Abstract
Phages (viruses that infect bacteria) play important roles in the gut ecosystem through infection of bacterial hosts, yet the gut virome remains poorly characterized. Mammalian gut viromes are dominated by double-stranded DNA (dsDNA) phages belonging to the order Caudovirales and single-stranded DNA (ssDNA) phages belonging to the family Microviridae. Since the relative proportion of each of these phage groups appears to correlate with age and health status in humans, it is critical to understand both ssDNA and dsDNA phages in the gut. Building upon prior research describing dsDNA viruses in the gut of Ciona robusta, a marine invertebrate model system used to study gut microbial interactions, this study investigated ssDNA phages found in the Ciona gut. We identified 258 Microviridae genomes, which were dominated by novel members of the Gokushovirinae subfamily, but also represented several proposed phylogenetic groups (Alpavirinae, Aravirinae, Group D, Parabacteroides prophages, and Pequeñovirus) and a novel group. Comparative analyses between Ciona specimens with full and cleared guts, as well as the surrounding water, indicated that Ciona retains a distinct and highly diverse community of ssDNA phages. This study significantly expands the known diversity within the Microviridae family and demonstrates the promise of Ciona as a model system for investigating their role in animal health.
Keywords: phage, ssDNA, Microviridae, virome, microbiome, gut, Ciona, invertebrate
1. Introduction
Recent studies of host-microbe interactions have recognized the importance of the holobiont, which acknowledges the complex partnerships between an animal and the entirety of its associated microbial communities [1]. The complex, dynamic relationship between an animal host and its microbial associates impacts many aspects of host physiology, including the procurement of nutrients and metabolic output [2,3]. In the nutrient- and mucus-rich gut of animals, the bacterial component of the microbiome can contribute orders of magnitude more gene products to host physiology than the host genome itself [4]. Although significant attention has been given to the metabolic contributions from the cellular component of the microbiome (i.e., bacteria, archaea, and fungi), it is now recognized that viruses also play important roles in the holobiont, both directly through infecting animal host cells, and indirectly through mediating microbiome dynamics.
Viruses present in gut communities are dominated by phages, which are viruses that infect bacteria [5,6,7,8,9,10,11]. The abundances of phages and bacteria are approximately equal in the human intestine [6,7,12], which contrasts with ratios observed in environmental samples where phages are typically ten times as abundant as bacteria [13]. Since phages can have dramatic influences on the structure and function of bacterial communities through host-specific lysis and horizontal gene transfer [13,14,15], phage-bacterial interactions appear crucial in all environments studied to date [15,16,17,18,19], including communities that are found in close association with animals [4,20,21,22,23,24]. While phage dynamics likely have dramatic influences on the physiology of the animal host, very little is known about the role of viromes (the cumulative viral community associated with a given host) or their impacts within the gut environment [6,8,9,10,25,26,27,28,29,30].
A variety of factors may influence the abundance, diversity, and ecological roles of phages in a complex ecosystem like the gut [31,32]. However, characterization of phage community composition is difficult due to a lack of universal gene markers within phage genomes [16]. Viral metagenomics, where the collective viral nucleic acids from a given sample are sequenced, is an efficient alternative approach for exploring the gut virome [5,12]. Although both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) phages have been identified in animal guts [27,33], the diversity of dsDNA phages remains the best characterized due to their large representation in culture collections and genomic sequence databases. In addition, standardly used library construction kits (e.g., the NexteraXT DNA library preparation kit, Illumina, San Diego, CA, USA) and viromic studies using linker-amplified shotgun sequencing approaches bias against ssDNA viruses [34]. However, the implementation of rolling circle amplification (RCA) to obtain enough DNA for sequencing has revealed a large diversity of genomes from ssDNA viruses in the guts of humans and other animals [6,7,12,33,35,36,37,38,39,40,41,42,43,44,45,46,47].
Studies investigating microbial communities in the human gut have shown that the microbiome structure seen in healthy newborns develops and changes dramatically within the first 2–3 years of life from a nearly sterile gut environment to a dynamic community maintained throughout adulthood [27]. Human gut phage communities are dominated by dsDNA phages from the order Caudovirales and members of the ssDNA phage family Microviridae [27,48,49]. Infant viromes appear to contain the highest phage richness at birth, which decreases with age [27,50]. While Caudovirales dominate in the earliest samples, the composition of the virome changes over time, with an increased relative abundance and richness of Microviridae by 2 years of age [27]. Gut phage communities often change in composition during disease states, such as inflammatory bowel disease, where the Microviridae:Caudovirales ratio decreases [51]. Despite the fact that several recent studies have highlighted phages belonging to the Microviridae as important gut virome components associated with an individual’s health state [12,27,33,37,51,52], little is known about the roles and dynamics of Microviridae in this complex ecosystem.
Studies leveraging simpler model systems to study the gut virome can enable hypothesis-driven experimental approaches to dissect these multifaceted biological and ecological processes. We have been developing a marine cosmopolitan sea squirt species, Ciona robusta (formerly Ciona intestinalis subtype A), in efforts to interrogate gut microbiome dynamics. This sessile, invertebrate chordate is a well-studied developmental model [53] with a sequenced genome [54]. Because Ciona is a filter feeding organism, its gut is a microcosm of microbial interactions that experience vast and continuous exposure to the large microbial and viral diversity found in seawater. Previous efforts have identified remarkable stability among some elements of the microbiome, which are distinct from surrounding seawater [20,55]. Ciona has structurally distinct gut compartments (stomach, midgut, hindgut) and maintains core bacterial species, some of which exhibit compartmentalization [20]. A recent effort characterizing the dsDNA Ciona gut virome revealed the dominance of tailed phages belonging to the order Caudovirales, as well as a diversity of eukaryotic dsDNA viruses. There was also evidence for compartmentalization of some components of the dsDNA virome, although to a lesser extent than seen for the corresponding bacterial communities [55]. Together, these findings suggest that strong selective pressures operate within the gut of this simple model organism. However, additional viral groups such as ssDNA viruses have yet to be characterized within Ciona, a finding that would support the use of this model system in studies to investigate the role of phages in animal guts.
The main objectives of the current study were to: (1) characterize the diversity of complete circular genomes related to the Microviridae, the most widely detected ssDNA phages among animals, from the Ciona gut, (2) evaluate if the identified ssDNA phages were unique to Ciona or if they were also found in water samples, and (3) determine if different gut compartments contained distinct ssDNA phage assemblages. We show that Ciona harbors a highly diverse community of ssDNA phages that is distinct from the water column and demonstrates some compartmentalization within the gut, but with the majority of genomes found in multiple gut compartments. The novel viral genomes reported here significantly broaden the known diversity of the Microviridae family, with a particular expansion in the subfamily Gokushovirinae and detection of a novel phylogenetic group.
