Abstract
To date, most transcriptome studies of schizophrenia focus on the analysis of protein-coding genes. Long noncoding RNAs (lncRNAs) are emerging as key tissue-specific regulators of cellular and disease processes. The amygdala brain region has been implicated in the pathophysiology of schizophrenia. We performed unbiased whole transcriptome profiling of amygdala tissues from 22 schizophrenia patients and 24 non-psychiatric controls using RNA-seq. We reconstructed amygdala transcriptome and employed systems biology approaches to annotating the functional roles of lncRNAs. As a result, we identified 839 novel lncRNAs in amygdala. We found in amygdala lncRNAs are more subtype-specific than protein-coding genes. We identified functional modules associated with “synaptic transmission”, “ribosome”, and “immune responses” which were related to schizophrenia pathophysiology that involved lncRNAs. Integrative functional analyses associating individual lncRNAs with specific pathways and functions further show that amygdala lncRNAs are connected with all of these pathways. Our study presents the first systematic landscape of lncRNAs in amygdala tissue from schizophrenia cases.
Abbreviation: mRNA, Messenger RNA; lncRNA, Long noncoding RNA; SCZ, Schizophrenia
Keywords: Schizophrenia, Next-generation sequencing, Transcriptome, Systems biology
Research in Context.
Evidence Before this Study
The etiology of schizophrenia remains unknown. Most previous studies focus on the roles of protein-coding genes (mRNAs) in the disease. In the recent advance, long non-coding RNAs (lncRNAs), a kind of transcripts larger than 200 nt but without protein-coding potential, have been illustrated to play important biological roles, especially in the neural system. Some lncRNAs such as Gomafu are found to contribute to schizophrenia. The amygdala is the region of the brain that has been considered to play a primary role in the processing of emotional response, and pathophysiology of schizophrenia.
Added Value of this Study
To identify amygdala specific lncRNAs relevant to schizophrenia, we conducted a systematic analysis of lncRNAs in amygdala by using RNA-seq data with high sequencing depth and characterized their functions and roles in schizophrenia. We constructed novel amygdala lncRNAs, and amygdala lncRNAs are found to be more subtype-specific than protein-coding genes. We annotated functional roles of lncRNAs in amygdala using a systems biology approach. We demonstrated that lncRNA are involved in dysregulated gene pathways “synaptic transmission”, “ribosome”, and “immune responses” in schizophrenia.
Implications of All the Available Evidence
Our study presented the first systematic landscape of lncRNAs in amygdala tissue from schizophrenia cases, and proposed biomarkers of lncRNAs in schizophrenia that are worthy experimental validation in the future.
Alt-text: Unlabelled Box
1. Introduction
Schizophrenia (SCZ) remains one of the most mysterious and costliest mental disorders in terms of human suffering and societal expenditure [1]. Thus, it's important to improve our understanding of its molecular mechanisms of schizophrenia for developing effective therapies. RNA-Seq has been widely used to profile schizophrenia transcriptome in recent years for this purpose. Most of these studies have focused on differential expression analysis of protein-coding genes [2, 3] and some have analyzed allele-specific expression in schizophrenia [4].
Long non-coding RNAs (lncRNAs), the transcripts of larger than 200 nt but without protein-coding potential [5, 6], have received much attention in recent years. Though not fully understood, lncRNAs are conceived to play important roles in a variety of biological functions through molecule functions such as signals, decoys, guides and scaffolds [7]. LncRNAs are generally expressed at low level, and many of them are unknown. This makes it difficult to detect and quantify these molecules. The emergence of RNA-Seq provides the exact technology as needed for studying lncRNAs. RNA-Seq has been used to investigate lncRNAs in neurons [8]. Previous studies have demonstrated that lncRNAs can have a profound impact on gene regulations, especially in neurons [[9], [10], [11], [12], [13], [14]]. However, limited information exists on the roles of lncRNAs in schizophrenia. In addition, many lncRNAs, especially tissue-specific lncRNAs, remain to be discovered or are involved with biological functions that are complex or poorly defined and remain to be annotated.
