Abstract
These data support the findings that dietary micronutrients influence the inflammatory responses and intestinal microbial community structure and function in a model of pouchitis-like small bowel inflammation reported in “Dietary Antioxidant Micronutrients Alter Mucosal Inflammatory Risk in a Murine Model of Genetic and Microbial Susceptibility” (Pierre et al., 2018) [1]. Briefly, wild-type and IL-10 deficient mice underwent surgical placement of small intestinal self-filling loops (SFL) and were subsequently fed purified control diet (CONT) or control diet supplemented with 4 micronutrients (AOX), retinoic acid, Vitamin C, Vitamin E, and selenium, for 14 days. These data include changes in host markers, such as body weight, mucosal levels of myeloperoxidase and syndecan-1, and luminal IgA and IgG levels. These data also include changes in the microbial compartment, including 16S community structure in the self-filling loop, conventionalized germ-free mice, and microbial substrate preference performed through anaerobic bacterial culturing of SLF CONT and AOX microbiota.
Specifications Table
| Subject area | Biology |
| More specific subject area | Gastroenterology |
| Type of data | Figures, Excel files |
| How data was acquired | Leica DM2500, Illumina MiSeq, Spectrophotometer |
| Data format | Processed |
| Experimental factors | Intestinal tissue and microbiome was examined following surgical procedures and dietary administration. |
| Experimental features | Immunofluorescences, ELISAs, Bacterial Assays, 16S sequencing |
| Data source location | Chicago, IL, USA |
| Data accessibility | Data is with this article |
| Related research article | Pierre JF, Hinterleitner R, Bouziat R, Hubert NA, Leone V, Miyoshi J, Jabri B, Chang EB. Dietary Antioxidant Micronutrients Alter Mucosal Inflammatory Risk in a Murine Model of Genetic and Microbial Susceptibility. Journal of Nutritional Biochemistry. 2017. In Press [1]. |
Value of the data
-
•
These data are valuable to nutrition and microbiome researchers interested in understanding the role dietary micronutrients can influence the structure and function of the intestinal microbiome.
-
•
These data can be used by inflammatory bowel disease and gastroenterology researchers interested in investigating intestinal inflammatory responses in genetically susceptible hosts under various dietary settings.
-
•
The 16S microbiome datasets provided here can be used to further examine how dietary micronutrients influence the structure of the intestinal microbiome at the genera levels.
1. Data
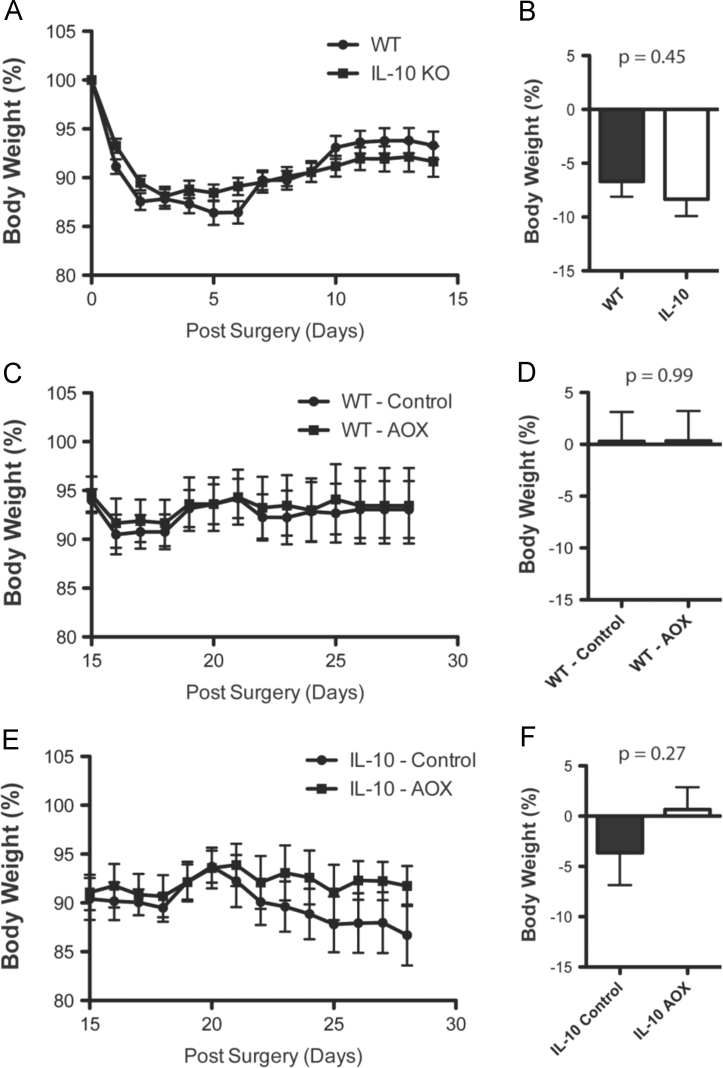
Fig. 1 shows the data on changes to body weight in WT and IL-10 KO animals following SFL surgery (A,B) and during subsequent feeding of CONT and AOX diet (C-F).