2. Materials and Methods
2.1. Sample Collections and Library Preparations
Sequences described in this manuscript are derived from viromes generated by Leigh et al. (2018), which focused on analysis of dsDNA viral diversity. Animal guts were sampled from Ciona robusta harvested near San Diego, CA (M-Rep, Carlsbad, CA, USA). Ciona robusta is an invertebrate that is not regulated or protected by environmental agencies in the USA. M-Rep maintains a valid collection license through the state of California that allows them to ship to research institutions throughout the USA. Research at the University of South Florida was approved by biosafety protocols 1199 IA and 1351E. Upon sample arrival in Florida, ten animals were selected at random; five specimens were placed into virus-free 100 kD-filtered artificial seawater [56] to clear guts of dietary contents (water changed every 4 h for 24 h). These samples are referred to as ‘cleared guts.’ The remaining five animals were dissected with full gut contents. All animal guts, full (F) or cleared (C), were tri-sected (stomach (S), midgut (M), hindgut (H)) and snap-frozen in liquid nitrogen. Collected tissues from each gut type (n = 5) were disrupted in 3 mL of sterile suspension buffer using the GentleMACS dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Tissue fragments were pelleted (6000× g for 10 min), the supernatant was filtered through a 0.22 μm pore size Sterivex filter (Merck Millipore, Burlington, MA, USA), and the filtrate containing virus-like particles (VLPs) was collected. In addition to the viral fraction from Ciona guts, viruses from surrounding seawater in Mission Bay (MB) where the animals were collected, as well as the flow-through holding-tank water (CB), were processed to determine if viruses detected in Ciona guts could be detected in the surrounding water. For this purpose, one liter of seawater was filtered through a 0.22 μm pore size Sterivex filter and VLPs in the filtrate were concentrated to 1 mL using a 100 kDa Amicon centrifugal filter (EMD, Merck Millipore). VLPs were purified and further concentrated via cesium chloride (CsCl) gradient centrifugation [56] and collecting the 1.2–1.5 g/mL fraction with a sterile syringe and needle into a 2 mL sterile tube. To remove potential bacteria or extracellular vesicles still present in the sample, chloroform (final concentration 20% v/v) was added to the viral fraction and incubated at room temperature for 10 min. Samples were then centrifuged for 30 s at maximum speed (20,000× g) and the top aqueous layer was recovered. Unencapsidated, free nucleic acids were then removed by treating with DNase I (2.5 U/μL final concentration) for 3 h at 37 °C with frequent vortexing; the nuclease was inactivated by treating with 20 mM final concentration of EDTA pH 8.0. Purified VLP samples were tested to rule out bacterial contamination by PCR amplification of the 16S rRNA gene using primers 27F and 1492R [57] and epifluorescence microscopy. Viral DNA was then extracted from 200 µL of the viral concentrate using the Qiagen MinElute Virus Spin Kit (Qiagen, Inc., Valencia, CA, USA) and amplified via rolling circle amplification (RCA) using the GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences, Pittsburgh, PA, USA), resulting in ~1 μg DNA per sample. Three identical RCA reactions per sample were prepared and pooled for sequencing. Qubit (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine the DNA concentration and amplification was verified with 1% agarose gel electrophoresis. Final, amplified products were cleaned via the MinElute PCR Purification Kit (Qiagen, Inc.). DNA quality and quantity were assessed using the BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed on an Illumina MiSeq platform generating mate-pair (2 × 250 bp) libraries (Operon, Eurofins MWG Operon LLC, Huntsville, AL, USA). Sequences were analyzed in the CyVerse Cyberinfrastructure using different bioinformatics applications (Apps) [58]. Briefly, raw sequences were trimmed based on quality scores using the Trimmomatic App version 0.35.0 [59] and quality-filtered sequences were then assembled using the SPAdes App version 3.6.0 [60]. Contigs were screened with the VirSorter App to detect potential viral sequences. VirSorter outputs were uploaded to MetaVir [61] (IDs 7811 (SF), 8143 (SC), 7815 (MF), 7814 (MC), 7812 (HF), 7910 (HC), 7816 (CB), 7819 (MB)).
2.2. Identification and Annotation of Microviridae Genomes
Since members of the Microviridae family have circular genomes, circular contig sequences were identified using MetaVir [62]. Circular contig sequences ranging from 1 kb to 8 kb in length were compared (BLASTx, e-value < 0.0001) against a curated database of 4120 Microviridae major capsid protein (MCP) amino acid sequences (Jonathan Vincent and Francois Enault, Université Clermont Auvergne). Contig sequences with significant matches in the Microviridae MCP database were then compared (BLASTx, e-value < 0.001) against the Genbank non-redundant (nr) database to eliminate contig sequences that had better matches to cellular organisms (i.e., false positives). BLASTx outputs were explored using the MEGAN community edition software v6.8.9 [63] to identify sequences related to the Microviridae. Microviridae-related sequences were then manually trimmed to unit length genomes by identifying repeated sequences, and unit-length genomes were annotated using Geneious v10.1.3 [64]. For this purpose, open reading frames (ORFs) encoding putative proteins >80 amino acids (aa) were compared against the Genbank nr database using BLASTp (e-value < 0.001). Whenever possible, ORFs were annotated based on phage proteins (PHA source database) found in the NCBI conserved domain database with nomenclature following Cherwa and Fane [65]. All genomes were manually edited to begin at the start codon of the MCP. Genome-wide and MCP pairwise identities were calculated using the sequence demarcation tool (SDT) v1.2 [66]. Genomes were clustered at 95% nucleic acid identity based on current species demarcation cutoffs for phages [67]. Complete genome sequences are available under Genbank Accession Numbers MH572269–MH572526.
2.3. Genome Comparisons and Phylogenetic Analysis
Reference MCP amino acid sequences, including VP1 (subfamily Gokushovirinae) and Protein F (subfamily Bullavirinae), were collected from GenBank. These reference sequences also contained select sequences related to Microviridae that were identified from metagenomes [37,68,69,70,71,72,73,74,75,76] and those integrated into bacterial genomes [36] (see Supplementary Table S1 for details). Reference MCP sequences were aligned with sequences identified in Ciona guts using MUSCLE [77] as implemented in Geneious v10.1.3 [64]. A maximum likelihood tree was then created using PhyML with aLRT-like probabilities for branch support [78] and visualized with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Branches with probability values less than 0.70 were collapsed via TreeGraph 2 [79].
To evaluate the diversity of Microviridae in different Ciona gut compartments, a recruitment analysis was performed following the pipeline suggested for analysis of viral abundance and distribution through iVirus as implemented in the CyVerse Cyberinfrastructure [80]. For this purpose, Bowtiebatch v1.0.0 and Read2Ref v1.0.1 were used with default parameters to map reads from each gut compartment against the 258 Microviridae genomes, requiring reads to cover >75% of the genome to consider a genome present in a compartment [80]. A binary matrix (presence/absence) was used to assess the number of shared Microviridae genomes in the different compartments. This information was summarized using the Venn Diagram package [81] with community indexes visualized as a dendrogram using the Vegan package [82] created in R v3.3.2 [83]. The binary data was analyzed with three methods (Bray-Curtis index, Jaccard index, Euclidian dissimilarity), each resulting in congruent tree topologies. Note that the number of reads recruiting to a given Microviridae genome was not considered either in this study or in comparing these genomes to the dsDNA phages [55], since there are known biases created by RCA that lead to overrepresentation of ssDNA circular genomes [34,84,85].
3. Results
3.1. Diversity of Microviridae in Ciona Gut Compartments
Analysis of the Ciona gut viromes, which included six libraries representing viral sequences from three gut compartments (stomach, midgut, and hindgut) from cleared and full guts, revealed 488 circular contig sequences (1–8 kb in length) with BLAST similarity to members of the Microviridae. After clustering these genomes based on 95% genome-wide pairwise identity, a total of 258 genomes were identified and named with the prefix, Ciona gut microphage (CGM), followed by a number (Supplementary Table S2). The average size of the identified CGM genomes was ~4.3 kb with a range from 3.9 kb to 5.8 kb, which is consistent with previously described members of the family Microviridae [65,86]. Coverage of the genomes in the compartment from which they were originally assembled ranged from 2× to 1167×, with a mean coverage of 35× (Supplementary Table S2).
