Abstract
Insertional mutagenesis is an important risk with all genetically modified cell therapies, including chimeric antigen receptor (CAR)-T cell therapy used for hematological malignancies. Here we describe a new tagmentation-assisted PCR (tag-PCR) system that can determine the integration sites of transgenes without using restriction enzyme digestion (which can potentially bias the detection) and allows library preparation in fewer steps than with other methods. Using this system, we compared the integration sites of CD19-specific CAR genes in final T cell products generated by retrovirus-based and lentivirus-based gene transfer and by the piggyBac transposon system. The piggyBac system demonstrated lower preference than the retroviral system for integration near transcriptional start sites and CpG islands and higher preference than the lentiviral system for integration into genomic safe harbors. Integration into or near proto-oncogenes was similar in all three systems. Tag-PCR mapping is a useful technique for assessing the risk of insertional mutagenesis.
Keywords: CD19 CAR-T cell, piggyBac transposon, Integration site mapping, Tag-PCR
Highlights
-
•
We developed a new tagmentation-assisted PCR technique (tag-PCR) to map the integration sites of genetically modified cells.
-
•
Tag-PCR enables a fast, simple, and comprehensive integration site mapping of CD19 CAR-T cells.
-
•
The insertional mutagenesis risk of piggyBac CAR-T cells appeared to be comparable to that of retroviral vector systems.
1. Introduction
The first successful application of gene therapy, retroviral gene therapy for patients with X-linked severe combined immunodeficiency, resulted in sustained reconstitution of the T cell pool and protection from infections [10]; however, the risk of development of T cell acute lymphoblastic leukemia raised concerns about the safety of gene therapy [11]. A number of approaches have been developed to reduce the potential risk associated with insertional mutagenesis, including systems based on the self-inactivating (SIN) retroviral vector [28] and the SIN lentiviral vector [3], and on non-virus vectors such as the Sleeping Beauty [7] and piggyBac [31] transposon systems. It is expected that these will provide safer gene transfer for clinical use.
During gene therapy for primary immunodeficiencies, if a vector becomes integrated into a part of the genome that contains a proto-oncogene such as LMO2, this can result in the development of leukemia [11]. Because of this, it is important to perform an integration site analysis to assess the potential insertional mutagenesis of a gene therapy. Various polymerase chain reaction (PCR)-based technologies have been developed to determine gene sequences in unknown DNA regions that flank a known sequence; these include inverse PCR [23], ligation-mediated PCR [21], and linear amplification-mediated PCR (LAM-PCR) [27]. However, because these methods use restriction enzymes to fragment the DNA, they can retrieve only a fraction of all the genomic integrations [12]. Nonrestrictive LAM-PCR (nrLAM-PCR), a high-throughput technique that does not involve restriction enzymes, was developed to overcome this drawback; it has been reported to be a comprehensive vector integration mapping method [25]. Target capture sequencing has also been reported to be an accurate, time-saving, and cost-effective method to determine the integration sites of viral vectors in human genes [29]. However, despite their improvements, these methods remain time-consuming. Therefore, we developed a new tagmentation-assisted PCR (tag-PCR) method to analyze the integration sites of genetically modified cells, which can analyze the integration sites of transgenes accurately and more comprehensively than restriction enzyme-based methods, and takes smaller number of steps to prepare next-generation sequencing (NGS) libraries because of the small number of sample preparation steps involved.
Therapy using genetically modified CD19-specific chimeric antigen receptor (CAR)-T cells (CD19 CAR-T cells) has been reported as a breakthrough treatment for CD19-positive B-cell lineage hematological malignancies. The initial clinical successes of CD19 CAR-T cell therapy have been achieved by SIN retroviral [2] and SIN lentiviral vector systems [9], but CD19 CAR-T cell engineering using non-viral vectors has also been developed in pre-clinical studies and phase I clinical trials [1,16,19].
In the present study, we applied the new tag-PCR method to the integration mapping of CD19 CAR-T cells to compare the cells produced by a piggyBac-mediated system with those from retroviral vector and lentivirus vector systems.
2. Materials and Methods
2.1. Vector Construction
The piggyBac transposase plasmid (pCMV-piggyBac) has been previously described [14,22,31]. The transposon plasmid for CD19-specific chimeric antigen receptor (pIRII-CAR.CD19) was produced by subcloning FMC63-28z receptor protein gene [17] into EcoRI and KpnI-digested pIRII-piggyBac transposon vector backbone [22]. pIRII-CAR.CD19 encodes CD19 CAR (FMC63-28z) comprising a single-chain variable fragment from an anti-CD19 antibody derived from the FMC63 mouse hybridoma, a portion of the human CD28 molecule, and the intracellular component of the human T cell receptor ζ molecule. Both vectors are transcriptionally regulated by the cytomegalovirus (CMV) immediate early gene enhancer/promoter sequence.
The CD19-encoding CAR retroviral and lentiviral vectors were produced by subcloning FMC63-28z into SFG retroviral [30] and CSII-CMV-MCS lentiviral (provided by Dr. Hiroyuki Miyoshi, RIKEN BioResource Center, Japan) backbones, respectively. The transfer vectors were co-transfected into the HEK293T cells using FuGENE HD (Promega, Madison, WI, USA) with packaging and envelope plasmids, according to the manufacturer's instructions. Further, supernatant was collected following 72-hour incubation, and aliquots were stored at −80 °C for further use. The schemata of the constructed vectors are illustrated in Supplementary Fig. 1.
