Abstract
Leucine can affect intestinal protein expressions, and improve mucosal immune function. However, little study has been conducted to determine the change of protein component by leucine treatment in intestine epithelial cells. The present study was to cover the key proteins and cell pathways that could be regulated by leucine treatment in porcine intestinal epithelial cell line (IPEC-J2) cells with the approach of proteome analysis. A total number of 3,211 proteins were identified in our approach by searching the database of Uniprot sus scrofa. Among identified proteins, there were 101 proteins expressed differently between control group and leucine group. Compared with the control group, there were 50 up-regulated proteins and 51 down-regulated proteins in leucine group. In these proteins, leucine treatment decreased the expression of some proteins including pyruvate kinase, glyceraldehyde-3-phosphate dehydrogenase, E3 ubiquitin ligase, cathepsin D, caspase 3 and caspase 6, and increased the levels of some proteins, such as some eukaryotic translation initiation factors, ubiquitin carboxyl-terminal hydrolase, DNA-related RNA polymerase II, urokinase plasminogen activator, cyclin-dependent kinase inhibitor 2b, MutL homolog 1, 5-methylcytosine binding domain 4, polymerase δ, α-tubulin, syntaxin 18, Ras homolog D, actin related protein 2/3 complex and cofilin. Via the analysis of Gene Ontology and pathways, these proteins in IPEC-J2 cells were related with some physiological functions, such as protein metabolism, glycolysis, cell proliferation, apoptosis and phagocytosis. Thus, these results suggest that leucine affects gut barrier function possibly via regulating cell proliferation and apoptosis, metabolism and phagocytosis.
Keywords: Leucine, IPEC-J2 cells, Proteome, Phagocytosis, Cell proliferation, Apoptosis and metabolism
1. Introduction
As a functional amino acid, leucine may regulate many important physiological functions, including protein and energy metabolism, cell proliferation and apoptosis, in different tissues and cells (Mao et al., 2013a, Zeanandin et al., 2012, Coëffier et al., 2011, Xiao et al., 2016, Toneto et al., 2016). Recently, the effect of leucine on gut function has gradually been focused (Zhang et al., 2014, Ren et al., 2016, Goichon et al., 2013). In our recent studies, leucine treatment can increase the expression of some specific proteins in LS174T cells (Mao et al., 2016), and dietary leucine supplementation can improve intestinal morphology, and alleviate the mucosal dysfunction of weaned pigs challenged by porcine rotavirus (PRV) (Mao et al., 2015).
In mammalian tissues and cells, leucine not only acts as the supply of substrate and energy for functional protein synthesis, and it also regulates some physiological functions by affecting intracellular signaling pathways, such as mammalian target of rapamycin (mTOR), adenosine 5′-monophosphate-activated protein kinase (AMPK) and extracellular regulated protein kinases (ERK) signaling pathway (Zhang et al., 2014, Ren et al., 2016, Dodd and Tee, 2012, Mao et al., 2011). The previous studies have shown that, by the proteomic or phospho-proteomic approach with two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), leucine supplementation regulates energy metabolism in human duodenal mucosa, and leucine deprivation affects the phosphorylation of translation-relative factors in muscle cells (Goichon et al., 2013, Talvas et al., 2008). However, these cannot completely explain our previous results that leucine supplementation can alleviate the mucosal dysfunction of weaned pigs challenged by PRV.
The porcine intestinal epithelial cell line (IPEC-J2), as a non-transformed intestinal cell line originally derived from jejunal epithelium of the neonatal piglet (Schierack et al., 2006), maintains some native characteristics including the expression of structural and functional proteins (Mao et al., 2013b, Zeng et al., 2013). The IPEC-J2 cells have often been utilized as the in vitro model to investigate the effect of nutrients on the intestinal function.
Therefore, the present study was conducted to determine the key proteins and cell pathways related to leucine treatment in IPEC-J2 cells through the proteomic approach with isobaric tags for relative and absolute quantitation (iTRAQ).
2. Material and methods
2.1. Cell culture and treatment
The porcine intestinal epithelial cell line (originally derived from jejunal epithelium of the neonatal piglet) was kindly provided by Professor Guoyao Wu (Texas A&M University, USA). Cell monolayers were cultured in plates at a humidified atmosphere with 5% CO2 at 37 °C. Standard media for feeding cells consisted of Dulbecco's Modified Eagle's Medium/F12 (DMEM/F12, Hyclone Laboratories Inc., Logan, UT, USA), 5% fetal calf serum (Gibco Laboratories Life Technologies Inc., Grand Island, NY, USA), 1% insulin/transferrin/sodium selenite (Invitrogen Corp., Carlsbad, CA, USA), 5 ng/mL epidermal growth factor (Invitrogen Corp., Carlsbad, CA, USA), and antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin; Hyclone Laboratories Inc., Logan, UT, USA). After cells were grown to 90% confluence, they were starved for 12 h in DMEM/F12 media. Then, cells were treated with 5 mmol/L L-leucine for 2 h, and then collected for proteomics assay.