Fig. 1.
(A,B) Body weight changes (% of pre-surgical baseline) during and after surgical recovery between WT and IL-10−/− animals. Body weight changes during and after CONT and AOX diet in WT (C,D) and IL-10−/− (D,F) animals. P values displayed.
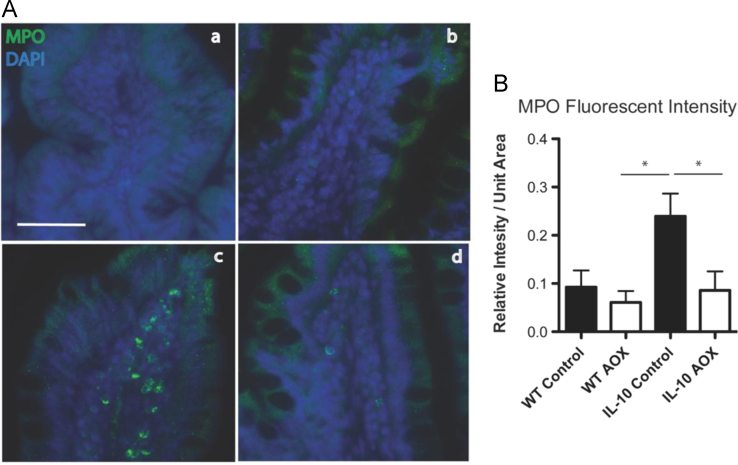
Fig. 2 shows the data on Myeloperoxidase levels in WT and IL-10 KO animals fed CONT or AOX diet.
Fig. 2.
(A) Immunofluorescence of Myeloperoxidase (MPO; green) and DAPI (blue) staining in self-filling loop mucosa of (a) WT CONT, (b) WT AOX (c), IL-10−/− CONT and (d) IL-10−/− AOX animals. (B) MPO fluorescent intensity displayed for each group. * P < 0.01. n = 5/group. Scale bar indicates 20 μm.
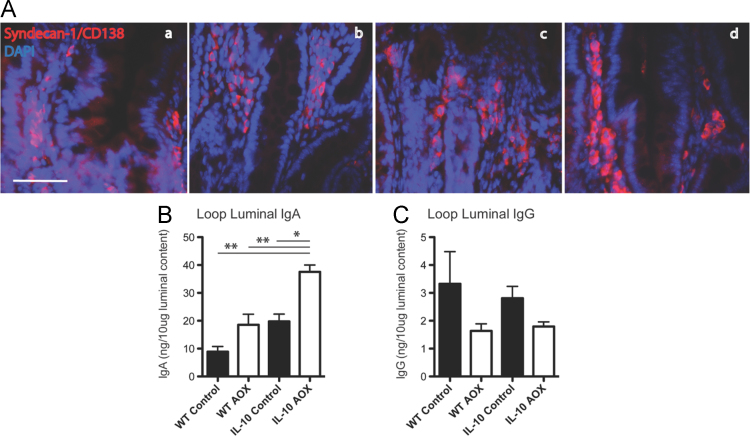
Fig. 3 shows the data on representative changes to Syndecan-1/CD138, which is a surface cell markers of plasma cells, following CONT and AOX diet (A). Fig. 3 also includes changes to luminal levels of IgA and IgG from the SFL following CONT and AOX diet.