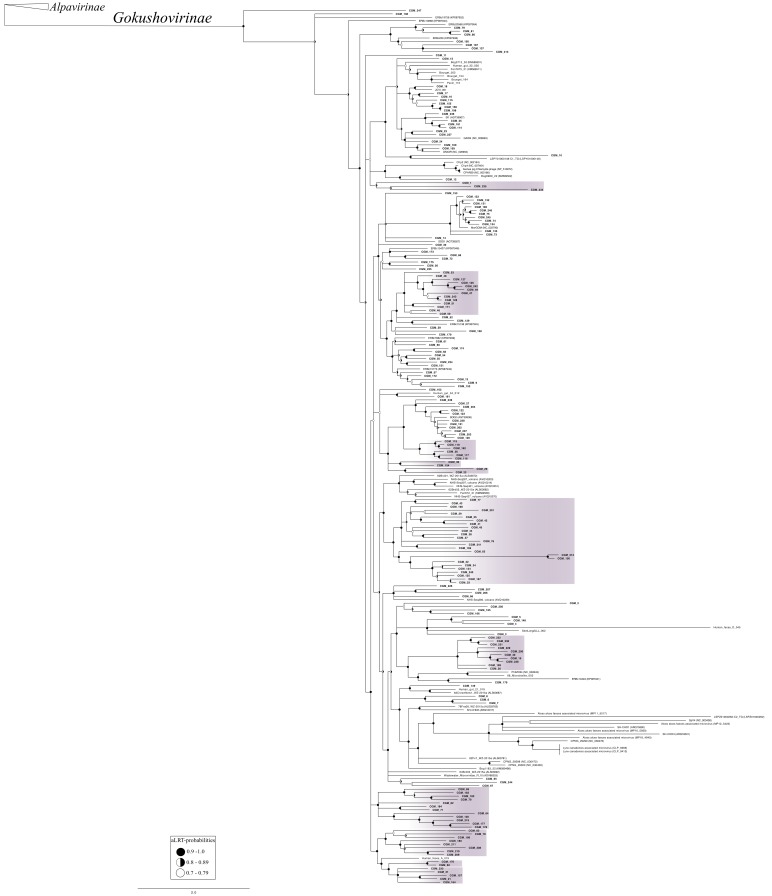
To assess diversity, each CGM genome was annotated and MCP amino acid sequences were used for phylogenetic analysis (Figure 1). Based on this analysis, the vast majority (n = 188) of CGM sequences grouped with the established subfamily Gokushovirinae [87], followed by sequences closely related to the proposed Group D microviruses (n = 33) [69] and the proposed subfamily Pichovirinae (n = 20) [73]. A smaller proportion of CGM sequences were related to Parabacteroides prophages (n = 7) [69,88], and members of the proposed Alpavirinae (n = 2) subfamily [36,37]. Six of the Ciona gut MCP sequences (CGM_251, CGM_223, CGM_252, CGM_222, CGM_249, CGM_250) form a distinct group from all previously described sequences and are referred to here as “Novel CGM Group”. None of the CGM MCP sequences grouped with the established Bullavirinae subfamily [87] or the proposed Aravirinae [68], Stokavirinae [68], or Sukshmavirinae [41] groups. However, two CGM sequences were most closely related to the pequeñoviruses, a proposed sister clade of the Bullavirinae [69]. Since 73% of the CGM genomes group within the Gokushovirinae subfamily, this subfamily is presented in a separate tree (Figure 2). Almost half (47%, n = 88) of the CGM MCP sequences within the Gokushovirinae subfamily do not group with previously reported sequences and share less than 70% aa identity with known Gokushovirinae MCP sequences.
Figure 1.
Maximum likelihood phylogenetic tree of predicted major capsid protein (MCP) sequences from the Ciona gut Microviridae (CGM, n = 258) along with representative sequences from previously proposed subfamilies (n = 96). The tree was created using PhyML with aLRT-probabilities; the scale bar represents the number of amino acid substitutions per site. Branches with probability values less than 0.7 were collapsed. Values greater than 0.7 are indicated at nodes. Suggested subfamily demarcations are delineated with dashed lines and colors based on previously classified sequences. Subfamilies for which CGM sequences were not identified are highlighted in grey color. Note: the Gokushovirinae sub-tree is displayed in Figure 2. Accession numbers for sequences used in this analysis are listed in Supplementary Tables S1 and S2.
Figure 2.
Maximum likelihood phylogenetic tree of predicted major capsid protein (MCP) sequences from the Ciona gut Microviridae (CGM, n = 188) that clustered within the established Gokushovirinae subfamily. MCP sequences representing Alpavirinae were used as an outgroup. The tree was created via PhyML with aLRT-probabilities; the scale bar represents the number of amino acid substitutions per site. Branches with probability values less than 0.7 were collapsed. Values greater than 0.7 are indicated at nodes. Clades highlighted in purple represent those where CGM sequences do not group with any previously described MCP sequences. Note: Accession numbers for sequences used in this analysis are listed in Supplementary Tables S1 and S2.
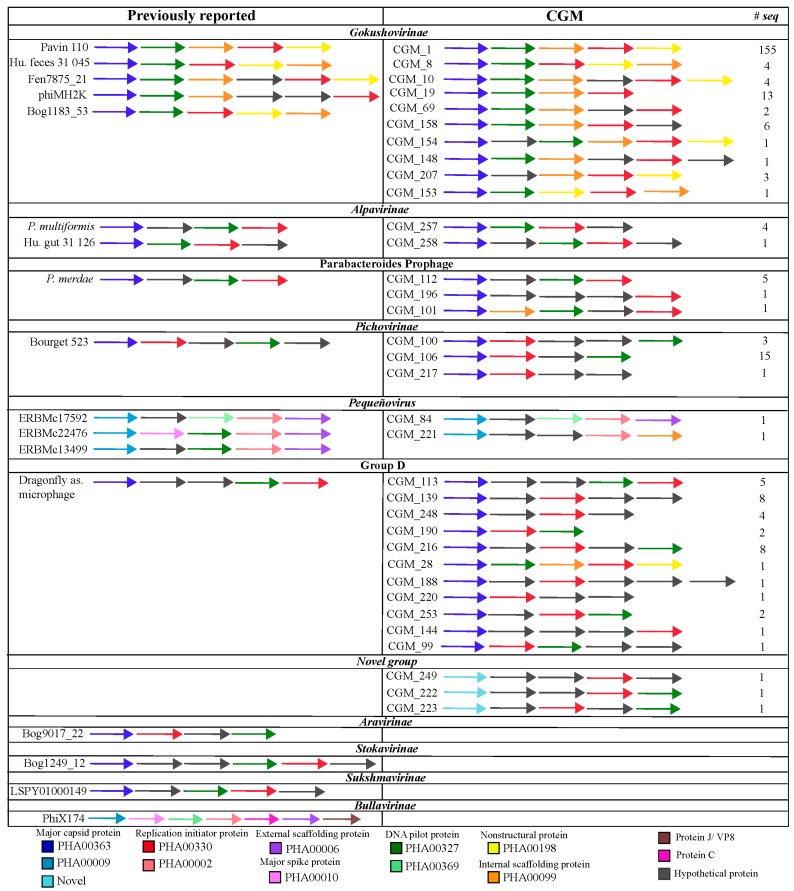
The gene synteny was compared between CGM genomes and previously reported Microviridae to evaluate if CGM genomes possess novel genome organizations (Figure 3). The CGM genomes expand upon previously identified gene organizations, yet many are syntenous with representative published sequences. The most diverse group, in terms of genome organization, was Group D, with 11 different patterns of gene organization.