2.2. Proteomics assays of IPEC-J2 cells
Protein extraction in IPEC-J2 cells was conducted as described previously with some modification (Zi et al., 2013). The collected IPEC-J2 cells were dissolved in the lysis buffer (8 mol/L urea, 30 mmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 1 mmol/L phenylmethanesulfonyl fluoride, 2 mmol/L ethylenediaminetetraacetic acid and 10 mmol/L DL-dithiothreitol [DTT]) with sonication. Following centrifugation at 20,000 × g for 30 min, the protein in the supernatant was reduced (10 mmol/L DTT, 56 °C for 60 min), alkylated (55 mmol/L iodoacetamide, room temperature for 45 min), and precipitated by precooled acetone at −20 °C for 30 min. After centrifugation at 20,000 × g for 30 min, the pellet was dissolved in 0.5 mol/L triethylamine borane buffer with 0.1% sodium dodecyl sulfate with sonication, and then centrifuged at 20,000 × g for 30 min. The supernatant was used for digestion. The protein concentration was determined by the Bradford assay.
Protein digestion, iTRAQ labeling, and labeled peptide analysis of strong cation exchange fractionation and reverse-phase nano-liquid chromatography/tandem mass spectrometer were conducted as described previously with some modification (Zi et al., 2013).
For protein identification, the data files of each fraction were combined together to perform searching against Uniprot sus scrofa protein database. Only unique peptides were contained for iTRAQ labeling quantification, and peptides with global false discovery rate values from fit less than 1% were considered for further analysis. And then, bioinformatics analysis (gene ontology [GO] and Kyoto Encyclopedia of Genes and Genomes [KEGG]) was conducted as described previously (Qin et al., 2013).
2.3. Western blot analysis
The antibody against pyruvate kinase (PYK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), E3 ubiquitin ligase (ITCH), cathepsin D, caspase 3, eukaryotic translation initiation factor 4A (eIF4A), ubiquitin carboxyl-terminal hydrolase (USP19), urokinase plasminogen activator (PLAU), 5-methylcytosine binding domain 4 (MBD4), α-tubulin (TUBA), Ras homolog D (RhoD), cofilin and β-actin were purchased from Cell Signaling (Davers, MA, USA), Abcam (Cambridge, MA, USA), Invitrogen Corp. (Carlsbad, CA, USA), LifeSpan BioSciences (Seattle, WA, USA) and Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA), respectively. Protein levels for the PYK, GAPDH, ITCH, cathepsin D, caspase 3, eIF4A, USP19, PLAU, MBD4, TUBA, RhoD, cofilin and β-actin in IPEC-J2 cells were determined by Western Blot analysis as described previously (Mao et al., 2011, Mao et al., 2017).
2.4. Statistical analysis
Within the iTRAQ analysis, differentially expressed proteins were determined based on the ratios of differently labeled proteins and P-values provided by Protein Pilot. Compared with the control group, proteins with values of P < 0.05 and changes of ≥1.2-fold were considered as significant differences. The data of Western Blot analysis were analyzed with unpaired t-test by SAS (version 8.1; SAS institute, Cary, NC, USA), and were indicated as means with their standard errors. P < 0.05 was considered statistically significant.