Fig. 3.
(A) Immunofluorescence of Syndecan-1/CD139 (red) and DAPI (blue) staining in self-filling loop mucosa of (a) WT CONT, (b) WT AOX (c), IL-10−/− CONT and (d) IL-10−/− AOX animals. Loop luminal levels of IgA (B) and IgG (C) and fluorescent intensity displayed for each group. * P < 0.01. n = 5/group. Scale bar indicates 20 μm.
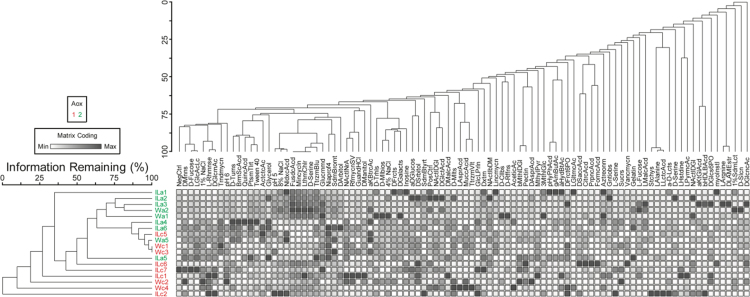
Fig. 4 shows the data on microbial community substrate preferences from communities collected from SFLs fed CONT or AOX diet in either genotype. Clustering of communities is shown by the left dendrogram by color.
Fig. 4.
Biolog Assay of Microbial Function. To screen microbial substrate utilization and tolerance, loop microbiota samples were inoculated in 96 well Biolog™ plates containing different metabolic substrates (sugars, amino acids) or environmental stressors (salts, pH, antibiotics). After 24 h, OD590 were recorded for plate well and Bray-Curtis dissimilarity was used to compare individual sample signatures. Two-way cluster dendrograms are displayed, which show microbiota in WT (w) and IL-10−/− (IL) on AOX (a) and CONT (c) diet cluster similarly, regardless of host genotype, suggesting diet is the primary driver of microbial function.
2. Experimental design, materials and methods
2.1. Animals
The use of all animals was approved by the IACUC at the University of Chicago. Wild-type (WT) or IL-10−/− gene-deficient mice on C57Bl/6 background were bred and housed under standard 12:12 light/dark conditions. Female mice (6–8 weeks old) were fed ad libitum gel diet 76A (Cat# 72-07-5022, Clear H20, Portland, ME) for 5-days before surgery to reduce the risk of intestinal obstruction at the anastomotic site. Females were chosen for this study since pilot data demonstrated higher survival rate post-operatively compared with male littermates. Prior to surgery, mice were anesthetized with ketamine/xylazine and an aseptic laparotomy was performed to resect 2.5 cm of ileum 3 cm proximal to the ileal-cecal value with anastomosis back to the distal ileum using 8-0 suture [1]. Compared with human surgery, the mouse model differs since colons remain in the animal. Removal of the colon in mice would dramatically complicate the surgical procedure, extend the surgical recovery period, and decrease overall animal survival. Following intestinal anastomosis, the abdomen was closed with interrupted 4-0 silk sutures and skin closured was performed with staples. Analgesics (buprenorphine 20 mg/kg BW) were provided to all animals post-operatively. Mice remained on gel diet 76A for 12 days following surgery and then were fed a purified Control diet (CONT) or Antioxidant diet (AOX) Pierre et al. [1] for 16 days. The antioxidant diet was formulated to be identical to CONT but with pharmacologically relevant levels of 4 micronutrients: retinoic acid, vitamin C, vitamin E, and selenium. Mice were weighted daily throughout the experiment. Animals were humanely euthanized and intestinal loop and loop mucosal scrapings were collected. Mesenteric lymph nodes were weighed and normalized to total body weight. Loop contents were snap frozen at − 80 °C for microbiota analysis, Biolog assay, and IgA/IgG measurements.