2.2. Generation of CD19 CAR-T Cells
We produced CD19 CAR-T cells from peripheral blood mononuclear cells (PBMC) obtained from three healthy volunteers. The Institutional Review Board of Nagoya University Graduate School of Medicine (Nagoya, Japan) approved the study protocol.
To produce piggyBac-CD19 CAR-T cells, we used 4D-Nucleofector device and P3 Primary Cell 4D-Nucleofector X Kit (Lonza, Basel, Switzerland) for gene transfer. Briefly, we used program EO-115 to electroporate 1 × 107 PBMC with 5 μg each of pIRII-CAR.CD19 transposon plasmid and pCMV-piggyBac transposase plasmid. Further, the transfected cells were co-cultured with irradiated autologous activated T cells (ATCs, stimulated with OKT3 and anti-CD28 antibody and cultured with IL-15 for 3 days) as feeder cells that were pulsed with four viral peptide pools (MACS GMP PepTivator; AdV5 Hexon, CMV pp65, EBV EBNA-1, and EBV BZLF1) (Miltenyi Biotec, Auburn, CA, USA), as previously reported [20]. Following stimulation, the cells were cultured in TexMACS Medium (Miltenyi Biotec) supplemented with human interleukin (IL)-7 (10 ng/mL, Miltenyi Biotec) and IL-15 (5 ng/mL, Miltenyi Biotec) in 24-well plates at 37 °C in a humidified 5% CO2 incubator. On day 7, the cells were transferred into G-Rex10 culture flasks (Wilson Wolf Manufacturing Inc., New Brighton, MN, USA) and co-cultured with four viral peptides-pulsed irradiated ATCs in the same manner as described above. Then, the product was cryopreserved at −80 °C for further use. Retroviral-CD19 CAR- and lentivirus-CD19 CAR-T cells were produced using MoMLV- and HIV-based systems, respectively [30]. Briefly, PBMC activated with anti-CD3 antibody (clone OKT3, Ortho Biotech, Bridgewater, NJ, USA), anti-CD28 antibody (BD Pharmingen, Franklin Lakes, NJ, USA), human IL-7 (10 ng/mL), and IL-15 (5 ng/mL) were transduced with viral supernatant using precoated plates with a recombinant fibronectin fragment (RetroNectin, Takara Bio, Otsu, Japan). The cells were cultured in complete media supplemented with IL-7 (10 ng/mL) and IL-15 (5 ng/mL) for 14 days. Following this, the products were cryopreserved at −80 °C for further use.
2.3. Flow Cytometric Analysis
T cell products were stained with a monoclonal antibody directed against the single-chain variable fragment (scFv) of the CD19 CAR generously provided by Dr. Gianpietro Dotti (University of North Carolina). CAR expression was examined by staining with allophycocyanin (APC)-conjugated mouse anti-human CD3 monoclonal antibody (mAb) and phycoerythrin (PE)-conjugated goat anti-mouse IgG (H + L) mAb (BD Pharmingen, Franklin Lakes, NJ, USA). Then, the cells were analyzed by BD FACSCalibur with BD Cell Quest Pro software (Becton Dickinson, Franklin Lakes, NJ, USA).
2.4. Tagmentation-Mediated PCR (Tag-PCR)
Genomic DNA was extracted from CAR-T cells using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). We performed two-step measurement of DNA using Qubit 2.0 Fluorometer (Life Technologies, CA, USA) to ensure accuracy. In addition, we routinely measured A260/A280 ratio using NanoDrop (Life Technologies) to assess protein contamination and diluted target DNA to ensure the concentration of EDTA in DNA was <0.1 mM. Further, 50 ng DNA was applied to tagmentation reaction for 5 min at 58 °C using the transposase and transposon complex provided in Illumina Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA). In this reaction, the DNA was fragmented at random positions and added with adapter sequences on both ends, serving as a scaffold for subsequent PCR reactions [4]. After purification with AMPure XP beads (Beckman Coulter, Brea, CA, USA), the tagmentation product was applied to the following two-step PCR.
In the first step of PCR, we performed 40-cycle PCR to amplify vector-genome junctions using PrimeSTAR GXL DNA Polymerase (TaKaRa Bio, Ohtsu, Japan), a Mastercycler pro S (Eppendorf, Hamburg, Germany), and primers, which were specific to an end of the transgene and adapter sequence, according to the manufacturers' instructions. We performed six patterns of PCR in parallel using 2 ng DNA for each reaction. The transgene-specific primer was tagged with an adapter sequence on its 5′ end, which served as a scaffold for the second step of PCR. In this step, platform-specific adapter sequences and individual indices for next-generation sequencer were attached to the first PCR product in the 14 cycles of thermal cycling. We used 1 μL of the first PCR product for the second step of PCR without any purification. The sequences of primers are listed in Supplementary Table 1, whereas paring of primers for the first PCR are summarized in Supplementary Table 2. In the second step of PCR, index numbers N723–N726, N716–N719, and N720–N722 were used for piggyBac, retrovirus, and lentivirus, respectively.
Lastly, the second PCR product was purified with AMPure XP beads, quantified, and subjected to sequencing on HiSeq2500 next sequencing platform with 2- × 150-bp paired end-reads (Illumina).
Three separate experiments using three different donors' PBMC were performed for each gene transfer system. After confirming that the profiles of integration sites did not significantly differ among the donors (Supplementary Fig. 2), we pooled the data for each gene transfer system.