3. Results
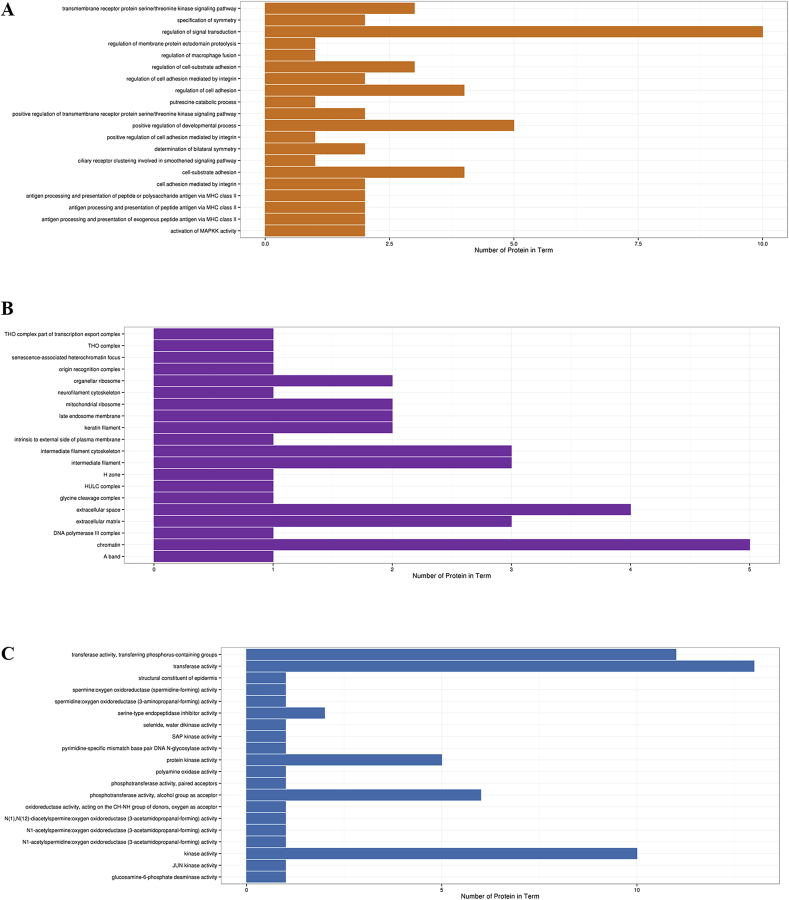
A total number of 3,211 proteins were identified and quantified in this study by searching the database of Uniprot sus scrofa. Among these identified proteins, there were 101 proteins shown ≥1.2-fold difference between control and leucine group (P < 0.05). Compared with control group, there were 50 up-regulated proteins and 51 down-regulated proteins in leucine group. Based on the GO enrichment analysis, the differentially expressed proteins in IPEC-J2 cells were mainly involved into 15 biological processes, 9 cellular components, and 6 molecular functions (Fig. 1).
Fig. 1.
Gene ontology (GO) classification of the differentially expressed proteins related (A) biological process, (B) cellular component and (C) molecular function in IPEC-J2 cells treated by leucine.
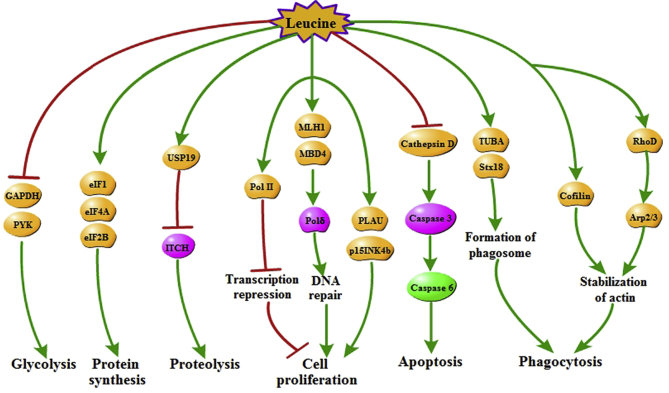
In addition, on the basis of pathways (KEGG) analysis, compared with the control group, a total of 21 differentially expressed pathway proteins showed ≥ 1.2-fold changes in leucine group (P < 0.05). More concretely, L-leucine treatment inhibited the expressions of PYK, GAPDH, ITCH, cathepsin D, caspase 3 and caspase 6, and increased the levels of proteins including some eukaryotic translation initiation factors (eIF1, eIF4A and eIF2B), USP19, DNA-related RNA polymerase II (Pol II), PLAU, cyclin-dependent kinase inhibitor 2b (p15INK4b), MutL homolog 1 (MLH1), MBD4, polymerase δ (Pol δ), TUBA, syntaxin 18 (Stx18), RhoD, actin related protein 2/3 complex (Arp2/3), and cofilin in IPEC-J2 cells. These proteins were primarily related to some physiological functions, such as cell proliferation, protein synthesis, proteolysis, glycolysis, apoptosis and phagocytosis (Fig. 2).
Fig. 2.
Analysis of cell signaling pathways regulated by leucine treatment in IPEC-J2 cells were summarized from Kyoto Encyclopedia of Genes and Genomes database. PYK = pyruvate kinase; GAPDH = glyceraldehyde-3-phosphate dehydrogenase; ITCH = E3 ubiquitin ligase; eIF1 = eukaryotic translation initiation factor 1; eIF4A = eukaryotic translation initiation factor 4A; eIF2B = eukaryotic translation initiation factor 2B; USP19 = ubiquitin carboxyl-terminal hydrolase; Pol II = DNA-related RNA polymerase II; PLAU = urokinase plasminogen activator; p15INK4b = cyclin-dependent kinase inhibitor 2b; MLH1 = MutL homolog 1; MBD4 = 5-methylcytosine binding domain 4; Pol δ = polymerase δ; TUBA = α-tubulin; Stx18 = syntaxin 18; RhoD = Ras homolog D; Arp2/3 = actin related protein 2/3 complex.