The main obstacle to characterize lncRNAs is their low expression levels. It requires very high sequencing depth for RNA-Seq, which is generally lacking in many previous transcriptome studies. Consequently, although many transcriptome studies of schizophrenia have been conducted, most of them don't investigate lncRNAs, partially due to the high sequencing depth requirement and the poor annotation of lncRNAs currently available [2, 3, [15], [16], [17], [18], [19], [20], [21], [22]]. Liu et al. demonstrated dysregulation of non-coding RNA in the amygdala region of schizophrenia patients [23], but a systematical demonstration of functional roles of lncRNAs in schizophrenia is lacking. Through its important role in the processing of emotions, amygdala region has been involved in the pathophysiology of schizophrenia. Currently, emotional perturbations are common symptoms of schizophrenia, suggesting that dysfunction in neurons within the amygdala region may contribute to the dysregulation and altered behavior of schizophrenia patients. To identify amygdala specific lncRNAs relevant to schizophrenia, we performed unbiased whole transcriptome profiling of human amygdala region using RNA-seq with high sequencing depth. We report the systematic identification of lncRNAs, and characterize their functions and roles in schizophrenia via systems biology approaches.
According to DSM-IV, schizophrenia can be divided into five subtypes: paranoid, disorganized, catatonic, undifferentiated and residual schizophrenia [24]. The clinical characteristics of each subtype can vary. It is interesting to investigate gene dysregulations in different subtypes that account for their distinct diversities. Unbiased whole transcriptome profiling would help to identify these dysregulations and interplay of these abnormally expressed genes systematically. In this study, we used amygdala tissue from 22 schizophrenia patients including three subtypes: paranoid, undifferentiated and disorganized, and 24 non-psychiatric controls. We profiled their amygdala whole transcriptomes with ~100 millions of 100 nt short reads per sample on average. To our knowledge, this is the first comprehensive characterization of lncRNAs in amygdala tissue from schizophrenia patients. The lncRNA landscape characterized here provides novel insights into the transcriptomic variations seen in the amygdala across subtypes of schizophrenia patients.
2. Materials and Methods
2.1. Experimental Design
We obtained 45 amygdala samples from postmortem brain from the Lieber Brain bank (http://www.libd.org), including 9 undifferentiated, 7 disorganized, 5 paranoid, and 24 controls without psychiatric diagnoses. The PMI, age and gender of cases and controls were well matched (Supplementary Table S1), as indicated by their no association with the phenotype (non-parametric Kruskal-Wallis rank sum test p-value of 0.2, 0.5 for PMI and age respectively; Chi-square test p-value of 0.67 for gender). The sample description and RNA-Seq data are available at https://www.ncbi.nlm.nih.gov/bioproject/379666.
2.2. RNA-Seq of Amygdala Tissue
RNA-Seq libraries were constructed using Illumina (RS-122-2001) TruSeq RNA sample Prep Kit following the manufacture instruction. The poly-A containing mRNA molecules were purified from 300 to 500 ng DNAse treated total RNA using oligo (dT) beads. Following the purification, the mRNA was fragmented into small pieces using divalent cations under elevated temperature (94 degree) for 2 min. Under this condition, the range of the fragments length is from 130 to 290 bp with a median length of 185 bp. Reverse transcriptase and random primers were used to generate the first strand cDNA from the cleaved RNA fragments. The second strand DNA was synthesized using DNA Polymerase I and RNaseH. These cDNA fragments then went through an end repair process using T4 DNA polymerase, T4 PNK and Klenow DNA polymerase, and the addition of a single ‘A’ base using Klenow exo (3′ to 5′ exo minus), then ligation of the illumine PE adapters using T4 DNA Ligase. An index was inserted into Illumina adapters so that multiple samples can be sequenced in a single lane. These products were then purified and enriched with PCR to create the final cDNA library for high throughput DNA sequencing using Highseq2000. The concentration of RNA-seq libraries was measured by Qubit (Invitrogen, CA) and quantified with qPCR. The quality of RNA-seq library was measured by LabChipGX (Caliper, MA) using HT DNA 1K/12K/Hi-sensitivity LabChip. The libraries are multiplexed and loaded on a flowcell for cluster generation on cBot (Illumina). While on sequencing run, the Illumina Real Time Analysis (RTA) module was used to perform image analysis, base calling, and the BCL Converter (CASAVA v1.8.2) were followed to generate FASTQ files which contain the sequence reads. The current sequencing depth is over 80 million (2x100bp 40 million paired-end) mappable sequencing reads.
