Fig. 4.
Chem-iLTP induces CaMKII–dependent PX-RICS phosphorylation to promote the assembly of PX-RICS and 14-3-3 and cell surface transport of GABAARs.
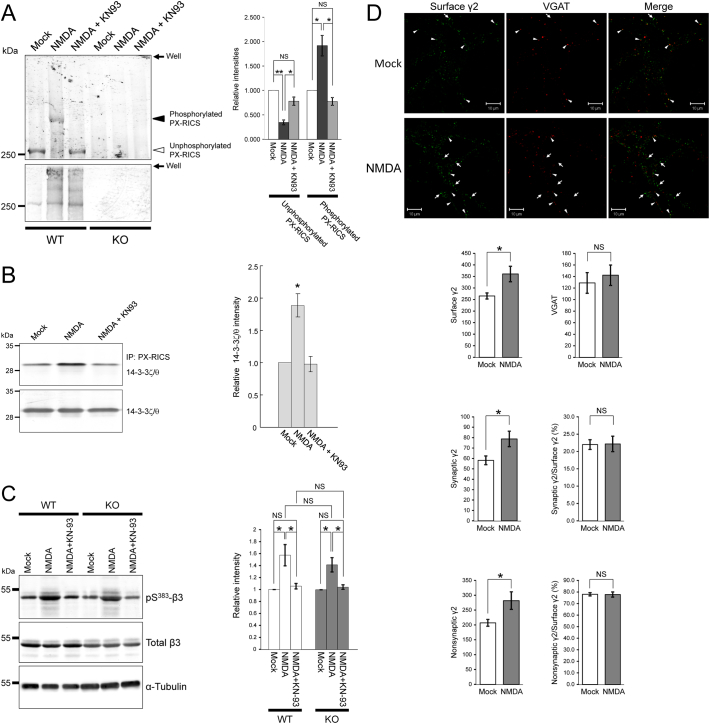
(A) WT and KO hippocampal neurons were moderately stimulated with NMDA in the presence or absence of KN93. The neuronal lysates were immunoprecipitated with an anti–PX-RICS antibody, and half of the immunoprecipitate was analyzed by Phos–tag (upper) or standard (lower) SDS-PAGE and immunoblotting. The intensities of unphosphorylated and phosphorylated PX-RICS bands (open and filled arrowheads, respectively) were corrected using those of total PX-RICS and the data were expressed as relative values with respect to the mock control (right panel).
(B) WT hippocampal neurons were moderately stimulated with NMDA, and the lysates were analyzed by immunoprecipitation with an anti–PX-RICS antibody followed by immunoblotting with anti–14-3-3ζ and anti–14-3-3θ antibodies (upper panel). Total 14–3-3ζ/θ protein in the neuronal lysates was assessed by immunoblotting with anti–14-3-3ζ and anti–14-3-3θ antibodies (lower panel). Quantitative analysis of 14-3-3ζ/θ bound to PX-RICS (right panel). The ratio of the band intensity of 14-3-3ζ/θ bound to PX-RICS to that of total 14-3-3ζ/θ was assessed, and the data were expressed as relative values with respect to mock control.
(C) Lysates of WT and KO hippocampal neurons modestly stimulated with NMDA in the presence or absence of KN93 were analyzed by immunoblotting with anti–β3 and anti–phospho-Ser383 β3 antibodies. α-tubulin was used as an internal control for the quantitative analysis.
(D) WT hippocampal neurons were modestly stimulated with NMDA and subjected to live cell labeling of the surface γ2, followed by co-staining with anti-VGAT antibody to identify synaptically localized GABAARs (arrowheads). Arrows indicate large or elongated puncta of nonsynaptic γ2 often observed in NMDA-treated neurons. Scale bars, 10 μm. n = 5 (mock) and 5 (NMDA) across two independent experiments.
Data are shown as the mean ± SEM from three (A, B, C) or two (D) independent experiments. NS, not significant; *P < 0.05; **P < 0.01 (Student's t-test).