Figure 3.
Gene synteny comparisons between previously described Microviridae genomes (left) and those discovered in the Ciona gut (right; CGM). All genomes were manually annotated to start at the major capsid protein (MCP) and open reading frames (ORFs) >80 aa are shown in linear fashion (i.e., overlapping genes are shown in order based on the position of the start codon). ORFs are color-coded based on PHA numbers (the phage protein subset of the Entrez protein cluster (PRK) database). One representative of each gene order known to exist within a given (proposed) subfamily is shown, and the numbers of CGM genomes containing a particular gene order are specified on the far right. The novel CGM group does not have representatives in the database, while the Aravirinae, Stokavirinae, Sukshmavirinae, and Bullavirinae were not detected among the CGM sequences. Details on the gene order for each CGM genome are available in Supplementary Table S2.
3.2. Structure of Microviridae Communities
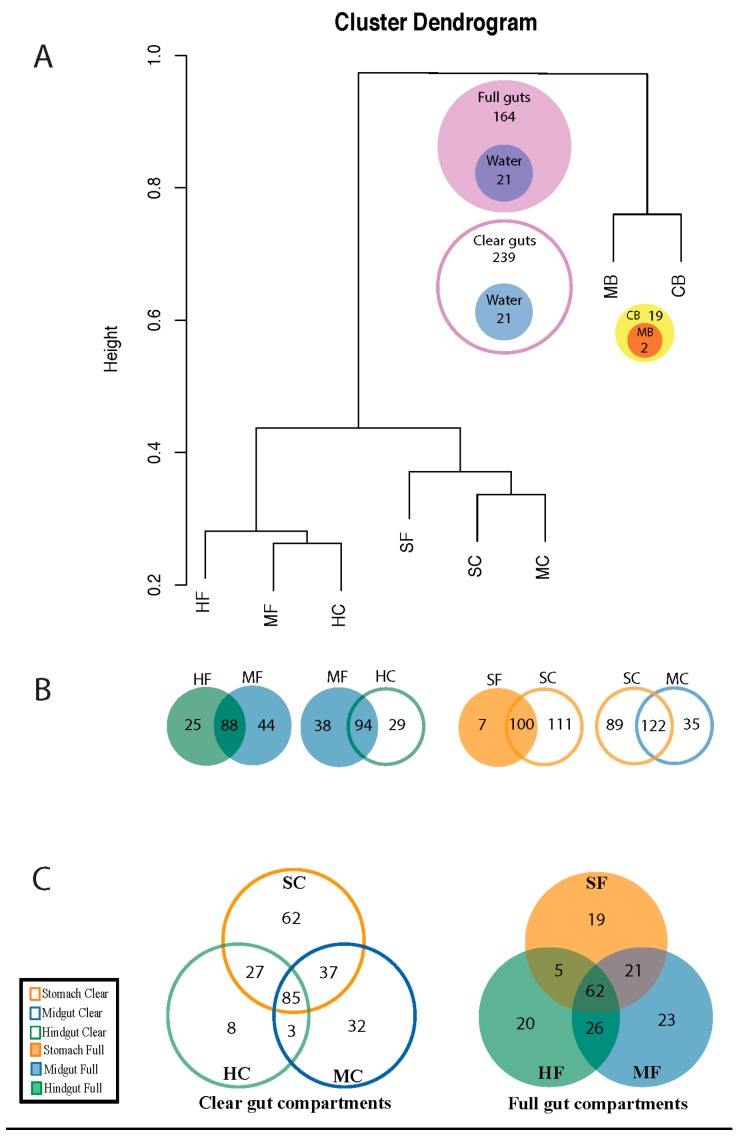
Richness of Microviridae genomes (i.e., the number of genomes that are <95% identical to each other) within gut compartments was compared among animals with either full or cleared guts. Stomach clear (SC) and midgut clear (MC) contained the highest number of genomes within the gut; these two compartments also share the largest number of genomes (Figure 4). Both water samples (MB & CB) group separately from the gut compartments (Figure 4), but all CGM sequences found in the water samples are also found throughout the gut compartments and all belong to the Gokushovirinae subgroup (Supplementary Table S2). Both the full and clear hindgut samples (HF and HC) share similarity with the midgut full (MF); however, the HF and MF share 88 genomes, the highest degree of overlap between the full gut compartments. The SC contained 211 Microviridae genomes; the largest number seen in any cleared gut compartment, while the HC had the lowest number of genomes (n = 123) of the cleared compartments, only 8 of which were unique to that compartment. Interestingly, despite being full of dietary material, the full gut compartments have a lower overall richness than the cleared ones. Four of the six sequences (CGM_251, CGM_223, CGM_250, CGM_257) belonging to the novel CGM group were only found in the hindgut full (Supplementary Table S2), and not seen in any other gut compartments. MF had the highest richness within the full gut, with a total of 132 genomes and 23 unique to that compartment. The lowest richness among the full compartments was found in the stomach, with 107 genomes and only 19 unique to SF.
Figure 4.
(A) Cluster dendrogram showing the relatedness among the CGM communities in the Ciona gut compartments and the surrounding water. (B) Venn diagrams showing comparisons between the closest groups on the dendrogram. (C) The three-way Venn diagrams specify shared and unique genomes detected in each of the compared groups. All diagrams were created based on the presence/absence of CGM genomes alone. The dendrogram was created using the Bray-Curtis dissimilarity index and the scale bar represents the dissimilarity values. Details on which CGM genomes were found in each compartment or water sample are available in Supplementary Table S2.
4. Discussion
Most microbiome research to date has focused on bacterial communities, with descriptions of the virome only recently gaining traction [25,27,89]. Understanding the gut virome is relevant to both the host and the cellular microbiome because viruses, whether infecting eukaryotic, bacterial or archaeal hosts, can have profound influences in shaping gut homeostasis [18,26,90]. Here, we described ssDNA phages found in the Ciona gut in an effort to further characterize the virome of this invertebrate model organism.
Recent applications of RCA in virome studies has dramatically increased our discovery of small, ssDNA viruses including phages belonging to the family Microviridae. These phages have now been described in a variety of habitats, including non-human animal guts [40,41,42,44,45,46,71,74,91], human guts [6,7,12,27,33,35,38,39,49,92,93,94], reclaimed water [95], sewage [96,97], fresh water systems [73,85,96,98,99,100], marine systems [24,100,101,102,103,104,105,106,107,108], methane seeps [70], modern stromatolites [109], confined aquifers [110], sediments [76,85,108,111,112,113], dragonflies [72], and fruit trees [114]. Despite this rapid increase in sequence information for ssDNA phages, their identification in such diverse environments has yet to reveal information about their hosts or functions. Now that a diversity of phages related to the Microviridae has been identified in Ciona, future experiments using this model organism can aim to define the role of these viruses in an animal gut.