2.3. Transcriptome Assembly and Novel lncRNAs Discovery
We followed the standard protocol of the transcript-level expression analysis of RNA-seq experiments [25]. We obtained an average read depth of 116M reads per sample (Supplementary Table S8). The first step is quality and adapter trimming. Trimming was done by Trim Galore - a wrapper script to automate quality and adapter trimming as well as quality control (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Adapter sequences and fragments with less than quality score 20 in raw reads were removed. The processed reads were then aligned to human reference genome version hg38 (reference chromosomes only) by HISAT2 [26]. Common SNPs recorded in dbSNP144 and splice sites from Ensemble (download from HISAT2 website) were taken into account to improve alignment accuracy. Only concordant read pairs were set to be reported. The output SAM files were converted to BAM files, then sorted and indexed by SAMtools [27]. Following alignment and filtering an average of 108 million reads were obtained per sample with an average alignment rate of 95% (Supplementary Table S8). After reads alignment, we applied the median transcript integrity number (medTIN) [28] score to assess the RNA quality of each sample. As a result, we obtained the medTINs in the range of 47.4 to 78.3, with a median of 70.8, which indicated a high level of RNA integrity (Supplementary Table S8). Obtained BAM files were used as input by assembler StringTie [29] (reference annotation was GENCODE v24 [30] + stringent Human Body Map lncRNAs set from Broad Institute [31]), default parameters were used, that means genomic regions with at least reads coverage 2.5 will be considered as transcripts. The resulting GTF files were combined by StringTie with –merge mode with GENCODE v24 + stringent Human Body Map lncRNAs as reference annotation. The merging took a consensus of individual transcriptomes, that could improve the reliability of novel loci and isoforms. After obtaining merged GTF file, we compared it with annotation of GENCODE v24 + stringent Human Body Map lncRNAs, by using the gffcompare tool from GFF utilities (https://ccb.jhu.edu/software/stringtie/gff.shtml), to identify novel transcripts (whose class code were labeled as “i”, “u” and “x” by gffcompare). As a result, we obtained 5924 novel transcripts. Then these novel transcripts were used to discover novel lncRNAs as the input of slncky software [32] to filter out potential unknown protein-coding genes or gene duplication events. We used slncky's novel lncRNAs report as our novel lncRNAs set. Then we added these lncRNAs annotation to the GENCODE v24 + stringent Human Body Map lncRNAs to do the following analysis.
2.4. Differential Expression and Pathway Enrichment Analysis
Differential expression analysis and summarization of expression levels were done based on read counts. All BAM files were first processed by the “prepDE.py” (https://ccb.jhu.edu/software/stringtie/dl/prepDE.py) script from stringTie to extract read count information in each RNA-Seq sample. The reference annotation GTF file we used was GENCODE v24 + stringent Human Body Map lncRNAs + our novel identified lncRNAs. After obtaining the read counts table, we conducted differential expression analysis and produced normalized counts using DESeq2 [33]. Reads Per Kilobase Million (RPKMs) were obtained by the normalized counts divided by gene lengths. Only expressed genes (i.e. the genes' 50th-percentile RPKM value is larger than 0, and the genes' 95th-percentile RPKM value is larger than 0.05) were selected to conduct differential expression analysis. P-values of DESeq2 were corrected using the Benjamini-Hochberg procedure [34] for multiple testing adjustment. To do pathway enrichment analysis, we calculated z-scores by using p-values and signs of log fold change from the output of DESeq2. These z-scores were used to rank genes, followed by pre-ranked GSEA [35]. Significantly enriched pathways were selected based on GSEA FDR q-values.
2.5. Expression Profile in Subtypes
We used an extended method described by Cabili et al. [14, 31] to quantify the SCZ subtype specificity of each lncRNA, mRNA, and pseudogene. For each gene, let vij be the RPKM value of the gene in the ith sample of the jth subtype, where i ranges from 1 to nij, the number of samples in subtype j, and j ranges from 1 to J (J = 4 in this study) in this study (Here “subtype” also contains controls). We first transform expression level by log2 transformation: xij = log2(vij + 1). Then we took the average of gene's expression levels across subtype: . Ideally, if one gene was perfectly subtype-specific, then should be equal to zero in all subtypes except one. Next, we calculated an expression profile that normalizes the gene expression in various subtypes. Formally:
Each ej is the proportion of the total expression that in the jth subtype. Third, we defined the idea expression profile in each subtype:
That is to say, Ej represents that a gene only expressed in the jth subtype. Finally, we calculated the distance between observed gene expression profile with the idea expression profile Ej by the Jensen-Shannon divergence. The Jensen-Shannon divergence is defined as:
where H is the Shannon entropy. Then we defined our subtype-specific score as:
The larger score means more subtype-specificity observed, and a score 1 means the gene expressed in only one subtype.