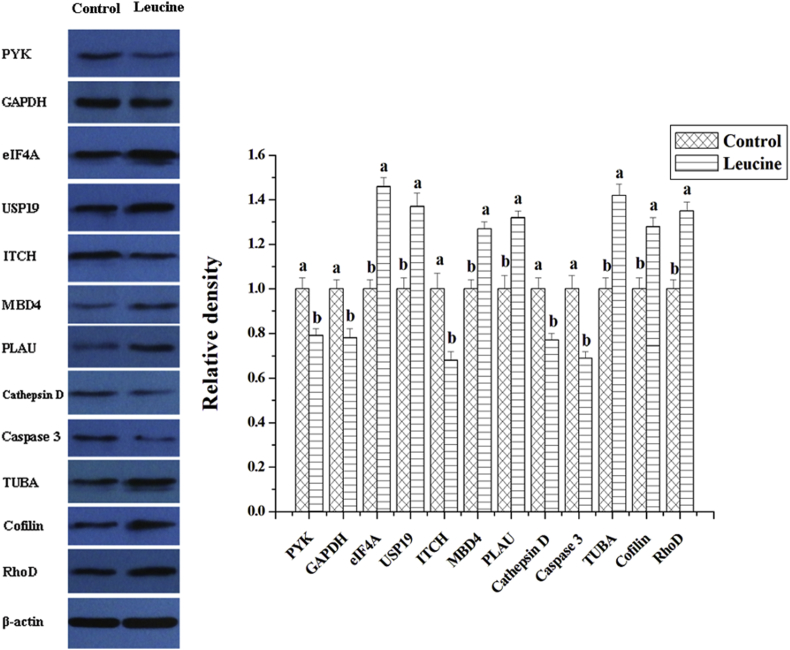
By Western Blot analysis, we measured the expressions of some key proteins that were affected by L-leucine treatment in IPEC-J2 cells. Following L-leucine treatment, the expressions of PYK, GAPDH, ITCH, cathepsin D and caspase 3 were inhibited (P < 0.05, Fig. 3), and the expressions of eIF4A, USP19, MBD4, PLAU, TUBA, cofilin and RhoD were stimulated (P < 0.05, Fig. 3).
Fig. 3.
The expressions of some key proteins affected by L-leucine treatment in IPEC-J2 cells. Representative western blots for these proteins in IPEC-J2 cells were shown. Results were expressed as the amount of these proteins to β-actin in each treatment as a ratio of L-leucine treatment to control. Values are means ± SE; n = 6. Values with different letters are significantly different (P < 0.05). PYK = pyruvate kinase; GAPDH = glyceraldehyde-3-phosphate dehydrogenase; ITCH = E3 ubiquitin ligase; eIF4A = eukaryotic translation initiation factor 4A; USP19 = ubiquitin carboxyl-terminal hydrolase; PLAU = urokinase plasminogen activator; MBD4 = 5-methylcytosine binding domain 4; TUBA = α-tubulin; RhoD = Ras homolog D.
4. Discussion
Intestinal epithelial cells may not only absorb kinds of nutrients, but also prevent the incursion of gut luminal hostile environment, including pathogens, as the first defense line of body. Thus, the functions of epithelial cells are critical for the health of animal and human. However, leucine could affect the gut function. Our previous studies have shown that leucine may alleviate the rotavirus-induced diarrhea via improving the intestinal mucosal barrier in piglets, and regulate the production of specific proteins, such as mucin 2, through stimulating phosphoinositide 3-kinase-protein kinase B-mTOR (PI3K-Akt-mTOR) signaling pathway in piglets and human colon epithelial cells (Mao et al., 2015, Mao et al., 2016). In addition, leucine treatment can also regulate the fatty acid β-oxidation in human duodenal mucosa (Goichon et al., 2013), increase the mRNA relative expression of several defensins in IPEC-J2 cells (Ren et al., 2016), and improve the ASCT2 amino acid transporter expression in IPEC-J2 cells via regulating PI3K-Akt-mTOR and ERK signaling pathways (Zhang et al., 2014). The present study showed that, in IPEC-J2 cells, leucine could affect the expression of 21 proteins that were related to cell proliferation, apoptosis, protein metabolism, glycolysis and phagocytosis by using proteome analysis.