In characterizing the diversity of Microviridae, we found 258 viral genomes within the Ciona gut, vastly outnumbering the total number of Microviridae genomes reported from any single animal gut study [6,33,35,37,38,41,42,74,94]. Although previous studies frequently identify sequences related to the Microviridae (most often belonging to the subfamily Gokushovirinae), detailed analysis of the genomes is rarely reported and contig sequences are often not available. Based on analysis of the MCP, which is typically used as a phylogenetic marker for this phage group [37,68,96,109], we found a variety of novel Microviridae groups. The CGM MCP sequence diversity encompassed 6 of the 10 established or proposed Microviridae groups in the literature, and a novel CGM clade was identified (Figure 1). Sequences belonging to the Gokushovirinae dominated in the Ciona gut, with 47% of the CGM gokushovirus MCP sequences forming clades without representatives from the literature (Figure 2). This finding adds to the growing body of literature stressing the predominance of gokushoviruses throughout numerous environments [12,33,35,37,38,70,98,105,107].
Interestingly, the only definitive hosts for Gokushovirinae are obligate parasitic or predatory bacteria [65] and laboratory isolates infect Bdellovibrio, Chlamydia, and Spiroplasma [86]. Although the hosts for the gokushoviruses identified in this study are not known, it should be noted that 16S ribosomal RNA gene data from a previous study of the Ciona gut [20] found representatives from each of these three known bacterial host groups. In addition, it is possible that gokushoviruses infect other types of bacteria with intracellular life stages. Other potential hosts for the gokushoviruses are the symbiotic bacteria belonging to the genus Endozoicomonas, which are dominant members of the core Ciona microbiome [20] and commonly associated with tunicates and other marine invertebrates from which gokushoviruses are frequently detected [24,115,116]. Finally, it is notable that Rickettsia were prevalent in the animals processed for this study [55], serving as another possible obligate intracellular bacterial host for the identified gokushoviruses.
Many of the CGMs were found to share a significant degree of synteny with previously observed members of the Microviridae (Figure 3). However, unique genome organizations were observed for several of the CGM genomes. The largest number of unique genome organizations was noted for Gokushovirinae and Group D viruses. The genomic features distinguishing the two established Microviridae subfamilies (Bullavirinae and Gokushovirinae), namely the MCP and scaffolding proteins, were conserved [65,86]. Based on these features, the vast majority of the CGM genomes are consistent with the Gokushovirinae subfamily, with the exception being the two CGM genomes grouping with the Bullavirinae sister clade, Pequeñovirus, which have an external scaffolding protein [70].
Surprisingly, the large diversity of Microviridae observed in the Ciona gut was mostly absent from the surrounding seawater. The MB and CB water samples only contained 2 and 19 ssDNA phage genomes, respectively, all of which were present in the Ciona gut. The CB water originates from the holding tanks where the animals are placed between field collection and shipping. Though the animals spend less than 8 h in these waters, they are passing water through their siphons, feeding, and releasing feces, which could potentially contribute to the increased number of CGM genomes seen in this sample compared to MB, where the animals were originally collected. It is possible that some Microviridae virions are too small to be captured on a 100 kDa filter; however, the more likely explanation for the lack of overlap between the water and animal gut samples is that these viruses are less prevalent in seawater than the Ciona guts, especially if their hosts are intracellular bacteria. Nearly all of the CGM genomes (237 viral genomes) were unique to the Ciona gut (Figure 4), which parallels previous findings that dsDNA phages from the Ciona gut were also significantly different from those in the water column [55]. The fact that gokushovirus richness was much higher in the Ciona gut than in surrounding marine environments suggests that Ciona could be concentrating hosts for these phages, providing a model for further studies into their host range and infection dynamics.
No significant correlation was noted between taxonomic classification of Microviridae and gut compartmentalization in this study; diverse environmental factors may influence the structure of these systems, but none appear to influence how these phages are dispersed. However, distinct viral signatures are still found among the stomach, midgut and hindgut compartments, which can inform us of similarity among these niches and may provide clues as to how some of these specific viruses and/or their hosts are distributed. For example, while a large number of phages are predominately found in the stomach, the midgut clear compartment is more closely related to the stomach (clear and full) while the midgut full more closely resembles the hindgut (clear and full) (Figure 4). These findings suggest that the midgut is likely an intermediate reservoir of phages and that some level of compartmentalization exists among a portion of the viral communities.
Clearing of animals is a process used to void the gut of dietary and fecal material, but this process is inherently stressful to a filter feeder because food is restricted from their diet. This stress could, in part, account for the higher number of diverse viral types recovered from the SC, if the clearing process liberates viruses from the mucosal lining of the gut that otherwise would be under-sampled when the gut is full of dietary material. Retention seems to vary from the stomach to the hindgut as the Microviridae richness diminishes towards the most distal areas of the gut. This trend is not seen within the full compartments, where the rapid transit of dietary and fecal material through the gut likely impacts the distribution and/or compartmentalization of some viruses. For example, laboratory feeding experiments performed with fluorescently tagged food particles and/or bacteria in the Dishaw lab have revealed food pellets exiting the animals within 45 to 60 min after feeding. This rapid transit is hypothesized to impact the stability of some of these niches and likely diminishes compartmentalization of viral communities. The stress of clearing could also induce prophages and Leigh et al. [55] hypothesized that an increased prevalence of temperate phages may be due to prophage induction caused by the stress of clearing. Although originally thought to be strictly lytic, Microviridae sequences have been discovered as prophages within the genomes of Bacteroidetes and Parabacteroides species common in the human mouth and gut [36,69]. Seven CGM genomes with MCPs most similar to the Parabacteroides prophages were identified, suggesting the possibility that temperate ssDNA phages exist in this system. Although these sequences are in the minority in regards to the overall CGM diversity, it supports prior evidence that lysogeny is common in the Ciona gut [55].
This single study of the Ciona gut revealed more complete Microviridae genomes than any environmental [68,69,70,96,98,104,105,106,107] or gut study [6,27,33,37,38,39,41,42,94,117] to date. Many prior studies examining animal guts have defined the structure of the viral community but have not probed the diversity of ssDNA phages present. Studies reporting ssDNA phage genomes have identified low diversity in comparison to the CGM communities. As an example, one study examining the feces of patients with coronary heart disease found 12 Gokushovirinae genomes and 2 Microviridae genomes that did not group with any known subfamily [35]. Another study focusing on the guts of termites found 12 Microviridae genomes, 2 of which were Gokushovirinae, 3 that did not group with any reference genomes, and 7 that they proposed to belong to a new subfamily Sukshmavirinae [41]. In comparison with prior studies, the remarkable Microviridae diversity (258 genomes) described in this study originate from the guts of only 10 Ciona individuals. These findings suggest that these phages, some of which may be infecting intracellular bacteria, likely infect hosts that are concentrated in or colonize the Ciona gut. As a filter-feeding organism that concentrates organic material from seawater, Ciona provides unique opportunities to explore questions about Microviridae within the gut environment. This is particularly true as the Ciona microbiome [20] can be manipulated and tightly controlled by rearing the animals germ-free [118]. Juvenile Ciona are small enough that dozens to hundreds of transparent juveniles can be reared on small tissue culture dishes, facilitating experimental manipulations. Therefore, the Ciona system affords many opportunities to address hypothesis-driven questions, possibly resulting in development of the first model for understanding the host range and biology of the Microviridae in animal guts.