2.6. Quantification of lncRNAs and lncRNA-Pathway Association
First, RPKMs were obtained by DESeq2. RPKMs were calculated by the normalized counts divided by gene length. In order to make the result more reliable, we only selected highly expressed genes, namely, the genes with 50th-percentile RPKM values larger than 0 and 95th-percentile RPKM values >0.25. Then RPKM value of each lncRNA locus was correlated with all highly expressed mRNAs loci (log2 transformed: log2 (RPKM+1)). All 45 samples were used to calculate correlations. Then for each lncRNA, a list of correlation-based (Pearson correlation) ranked mRNAs was constructed and subject to GSEA to do pre-ranked analysis [35]. An association matrix of lncRNAs and GO terms was made: if a GO term was enriched in one lncRNA with FDR threshold 0.01, then we would assign 1 or −1 based on positive or negative enriched, otherwise 0 will be assigned.
Heatmap was generated based on the association of lncRNAs with GO terms, and hierarchical clustering was applied to both rows and columns. We cut lncRNAs into 10 clusters based on hierarchical clustering. Then we used Fisher exact test to rank GO terms in each cluster to determine the enrichment level of associated GO terms of each cluster with respect to other clusters.
2.7. Gene Co-Expression Network Analysis
Co-expression networks of highly expressed genes were identified by WGCNA according to the methods described previously [36, 37]. All 45 samples were used to construct WGCNA network. First, normalized RPKM values of highly expressed genes were quantified by DESeq2. Then a matrix of correlation (Pearson correlation) between all pairs of highly expressed genes was generated and further converted to an adjacency matrix with a power value β. The value of power value β was 20, as determined by the function pickSoftThreshold provided by WGCNA that met the scale-free fitting threshold of 0.9. Co-expression modules were found by the dynamic hybrid tree cut algorithm, with the parameter settings: height cutoff = 0.95, deepSplit = 4 and minimum cluster size = 20. If the correlation between the eigenvalues of two modules is >0.85, the two modules were considered as so similar that they were merged. The module-trait correlation was defined as the correlation between the module eigenvalue and the trait.
After identifying co-expression networks modules, we exported the modules, which were used as input to Cytoscape [38] for visualization. ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks) [39] was applied to identify significant interactions between genes in each module based on their mutual information and remove indirect interactions through data processing inequality (DPI). The arguments for ARACNE were set to be DPI tolerance of 0 and mutual information threshold of 0.5.
3. Results
3.1. Identification of a Stringent Set of Novel Amygdala lncRNAs
To discover novel lncRNAs, we first assembled transcriptome from RNA-Seq data of all 45 samples (clinical information of samples was summarized in Supplementary Table S1), followed by a stringent pipeline to remove transcripts with protein-coding potential or low-quality data (Method). We identified a set of 839 novel lncRNA genes with high quality (Supplementary Table S2, the names of novel lncRNAs begin with “MSTRG”). We found that a dominant majority of these novel lncRNAs (>80%) were intergenic, falling entirely in intergenic regions. About 10% were divergent, being transcribed in the opposite orientation of a coding gene with which they share a promoter. We found very few lncRNAs that were miRNA host genes or snoRNA host genes. To further evaluate the reliability of these novel lncRNAs, we applied the same protocol to assemble transcriptome from the RNA-Seq dataset of 79 GTEx normal amygdala tissues [40]. As a result, we recovered 215 of the novel lncRNAs completely, namely, with all exons found in the GTEx transcriptomes, and 106 of the novel lncRNAs recovered partially in GTEx dataset (Supplementary Table S2). We applied the RPKM saturation module from RSeQC [41] to evaluate that the current sequencing depth was saturated for these 839 novel lncRNAs (Fig. S1).