2.2. Conventionalization of germ-free IL-10 mice
Germ-free animals were individually housed and maintained in flexible film positive pressure isolators (CBC, Madison, WI). Gnotobiotic diets were autoclaved and culture and PCR screening was performed weekly to ensure sterility. For conventionalization of germ-free IL-10 animals, 1 g of luminal contents was obtained from wild-type CONT and AOX loops, resuspended in 5 mL sterile PBS, and vigorously resuspended. Particulate matter was centrifuged (1000g for 10 s) and 200 µL of microbial supernatant was administered in each mouse via oral gavage. To reinforce colonization, this process was repeated 7 days later. Donor contents and colonized intestinal microbiome were analyzed by 16S rRNA gene sequencing 3 weeks following conventionalization.
2.3. RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA isolation was performed on SFL mucosal scrapings with the Trizol (Ambion) and chloroform method as described previously [1]. Gene-specific primers from murine GAPDH, TNFα, IFNγ, IL-17α, IL-10, CD103, and CCR9 are shown in reference Table 1. All measurements were normalized by the expression of GAPDH gene, considered as a stable housekeeping gene. Gene expression was determined using the delta-delta Ct method: 2−▲▲CT(▲▲CT = [Ct(target gene) – Ct(GAPDH)]patient – [Ct(target gene) – Ct(GAPDH)]control). Real-time data were analyzed using the Roche LightCycle® (Roche Applied Science).
Table 1.
Study primers.
|
Mouse Primers | ||
|---|---|---|
| Gene name | Forward Primer | Reverse Primer |
| GAPDH(NM_001289726.1) | 5′-AACCCTTAAGAGGGATGCTG-3′ | 5′-CATTTTGTCTACGGGACGAG-3′ |
| TNFα (NM_013693.3) | 5′-GCCTCCCTCTCATCAGTTCT-3′ | 5′-CACTTGGTGGTTTGCTACGA-3′ |
| IFNγ (NM_008337.4) | 5′-CACGGCACAGTCATTGAAAG-3′ | 5′-TTTTGCCAGTTCCTCCAGAT-3′ |
| IL-17α (NM_010552.3) | 5′-ATTCTGTTCTCATCCAGCAA-3′ | 5′-CATCTTCTCGACCCTGAAAG-3′ |
| IL-10 (NM_010548.2) | 5′-TACTGCTAACCGACTCCTTA-3′ | 5′-GGATCATTTCCGATAAGGCT-3′ |
| CD103 (NM_008399.3) | 5′-ATCTGACAAAGACTAAGGAC-3′ | 5′-GATAGCACAGACCACTGAAT-3′ |
| CCR9 (NM_001166625.1) | 5′-GTACTGGCTTGTGTTCATTG-3′ | 5′-GTCATGGTCTTCACTCTTGT-3′ |
| 16S Sequencing Primers | ||
| 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ | ||
| 338F: 5′-GTGCCAGCMGCCGCGGTAA-3′ | ||
| 806R: 5′-GGACTACHVGGGTWTCTAAT-3′ | ||
| 1492R: 5′-GGTTACCTTGTTACGACT-3′ | ||
2.4. Immunofluorescence (IF), and histopathological scoring
SFL samples were fixed in 4% formalin/PBS overnight at room temperature. Five-micron sections were cut, deparaffinized, and stained via immunofluorescence (IF) with Anti-MPO and Anti-Syndecan. Antigen retrieval utilized sodium citrate (pH 6.0) with blocking in 10% BSA. Primary antibodies were incubated on samples (1:200 rabbit anti-MPO; 1:1000 rabbit anti-syndecan) overnight at 4 °C. Samples were rinsed and incubated with Alexa Fluor conjugated secondary antibody (555 goat anti-rabbit, Invitrogen life science). Slides were counterstained with DAPI and visualized on a Leica DM2500 microscope (Leica Microsystems, Wetzlar, Germany) using Image Pro-Plus software (Media Cybernetics, Silver Springs, MD, USA).