Acknowledgments
The authors would like to thank Anni Djurhuus for assistance with R-code, Arvind Varsani for assistance with genome annotation, and Jonathan Vincent and Francois Enault for providing Microviridae MCP query sequences.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4915/10/8/404/s1, Table S1: Reference sequences used in constructing major capsid protein phylogenetic trees, Table S2: Details for each of the CGM phage genomes, including coverage, which compartment(s) they were detected in, and gene annotations.
Author Contributions
Conceptualization, K.R., L.J.D., M.B.; Methodology, A.C., K.R., B.A.L.; Formal Analysis, A.C. and K.R.; Resources, L.J.D. and M.B.; Writing-Original Draft Preparation, A.C.; Writing-Review & Editing, A.C., K.R., B.A.L., L.J.D., M.B.; Visualization, A.C.; Supervision, K.R., L.J.D., M.B.; Project Administration, L.J.D. and M.B.; Funding Acquisition, L.J.D. and M.B.
Funding
This research was funded by National Science Foundation grant numbers IOS-1456301 and DEB-1555854. The APC was funded by National Science Foundation grant number IOS-1456301. A.C. was funded by the University of South Florida College of Marine Science’s Endowed Southern Kingfish Association Fellowship and George Lorton Fellowship in Marine Science. B.A.L. was funded by National Science Foundation Graduate Research Fellowship award number 1144244 and the University of South Florida College of Marine Science’s Endowed Mahaffey Family Graduate Fellowship in Marine Science.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Theis K.R., Dheilly N.M., Klassen J.L., Brucker R.M., Baines J.F., Bosch T.C., Cryan J.F., Gilbert S.F., Goodnight C.J., Lloyd E.A. Getting the hologenome concept right: An eco-evolutionary framework for hosts and their microbiomes. mSystems. 2016;1:e00028-16. doi: 10.1128/mSystems.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordenstein S.R., Theis K.R. Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLoS Biol. 2015;13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitbart M., Hewson I., Felts B., Mahaffy J.M., Nulton J., Salamon P., Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F., Gordon J.I. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minot S., Sinha R., Chen J., Li H., Keilbaugh S.A., Wu G.D., Lewis J.D., Bushman F.D. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirzaei M.K., Maurice C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 9.Manrique P., Dills M., Young M.J. The human gut phage community and its implications for health and disease. Viruses. 2017;9:141. doi: 10.3390/v9060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manrique P., Bolduc B., Walk S.T., van der Oost J., de Vos W.M., Young M.J. Healthy human gut phageome. Proc. Natl. Acad. Sci. USA. 2016;113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yutin N., Makarova K.S., Gussow A.B., Krupovic M., Segall A., Edwards R.A., Koonin E.V. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 2018;3:38–46. doi: 10.1038/s41564-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M.-S., Park E.-J., Roh S.W., Bae J.-W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 2011;77:8062–8070. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suttle C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 14.Koskella B., Brockhurst M.A. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro O.H., Kushmaro A., Brenner A. Bacteriophage predation regulates microbial abundance and diversity in a full-scale bioreactor treating industrial wastewater. ISME J. 2010;4:327–336. doi: 10.1038/ismej.2009.118. [DOI] [PubMed] [Google Scholar]
- 16.Rohwer F., Thurber R.V. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 17.Breitbart M. Marine viruses: Truth or dare. Annu. Rev. Mar. Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 18.Scarpellini E., Ianiro G., Attili F., Bassanelli C., De Santis A., Gasbarrini A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015;47:1007–1012. doi: 10.1016/j.dld.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thurber R.V., Payet J.P., Thurber A.R., Correa A.M. Virus–host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 2017;15:205–216. doi: 10.1038/nrmicro.2016.176. [DOI] [PubMed] [Google Scholar]
- 20.Dishaw L.J., Flores-Torres J., Lax S., Gemayel K., Leigh B., Melillo D., Mueller M.G., Natale L., Zucchetti I., De Santis R., et al. The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS ONE. 2014;9:e93386. doi: 10.1371/journal.pone.0093386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders J.G., Beichman A.C., Roman J., Scott J.J., Emerson D., McCarthy J.J., Girguis P.R. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soverini M., Quercia S., Biancani B., Furlati S., Turroni S., Biagi E., Consolandi C., Peano C., Severgnini M., Rampelli S. The bottlenose dolphin (Tursiops truncatus) faecal microbiota. FEMS Microbiol. Ecol. 2016;92:fiw055. doi: 10.1093/femsec/fiw055. [DOI] [PubMed] [Google Scholar]
- 23.Isaacson R., Kim H.B. The intestinal microbiome of the pig. Anim. Health Res. Rev. 2012;13:100–109. doi: 10.1017/S1466252312000084. [DOI] [PubMed] [Google Scholar]
- 24.Laffy P.W., Wood-Charlson E.M., Turaev D., Jutz S., Pascelli C., Botté E.S., Bell S.C., Peirce T.E., Weynberg K.D., van Oppen M.J. Reef invertebrate viromics: Diversity, host specificity and functional capacity. Environ. Microbiol. 2018 doi: 10.1111/1462-2920.14110. [DOI] [PubMed] [Google Scholar]
- 25.Ogilvie L.A., Jones B.V. The human gut virome: A multifaceted majority. Front. Microbiol. 2015;6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carding S.R., Davis N., Hoyles L. The human intestinal virome in health and disease. Aliment. Pharmacol. Ther. 2017;46:800–815. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim E.S., Zhou Y., Zhao G., Bauer I.K., Droit L., Ndao I.M., Warner B.B., Tarr P.I., Wang D., Holtz L.R. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills S., Shanahan F., Stanton C., Hill C., Coffey A., Ross R.P. Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzger R.N., Krug A.B., Eisenächer K. Enteric virome sensing—Its role in intestinal homeostasis and immunity. Viruses. 2018;10:146. doi: 10.3390/v10040146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keen E.C., Dantas G. Close encounters of three kinds: Bacteriophages, commensal bacteria, and host immunity. Trends Microbiol. 2018;10 doi: 10.1016/j.tim.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Toole P.W., Jeffery I.B. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 33.Reyes A., Blanton L.V., Cao S., Zhao G., Manary M., Trehan I., Smith M.I., Wang D., Virgin H.W., Rohwer F., et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. USA. 2015;112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux S., Solonenko N.E., Dang V.T., Poulos B.T., Schwenck S.M., Goldsmith D.B., Coleman M.L., Breitbart M., Sullivan M.B. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ. 2016;4:e2777. doi: 10.7717/peerj.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo L., Hua X., Zhang W., Yang S., Shen Q., Hu H., Li J., Liu Z., Wang X., Wang H., et al. Viral metagenomics analysis of feces from coronary heart disease patients reveals the genetic diversity of the Microviridae. Virol. Sin. 2017;2:130–138. doi: 10.1007/s12250-016-3896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krupovic M., Forterre P. Microviridae goes temperate: Microvirus-related proviruses reside in the genomes of Bacteroidetes. PLoS ONE. 2011;6:e19893. doi: 10.1371/journal.pone.0019893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roux S., Krupovic M., Poulet A., Debroas D., Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS ONE. 2012;7:e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waller A.S., Yamada T., Kristensen D.M., Kultima J.R., Sunagawa S., Koonin E.V., Bork P. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–1402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes A., Wu M., McNulty N.P., Rohwer F.L., Gordon J.I. Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. USA. 2013;110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno P.S., Wagner J., Mansfield C.S., Stevens M., Gilkerson J.R., Kirkwood C.D. Characterisation of the canine faecal virome in healthy dogs and dogs with acute diarrhoea using shotgun metagenomics. PLoS ONE. 2017;12:e0178433. doi: 10.1371/journal.pone.0178433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tikhe C.V., Husseneder C. Metavirome sequencing of the termite gut reveals the presence of an unexplored bacteriophage community. Front. Microbiol. 2018;8:2548. doi: 10.3389/fmicb.2017.02548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’arc M., Furtado C., Siqueira J.D., Seuánez H.N., Ayouba A., Peeters M., Soares M.A. Assessment of the gorilla gut virome in association with natural simian immunodeficiency virus infection. Retrovirology. 2018;15:19. doi: 10.1186/s12977-018-0402-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Székely A.J., Breitbart M. Single-stranded DNA phages: From early molecular biology tools to recent revolutions in environmental microbiology. FEMS Microbiol. Lett. 2016;363:fnw027. doi: 10.1093/femsle/fnw027. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W., Li L., Deng X., Kapusinszky B., Pesavento P.A., Delwart E. Faecal virome of cats in an animal shelter. J. Gen. Virol. 2014;95:2553–2564. doi: 10.1099/vir.0.069674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smits S.L., Raj V.S., Oduber M.D., Schapendonk C.M., Bodewes R., Provacia L., Stittelaar K.J., Osterhaus A.D., Haagmans B.L. Metagenomic analysis of the ferret fecal viral flora. PLoS ONE. 2013;8:e71595. doi: 10.1371/journal.pone.0071595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Shan T., Wang C., Côté C., Kolman J., Onions D., Gulland F.M., Delwart E. The fecal viral flora of California sea lions. J. Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van den Brand J.M., van Leeuwen M., Schapendonk C.M., Simon J.H., Haagmans B.L., Osterhaus A.D., Smits S.L. Metagenomic analysis of the viral flora of pine marten and European badger feces. J. Virol. 2011;86:2360–2365. doi: 10.1128/JVI.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minot S., Bryson A., Chehoud C., Wu G.D., Lewis J.D., Bushman F.D. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breitbart M., Haynes M., Kelley S., Angly F., Edwards R.A., Felts B., Mahaffy J.M., Mueller J., Nulton J., Rayhawk S., et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Lim E.S., Wang D., Holtz L.R. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016;24:801–810. doi: 10.1016/j.tim.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Norman J., Handley S., Baldridge M., Droit L., Liu C., Keller B., Kambal A., Monaco C., Zhao G., Fleshner P., et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuo T., Wong S.H., Lam K., Lui R., Cheung K., Tang W., Ching J.Y., Chan P.K., Chan M.C., Wu J.C. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh N., Satou Y., Davidson B., Levine M. Ciona intestinalis: An emerging model for whole-genome analyses. Trends Genet. 2003;19:376–381. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 54.Dehal P., Satou Y., Campbell R., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 55.Leigh B.A., Djurhuus A., Breitbart M., Dishaw L.J. The gut virome of the protochordate model organism, Ciona intestinalis subtype A. Virus Res. 2018;244:137–146. doi: 10.1016/j.virusres.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 56.Thurber R.V., Haynes M., Breitbart M., Wegley L., Rohwer F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 57.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goff S.A., Vaughn M., McKay S., Lyons E., Stapleton A.E., Gessler D., Matasci N., Wang L., Hanlon M., Lenards A., et al. The iPlant collaborative: Cyberinfrastructure for plant biology. Front. Plant Sci. 2011;2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roux S., Tournayre J., Mahul A., Debroas D., Enault F. Metavir 2: New tools for viral metagenome comparison and assembled virome analysis. BMC Bioinform. 2014;15:76. doi: 10.1186/1471-2105-15-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roux S., Faubladier M., Mahul A., Paulhe N., Bernard A., Debroas D., Enault F. Metavir: A web server dedicated to virome analysis. Bioinformatics. 2011;27:3074–3075. doi: 10.1093/bioinformatics/btr519. [DOI] [PubMed] [Google Scholar]
- 63.Huson D.H., Beier S., Flade I., Górska A., El-Hadidi M., Mitra S., Ruscheweyh H.-J., Tappu R. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cherwa J.E., Fane B.A. eLS. John Wiley & Sons, Ltd.; Chichester, UK: 2001. Microviridae: Microviruses and Gokushoviruses. [DOI] [Google Scholar]
- 66.Muhire B.M., Varsani A., Martin D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adriaenssens E., Brister J.R. How to name and classify your phage: An informal guide. Viruses. 2017;9:70. doi: 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Labonté J.M., Suttle C.A. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 2013;7:2169–2177. doi: 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quaiser A., Dufresne A., Ballaud F., Roux S., Zivanovic Y., Colombet J., Sime-Ngando T., Francez A.-J. Diversity and comparative genomics of Microviridae in Sphagnum-dominated peatlands. Front. Microbiol. 2015;6:375. doi: 10.3389/fmicb.2015.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bryson S.J., Thurber A.R., Correa A., Orphan V.J., Vega Thurber R. A novel sister clade to the enterobacteria microviruses (family Microviridae) identified in methane seep sediments. Environ. Microbiol. 2015;17:3708–3721. doi: 10.1111/1462-2920.12758. [DOI] [PubMed] [Google Scholar]
- 71.Walters M., Bawuro M., Christopher A., Knight A., Kraberger S., Stainton D., Chapman H., Varsani A. Novel single-stranded DNA virus genomes recovered from chimpanzee feces sampled from the Mambilla Plateau in Nigeria. Genome Announc. 2017;5:e01715–e01716. doi: 10.1128/genomeA.01715-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosario K., Dayaram A., Marinov M., Ware J., Kraberger S., Stainton D., Breitbart M., Varsani A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta) J. Gen. Virol. 2012;93:2668–2681. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- 73.Roux S., Enault F., Robin A., Ravet V., Personnic S., Theil S., Colombet J., Sime-Ngando T., Debroas D. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS ONE. 2012;7:e33641. doi: 10.1371/journal.pone.0033641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kraberger S., Waits K., Ivan J., Newkirk E., VandeWoude S., Varsani A. Identification of circular single-stranded DNA viruses in faecal samples of Canada lynx (Lynx canadensis), moose (Alces alces) and snowshoe hare (Lepus americanus) inhabiting the Colorado San Juan Mountains. Infect. Genet. Evol. 2018;64 doi: 10.1016/j.meegid.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 75.Shu P., Butt A.M., Mi Z., Wang W., An X., Pei G., Zhang Z., Huang Y., Zhang X., Shi T. Identification and genomic analysis of a novel member of Microviridae, IME-16, through high-throughput sequencing. Virol. Sin. 2015;30:301–304. doi: 10.1007/s12250-015-3596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu D.-T., He J.-Z., Zhang L.-M., Han L.-L. Viral metagenomics analysis and eight novel viral genomes identified from the Dushanzi mud volcanic soil in Xinjiang, China. J. Soils Sediments. 2018 doi: 10.1007/s11368-018-2045-9. [DOI] [Google Scholar]
