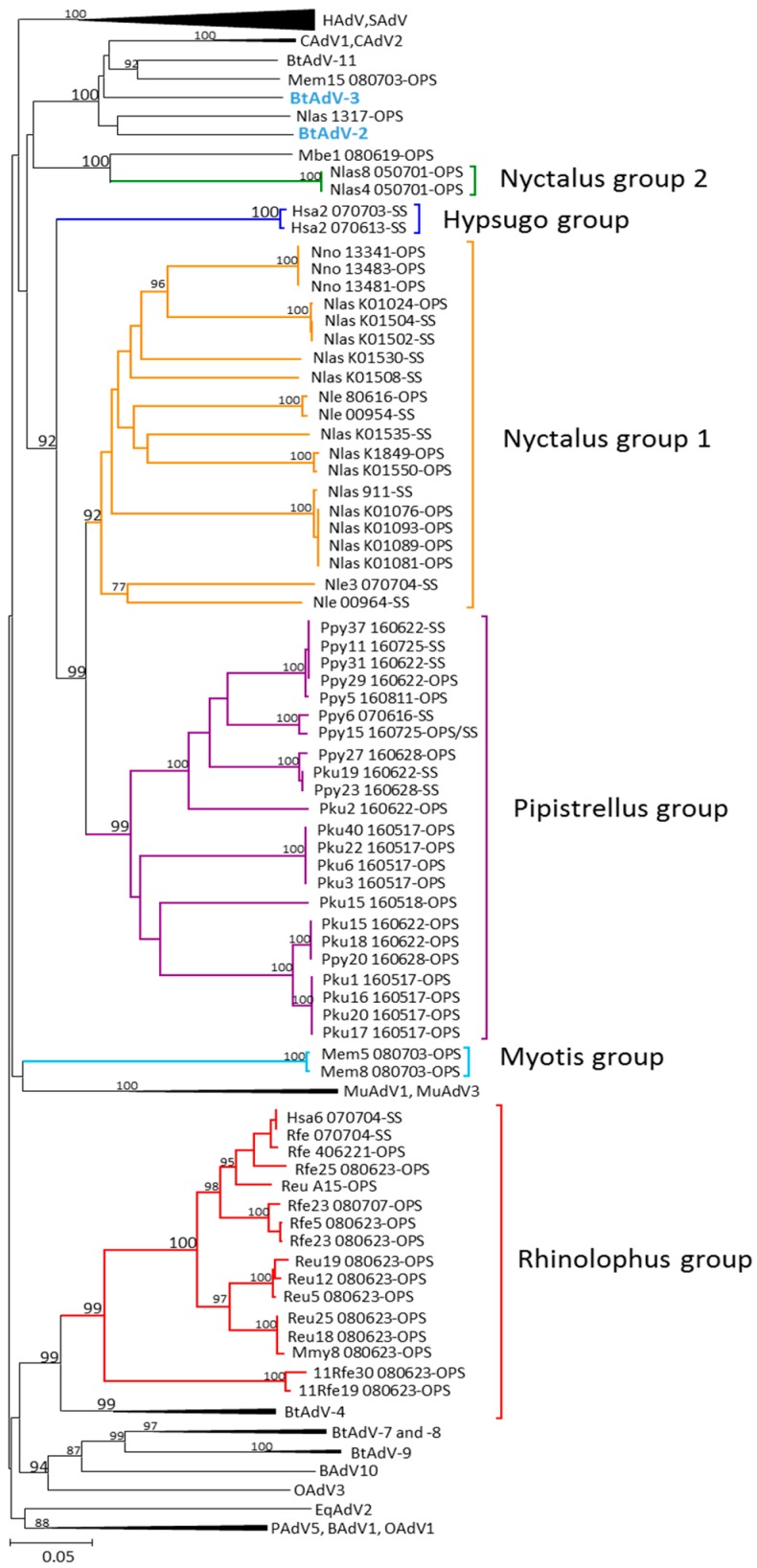
Figure 2.
Phylogenetic tree based on the analysis of the hexon partial gene. Trees were estimated with MEGA 5.2 software by using the neighbour-joining method on Tamura 3 parameters model. A bootstrap test was replicated for 5000 times. Numbers represent percentage bootstrap support. GenBank accession numbers for the sequences included in the tree are as follows: BtAdV-3 (strain TJM, GU226970, Bat mastadenovirus A), BtAdV-2 (strain PPV1, JN252129, Bat mastadenovirus B), human AdVs: type 1 (AF534906), type 2 (J01917), type 3 (DQ086466), type 4 (AY594254), D8 strain Ger/Berlin/04_2003 (KT862545), type 9 (AJ854486), type 12 (X73487), type 14 (FJ841902), type 16 (X74662), type 21 (KF528688), type 24 (JN226751), type 27 (JN226753), type 42 (JN226761), type 45 (JN226764), simian AdVs: type 1 (AY771780), type 4 (KP853121), ovine AdVs: type 1 (DQ630754), type 3 strain (DQ630756), porcine AdV 5 (AF289262), murine AdVs: type 1 (M81889), type 3 (EU835513), bovine AdVs: type 1 (DQ630761), type 10 (AF282774), canine AdVs: type 1 (KX545420), type 2 (U77082), equine AdV type 2 (L80007), bat mastadenovirus: BtAdV-7 (strain WIV12, KT698856, Bat mastadenovirus D), BtAdV-8 (strainWIV13, KT698852, Bat mastadenovirus E), BtAdV-9 (strain WIV17, KX961095, Bat mastadenovirus F), Rousettus leschenaultii WIV18 (NC_035072), BtAdV-4 (strain WIV9, KT698853, Bat mastadenovirus C), Rhinolophus sinicus WIV10 (NC_029899), R. sinicus WIV11 (NC_029902), BtAdV-11 (strain 250-A, KX871230, Bat mastadenovirus G).