3.2. Amygdala lncRNAs are Shorter, Less Complex, and Expressed at Low Levels
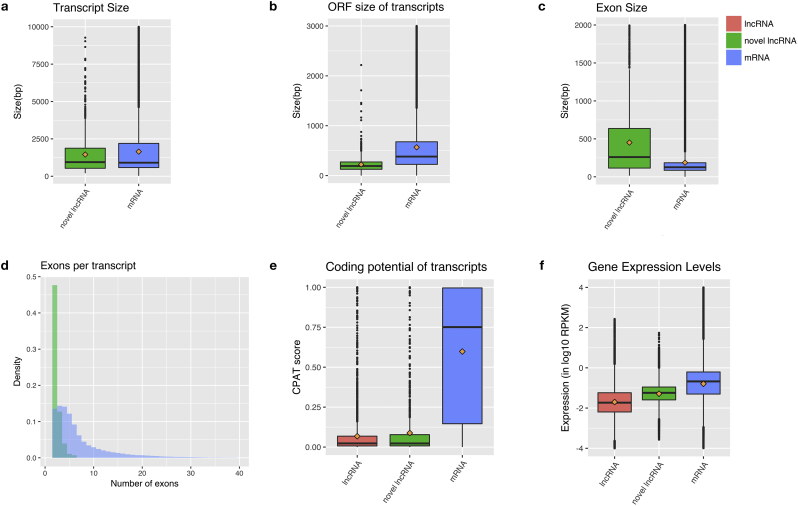
We first compared the structure, expression level and coding potentials of our assembled transcriptome (Fig. 1). All comparisons were done using one-sided Wilcoxon test. We found that novel lncRNAs, compared with protein-coding genes (we use the term mRNA to indicate protein-coding gene in this paper), have significantly shorter transcript length (Fig. 1a, 1477 bp vs. 1727 bp, p-value = 7.566e-06), shorter ORFs (Fig. 1b, 192 bp vs. 390 bp, p-value < 2.2e-16), and longer exons (Fig. 1c, 766 bp vs 238 bp, p-value < 2.2e-16). These properties were consistent with the lower estimated number of exons for lncRNAs compared with mRNAs (Fig. 1d, 2 vs. 5, p-value < 2.2e-16). Taken together, we conclude that amygdala lncRNAs are shorter and less complex than mRNAs.
Fig. 1.
Genomic features of novel lncRNAs. (a) Transcript sizes of lncRNAs and mRNAs. (b) Open reading frame (ORF) sizes of lncRNA and mRNAs. (c) Exon sizes of lncRNAs and mRNAs. (d) Numbers of exons per lncRNA and mRNAs. (e) Coding potential (CPAT scores) of known lncRNAs, novel lncRNAs, and mRNAs. (f) Expression levels (RPKM values) of known lncRNAs, novel lncRNAs, and mRNAs. (a), (b), (c), (e), (f) are standard boxplots, which display the distribution of data by presenting the inner fence (the whisker, taken to 1.5× the Inter Quartile range, or IQR, from the quartile), first quartile, median, third quartile and outliers. The means are marked as tan diamonds.
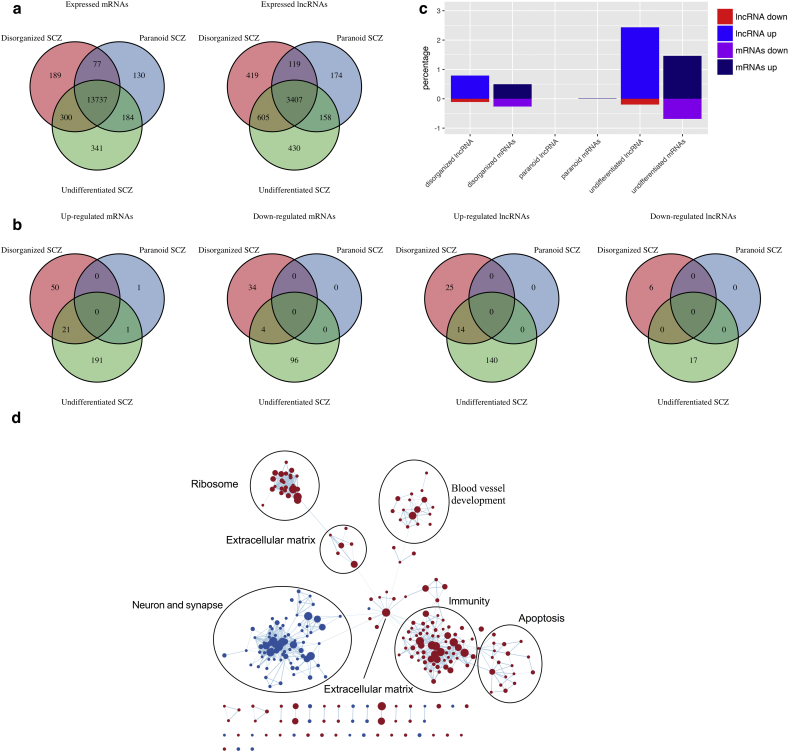
Fig. 2.