- 77.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lefort V., Longueville J.-E., Gascuel O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stöver B.C., Müller K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bolduc B., Youens-Clark K., Roux S., Hurwitz B.L., Sullivan M.B. iVirus: Facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 2017;11:7–14. doi: 10.1038/ismej.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen H., Boutros P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oksanen J., Blanchet F.G., Kindt R., Legendre P. Vegan: Community Ecology. 2013. R Package Version 1.18-28/r1569. [Google Scholar]
- 83.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 84.Kim K.-H., Bae J.-W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 2011;77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim K.-H., Chang H.-W., Nam Y.-D., Roh S.W., Kim M.-S., Sung Y., Jeon C.O., Oh H.-M., Bae J.-W. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doore S.M., Fane B.A. The Microviridae: Diversity, assembly, and experimental evolution. Virology. 2016;491:45–55. doi: 10.1016/j.virol.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 87.Lefkowitz E.J., Dempsey D.M., Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sakamoto M., Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 2006;56:1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 89.Reyes A., Semenkovich N.P., Whiteson K., Rohwer F., Gordon J.I. Going viral: Next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dahiya D., Renuka K. The gut virome: A neglected actor in colon cancer pathogenesis. Futur. Med. 2017;12:1345–1348. doi: 10.2217/fmb-2017-0159. [DOI] [PubMed] [Google Scholar]
- 91.Jørgensen T.S., Xu Z., Hansen M.A., Sørensen S.J., Hansen L.H. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS ONE. 2014;9:e87924. doi: 10.1371/journal.pone.0087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santiago-Rodriguez T.M., Fornaciari G., Luciani S., Dowd S.E., Toranzos G.A., Marota I., Cano R.J. Natural mummification of the human gut preserves bacteriophage DNA. FEMS Microbiol. Lett. 2015;363:fnv219. doi: 10.1093/femsle/fnv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCann A., Ryan F.J., Stockdale S.R., Dalmasso M., Blake T., Ryan C.A., Stanton C., Mills S., Ross P.R., Hill C. Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ. 2018;6:e4694. doi: 10.7717/peerj.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosario K., Nilsson C., Lim Y.W., Ruan Y., Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 96.Hopkins M., Kailasan S., Cohen A., Roux S., Tucker K.P., Shevenell A., Agbandje-McKenna M., Breitbart M. Diversity of environmental single-stranded DNA phages revealed by PCR amplification of the partial major capsid protein. ISME J. 2014;8:2093–2103. doi: 10.1038/ismej.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pearson V.M., Caudle S.B., Rokyta D.R. Viral recombination blurs taxonomic lines: Examination of single-stranded DNA viruses in a wastewater treatment plant. PeerJ. 2016;4:e2585. doi: 10.7717/peerj.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong X., Guidoni B., Jacas L., Jacquet S. Structure and diversity of ssDNA Microviridae viruses in two peri-alpine lakes (Annecy and Bourget, France) Res. Microbiol. 2015;166:644–654. doi: 10.1016/j.resmic.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 99.López-Bueno A., Tamames J., Velázquez D., Moya A., Quesada A., Alcamí A. High diversity of the viral community from an Antarctic lake. Science. 2009;326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 100.Kim Y., Van Bonn W., Aw T.G., Rose J.B. Aquarium viromes: Viromes of human-managed aquatic systems. Front. Microbiol. 2017;8:1231. doi: 10.3389/fmicb.2017.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Breitbart M., Salamon P., Andresen B., Mahaffy J.M., Segall A.M., Mead D., Azam F., Rohwer F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bench S.R., Hanson T.E., Williamson K.E., Ghosh D., Radosovich M., Wang K., Wommack K.E. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Labonté J., Suttle C. Metagenomic and whole-genome analysis reveals new lineages of gokushoviruses and biogeographic separation in the sea. Front. Microbiol. 2013;4:404. doi: 10.3389/fmicb.2013.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Labonté J.M., Hallam S.J., Suttle C.A. Previously unknown evolutionary groups dominate the ssDNA gokushoviruses in oxic and anoxic waters of a coastal marine environment. Front. Microbiol. 2015;6:315. doi: 10.3389/fmicb.2015.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tucker K.P., Parsons R., Symonds E.M., Breitbart M. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 2011;5:822–830. doi: 10.1038/ismej.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu D.-T., Han L.-L., Zhang L.-M., He J.-Z. Diversity and distribution characteristics of viruses in soils of a marine-terrestrial ecotone in East China. Microb. Ecol. 2018;75:375–386. doi: 10.1007/s00248-017-1049-0. [DOI] [PubMed] [Google Scholar]
- 108.Yoshida M., Mochizuki T., Urayama S.-I., Yoshida-Takashima Y., Nishi S., Hirai M., Nomaki H., Takaki Y., Nunoura T., Takai K. Quantitative viral community DNA analysis reveals the dominance of single-stranded DNA viruses in offshore upper bathyal sediment from Tohoku, Japan. Front. Microbiol. 2018;9:75. doi: 10.3389/fmicb.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Desnues C., Rodriguez-Brito B., Rayhawk S., Kelley S., Tran T., Haynes M., Liu H., Furlan M., Wegley L., Chau B., et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 110.Smith R.J., Jeffries T.C., Roudnew B., Seymour J.R., Fitch A.J., Simons K.L., Speck P.G., Newton K., Brown M.H., Mitchell J.G. Confined aquifers as viral reservoirs. Environ. Microbiol. Rep. 2013;5:725–730. doi: 10.1111/1758-2229.12072. [DOI] [PubMed] [Google Scholar]
- 111.Yoshida M., Takaki Y., Eitoku M., Nunoura T., Takai K. Metagenomic analysis of viral communities in (hado) pelagic sediments. PLoS ONE. 2013;8:e57271. doi: 10.1371/journal.pone.0057271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han L.-L., Yu D.-T., Zhang L.-M., Shen J.-P., He J.-Z. Genetic and functional diversity of ubiquitous DNA viruses in selected Chinese agricultural soils. Sci. Rep. 2017;7:45142. doi: 10.1038/srep45142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reavy B., Swanson M.M., Cock P.J., Dawson L., Freitag T.E., Singh B.K., Torrance L., Mushegian A.R., Taliansky M. Distinct circular single-stranded DNA viruses exist in different soil types. Appl. Environ. Microbiol. 2015;81:3934–3945. doi: 10.1128/AEM.03878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Basso M.F., da Silva J.C.F., Fajardo T.V.M., Fontes E.P.B., Zerbini F.M. A novel, highly divergent ssDNA virus identified in Brazil infecting apple, pear and grapevine. Virus Res. 2015;210:27–33. doi: 10.1016/j.virusres.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 115.Neave M.J., Apprill A., Ferrier-Pagès C., Voolstra C.R. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 2016;100:8315–8324. doi: 10.1007/s00253-016-7777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schreiber L., Kjeldsen K.U., Funch P., Jensen J., Obst M., López-Legentil S., Schramm A. Endozoicomonas are specific, facultative symbionts of sea squirts. Front. Microbiol. 2016;7:1042. doi: 10.3389/fmicb.2016.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krupovic M., Prangishvili D., Hendrix R.W., Bamford D.H. Genomics of bacterial and archaeal viruses: Dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leigh B.A., Liberti A., Dishaw L.J. Generation of germ-free Ciona intestinalis for studies of gut-microbe interactions. Front. Microbiol. 2016;7:2092. doi: 10.3389/fmicb.2016.02092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.