2.5. Analysis of Sequencing Data for Integration Mapping
In the data of high-throughput sequencing, we selected reads containing vector–genomic DNA junctions using the complete match of a 15-bp specific sequence on the vector. Following the removal of vector-derived part of the sequences, such reads were aligned to the hg19 human genome using Burrows–Wheeler aligner with a-MEM option [18]. We considered the alignment valid if (i) there was no inserted base between the vector and genome sequences; (ii) the aligned read was ≥20 bp; (iii) mapping quality was ≥30; and (iv) identity with reference genome was ≥95%.
We evaluated the site of genomic integration for the presence of RefSeq genes, CpG islands, and transcription start sites (TSSs). Integration into a RefSeq gene (intragene) was defined as insertion between the transcriptional start and stop sites of a gene. We defined 572 proto-oncogenes based on Sanger Institute Cancer Gene Census Table (http://www.sanger.ac.uk/genetics/CGP/Census/; as of July 4, 2016).
To assess the frequency of integration into genomic safe harbors (GSHs), we used the previously proposed criteria: (i) a minimum distance of 50 kb from the 5′ end of any gene; (ii) a minimum distance of 300 kb from any cancer-related gene; (iii) a minimum distance of 300 kb from any microRNA (miRNA); (iv) located outside a transcription unit; and (v) located outside ultraconserved regions of the human genome [24,26]. The positions of miRNA were obtained from miRBase11 as of April 17, 2017. We defined the ultraconserved region using the phyloP conservation score of ≥1.0, which was deposited in UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTables).
We simulated random integration sites by generating random numbers across the hg19 co-ordinate using Mersenne twister pseudorandom number generator.
2.6. Validation of the Integration Sites Generated by Tag-PCR
To validate the integration sites determined by tag-PCR, we detected the integration sites of piggyBac-CAR-T cells using target capture sequencing and compared them with those obtained from tag-PCR. In brief, we designed custom SureSelect bait capturing the whole part of pIRII-CAR.CD19 vector (Agilent Technologies, Santa Clara, CA, USA). Further, we performed target enrichment and sequencing according to the manufacturers' instructions. Using in-house programs, we detected sequence reads containing vector–genome junctions and performed subsequent analyses as in the case of tag-PCR.
2.7. Sequence Logo Analysis
We used WebLogo [5] to analyze integration sites determined by our study to evaluate and visualize consensus sequence motifs. The standard logo plots revealed a possible consensus sequence, with the height of the nucleotide representing the level of conservation at that position.
2.8. Statistical Analysis
The Fisher's exact test was used to compare the integration frequencies. All p-values reported are two-sided, and the values <0.05 were considered statistically significant. All statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University), a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria) [15].
3. Results
3.1. Comparison of the Tag-PCR Procedure with those of the Other Site Mapping Methods
Fig. 1 provides a schematic overview of the tag-PCR (Fig. 1C) site integration mapping method in comparison to the LAM-PCR (Fig. 1A) and nrLAM-PCR (Fig. 1B) methods. The tagmentation reaction simultaneously fragments the DNA at random positions and adds adapter sequences on both sides of the fragments. Using the adapter sequences as scaffolds, PCR amplifies the vector–genome junctions. This procedure does not utilize restriction enzyme digestion and involves fewer processing steps than the other two methods.
Fig. 1.
Schematic outline of the tag-PCR method of integrated site mapping compared with standard LAM-PCR and nrLAM-PCR methods.
Linear amplification-mediated polymerase chain reaction (LAM-PCR) (A), and non-restriction LAM-PCR (nrLAM-PCR) (B) start with the amplification of vector–genome junctions using biotinylated primers, hybridizing close to the end of the known vector DNA sequence. Biotinylated single strand (ss)DNA is captured on magnetic particles. LAM-PCR then uses restriction enzyme digestion and linker ligation after double strand (ds)DNA synthesis. The product is amplified by nested PCR with linker- and vector-specific primers. In nrLAM-PCR, the ssDNA linker sequence is directly ligated to the unknown end of the pre-amplified ssDNA following amplification by nested PCR with linker- and vector-specific primers. Additional PCR is performed to add sequencing specific adapters to these (nr)LAM-PCR products. In tag-PCR (C), the tagmentation enzyme randomly fragments genomic DNA and tags the fragments with adapters at both ends. DNA fragments containing the vector–genome junction are amplified with a primer that is complementary to the adapter sequence and the other primer that is complementary to the vector sequence and tagged with the additional adapter sequence. A second PCR is performed to add the complete adapter sequences necessary for sequencing. Finally, the libraries are analyzed by a next-generation sequencer. Estimated durations are indicated alongside each step. IR, inverted repeat; LC, linker cassette; ssLC, single strand linker cassette.
