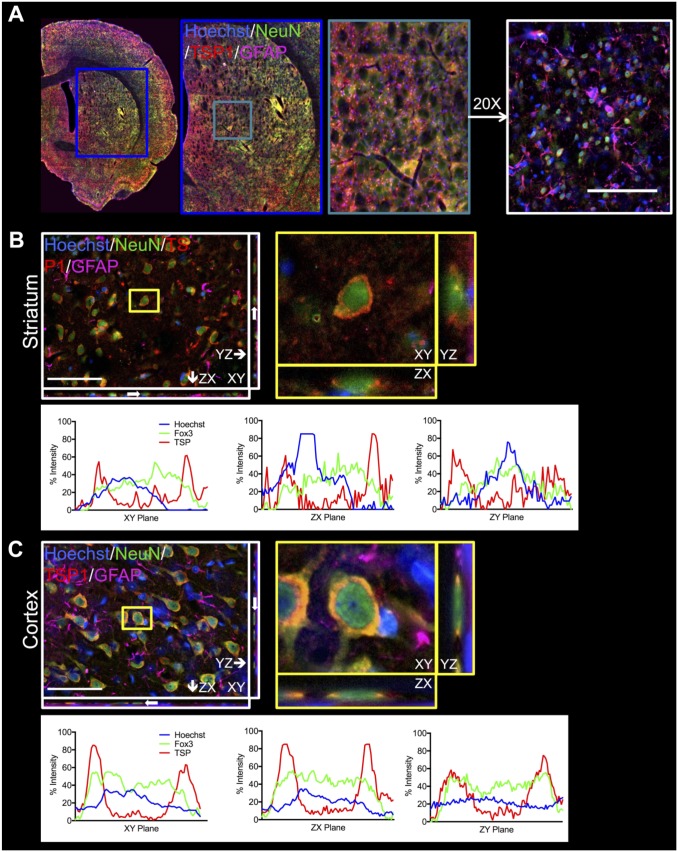
Images were processed using a pipeline of modules developed in the biological image analysis software, CellProfiler (2.1.1 rev. 6c2d896 for Mac; see http://cellprofiler.org/).24 The pipeline generated automated counts of cell phenotype populations and their association with TSP1, which was then used to calculate the percentages of specific cell phenotypes that associate with TSP1. Images are first taken based upon region—either the striatum or cortex—and then acquired with consistent acquisition settings (Table 2) before being processed by the pipeline (Fig. 2A). Detailed settings are summarized in Table 3. Staining of extracellular molecules was shown to be extracellular to the cells, shown in Fig. 2B and C for neurons in the striatum and cortex, respectively. Orthogonal views obtained from the z-stacks at higher magnification (40× oil immersion objective) show TSP1 is peripheral to the phenotype staining (NeuN, shown in Fig. 2B and C). Intensity profiles of these orthogonal view planes further confirm this, with the phenotype and nuclear stains localized in the center and the TSP1 stain localized outside of this center.
Figure 2.
Confirmation of the extracellular nature of TSP in the striatum and cortex. (A) Example of visualization of the striatum with 10× and then 20× objectives in a whole hemisphere stitched image. Qualitatively, the TSP marker appears extracellular. Scale bar is 200 μm. (B) z-stack with a 40× objective in the striatum. White arrows indicate the cell (neuron) highlighted in the yellow box for each orthogonal view. Intensity profiles are shown for each orthogonal view, demonstrating the nuclear colocalization of Hoechst and the phenotype marker (NeuN, here) and the extracellular association of the cell with TSP. Scale bar is 100 μm. (C) z-stack with a 40× objective in the cortex. The same views are shown as in B, but for the cortex. Intensity profiles confirm the extracellular nature of the TSP marker. Scale bar is 100 µm. Abbreviations: GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Table 3.
Key Parameters for CellProfiler Modules.
| Module | Parameter | Value |
|
|---|---|---|---|
| Striatum | Cortex | ||
| Background removal—Hoechst | |||
| EnhanceOrSuppressFeatures | Feature type | Speckles | Speckles |
| Feature size | 50 | 50 | |
| ApplyThreshold | Threshold strategy | Adaptive | Adaptive |
| Threshold method | Robust background | Robust background | |
| Smoothing | Automatic | Automatic | |
| Threshold correction | 0.99 | 0.99 | |
| Size of adaptive window | 100 | 100 | |
| Background removal—NeuN | |||
| EnhanceOrSuppressFeatures | Feature type | Speckles | Speckles |
| Feature size | 40 | 100 | |
| ApplyThreshold | Threshold strategy | Adaptive | Adaptive |
| Threshold method | Robust background | Robust background | |
| Smoothing | Manual: 1 | Manual: 1 | |
| Threshold correction | 1.00 | 1.00 | |
| Size of adaptive window | 325 | 300 | |
| Background removal—GFAP | |||
| EnhanceOrSuppressFeatures | Feature type | Speckles | Speckles |
| Feature size | 100 | 100 | |
| ApplyThreshold | Threshold strategy | Adaptive | Adaptive |
| Threshold method | Robust background | Robust background | |
| Smoothing | Manual: 1 | Manual: 1 | |
| Threshold correction | 0.70 | 0.80 | |
| Size of adaptive window | 100 | 500 | |
| Background removal—TSP1 | |||
| EnhanceOrSuppressFeatures | Feature type | Speckles | Speckles |
| Feature size | 50 | 50 | |
| ApplyThreshold | Threshold strategy | Adaptive | Adaptive |
| Threshold method | Robust background | Robust background | |
| Smoothing | Manual: 1 | Manual: 1 | |
| Threshold correction | 0.40 | 0.50 | |
| Size of adaptive window | 325 | 325 | |
| Direct object identification—Nuclei | |||
| IdentifyPrimaryObjects | Typical diameter range | 10–50 | 10–50 |
| Threshold strategy | Global | Global | |
| Threshold method | Otsu—Three classes | Otsu—Three classes | |
| Threshold smoothing | Manual: 3 | Manual: 3 | |
| Threshold correction | 1.0 | 1.0 | |
| Bounds on threshold | 0.02–1.00 | 0.02–1.00 | |
| Declumping smoothing | Manual: 5 | Manual: 5 | |
| Distance to suppress local maxima | 10 | 10 | |
| Direct object identification—Filtered neurons | |||
| IdentifyPrimaryObjects | Typical diameter range | 13–50 | 20–80 |
| Threshold strategy | Global | Global | |
| Threshold method | Otsu—Three classes | Otsu—Three classes | |