Many studies have shown that leucine treatment can increase the protein synthesis in different tissues and cells, which could be derived from stimulation of the intracellular translation initiation and elongation via regulating eIF4E, eIF2α, eIF2B and eEF2 (Dodd and Tee, 2012, Pedroso et al., 2015, Suryawan et al., 2011). However, leucine deprivation also decreases the phosphorylation of eIF4B in C2C12 myoblasts by the phospho-proteomic analysis (Talvas et al., 2008). In addition, eIF1 plays a central role in ensuring the fidelity of codon selection start (Ivanov et al., 2010), and eIF2B maintains eIF2 recycling and translation initiation rates (Jennings and Pavitt, 2014), and eIF4A, the component of eIF4F, plays a role in delivery of RNA helicase to the 5′ region (Gingras et al., 1999). These factors directly regulate the translation initiation of protein synthesis. Through the proteome analysis, this study also showed that leucine treatment increased eIF1, eIF2B, and eIF4A expression in IPEC-J2 cells.
Besides protein synthesis, leucine treatment can also decrease the proteolysis in tissues and cells, especially skeletal muscles, which was mainly regulated by ubiquitin-proteasome signaling pathway (Nakashima et al., 2005, Sadiq et al., 2007). Ubiquitin carboxyl-terminal hydrolase, known as a kind of ubiquitin carboxyl-terminal hydrolase, can remove ubiquitin conjugates from substrates and disassemble them (Lee et al., 2014). E3 ubiquitin ligase, a novel E3 ubiquitin-protein ligase, is responsible for recognizing substrate and promoting ubiquitin ligation to substrate (Qiu et al., 2000). In this study, leucine supplementation could enhance the USP19 expression, and reduce the ITCH expression in IPEC-J2 cells, which further demonstrated that leucine inhibited the proteolysis by regulating ubiquitin-proteasome signaling pathway.
MutL homolog 1, one of the MutL homologs, plays a role in nuclear mismatch repair through combination with the other MutL homologs (Bronner et al., 1994). 5-methylcytosine binding domain 4, also known as MED1, is a novel DNA repair protein, and acts as a mismatch-specific glycosylase, which can interact with MLH1 (Petronzelli et al., 2000). Polymerase δ is the key enzyme that plays a critical role in the elongation of both the leading and lagging strands of DNA, and regulates DNA mismatch repair (Li et al., 2006). Our study showed that leucine increased expressions of MLH1, MBD4 and Pol δ, which could lead to the improvement of DNA repair. DNA-related RNA polymerase II can inhibit indirectly the transcription repression by recruiting factors that modify chromatin structures and interacting with components of the transcription machinery, and may modulate transcription of epithelial cells (Hahn, 2004). DNA-related RNA polymerase II was up-expressed by leucine treatment, which could enhance the transcription in IPEC-J2. In addition, PLAU has been implicated in remodeling of the extracellular matrix, enhancing cell proliferation and migration, and modulating cell adhesion (Duffy, 2004). The p15INK4b, one of the cyclin-dependent kinase inhibitors, modulates cell growth via the change in cell-ECM contact and the increase in G0/G1 (Wall et al., 2007). Leucine could also increase the levers of PLAU and p15INK4b in IPEC-J2 cells. Thus, recent studies have shown that leucine supplementation or deprivation regulates cell proliferation in different tissues (including gut), which could be derived from leucine affecting the expressions of these proteins.
Cathepsin D, a soluble lysosomal aspartic endopeptidase synthesized in rough endoplasmic reticulum, cleaves and activates BH3 interacting domain death agonist, and causes mitochondrial dysfunction, which will further stimulate caspases (Benes et al., 2008, Brentnall et al., 2013, LeBlanc et al., 1999). This is the classic caspase-dependent pathways of apoptosis. Moreover, GAPDH can also regulate the apoptosis (Sirover, 2011). In the present study, leucine treatment could decrease the expressions of cathepsin D, caspase 3, capase 6 and GAPDH, which demonstrated that leucine inhibited the apoptosis in IPEC-J2 cells. Toneto et al. (2016) reported that dietary leucine supplementation may attenuate apoptosis in tumour-bearing rats. In addition, our previous study has shown that dietary leucine supplementation may improve the effects of rotavirus infection on mucosal morphology and goblet cell numbers in the jejunal mucosa of pigs (Mao et al., 2015), which can be relative to leucine improving cell proliferation and apoptosis via regulating the related-protein expression.