3.2. Transduction Efficiency of piggyBac-, Retrovirus-, and Lentivirus-CD19 CAR-T Cells
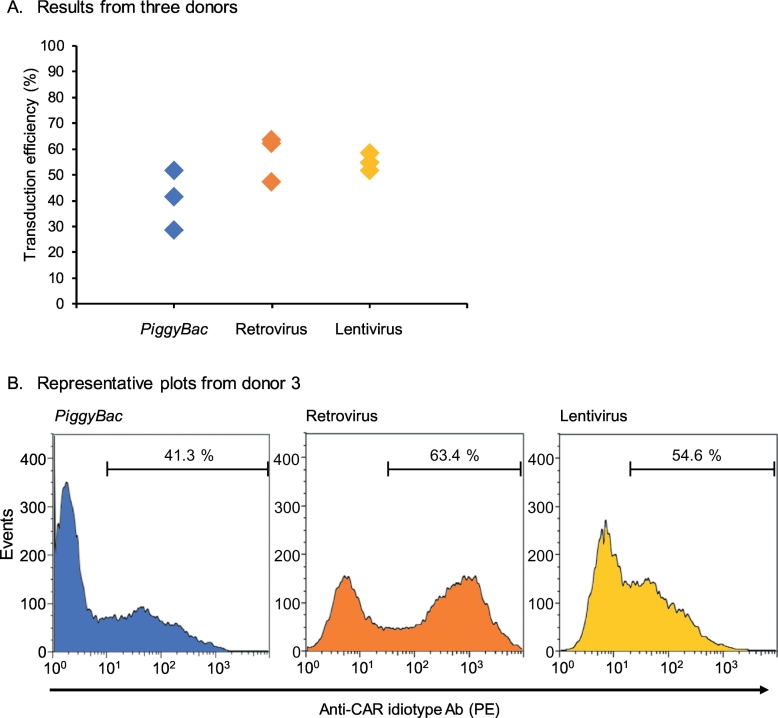
We generated CD19 CAR-T cells using three methods of transduction: piggyBac transposon, retrovirus, and lentivirus. The cells were cultured for 14 days and the transduction efficiency of the CD19 CAR gene (FMC63-28z) was measured by flow cytometry using an anti-idiotype antibody (Fig. 2). There was no statistical difference in the transduction efficiency of the cells produced by the piggyBac transposon, retrovirus, and lentivirus systems (28.4%–51.4%, 47.1%–63.4%, and 51.4%–58.4%, respectively). This result indicates that the piggyBac transposon system had comparable transduction efficiency to those of the retrovirus and lentivirus vectors.
Fig. 2.
Chimeric antigen receptor expression.
The frequency of chimeric antigen receptor (CAR) expression was measured by flow cytometry using anti-idiotype antibodies after CAR-T cells had been cultured for 14 days. (A) CAR expression for the piggyBac, retrovirus, and lentivirus vectors obtained from three healthy donors. Each donor's CAR expression is shown by a single data point. (B) Representative results of CAR expression in primary T cells obtained from donor 3. The x- and y-axes indicate the fluorescence intensity and frequency of events, respectively.
3.3. Mapping of Genomic Integration Sites
Using tag-PCR to identify the integration sites, we mapped 602, 815, and 1609 integration sites within the human genome for the piggyBac-, retrovirus-, and lentivirus-CAR-T cells, respectively. The sites are listed in Supplementary Table 3. Consensus sequencing around the integration sites confirmed that the piggyBac transposon system integrated CD19 CAR genes into TTAA sequences in the human genome, and that there was palindromic preference for upstream and downstream repetitive A or T sequences surrounding the central TTAA nucleotide (Supplementary Fig. 3A), as noted previously [8]. In contrast, the retrovirus and lentivirus systems displayed weak preferences for integration at the primary DNA sequence level (Supplementary Fig. 3B and C).
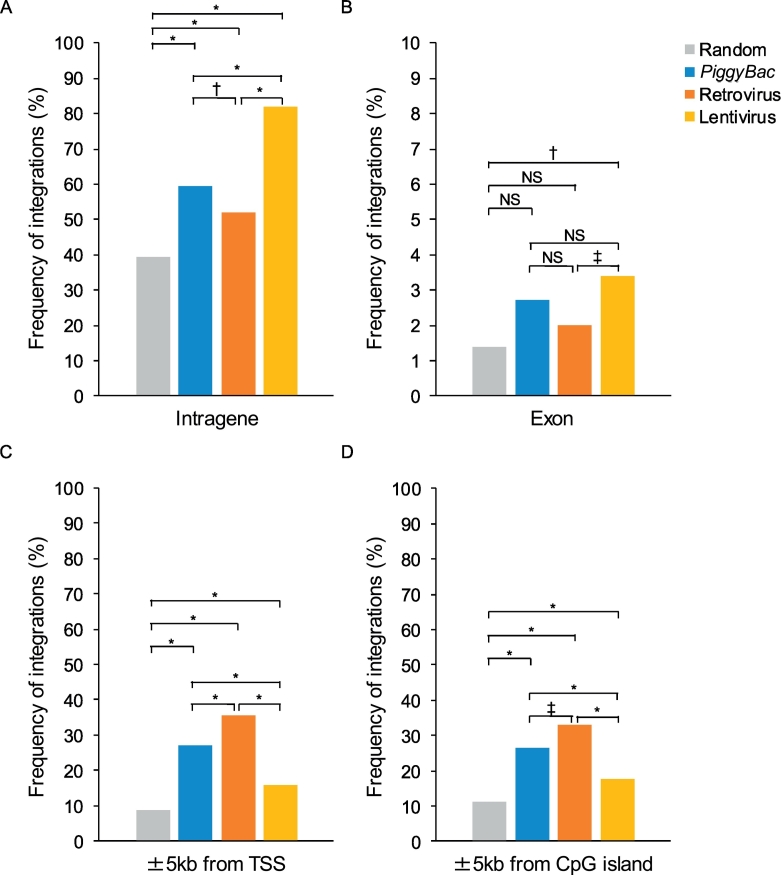
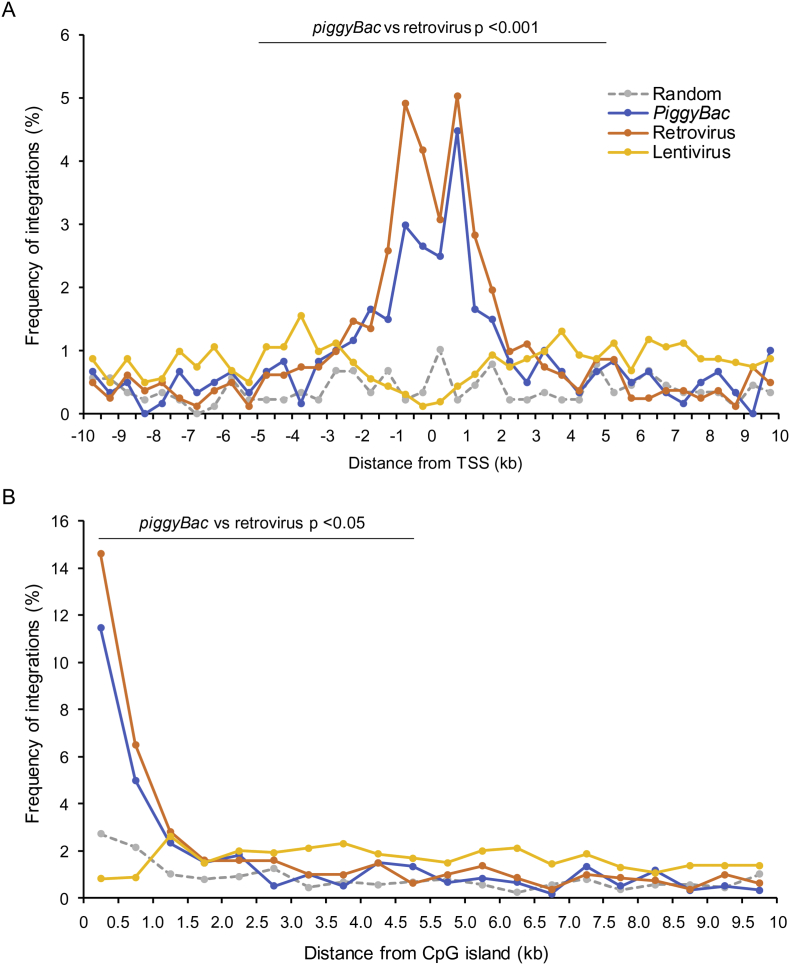