| Threshold smoothing | Manual: 3 | Manual: 3 | |
| Threshold correction | 0.9 | 0.9 | |
| Bounds on threshold | 0.02–1.00 | 0.02–1.00 | |
| Declumping smoothing | Manual: 5 | Manual: 5 | |
| Distance to suppress local maxima | 10 | 10 | |
| MeasureObjectNeighbors on neuron objects | Method to determine neighbors | Adjacent | Adjacent |
| FilterObjects on neuron objects | Filtering mode | Measurements | Measurements |
| Filtering method | Limits | Limits | |
| Measurement filter | Neighbors > Percent Touching > Adjacent | Neighbors > Percent Touching > Adjacent | |
| Filter using maximum value | 35 | 40 | |
| Association—Filtered astrocyte nuclei | |||
| ApplyThreshold on background removed GFAP stain | Output image type | Binary | Binary |
| Threshold strategy | Manual: 0.07 | Manual: 0.07 | |
| MaskObjects (objects: nuclei, mask: binary GFAP) | Remove based on fractional overlap | 0.1 | 0.16 |
| ApplyThreshold on background removed NeuN stain | Output image type | Binary | Binary |
| Threshold strategy | Manual: 0.05 | Manual: 0.05 | |
| MaskObjects (objects: astrocyte nuclei, mask: binary NeuN) | Remove based on fractional overlap with inverted mask | 0.9 | 0.9 |
| Association—Neurons and astrocytes that associate with TSP1 | |||
| ApplyThreshold on background removed TSP1 stain | Output image type | Binary | Binary |
| Threshold strategy | Manual: 0.09 | Manual: 0.07 | |
| MaskObjects (objects: filtered neurons, mask: binary TSP1) | Remove based on fractional overlap | 0.05 | 0.07 |
| MaskObjects (objects: filtered astrocyte nuclei, mask: binary TSP1) | Remove based on fractional overlap | 0.15 | 0.1 |
Abbreviations: GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Iterative Pipeline Development and Validation
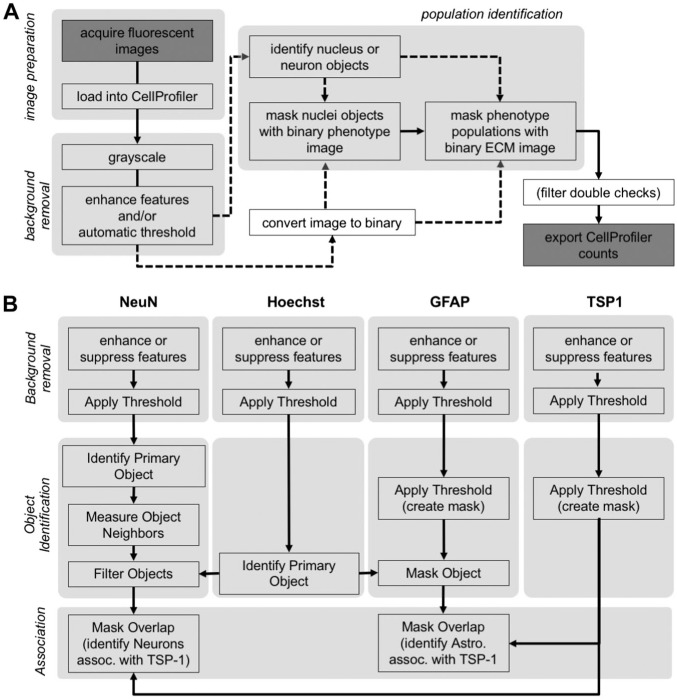
A conceptual pipeline was developed to account for image acquisition, background removal and cell and TSP1 identification, as well exporting of the data for analysis (Fig. 3A). Specific module combinations were made from various CellProfiler image-processing tools to achieve removal of background in individual channels, to identify individual phenotype populations colocalizing with cell nuclei, and to define cell phenotype pericellular associations with TSP1 (Fig. 3B). It is important to note that the nucleus or cell body is being digitally identified as an object that does not represent the entire cell. For example, in identifying neurons, it is not the entire neuron (including the cell body, axon, etc.) that is being identified, but a localized area where the phenotype stain NeuN has visualized the cell body. Relying on the immunostaining, this identification can provide a definition of the area occupied by the cell. To validate the combination of modules and chosen settings, each module combination was tested for accuracy/trueness by comparing the manual counts done on the images before pipeline development to the automated counts generated by CellProfiler for a set of 10 images. The set of 10 images were chosen to best represent the possible variability within each stain to produce a pipeline that is as robust as possible. We considered the precision of measurements to be affected by random errors reflected in the variance of measuring the same image multiple times. Accuracy, that is, a description of systematic errors that defines the proximity of measurements results to the true value (as defined by ISO5725-1),30 was determined by calculating Pearson’s r for a correlation between the two sets of counts for the same 10 images. In order for a module combination to be accepted for the final pipeline, a Pearson’s r > 0.90 was required with a p value<0.05 (Type I error, that is, a cell being identified as belonging to a certain population when it does not). A power of >80% was considered sufficient to avoid Type II errors, that is, a cell that belongs to a certain population not being identified as such. If a module combination did not pass this threshold, iterative modifications and retesting were performed until the criteria were reached. The pipeline for analyzing images from the striatum was developed first and was then adapted to analyze images from the cortex. The same iterative method of development was used to adapt the pipeline settings to analyze the cortex images and no new modules were added. Pipelines are available from the authors upon request.
Figure 3.