2.5. Loop content IgA and IgG enzyme-linked immunosorbent assay
Loop contents were normalized by weight and resuspended in PBS + protease inhibitor. Samples were vortexed for 20 min, spun down for 2 min at 1000×g, and diluted 1:100. IgA and IgG ELISAs were carried out according to manufacturer instructions (Cat# 88–50450, Cat# 88–50400; eBiosciences, San Diego, CA).
2.6. 16S DNA isolation
Intestinal contents were homogenized in 1 mL extraction buffer [50 mM Tris (pH 7.4), 100 mM EDTA (pH 8.0), 400 mM NaCl, 0.5% SDS] containing 20 uL proteinase K (20 mg/ml) as previously described [1]. 0.1-mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK, USA) were added to the extraction tubes and physical disruption was carried out for 2 × 1 min to lyse microbial cells. An overnight incubation at 55 °C was carried out with agitation,followed by extraction with phenol:chloroform:isoamyl alcohol, and precipitation with ethanol. Isolated DNA was resuspended in nuclease-free water and frozen at − 80 °C prior to sequencing.
2.7. 16S rRNA-based polymerase chain reaction, Illumina library preparation, and data analysis
The 16S gene was amplified by PCR as previously described [2]. Next, PCR primers targeted the 338–806 bp region of the 16S rRNA encoding gene (Table 1) that also contained Illumina 3′ adapter sequences as and 12-bp barcodes. Sequencing was carried out on an Illumina MiSeq DNA sequencer at Argonne National Laboratory. Sequences were trimmed and classified with the QIIME toolkit (version 1.8.0). Using the QIIME wrappers, OTUs were picked at 97% sequence identity for each OTU. Representative sequences were aligned using PyNAST and taxonomy assigned using uclust. The PyNAST-aligned sequences were utilized for phylogenetic analysis and weighted UniFrac distances.
2.8. Biolog assay
To determine substrate utilization of loop microbial communities shaped under the SFL conditions on each diet, biolog assays was carried out as previously described [1]. Briefly, an anaerobic chamber was used to resuspend homogenized loop contents (~ 10 mg) in Biolog Inoculum Fluid-A (IF-A; Cat #72401, Biolog, Heyward, CA) containing tetrazolium indicator dye that allows quantification of metabolic redox. After inoculation in the anaerobic chamber, plates were incubated overnight at 37 °C and monitored continuously for optical densities at 590 nm. After 24 h, Bray-Curtis dissimilarity analysis was applied to samples to compare experimental groups using Two-way cluster dendrograms and Indicator analysis [3].
2.9. Statistics
Data from the in vivo studies are presented as mean ± SEM; statistical significance was analyzed by the Analysis of Variance test (ANOVA) for multiple groups followed by analysis between two groups (Tukey-Frame׳s multiple comparisons test) or T-test. P <0.05 was considered statistically significant, unless otherwise stated.
Footnotes
Transparency document associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.08.026.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.08.026.
Transparency document. Supplementary material
Transparency document
Appendix A. Supplementary material
Table 2. Genera level data of 16S taxonomy of self-filling loops.
Table 3. Genera of data of 16S taxonomy of conventionalized animals.
References
- 1.Pierre J.F., Hinterleitner R., Bouziat R., Hubert N.A., Leone V., Miyoshi J., Jabri B., Chang E.B. Dietary antioxidant micronutrients alter mucosal inflammatory risk in a murine model of genetic and microbial susceptibility. J. Nutr. Biochem. 2018;54:95–104. doi: 10.1016/j.jnutbio.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierre J.F. Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. - Gastrointest. Liver Physiol. 2016;311(2):G286–G304. doi: 10.1152/ajpgi.00202.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.B. McCune, M. Mefford, Multivariate Analysis of Ecological Data, in: Software Design, Gleneden Beach, Oregon, USA, 1999.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Table 2. Genera level data of 16S taxonomy of self-filling loops.
Table 3. Genera of data of 16S taxonomy of conventionalized animals.