The GAPDH and PYK are the key enzymes of glycolysis. The GAPDH catalyzes the oxidative phosphorylation of glyceraldehyde 3-phosphate to 1,3-diphosphoglycerate (Sirover, 2011), and PYK is to catalyze the transphosphorylation from phosphoenolpyruvate to ADP as the last step of glycolysis (Israelsen and Vander Heiden, 2015). In this study, GAPDH and PYK in IPEC-J2 cells were decreased by leucine, which illustrated that glycolysis was reduced, and energy derived from carbohydrate was decreased. Thus, supplementing leucine in the media changed the energy metabolism and utilization of IPEC-J2 cells.
Actin related protein 2/3 complex is the only known nucleator of branched F-actin filaments, and plays essential roles in vesicle trafficking (Zhuo et al., 2013). Loss of Arp2/3 complex activity resulted in unexpected phenotypes of the intestinal epithelium, including tight junction defects (Zhuo et al., 2015). RhoD partially coordinates Arp2/3-dependent mechanism to control the actin filament system (Gad et al., 2012). Cofilin has a central role for controlling actin dynamics by regulating actin polymerization and depolymerization, as well as by inducing dendritic nucleation, which is required for the early polymerization response to growth factor stimulation and the formation of protrusions (Hu et al., 2016). The TUBA polypeptide is the component of microtubule subunits (Tang et al., 2016). Syntaxin 18 can regulate the specific and direct fusion of endoplasmic reticulum, plasma or phagosomal membranes (Hatsuzawa et al., 2006). In this study, leucine treatment could increase the expressions of Arp2/3, RhoD, cofilin, TUBA and Stx18 in IPEC-J2 cells, which would strengthen the phagocytosis via regulating phagosome formation and actin stabilization. In addition, the phagocytosis plays an important role for the host defense to pathogens (Greenberg and Grinstein, 2002). Thus, our previous study demonstrated that dietary leucine supplementation alleviated the rotavirus-induced diarrhea in piglets (Mao et al., 2015), which could be related to the improvement of phagocytosis in gut epithelial cells.
5. Conclusions
In summary, based on the proteome analysis (iTRAQ), the expressions of 101 proteins were regulated by leucine treatment in IPEC-J2 cells. Some of these proteins in IPEC-J2 cells were related with some physiological functions. Besides protein metabolism, cell proliferation and apoptosis that have been reported in recent studies, we also found that leucine treatment could regulate glycolysis and phagocytosis. These verified the results of our previous study in piglets (Mao et al., 2015). In addition, with the porcine model being one of the best models for studying human gastrointestinal function (Meurens et al., 2012, Zhang et al., 2013), our findings will also be able to further reveal the possible function of leucine on gut health in human.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was financially supported by the grant from the National Basic Research Program (973 Program) of China (2013CB127306), the grant from National Natural Science Foundation of China (31201812), and the earmarked fund for the China Agriculture Research System (CARS-35). Special thanks to Professor De Wu from Sichuan Agricultural University for editing the manuscript.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Contributor Information
Xiangbing Mao, Email: acatmxb2003@163.com.
Lianqiang Che, Email: clianqiang@hotmail.com.
References
- Benes P., Vetvicka V., Fusek M. Cathepsin D – many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentnall M., Rodriguez-Menocal L., De Guevara R.L., Cepero E., Boise L.H. Caspase-9, caspase-3 and caspasse-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14:32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner E.C., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Coëffier M., Claeyssens S., Bensifi M., Lecleire S., Boukhettala N., Maurer B. Influence of leucine on protein metabolism, phosphokinase expression, and cell proliferation in human duodenum. Am J Clin Nutr. 2011;93:1255–1262. doi: 10.3945/ajcn.111.013649. [DOI] [PubMed] [Google Scholar]
- Dodd K.M., Tee A.R. Leucine and mTORC1: a complex relationship. Am J Physiol Endocrinol Metab. 2012;302:E1329–E1342. doi: 10.1152/ajpendo.00525.2011. [DOI] [PubMed] [Google Scholar]