(A) Flowchart of image acquisition through processing to acquiring the final counts. Images are acquired after immunohistochemistry and are loaded into CellProfiler before undergoing background removal. A conceptual overview of image processing is presented. (B). Images are processed through several routes, depending on which stain is in the image. For example, Hoechst is used to identify nuclei objects before being associated with a phenotype stain (GFAP) to identify a phenotype population (astrocytes). This phenotype population is then masked with the binary version of the ECM molecule (TSP) which has gone through background removal, to identify the amount of that phenotype population that associates with the ECM molecule (astrocytes associating with TSP). There may be double checks, if possible, to exclude incorrectly identified objects. For astrocytes, identified astrocyte nuclei were masked with the stain for neurons (NeuN) to ensure that no identified objects were also being identified as neurons. The final counts were then exported. Abbreviations: ECM, extracellular matrix; GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Individual Image Processing and Analysis Modules
Background Removal
General background removal consisted of three modules to convert the individual 24 bit red, green, blue (RGB) channel images to 8 bit grayscale images, enhance features, and to perform an automatic threshold. Images from individual fluorescent channels are converted to grayscale to be processed by subsequent modules. Features of the image (features meaning specific staining within the image) are treated as Speckle type features and enhanced to increase the contrast between specific staining and background, without losing image integrity. Remaining background with nonzero intensity was removed using the ApplyThreshold module. Background was removed on a tile basis, where the threshold is calculated and applied individually for tiles of the image (the size of the tile being customizable), rather than a single threshold calculated and applied to the whole image, using the Adaptive strategy. The threshold was calculated using the RobustBackground method, which assumes that the background intensity distribution approximates a Gaussian normal distribution. The top and bottom 5% of pixel intensities were trimmed before calculating the mean and standard deviation of the rest of the image. The threshold was determined to be the mean plus double the standard deviation.
Direct Object (Cell) Identification
For nuclear stains (e.g., Hoechst) or stains that are oval shaped and encompass the nucleus and some of the cytoplasm (e.g., NeuN for neurons), cell populations can be directly identified from the histology images. Once background was removed, images were processed by a single module (IdentifyPrimaryObjects) that identified primary objects (objects with no current relationship to other objects) based on intensity distributions. For this type of identification, the global threshold strategy (calculating a single threshold value based on the whole image) with a three-class Otsu thresholding method was used. This method calculates the threshold based on separating pixel intensities into three classes, minimizing the variance within each, and determines the threshold that separates the lower two classes. For molecules whose histological visualization is not localized akin to nuclear or nuclear-like stains, direct object identification cannot be used. For example, GFAP visualizes intermediate filaments of astrocytes and each identified astrocyte may have several immediate filaments protruding from the nuclear area stained. Use of the direct object identification method on GFAP would identify many objects along the stained intermediate filaments as the stain intensity varies along the filaments and is not localized to a small area, creating vast inaccuracies. Thus, phenotypes whose histological visualization is not nuclear, or localized to the nucleus, and identifiable by direct object identification must be identified using the association method.
Association
Association of a visualized molecule with previously identified cell objects was achieved through masking with a binary image and based on an overlap of the molecule with an object. Following background removal and direct object identification of the objects to associate with the molecule, the histological image was converted to binary using a manual threshold. The binary image can then be used as a mask over the desired objects. Based on a user specified fractional overlap (Table 3) with the mask (stain) image, objects are accepted (or rejected) as objects that associate with the stain and identified as a subpopulation. This can be used to both identify phenotype populations as subpopulations of all nuclei and populations of a certain phenotype that associate with TSP1 as subpopulations of the phenotype population.
False Positive Corrections
For both neuron and astrocyte populations, additional validations to ensure that identified objects were indeed of the intended phenotype were performed. Clusters of many cells are not common in healthy striatum and cortex, but were identified from the NeuN stain. To correct for this, two modules were used to identify objects that were too close to each other for each to be a cell. The first module measured the amount of perimeter that objects were touching neighboring objects, while the second excluded objects whose perimeters were in contact more than 35% (striatum) or 40% (cortex) with neighboring objects (these numbers were enough to reject typically clumped objects), as these were most likely not individual cell objects and rather wrongly identified from the staining background.
Phenotypes that were identified by the association method had a greater chance of measurement error as there were several more processing steps involved than in direct object identification. Each processing step introduces some amount of potential error, stemming from whether the processing step is robust enough to accurately process each image. Following this logic, if two phenotypes could be identified within the same round of histology (e.g., staining for NeuN and GFAP at the same time, in different channels) and phenotype 1 is identified using direct object identification while phenotype 2 is identified using the association method, then we can rely on phenotype 1 to check phenotype 2. Here, phenotype 1 are neurons and phenotype 2 are astrocyte nuclei. As there is a lower chance of error in identifying neurons, we can use the same steps as the association method to make sure that objects identified as astrocyte nuclei do not overlap with identified neuron objects. In this case, astrocyte nuclei were masked with a binary version of the NeuN stain and objects were rejected if the overlap was greater than 90% (Table 3).
Area Occupied by Immunohistochemical Markers
The area occupied by each stain (phenotypic and ECM) was also measured. This was done using the MeasureImageAreaOccupied module to measure the area occupied by the binary versions of the phenotype and ECM stains after background removal and the total area of the image (consistent for all images). Binary images were taken from previous modules that had created binary versions of each of the stains—the GFAP stain from the identification of astrocyte nuclei, the NeuN stain from filtering wrongly identified astrocyte nuclei, and the TSP1 stain from identifying neurons and astrocyte nuclei that associated with TSP1. Each of these binary images was produced by applying a threshold, as described in previous sections. The percent area occupied by TSP1 can also act as a check to ensure consistency with staining and image processing.
Data Export and Statistics
Counts for all identified populations were exported from CellProfiler and imported into Excel, where the percentages of phenotypes associating with TSP1 were calculated based on cell counts. This was achieved by simply dividing the number of identified phenotype objects that associated with TSP1 by the total number of identified phenotype objects before multiplying by 100. Arithmetic means were calculated for all images (n=15 per section per animal) in a given tissue section at the same Bregma position for each animal. Using Prism 6 (GraphPad), biological replicates (n=5) were graphed as the mean ± standard deviation as data was normally distributed. For accuracy counts, Pearson’s r correlation was calculated with significance set at p<0.05 and 1-β at 0.8. For association analysis, repeated measures two-way ANOVA with post hoc Sidak tests and t-tests were used with significances set at p<0.05.