We analyzed the frequencies of the three vectors' integration into known genomic elements, including genes (exons and introns), the regions surrounding (i.e. within ±5 kb of) TSSs, and regions surrounding CpG islands (with an absolute distance <5 kb, Fig. 3). The integration frequency into genes (intragenic regions) of piggyBac was 59.6%, which was significantly higher than that for the retrovirus system (51.9%, P = .0042) and lower than that for the lentivirus system (81.9%, P = 9.7 × 10−26; Fig. 3A). The frequency of insertions into exons was comparable for piggyBac (2.7%), retrovirus (2.0%), and lentivirus (3.4%; Fig. 3B). Consistent with previous reports [6], piggyBac exhibited a preference for gene integration around TSSs (±5 kb, 27.1%) and CpG islands (<5 kb, 26.6%), which was similar to, but weaker than, the pattern shown by the retrovirus vector (TSS, 35.7%, P = 6.7 × 10−4; CpG, 32.8%, P = .014; Fig. 3C and D and Fig. 4). Compared with these two systems, such preference was significantly weaker in the lentivirus system (TSS, 15.9% and CpG, 17.7%).
Fig. 3.
Frequency of integration into or around genomic elements.
Frequency of integration in genes (A), exons (B), 5 kb windows around transcriptional start sites (TSSs) (C), and 5 kb windows around CpG islands (D). Gray bars indicate the results from random simulation. * P < .001; † P < .01; ‡ P < .05; NS, not significant.
Fig. 4.
Integration frequency around transcriptional start sites and CpG islands.
(A) The frequency of insertion within a distance of 500 bp from the nearest transcriptional start site (TSS). (B) The frequency of insertion within a distance of 500 bp from the nearest CpG island. The frequencies for the piggyBac, retrovirus, and lentivirus vectors are illustrated in blue, orange, and yellow, respectively. The gray dotted lines represent simulated random integration events.
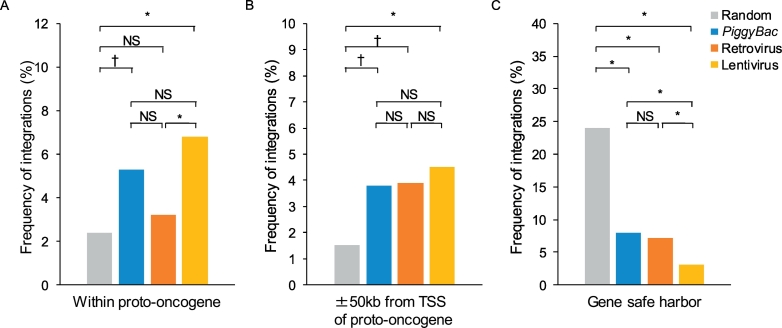
We analyzed the frequencies of integration of the three vectors into or around (±50 kb) the TSSs of known proto-oncogenes (Fig. 5). The frequency into known proto-oncogene for piggyBac (5.3%) was not significantly different from that for retrovirus (3.2%) and lentivirus (6.8%), nor around the TSSs of known proto-oncogene (piggyBac, 3.8%; retrovirus, 3.9%; lentivirus, 4.5%) (Fig. 5A and B). There was no integration of any of the three vectors into or within 50 kb of the TSSs of proto-oncogenes that have been reported to induce leukemogenesis in hematopoietic stem cell-based gene therapy, such as LMO2, BMI2, CCND2, MN1 or MECOM (MDS1-EVI1) (Supplementary Table 4).