- Duffy M.J. The urokinase plasminogen activator system: role in malignancy. Curr Pharmaceut Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- Gad A.K.B., Nehru V., Ruusala A., Aspenström P. RhoD regulates cytoskeletal dynamics via the actin nucleation-promoting factor WASp homologue associated with actin Golgi membranes and microtubules. Mol Biol Cell. 2012;23:4807–4819. doi: 10.1091/mbc.E12-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Goichon A., Chan P., Lecleire S., Coquard A., Cailleux A., Walrand S. An enteral leucine supply modulates human duodenal mucosal proteome and decreases the expression of enzymes involved in fatty acid beta-oxidation. J Proteomics. 2013;78:535–544. doi: 10.1016/j.jprot.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Greenberg S., Grinstein S. Phagocytosis and innate immunity. Curr Opin Immunol. 2002;14:136–145. doi: 10.1016/s0952-7915(01)00309-0. [DOI] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K., Tamura T., Hashimoto H., Hashimoto H., Yokoya S., Miura M. Involvement of syntaxin 18, an endoplasmic reticulum (ER)-localized SNARE protein, in ER-mediated phagocytosis. Mol Biol Cell. 2006;17:3964–3977. doi: 10.1091/mbc.E05-12-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zhu L., Yang X., Lin J., Yang Q. The epidermal growth factor receptor regulates cofilin activity and promotes transmissible gastroenteritis virus entry into intestinal epithelial cells. Oncotarget. 2016;7:12206–12221. doi: 10.18632/oncotarget.7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen W.J., Vander Heiden M.G. Pyruvate kinase: function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci U S A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M.D., Pavitt G.D. A new function and complexity for protein translation initiation factor eIF2B. Cell Cycle. 2014;13:2660–2665. doi: 10.4161/15384101.2014.948797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc A., Liu H., Goodyer C., Bergeron C., Hammond J. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer's disease. J Biol Chem. 1999;274:23426–23436. doi: 10.1074/jbc.274.33.23426. [DOI] [PubMed] [Google Scholar]
- Lee J., Kim W., Gygi S., Ye Y. Characterization of the deubiquitinating activity of USP19 and its role in endoplasmic reticulum-associated degradation. J Biol Chem. 2014;289:3510–3517. doi: 10.1074/jbc.M113.538934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xie B., Zhou Y., Rahmeh A., Trusa S., Zhang S. Functional roles of p12, the fourth subunit of human DNA polymerase δ. J Biol Chem. 2006;281:14748–14755. doi: 10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- Mao X., Hu H., Tang J., Chen D., Yu B. Leucine increases mucin 2 and occluding production in LS174T cells partially via PI3K-Akt-mTOR pathway. Anim Nutr. 2016;2:218–224. doi: 10.1016/j.aninu.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Liu M., Tang J., Chen H., Chen D., Yu B. Dietary leucine supplementation improves the mucin production in the jejunal mucosa of the weaned pigs challenged by porcine rotavirus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Qi S., Yu B., He J., Yu J., Chen D. Zn2+ and L-isoleucine induce the expression of porcine β-defensins in IPEC-J2 cells. Mol Biol Rep. 2013;40:1547–1552. doi: 10.1007/s11033-012-2200-0. [DOI] [PubMed] [Google Scholar]
- Mao X., Xiao X., Chen D., Yu B., He J., Chen H. Dietary apple pectic oligosaccharide improves gut barrier function of rotavirus-challenged weaned pigs by increasing antioxidant capacity of enterocytes. Oncotarget. 2017;8:92420–92430. doi: 10.18632/oncotarget.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Zeng X., Huang Z., Wang J., Qiao S. Leptin and leucine synergistically regulate protein metabolism in C2C12 myotubes and mouse skeletal muscles. Br J Nutr. 2013;110:256–264. doi: 10.1017/S0007114512004849. [DOI] [PubMed] [Google Scholar]
- Mao X., Zeng X., Wang J., Qiao S. Leucine promotes leptin receptor expression in mouse C2C12 myotubes through the mTOR pathway. Mol Biol Rep. 2011;38:3201–3206. doi: 10.1007/s11033-010-9992-6. [DOI] [PubMed] [Google Scholar]
- Meurens F., Summerfield A., Nauwynck H., Saif L., Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Ishida A., Yamazaki M., Abe H. Leucine suppresses myofibrillar proteolysis by down-regulating ubiquitin-proteasome pathway in chick skeletal muscles. Biochem Biophys Res Commun. 2005;336:660–666. doi: 10.1016/j.bbrc.2005.08.138. [DOI] [PubMed] [Google Scholar]