Results
Identification of Cell Nuclei and Phenotypes
To analyze the association of TSP1, phenotype populations must first be identified. Here, we identified neuron and astrocyte populations, as well as the total number of cells, in all acquired images. This analysis was applied to both the striatum and cortex, as there are notable differences in the phenotypic localization of TSP1.
Cell Nuclei
To account for the total number of cells present within individual images and yield percentages of the total amount of cells, the number of cells present needed to be measured. For this, cell nuclei were identified based on the nuclear stain Hoechst using direct object identification. Images of the Hoechst stain (blue channel) were first processed for background removal before nuclei were identified (Fig. 4A). An example of adapting the settings (such as threshold lenience, binary image threshold, fractional overlap, etc.) of specific modules to produce automated counts with a high enough correlation (i.e., iterative pipeline development) to the manual counts is changing the range of acceptable sizes for identified objects (Fig. 4B). Once objects are identified by direct object identification in the module IdentifyPrimaryObjects, they are accepted or rejected based on a user-defined size range. By changing the size range for identifying nuclei in an iterative manner, the required correlation coefficient with low error (<5%) was reached (Fig. 4B). An initial estimate of the average size of nuclei in pixels (1–30 pixels at this magnification and resolution), yielded an acceptable correlation, but it was visually clear that larger, correctly stained nuclei were not being accepted (Fig. 4B). Thus, further changes to the size range resulted in a correlation of 0.9565 (p<0.0001) with an average error of 3.28%.
Figure 4.
Process of identifying nuclei objects and an example of accuracy testing in the striatum are shown. (A) The fluorescent image of Hoechst is first converted to grayscale before the features are enhanced and background is thresholded. Then, objects are identified based on intensity distribution. The colors of the objects are used only to tell objects apart. The accuracy check is shown. (B) An example of how one parameter in direct object identification was changed to achieve accuracy is shown with the raw image and the manual counts (red dots) on the left. In IdentifyPrimaryObjects, a user-defined range of pixel lengths is used to accept or reject identified objects based on diameter. Size 1 is 1 to 30 pixels and it is clear that objects that are too large, but that are still correct cells, are being excluded. Despite Pearson’s r being >0.9 and p being <0.0001, it is evident that there is still error for several of the counts and that some automated counts being equally over- or underestimated has resulted in the high correlation. Increasing the lower and upper bounds of the range, as in Size 2 (20–50 pixels), then excludes smaller correctly identified cells and decreases Pearson’s r below the acceptable threshold. A final adjustment to Size 3 (10–50 pixels) increases correlation with the counts much closer to the trend line, indicating acceptable accuracy. Scale bars represent 200 µm.
Neuron Cell Identification
As with Hoechst, neuronal staining was nuclear and hence an adaption of this module combination afforded the quantification of phenotype stains with nuclear morphology. Neurons were identified via direct object identification (IdentifyPrimaryObjects) after background removal on the NeuN stain (Fig. 5A). Due to the sometimes significant areas of medium intensity background in the NeuN stain, cell counts were often an overestimation due to large cell clusters being identified from background, and not specific, staining. To correct for this, objects were removed based on the percentage that their object perimeter was in direct contact with a neighboring object, which signified a high likelihood of being in an extraneous cluster that was not common for neurons. This additional correction yielded a correlation of 0.9256 (p<0.0001) with an average error of 4.97%.
Figure 5.
Examples of phenotype population identification in the striatum by both direct object identification (neurons) and association identification (astrocytes). (A) The fluorescent NeuN stain was first converted to grayscale before the features were enhanced and the background was thresholded. As the NeuN stain is continuous and localized to the nucleus, neuron objects were identified by direct object identification. The NeuN stain, however, also has notable clouds of background staining that sometimes led to extraneous identification of clusters of cells. To exclude these extra objects, a double check was performed where the percent of the perimeters that are touching of two identified neighboring objects was measured. Objects were then filtered by this measurement and excluded from the final count if the percentage was above a user-defined threshold. (B) The fluorescent GFAP stain was first converted to grayscale before the features were enhanced and the background was thresholded. As the GFAP stain is not continuous and localized to the nucleus, astrocyte nuclei objects were identified by association of previously directly identified nuclei objects and the GFAP stain. After background removal, the GFAP stain was converted to binary and previously identified nuclei objects for that image were masked with the binary GFAP stain. Objects were accepted based on a user specified fractional overlap of the nuclei objects and the stain. The neuron counts were used as a double check on the astrocyte counts as these two populations should not overlap. A binary version of the processed NeuN stain was used as a mask over astrocyte nuclei objects where objects that overlapped beyond a user-defined threshold were excluded from the final astrocyte count. Scale bars are 200 µm. Abbreviation: GFAP, glial fibrially acid protein.
Astrocyte Cell Identification
Astrocytes were identified by the association approach due to a lack of a nuclear morphology (Fig. 5B). GFAP stains underwent background removal, reducing blur due to out of focus projections, and a user specified threshold was used to produce binary images. These binary images were then used to mask previously identified nuclear objects (identified by direct object identification). This resulted in a population of identified cell objects—nuclei—that colocalized with the phenotype histology—GFAP (Fig. 5B). For astrocytes, an additional correction was performed to exclude incorrectly identified astrocyte nuclei objects that were actually neuron nuclei. This was done by creating a binary version of the NeuN stain—a threshold was applied to the NeuN image that had undergone background removal. The initially identified astrocyte nuclei objects were then masked by the binary NeuN image and objects were removed if there was more than a 90% overlap between the objects and the NeuN binary image. Iterative pipeline development yielded a correlation of 0.9442 (p<0.0001) with an average error of 4.40%.