Fig. 5.
Frequency of integration into or around transcriptional start site of proto-oncogenes and gene safe harbors.
The frequency of integration within proto-oncogenes (A), within 50-kb windows around transcriptional start sites (TSSs) of proto-oncogenes (B), and gene safe harbors (C). Gray bars indicate results from random simulation. * P < .001; † P < .01; NS, not significant.
We also analyzed integration of the three vectors into GSHs (Fig. 5C). We defined these based on previously proposed criteria regarding their location [24,26]. All three vectors demonstrated a lower frequency of integration into GSHs than that of random simulation control. The frequency for piggyBac (8.3%) was not significantly different from that for retrovirus (7.2%) but was significantly higher than that for lentivirus (2.9%, P = 2.6 × 10−7) (Fig. 5C).
3.4. Validation of Tag-PCR-Based Integration Site Mapping with Target Capture Sequencing
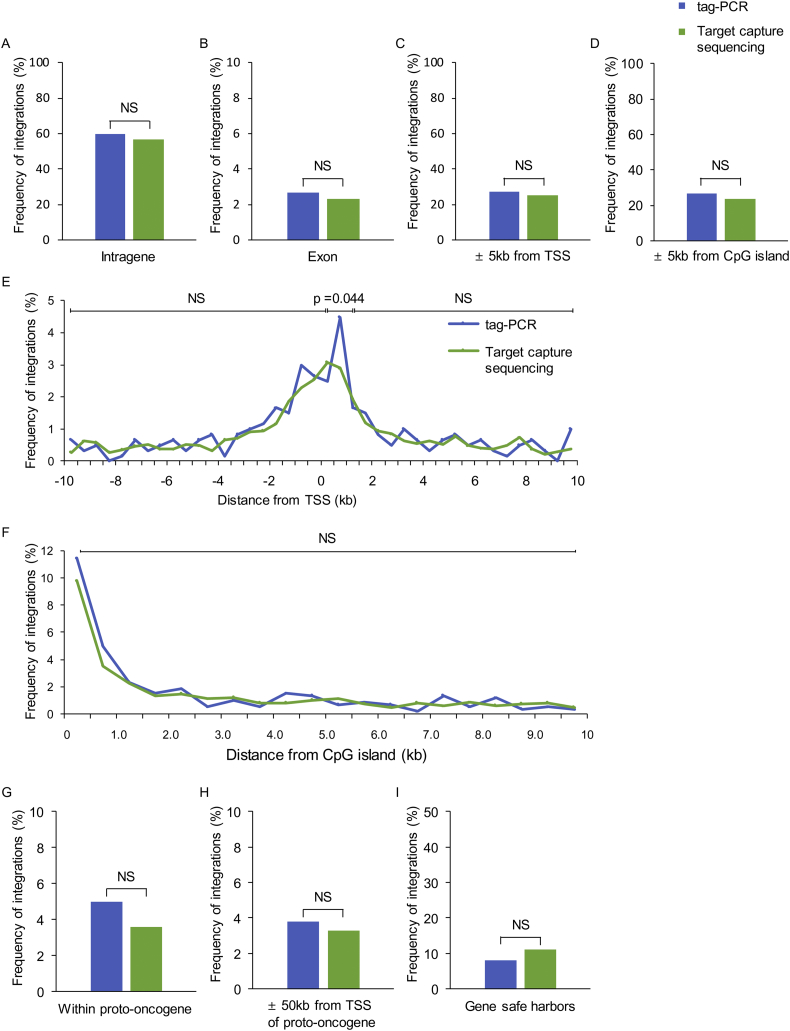
We compared various properties of the integration sites of piggyBac-CD19 CAR-T cells generated by tag-PCR and by target capture sequencing. Starting from the identical DNA specimen, only eight integration sites were identified in common in both tag-PCR and target capture sequencing, indicating that our piggyBac CAR-T cell products were highly polyclonal. The frequency of integration into each element of genes, the integration pattern around TSSs and CpG islands, the integration into or around TSSs of proto-oncogenes, and the integration into GSHs were not significantly different between the two methods (Fig. 6).
Fig. 6.
Validation of tag-PCR system by target capture sequencing. Comparison of integration site mapping of piggyBac CAR-T cell generated by tag-PCR and that by target capture sequencing. We calculated several parameters, including frequency of integration in genes (A), exons (B), 5-kb windows around transcriptional start sites (TSSs) (C), 5-kb windows from CpG islands (D), the patterns of insertions around TSSs (E), CpG islands (F), integration within proto-oncogenes (G), 50-kb windows around TSS of proto-oncogenes (H), and gene safe harbors (I) using data from both methods.