- Pedroso J.A.B., Zampieri T.T., Donato J. Reviewing the effects of L-leucine supplementation in the regulation of food intake, energy balance, and glucose homeostasis. Nutrients. 2015;7:3914–3937. doi: 10.3390/nu7053914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronzelli F., Riccio A., Markham G.D., Seeholzer S.H., Stoerker J., Genuardi M. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- Qin J., Gu F., Liu D., Yin C., Zhao S., Chen H. Proteomic analysis of elite soybean Jidou17 and its parents using iTRAQ-based quantitative approaches. Proteome Sci. 2013;11:12. doi: 10.1186/1477-5956-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Joazeiro C., Fang N., Wang H., Elly C., Altman Y. Recognition and ubiquitination of notch by itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Ren M., Zhang S., Liu X., Li S., Mao X., Zeng X. Different lipopolysaccharide branched-chain amino acids modulate porcine intestinal endogenous β-defensin expression through the Sirt1/ERK/90RSK pathway. J Agric Food Chem. 2016;64:3371–3379. doi: 10.1021/acs.jafc.6b00968. [DOI] [PubMed] [Google Scholar]
- Sadiq F., Hazlerigg D.G., Lomax M.A. Amino acids and insulin act additively to regulate components of the ubiquitin-proteasome pathway in C2C12 myotubes. BMC Mol Biol. 2007;8:23. doi: 10.1186/1471-2199-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierack P., Nordhoff M., Pollmann M., Weyrauch D.K., Amasheh S., Lodemann U. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem Cell Biol. 2006;125:293–305. doi: 10.1007/s00418-005-0067-z. [DOI] [PubMed] [Google Scholar]
- Sirover M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Suryawan A., Orellana R.A., Fiorotto M.L., Davis T.A. Leucine acts as a nutrient signal to stimulate protein synthesis in neonatal pigs. J Anim Sci. 2011;89:2004–2016. doi: 10.2527/jas.2010-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talvas J., Obled A., Sayd T., Chambon C., Mordier S., Fafournoux P. Phospho-proteomic approach to identify new targets of leucine deprivation in muscle cells. Anal Biochem. 2008;381:148–150. doi: 10.1016/j.ab.2008.05.038. [DOI] [PubMed] [Google Scholar]
- Tang E.I., Lee W.M., Cheng Y.C. Coordination of actin- and microtubule-based cytoskeletons supports transport of spermatids and residual bodies/phagosomes during spermatogenesis in the rat testis. Endocrinology. 2016;157:1644–1659. doi: 10.1210/en.2015-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneto A.T., Ramos L.A.F., Salomão E.M., Tomasin R., Aereas M.A., Gomes-marcondes M.C.C. Nutritional leucine supplementation attenuates cardiac failure in tumour-bearing cachectic animals. J Cachexia Sarcopenia Muscle. 2016;7:1–10. doi: 10.1002/jcsm.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall S.J., Zhong Z., DeClerk Y.A. The cyclin-dependent kinase inhibitors p15INK4b and p21CIP1 are critical regulators of fibrillar collagen-induced tumor cell cycle arrest. J Biol Chem. 2007;282:24471–24476. doi: 10.1074/jbc.M702697200. [DOI] [PubMed] [Google Scholar]
- Xiao F., Wang C., Yin H., Yu J., Chen S., Fang J. Leucine deprivation inhibits proliferation and induces apoptosis of human breast cancer cells via fatty acid synthase. Oncotarget. 2016;7:63679. doi: 10.18632/oncotarget.11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeanandin G., Balage M., Schneider S.M., Dupont J., Hébuterne X., Mothe-Satney I. Differential effect of long-term leucine supplementation on skeletal muscle and adipose tissue in old rats: an insulin signaling pathway approach. Age. 2012;34:371–387. doi: 10.1007/s11357-011-9246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Sumkara L.T., Jiang W., Bible M., Carter S., Ma X. Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Widmer G., Tzipori S. A pig model of the human gastrointestinal tract. Gut Microb. 2013;4:193–200. doi: 10.4161/gmic.23867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ren M., Zeng X., He P., Ma X., Qiao S. Leucine stimulates ASCT2 amino acid transporter expression in porcine jejunal epithelial cell line (IPEC-J2) through PI3K/Akt/mTOR and ERK signaling pathways. Amino Acids. 2014;46:2633–2642. doi: 10.1007/s00726-014-1809-9. [DOI] [PubMed] [Google Scholar]
- Zhuo K., Muroyama A., Underwood J., Leylek R., Ray S., Soderling S.H. Actin-related protein2/3 complex regulates tight junctions and terminal differentiation to promote epidermal barrier formation. Proc Natl Acad Sci U S A. 2013;110:E3820–E3829. doi: 10.1073/pnas.1308419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Sumigray K.D., Lechler T. The Arp2/3 complex has essential roles in vesicle trafficking and transcytosis in the mammalian small intestine. Mol Biol Cell. 2015;26:1995–2004. doi: 10.1091/mbc.E14-10-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J., Zhang J., Wang Q., Zhou B., Zhong J., Zhang C. Stress responsive proteins are actively regulated during rice (Oryza sativa) embryogenesis as indicated by quantitative proteomics analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074229. [DOI] [PMC free article] [PubMed] [Google Scholar]