Identification of TSP1 and Cellular Attribution
To identify populations of phenotypes that associate with TSP1, the association method was used similarly to how it was used to identify astrocyte nuclei objects. TSP1 images (red channel) underwent background removal before being converted to a binary image via applying a user-defined threshold (Fig. 6A). The binary image was used to quantify the relative percent area that TSP1 covers and this can also be used to confirm uniformity of the TSP1 stain. Over the set of test images (n=10), the area covered was on average 8.17% ± 2.73%. This binary TSP1 image was then used as a mask over the phenotype objects—those objects being nuclei, astrocyte nuclei, and neurons (identified previously through direct object identification or association; Fig. 6B). Based on a user specified fractional overlap between the TSP1 stain and the phenotype objects (astrocyte nuclei or neuron objects), objects were accepted or rejected as associating with TSP1 (Fig. 6B). Fractional overlaps were determined for neurons associating with TSP1 and for nuclei associating with TSP1 (astrocytes were identified by their nuclei, thus determining a fractional overlap for all nuclei provides a robust method to analyze any subset of nuclei). Iterative pipeline development yielded a correlation of 0.9538 (p<0.0001) and an average error of 5.72% for nuclei associating with TSP1 and a correlation of 0.9684 (p<0.0001) and an average error of 4.93% for neurons associating with TSP1.
Figure 6.
The processing of ECM molecule stains and association of the stain with phenotype populations is shown (in the striatum). (A) The fluorescent TSP1 stain was first converted to grayscale before the features were enhanced and the background was thresholded. The image was then converted to binary to use as a mask with identified phenotype populations. The percent area covered by the binary image was also measured as another way to observe the distribution of TSP, shown here for the test images (with error bars representing the standard deviation), averaging 8.17%. (B) Subpopulations of phenotypes that associated with TSP were identified by the association method. Here, the association of nuclei with TSP is shown. Nuclei objects were first identified from the fluorescent Hoechst image and the fluorescent TSP image was processed and converted to binary. The binary TSP image (white) was then used to mask identified nuclei objects (gray) and objects that passed a user specified fractional overlap were accepted into the final subpopulation as associating with TSP (yellow). Scale bars are 200 µm. Abbreviations: ECM, extracellular matrix; TSP1, thrombospondin 1.
Quantification of TSP Distribution and Association With Phenotypes
A homogeneous distribution throughout the anterior-posterior striatum and cortex is observed, though there is notably stronger staining in the cortex (Fig. 2C). Qualitatively, TSP1 associates more with neurons than with astrocytes. Morphologically TSP1 staining is present within/surrounding the cytoplasm and can be visualized as associating with different phenotypes (Fig. 7A).
Figure 7.
Quantitative results of the distribution and association of TSP1 in the striatum and cortex. (A). Examples (in the striatum) of correctly identified neurons and astrocytes that associate with TSP1. Scale bars are 200 µm. (B) Percentage of the total identified cell population that were classified as neurons (42.54%, 43.67%) and astrocytes (17.37%, 10.08%) in the striatum and the cortex, respectively. (C) Percent area covered by each phenotypic stain, NeuN (3.88%, 7.04%) and GFAP (4.69%, 3.83%) in the striatum and the cortex, respectively. The percent area covered by NeuN is significantly different in the striatum versus the cortex (p<0.0332). (D) Anterior to posterior distribution of the percent area covered by the TSP1 stain in the striatum and the cortex. Distributions were not significantly different both anterior to posterior and between regions. (E) Average of the percent area covered by TSP1 (8.42%, 7.40%) in the striatum and cortex, respectively. (F) Anterior to posterior distribution of the percentage of phenotypes that associated with TSP1 in both the striatum and the cortex. Distributions were not significantly different anterior to posterior. (G) Averages of the percentage of phenotypes, neurons (94.58%, 88.45%) and astrocytes (47.55%, 28.09%) that associated with TSP1 in the striatum and cortex, respectively. The percentage of astrocytes that associate with TSP1 is significantly different in the striatum versus the cortex (p<0.0332). All error bars represent the standard deviation. Abbreviations: GFAP, glial fibrially acid protein; TSP1, thrombospondin 1.
Phenotypes were first identified before association with TSP1. In the striatum, neurons and astrocytes accounted for 42.54% and 17.37% of the population, respectively, while in the cortex, they accounted for 43.67% and 10.08% of the population (Fig. 7B). Neither the percentage of neurons nor astrocytes were significantly different between the striatum and cortex, though they differed significantly from each other (p<0.0021). In addition to quantifying the number of each phenotype, the percent area covered by each of their histological stains was measured. Neuronal staining (NeuN) covered 3.88% and 7.04% of the total area in the striatum and cortex, respectively, while astrocyte staining (GFAP) covered 4.69% and 3.83% of the total area (Fig. 7C). Neurons and astrocytes in both regions did not differ significantly along the anterior-posterior axis, indicating that the staining and image-processing methods were consistent and uniform. Only neuronal staining was significantly different between the striatum and cortex and this was visually evident (p<0.0332). The percent area occupied by TSP1 was also calculated for all images. This percentage did not significantly differ anterior to posterior or between regions (Fig. 7D). On average, the percent area covered by the TSP1 stain was 8.85% in the striatum and 7.40% in the cortex (Fig. 7E).
The percentage of neurons and astrocytes that associated with TSP1 also did not significantly differ along the anterior to posterior axis. The percentage of astrocytes that associated with TSP1 did significantly differ between the striatum and cortex (p<0.0001), while the percentage of neurons that associated did not (Fig. 7F). On average, the percentage of neurons that associated with TSP1 was 94.58% in the striatum and 88.45% in the cortex (Fig. 7G). For astrocytes, the percentage that associated with TSP1 was significantly lower (p<0.0005) than that for neurons with 47.55% in the striatum and 28.09% in the cortex (Fig. 7G).