Dysregulation of expression of mRNAs, lncRNAs and gene pathways. (a) Venn diagrams of expressed protein-coding genes (left) and lncRNAs (right) in three subtypes. (b) Venn diagrams of upregulated/downregulated mRNAs and lncRNAs in three subtypes. (c) Percentages of dysregulated mRNAs and lncRNAs. (d) Enrichment map for significant GO terms in schizophrenia (all SCZs vs normal), nominal p < 0.005 and FDR q value <0.1. Nodes represent gene sets that are significantly up or down regulated, as determined by GSEA, where the node size corresponds to the size of the GO term. Edges indicate overlap between gene sets, where the thickness indicates the overlap coefficient. Red nodes indicate up-regulation and blue nodes indicate down-regulation.
We then measured protein-coding potential of lncRNAs and mRNAs using the CPAT [42], which was a logistic regression model built with four sequence features: open reading frame size, open reading frame coverage, Fickett TESTCODE statistic and hexamer usage bias. We found that both known lncRNAs and novel lncRNAs had comparable low protein-coding potential, which was significantly lower than that of mRNAs (Fig. 1e, median of 0.023, 0.023, 0.75 for known lncRNAs, novel lncRNAs and mRNAs, p-value < 2.2e-16 for both known lncRNA vs mRNAs and novel lncRNAs vs mRNAs). This suggests that our algorithm to identify novel lncRNAs is reliable. Notably, SCZ lncRNAs were expressed on average at about 10-fold lower levels than mRNAs, with the median expression level of 0.018, 0.057 and 0.21 RPKM for known lncRNAs, novel lncRNAs, and mRNAs, respectively (Fig. 1f, p-value < 2.2e-16 for both known lncRNAs vs mRNAs and novel lncRNAs vs mRNAs). Similar findings were observed in other human tissues [31, 43]. Novel amygdala lncRNAs exhibited similar characteristics as known lncRNAs. Therefore, in the following sections, we combined the novel and known lncRNAs for further functional analysis.
3.3. Identification of Differentially Expressed Genes and Pathways
After obtaining the assembled novel lncRNAs, we appended these lncRNAs definitions together with the Broad Institute's lncRNAs set to the GENCODE v24 annotation. As a result, we have 19,815 mRNAs and 18,805 lncRNAs. To rule out the potential unreliability of lowly expressed genes, we define a gene as expressed if, [1] the genes' 50th-percentile RPKM value is larger than 0, and [2] the genes' 95th-percentile RPKM value is larger than 0.05. Among all SCZ and control samples, there were 14,570 expressed mRNAs, and 4596 expressed lncRNAs (of which 697 were novel). For the three subtypes of SCZ samples, 14,331 mRNAs and 4336 lncRNAs on average were expressed. Of those, 13,737 mRNAs and 3407 lncRNAs were detected in all three subtypes of SCZs, respectively, suggesting a high level of diversity of lncRNA expression across the three subtypes (Fig. 2a).
To characterize schizophrenia-associated dysregulation of gene expression, we performed four differential expression analyses of the schizophrenia subtypes in comparison with normal control samples: (1) all SCZs vs control, (2) disorganized SCZs vs controls, (3) paranoid SCZs vs controls, and (4) undifferentiated SCZs vs controls, in search for both common SCZ related genes as well as SCZ subtypes specific genes (in the set of expressed genes). Under the cutoff of adjusted p-value < 0.05, there are 345, 182, 2 and 541 differentially expressed genes for these four comparisons, respectively (Supplementary Table S3). The comparison of differentially expressed mRNAs across 3 subtypes was summarized (Fig. 2b, c). Among them, we uncovered several protein-coding genes significantly differentially expressed in all SCZ samples that were reported in previous studies [18, 19, 21, 22]. These include HBA1, HBA2, HBB, IFITM1, GBP1, IFITM2, SERPINA3. These results were identical to the previous study focusing on protein-coding genes in the amygdala of schizophrenia patients [20].