4. Discussion
Deep evaluation of piggyBac transposon integrations is needed for consideration of a piggyBac-modified human T cell product for human therapeutic application. To this end, we developed a new genome-wide integration site mapping system with tag-PCR technique, which allows the non-selective assessment of the DNA sequences that flank the vector integration. Other conventional mapping methods are also able to identify the integration sites of gene transfer vectors, but tag-PCR-based mapping offers two main technological advantages: it can analyze the integration sites of transgenes accurately and more comprehensively than restriction enzyme-based methods, and it is simpler and faster technique for site mapping than the methods that do not use restriction enzymes. Inverse PCR [23] and LAM-PCR [27] are applied to genomic DNA that has been digested by specific restriction enzymes; thus, they analyze the junctions of the host DNA and transgenes that integrate near the cleavage sites of each restriction enzyme. In contrast, tagmentation enzymes cut the genomic DNA at random positions, so tag-PCR is able to analyze the sequences flanking a known DNA sequence more comprehensively, without missing any specific restriction fragments. Like tag-PCR, the recently developed nrLAM-PCR and target capture sequencing systems are high-throughput techniques that do not use restriction enzymes and can produce non-biased, comprehensive integration site mapping. However, tag-PCR is able to perform integration site mapping more simply and faster than these other systems, potentially saving time. With tag-PCR mapping, it takes only a single day to prepare NGS libraries because of the small number of sample preparation steps. However, validation of tag-PCR integration site mapping against target capture sequencing confirmed its equivalent performance, despite its speed and simplicity. Because of these advantages, tag-PCR will allow integration site mapping of all genetically modified cell products, as well as periodic monitoring of the clonality of infused cells in individual patients, thereby reinforcing the safety of genetically modified cell therapy in the clinical setting.
In this study, we used tag-PCR to evaluate the integration mapping of CD19 CAR-T cell products produced by piggyBac, retrovirus, and lentivirus systems. To our knowledge, this was the first study to analyze genome-wide insertion mapping of piggyBac-generated human CD19 CAR-T cell products. The piggyBac system demonstrated a lower preference than the retrovirus system and a higher preference than the lentivirus system to integrate near TSSs and CpG islands. Our results are in line with the results of a previous study evaluated green fluorescent protein transgene integrations, and not CD19 CAR transgene integrations, in human T cells assessed by insertional mapping with LAM-PCR system [6,8,13]. Although the integration mapping property cannot fully evaluate the safety of gene therapies, the insertional mutagenesis risk of piggyBac CAR-T cells seemed to be comparable to that of retroviral CAR-T cells which have demonstrated safety with regards to genotoxicity over a long period of time in humans.
In conclusion, we have developed a new integration mapping methodology based on tag-PCR, and we applied it to evaluate CD19 CAR-T cell products of piggyBac, retrovirus, and lentivirus systems. This demonstrated that tag-PCR mapping is a useful technique for assessing the risk of insertional mutagenesis and for clonality analysis of genetically modified cells. Given the reduced cost of vector production, piggyBac-modified CAR-T cells could be a feasible therapeutic approach for cancer and possibly other diseases.
Funding
This work was supported by Practical Research for Innovative Cancer Control provided by Ministry of Health, Labour and Welfare. (14526464)
Competing Interests
NN, DM, YN, and YT have patent applications related to this paper. The other authors declare no competing financial interests.
Acknowledgments
Acknowledgements
The authors would like to thank Dr. Gianpietro Dotti from University of North Carolina for his generous provision of a monoclonal antibody against CD19 CAR. The authors would also thank Ms. Yoshie Miura and Ms. Yinyan Xu for their valuable assistance. The authors acknowledge the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine for technical support for next-generation sequencing. The authors would like to thank Enago (www.enago.jp) for the English language review.
Author Contributions
M.H., N.N., Y.O., and H.M. designed and performed the research, analyzed the data, and wrote the paper. S.S., N.K., S.T., D.M, S.K., D.I., N.M., R.T., K.S., D.K., Y.S., E.N., and A.N. performed the research and analyzed the data. A.H., S.K., Y.N., and Y.T. designed the research and analyzed the data. M.W. provided materials and wrote the paper. All authors critically reviewed the content of the manuscript and agreed on the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.07.008.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Bishop D.C., Xu N., Tse B., O'Brien T.A., Gottlieb D.J., Dolnikov A., Micklethwaite K.P. Piggybac-engineered T cells expressing CD19-specific CARs that lack IgG1 Fc spacers have potent activity against B-ALL xenografts. Mol Ther. 2018;26(8) doi: 10.1016/j.ymthe.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brentjens R.J., Riviere I., Park J.H., Davila M.L., Wang X., Stefanski J., Taylor C., Yeh R., Bartido S., Borquez-Ojeda O., Olszewska M., Bernal Y., Pegram H., Przybylowski M., Hollyman D., Usachenko Y., Pirraglia D., Hosey J., Santos E., Halton E., Maslak P., Scheinberg D., Jurcic J., Heaney M., Heller G., Frattini M., Sadelain M. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118:4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbonaro D.A., Zhang L., Jin X., Montiel-Equihua C., Geiger S., Carmo M., Cooper A., Fairbanks L., Kaufman M.L., Sebire N.J., Hollis R.P., Blundell M.P., Senadheera S., Fu P.Y., Sahaghian A., Chan R.Y., Wang X., Cornetta K., Thrasher A.J., Kohn D.B., Gaspar H.B. Preclinical demonstration of lentiviral vector-mediated correction of immunological and metabolic abnormalities in models of adenosine deaminase deficiency. Mol Ther. 2014;22:607–622. doi: 10.1038/mt.2013.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 5.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galvan D.L., Nakazawa Y., Kaja A., Kettlun C., Cooper L.J., Rooney C.M., Wilson M.H. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geurts A.M., Yang Y., Clark K.J., Liu G., Cui Z., Dupuy A.J., Bell J.B., Largaespada D.A., Hackett P.B. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 8.Gogol-Doring A., Ammar I., Gupta S., Bunse M., Miskey C., Chen W., Uckert W., Schulz T.F., Izsvak Z., Ivics Z. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the piggyBac transposon in primary human CD4(+) T cells. Mol Ther. 2016;24:592–606. doi: 10.1038/mt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., Teachey D.T., Chew A., Hauck B., Wright J.F., Milone M.C., Levine B.L., June C.H. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S., Le Deist F., Carlier F., Bouneaud C., Hue C., De Villartay J.P., Thrasher A.J., Wulffraat N., Sorensen R., Dupuis-Girod S., Fischer A., Davies E.G., Kuis W., Leiva L., Cavazzana-Calvo M. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Sorensen R., Forster A., Fraser P., Cohen J.I., de Saint Basile G., Alexander I., Wintergerst U., Frebourg T., Aurias A., Stoppa-Lyonnet D., Romana S., Radford-Weiss I., Gross F., Valensi F., Delabesse E., Macintyre E., Sigaux F., Soulier J., Leiva L.E., Wissler M., Prinz C., Rabbitts T.H., Le Deist F., Fischer A., Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 12.Harkey M.A., Kaul R., Jacobs M.A., Kurre P., Bovee D., Levy R., Blau C.A. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cells Dev. 2007;16:381–392. doi: 10.1089/scd.2007.0015. [DOI] [PubMed] [Google Scholar]
- 13.Huang X., Guo H., Tammana S., Jung Y.C., Mellgren E., Bassi P., Cao Q., Tu Z.J., Kim Y.C., Ekker S.C., Wu X., Wang S.M., Zhou X. Gene transfer efficiency and genome-wide integration profiling of sleeping beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huye L.E., Nakazawa Y., Patel M.P., Yvon E., Sun J., Savoldo B., Wilson M.H., Dotti G., Rooney C.M. Combining mTor inhibitors with rapamycin-resistant T cells: a two-pronged approach to tumor elimination. Mol Ther. 2011;19:2239–2248. doi: 10.1038/mt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S., Maiti S., Dai J., Moriarity B., Forget M.A., Senyukov V., Orozco A., Liu T., McCarty J., Jackson R.N., Moyes J.S., Rondon G., Qazilbash M., Ciurea S., Alousi A., Nieto Y., Rezvani K., Marin D., Popat U., Hosing C., Shpall E.J., Kantarjian H., Keating M., Wierda W., Do K.A., Largaespada D.A., Lee D.A., Hackett P.B., Champlin R.E., Cooper L.J. Phase I trials using sleeping beauty to generate CD19-specific CAR T cells. J Clin Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochenderfer J.N., Feldman S.A., Zhao Y., Xu H., Black M.A., Morgan R.A., Wilson W.H., Rosenberg S.A. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother. 2009;32:689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manuri P.V., Wilson M.H., Maiti S.N., Mi T., Singh H., Olivares S., Dawson M.J., Huls H., Lee D.A., Rao P.H., Kaminski J.M., Nakazawa Y., Gottschalk S., Kebriaei P., Shpall E.J., Champlin R.E., Cooper L.J. piggyBac transposon/transposase system to generate CD19-specific T cells for the treatment of B-lineage malignancies. Hum Gene Ther. 2010;21:427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita D., Nishio N., Saito S., Tanaka M., Kawashima N., Okuno Y., Suzuki S., Matsuda K., Maeda Y., Wilson M.H., Dotti G., Rooney C.M., Takahashi Y., Nakazawa Y. Enhanced expression of anti-CD19 chimeric antigen receptor in piggyBac transposon-engineered T cells. Mol Ther Methods Clin Dev. 2018;8:131–140. doi: 10.1016/j.omtm.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller P.R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 22.Nakazawa Y., Huye L.E., Dotti G., Foster A.E., Vera J.F., Manuri P.R., June C.H., Rooney C.M., Wilson M.H. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochman H., Gerber A.S., Hartl D.L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A., Leslie C., Bushman F.D., Studer L., Sadelain M. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paruzynski A., Arens A., Gabriel R., Bartholomae C.C., Scholz S., Wang W., Wolf S., Glimm H., Schmidt M., von Kalle C. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 26.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 28.Thornhill S.I., Schambach A., Howe S.J., Ulaganathan M., Grassman E., Williams D., Schiedlmeier B., Sebire N.J., Gaspar H.B., Kinnon C., Baum C., Thrasher A.J. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ustek D., Sirma S., Gumus E., Arikan M., Cakiris A., Abaci N., Mathew J., Emrence Z., Azakli H., Cosan F., Cakar A., Parlak M., Kursun O. A genome-wide analysis of lentivector integration sites using targeted sequence capture and next generation sequencing technology. Infect Genet Evol. 2012;12:1349–1354. doi: 10.1016/j.meegid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Vera J., Savoldo B., Vigouroux S., Biagi E., Pule M., Rossig C., Wu J., Heslop H.E., Rooney C.M., Brenner M.K., Dotti G. T lymphocytes redirected against the kappa light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood. 2006;108:3890–3897. doi: 10.1182/blood-2006-04-017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson M.H., Coates C.J., George A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2