Discussion
Although the ECM is a major structural and signaling component of the brain, the distribution and localization of ECM molecules remains poorly defined. Elucidating the association of ECM molecules with individual cell phenotypes will lead to a more detailed understanding of their physiological role in situ. We here described an automated computerized method that affords a quantitative characterization of cell phenotypes and their association with ECM molecules that removes possible inter- and intra-user bias. Specifically, we investigated the association of TSP1 with neurons and astrocytes in the striatum and cortex, as well as the spatial distribution of TSP1 in both regions.
Automation and Validation of Image Analysis for Cells and ECM
In theory, completely automated quantification is the ideal route for quantitative analyses. Automation, as opposed to manual quantification, eliminates possible user bias, whether that is inter-user bias between different persons carrying out the quantification or intra-user bias within a single person carrying out the quantification at different times. Automated methods also open up the potential for large sample size processing and high-throughput approaches.22,31 With larger sample sizes and faster processing time, not only is the confidence in accuracy increased but the time to do so is also decreased. There are several programs that can give automated nuclei counts, such as Fiji (see https://fiji.sc/), however, nuclei counts only provide a general measure of cellularity of a tissue32 and more complex information is required to provide a more thorough characterization of tissues composition. For example, examining the association and distribution of functional tissue components, such as ECM and different phenotypes (not merely all cells or only a single phenotype).
There are some automated image analysis tools (e.g., CellProfiler, FARSIGHT) that afford a more detailed and complex investigation beyond nuclei counting. FARSIGHT combines several different segmentation models to identify individual cell nuclei, but also can determine the distance between these and other cells as well as classify specific phenotypes.22,32 CellProfiler, similarly, can identify different phenotypes, but employs a pipeline method. Different modules can be applied with various methods of completing each module available (e.g., different threshold methods, choices in user specified parameters). This framework was utilized, here, where identification of phenotypes was generalized into two classes—direct object identification and association. This allowed for the pipeline to potentially cover all phenotypes—those that could not be identified by direct object identification would be able to be identified by association.
In addition to the identification of cell phenotypes, we here needed to analyze the distribution of the ECM in a truly quantitative manner. Existing histological quantifications of the normal neural ECM are commonly semi-quantitative in that they assign grades to qualitative judgments (e.g., rating how intense staining appears) or in that they introduce manual error checks (where users flip through processed images and manually add/delete cells from identified populations).8,11,19,22,23,32 Although the manual error checks in some semiautomated analyses minimize error to nearly 0% (if error is being measured relative to manual counts), this loss in time efficiency is not always desirable. For the analysis of percentages, such as that presented here, while accuracy is still important, consistency is equally important and minor errors will not be detrimental. Here, by employing iterative pipeline testing, we designed a pipeline that is robust enough for percentage comparisons without the need for manual error checks. In addition, the average percent error had to be less than 10%, as it is possible to have high correlation between two sets of data, even if there is high percent error. These values were chosen based on biologically plausibility, what is commonly achieved in other automated image analysis methods,19 and how much error would not be negligible in the interpretation of results. With this semiautomated method, we decrease the possible intra- and inter-user bias and enable high-throughput processing.
Many existing quantifications of normal neural ECM based on Western Blots or PCR also lack information about their spatial distribution.7–9 By using histological images and automated background removal, we were able to acquire a measure of the amount of TSP1 that is visualized by immunostaining and, thus, an idea of the homogeneity of the distribution of TSP1. However, immunohistochemistry also imposes methodological limitations. With many variations in histology (different antibodies, various precision and accuracy of stains, staining methods) and image acquisition (microscope models, exposure times, gain), the intensity, texture, and quality of features and potential image artifacts demand analysis and validation specific to each acquisition.22 Although one method may work very well for nuclei segmentation it, for example, may not work sufficiently well for neuron or astrocyte segmentation. This makes the ability to customize pipelines attractive for image analysis involving histology.
TSP1 Associates With Most Neurons and Some Astrocytes in Normal Rat Brain
With the ability to analyze the percentages of cell phenotype populations that associate or associate with other stains, the distribution of the ECM can be analyzed. Here, we specifically investigated the spatial distribution and association of TSP1 with two neural phenotypes in healthy rat brain (striatum and cortex). TSP1 is evenly distributed throughout the striatum and cortex. Using the proposed automated approach, we can additionally note that TSP1 associates more often with neurons than astrocytes. The developed pipeline confirms this and allows us to perform additional analysis to validate these observations in a high-throughput fashion.
As each phenotype population is being determined from the different phenotypic stains and methods of identification (either direct object identification for neurons or association for astrocytes), we are able to investigate the phenotypes themselves. We found here that the pipeline determined that about 40% to 45% of the total population are NeuN+ neurons, while about 10% to 20% of the population are GFAP+ astrocytes. More astrocytes are present in the striatum than cortex, consistent with the literature.33 We also determined the percent area covered by each of the phenotypic stains, helping to ensure consistency in the images acquired and processed. All covered quite a low area (<10%) suggesting that phenotypic stains are quite localized to the nucleus and not diffuse. The higher percent area covered by the NeuN immunostaining in the cortex also confirms our visual inspection, as neurons stained in the cortex were often much larger than that in the striatum.
Analysis of the TSP1 stain resulted in the same observation that TSP1 appears homogenously distributed throughout the striatum and cortex and along the anterior-posterior axis, with no significant difference in coverage between these areas. Similar to the phenotypic stains, the area covered was under 10% in both the striatum and the cortex, again suggesting that there was uniform staining and consistent image processing. In addition, the overall low area covered by TSP1 supports previous Western Blots that showed that TSP1 levels are quite low or absent in the healthy adult brain.7
The percentage of neurons and astrocytes associating with TSP1 indeed reflects the functional roles of TSP1. As TSP1 plays both a role in crucial developmental events, such as presynaptic differentiation, and is produced by astrocytes, it was expected that there would be significant associations with both neurons and astrocytes. The difference in the amount of association between phenotypes, which did not significantly differ anterior to posterior, was very apparent. Although most, if not all, neurons were found to associate with TSP1, the percentage of astrocytes that were found to associate was dramatically lower in both the striatum and cortex, confirming earlier qualitative observations. This is supported by earlier studies that have shown that while TSP1 associates with astrocytes early in development, that percentage decreases as the brain matures.7,17 The nearly total association of TSP1 with neurons is to be expected, as TSP1 is involved in synapse formation and plays a role in synaptic plasticity. To our knowledge, however, this neuronal association has not been previously quantified in the brain. Although the percentage of neurons that associate with TSP1 did not significantly differ between the striatum and the cortex, the percentage of astrocytes that associated was approximately 20% more in the striatum than in the cortex. A more detailed assay would be needed to investigate why there are differences in the percentage of astrocytes that associate with TSP1 in the striatum versus the cortex. This information more thoroughly characterizes the presence of TSP1 in the normal brain and potentially gives insight into the components of the ECM that are most necessary in regenerating or repairing damaged brain tissue.
This method additionally eliminates the issues of inter- and intra-user bias and the limitations of using manual counting to quantify cellular associations. Enabling high-throughput approaches to be automated reduces bias, manual computation time, and allows for large sample sizes to be processed. Ultimately, this leads to more precise data and can give insight into how other ECM molecules are spatially organized. This becomes important in pathological conditions where ECM has been disrupted. In such conditions, such as stroke or other neural tissue disruptions, hydrogels and biomaterials are often of interest, in conjunction with neural stem cells, to help regenerate the lost or damaged tissue.34,35
Despite being a major functional component of the brain, the normal distribution and localization of the ECM remains poorly defined. We here demonstrated the use of a semiautomated image analysis approach to reliably quantify different cell phenotypes and their association with TSP1. Although most neurons associated with TSP1 in both the striatum and the cortex, only about half and about one third of astrocytes associated with TSP1 in the striatum and the cortex, respectively. We additionally found that TSP1 occupies, histologically, little space in the striatum (~9%) and cortex (~7%). These findings both support previously reported TSP1 protein levels in the adult brain and quantify the amount of cellular association of TSP1 in the normal brain. This same approach can be applied to characterize the associations of other ECM molecules with cellular phenotypes. Improving the description and quantification of the spatial distribution of ECM molecules and neural phenotypes will increase our understanding of the structural and functional roles these molecules play in health, disease, and tissue repair.
Supplemental Material
Supplemental material, JHC771677_Supplemental_Material for Quantification of the Extracellular Matrix Molecule Thrombospondin 1 and Its Pericellular Association in the Brain Using a Semiautomated Computerized Approach by Jessie R. Liu and Michel Modo in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors thank Ms. Danielle Stark for the help with immunohistochemistry and Mr. Harmanvir Ghuman with staining of the thrombospondin 1 (TSP1) knockout tissues. We are grateful to Drs. David Roberts (NCI) and John Sipes (NCI) for providing wild-type and TSP1 knockout liver tissue for validation of the TSP1 antibody.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JRL performed perfusion of animals, cutting of histological slices, immunohistochemistry, image acquisitions, analysis pipeline development, graphing and analysis of data as well as drafted the manuscript. MM conceived of the study, provided funding, edited the manuscript, and provided supervision to the experimental procedures. JRL and MM approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was funded in part by National Institute of Neurological Disorders and Stroke (NINDS; R01NS08226) and National Institute of Biomedical Imaging and Bioengineering (NIBIB; R01EB016629).
Contributor Information
Jessie R. Liu, Department of Bioengineering, University of Pittsburgh, Pittsburgh, Pennsylvania
Michel Modo, Department of Radiology, McGowan Institute for Regenerative Medicine and Centre for the Neural Basis of Cognition, University of Pittsburgh, Pittsburgh, Pennsylvania.
Literature Cited
- 1. Bruckner G, Hartig W, Kacza J, Seeger J, Welt K, Brauer K. Extracellular matrix organization in various regions of rat brain grey matter. J Neurocytol. 1996;25:333–46. [DOI] [PubMed] [Google Scholar]
- 2. Sykova E. The extracellular space in the CNS: its regulation, volume and geometry in normal and pathological neuronal function. Neuroscientist. 1997;3:28–41. [Google Scholar]
- 3. Dityatev A, Seidenbecher CI, Schachner M. Compart-mentalization from the outside: the extracellular matrix and functional microdomains in the brain. Trends Neurosci. 2010;33:503–12. [DOI] [PubMed] [Google Scholar]
- 4. Marcoli M, Agnati LF, Benedetti F, Genedani S, Guidolin D, Ferraro L, Maura G, Fuxe K. On the role of the extracellular space on the holistic behavior of the brain. Rev Neurosci. 2015;26:489–506. [DOI] [PubMed] [Google Scholar]
- 5. Bonneh-Barkay D, Wiley CA. Brain extracellular matrix in neurodegeneration. Brain Pathol. 2009;19:573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haylock-Jacobs S, Keough MB, Lau L, Yong VW. Chondroitin sulphate proteoglycans: extracellular matrix proteins that regulate immunity of the central nervous system. Autoimmun Rev. 2011;10:766–72. [DOI] [PubMed] [Google Scholar]
- 7. Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–33. [DOI] [PubMed] [Google Scholar]
- 8. Costa C, Tortosa R, Domenech A, Vidal E, Pumarola M, Bassols A. Mapping of aggrecan, hyaluronic acid, heparan sulphate proteoglycans and aquaporin 4 in the central nervous system of the mouse. J Chem Neuroanat. 2007;33:111–23. [DOI] [PubMed] [Google Scholar]
- 9. Deepa SS, Carulli D, Galtrey C, Rhodes K, Fukuda J, Mikami T, Sugahara K, Fawcett JW. Composition of perineuronal net extracellular matrix in rat brain: a different disaccharide composition for the net-associated proteoglycans. J Biol Chem. 2006;281:17789–800. [DOI] [PubMed] [Google Scholar]
- 10. Giamanco KA, Matthews RT. Deconstructing the perineuronal net: cellular contributions and molecular composition of the neuronal extracellular matrix. Neuroscience. 2012;218:367–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kecskes S, Gaal B, Racz E, Birinyi A, Hunyadi A, Matesz C. Extracellular matrix molecules exhibit unique expression pattern in the climbing fiber-generating precerebellar nucleus, the inferior olive. Neuroscience. 2015;284:412–21. [DOI] [PubMed] [Google Scholar]
- 12. Kwok JC, Carulli D, Fawcett JW. In vitro modeling of perineuronal nets: hyaluronan synthase and link protein are necessary for their formation and integrity. J Neurochem. 2010;114:1447–59. [DOI] [PubMed] [Google Scholar]
- 13. Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 2008;3:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adams JC, Watt FM. Regulation of development and differentiation by the extracellular matrix. Development. 1993;117:1183–98. [DOI] [PubMed] [Google Scholar]
- 15. Lin TN, Kim GM, Chen JJ, Cheung WM, He YY, Hsu CY. Differential regulation of thrombospondin-1 and thrombospondin-2 after focal cerebral ischemia/reperfusion. Stroke. 2003;34:177–86. [DOI] [PubMed] [Google Scholar]
- 16. Roll L, Faissner A. Influence of the extracellular matrix on endogenous and transplanted stem cells after brain damage. Front Cell Neurosci. 2014;8:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eroglu C. The role of astrocyte-secreted matricellular proteins in central nervous system development and function. J Cell Commun Signal. 2009;3:167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liauw J, Hoang S, Choi M, Eroglu C, Choi M, Sun GH, Percy M, Wildman-Tobriner B, Bliss T, Guzman RG, Barres BA, Steinberg GK. Thrombospondins 1 and 2 are necessary for synaptic plasticity and functional recovery after stroke. J Cereb Blood Flow Metab. 2008;28:1722–32. [DOI] [PubMed] [Google Scholar]
- 19. Bjornsson CS, Lin G, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B. Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue. J Neurosci Methods. 2008;170:165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meruvia-Pastor OE, Soh J, Schmidt EJ, Boughner JC, Xiao M, Jamniczky HA, Hallgrimsson B, Sensen CW. Estimating cell count and distribution in labeled histological samples using incremental cell search. Int J Biomed Imaging. 2011;2011:874702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmitz C, Eastwood BS, Tappan SJ, Glaser JR, Peterson DA, Hof PR. Current automated 3D cell detection methods are not a suitable replacement for manual stereologic cell counting. Front Neuroanat. 2014;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin G, Chawla MK, Olson K, Barnes CA, Guzowski JF, Bjornsson C, Shain W, Roysam B. A multi-model approach to simultaneous segmentation and classification of heterogeneous populations of cell nuclei in 3D confocal microscope images. Cytometry A. 2007;71:724–36. [DOI] [PubMed] [Google Scholar]
- 23. Weaver CM, Hof PR, Wearne SL, Lindquist WB. Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Comput. 2004;16:1353–83. [DOI] [PubMed] [Google Scholar]
- 24. Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–5. [DOI] [PubMed] [Google Scholar]
- 25. Modo M, Beech JS, Meade TJ, Williams SC, Price J. A chronic 1 year assessment of MRI contrast agent-labelled neural stem cell transplants in stroke. Neuroimage. 2009;47(Suppl 2):T133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Durrans A, Gao D, Gupta R, Fischer KR, Choi H, El Rayes T, Ryu S, Nasar A, Spinelli CF, Andrews W, Elemento O, Nolan D, Stiles B, Rafii S, Narula N, Davuluri R, Altorki NK, Mittal V. Identification of reprogrammed myeloid cell transcriptomes in NSCLC. PLoS ONE. 2015;10:e0129123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Labrousse-Arias D, Castillo-Gonzalez R, Rogers NM, Torres-Capelli M, Barreira B, Aragones J, Cogolludo A, Isenberg JS, Calzada MJ. HIF-2alpha-mediated induction of pulmonary thrombospondin-1 contributes to hypoxia-driven vascular remodelling and vasoconstriction. Cardiovasc Res. 2016;109:115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mandel ER, Dunford EC, Trifonova A, Abdifarkosh G, Teich T, Riddell MC, Haas TL. Prazosin can prevent glucocorticoid mediated capillary rarefaction. PLoS ONE. 2016;11:e0166899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim SK, Hayashi H, Ishikawa T, Shibata K, Shigetomi E, Shinozaki Y, Inada H, Roh SE, Kim SJ, Lee G, Bae H, Moorhouse AJ, Mikoshiba K, Fukazawa Y, Koizumi S, Nabekura J. Cortical astrocytes rewire somatosensory cortical circuits for peripheral neuropathic pain. J Clin Invest. 2016;126:1983–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. International Organization for Standardization. Accuracy (trueness and precision) of measurement methods and results—part 1: general principles and definitions. ISO, Switzerland; 1994. [Google Scholar]
- 31. Zhang Y, Zhou X, Degterev A, Lipinski M, Adjeroh D, Yuan J, Wong ST. A novel tracing algorithm for high throughput imaging Screening of neuron-based assays. J Neurosci Methods. 2007;160:149–62. [DOI] [PubMed] [Google Scholar]
- 32. Ross JD, Cullen DK, Harris JP, LaPlaca MC, DeWeerth SP. A three-dimensional image processing program for accurate, rapid, and semi-automated segmentation of neuronal somata with dense neurite outgrowth. Front Neuroanat. 2015;9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bandeira F, Lent R, Herculano-Houzel S. Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat. Proc Natl Acad Sci U S A. 2009;106:14108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ghuman H, Massensini AR, Donnelly J, Kim SM, Medberry CJ, Badylak SF, Modo M. ECM hydrogel for the treatment of stroke: characterization of the host cell infiltrate. Biomaterials. 2016;91:166–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moshayedi P, Nih LR, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST. Systematic optimization of an engineered hydrogel allows for selective control of human neural stem cell survival and differentiation after transplantation in the stroke brain. Biomaterials. 2016;105:145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, JHC771677_Supplemental_Material for Quantification of the Extracellular Matrix Molecule Thrombospondin 1 and Its Pericellular Association in the Brain Using a Semiautomated Computerized Approach by Jessie R. Liu and Michel Modo in Journal of Histochemistry & Cytochemistry