To determine which functional pathways are most robustly involved in schizophrenia pathogenesis, we performed gene set enrichment analysis (GSEA) for each subtype and all case samples combined. At FDR level of 0.1 (nominal p-value < 0.005), there were 178, 40, 355 and 123 GO terms positively enriched genes, and 94, 147, 4 and 107 GO terms negatively enriched genes in all SCZ, disorganized SCZ, paranoid SCZ, and undifferentiated SCZ, respectively. The three subtypes demonstrated enrichment in many unique gene sets, while some common ones were identified across all three subtypes (Supplementary Table S4). When considering all SCZ vs. normal, we found the dysregulated pathways came from four main categories: “immunity”, “blood vessel development” and “ribosome and protein synthesis” were up-regulated; and “neuron and synapse” were down-regulated (Fig. 2d). We also found the gene pathway from “immunity”, “ribosome”, and “neuron and synapse” categories in all three subtypes of SCZ (Supplementary Table S4). These results agreed with previous studies on dysregulation of functional pathways in SCZ especially in amygdala tissue [20]. We concluded that in the amygdala tissues of schizophrenia patients, gene pathways related to immune response, blood vessel development and ribosome were upregulated in expression, gene pathways related to synaptic transmission and behavior were suppressed. We want to further identify the potential roles of lncRNAs relating to these functional pathways in the following sections.
At FDR level of 0.05, we detected 110, 45, 0, 171 differentially expressed lncRNAs in all SCZ samples, disorganized SCZ samples, paranoid SCZ samples and undifferentiated SCZ samples, respectively (Supplementary Table S3). Venn diagrams and percentages of differentially expressed lncRNAs in the three subtypes were presented (Fig. 2b, c). Previous studies have reported lncRNAs association with schizophrenia, including MIAT, DLX6-AS1, and BDNF-AS [12, 44]. We didn't find these lncRNAs significantly differentially expressed in our dataset.
3.4. Expression Profiles of lncRNAs in Schizophrenia Subtypes
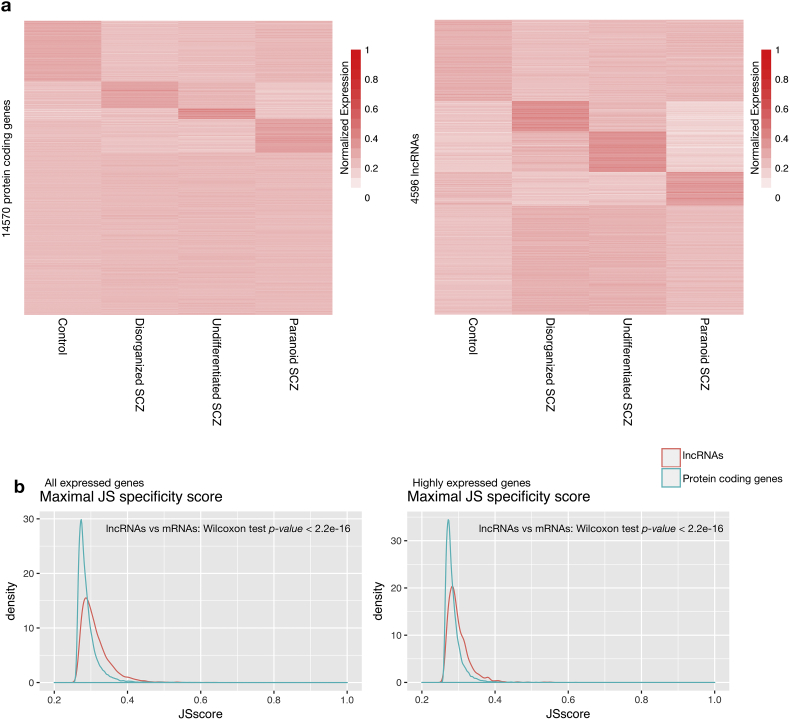
To determine whether amygdala lncRNAs are subtype-specific in schizophrenia, we characterized the expression profiles of lncRNAs and mRNAs across the three schizophrenia subtypes in comparison with controls. A larger number of lncRNAs exhibited subtype-specific expression patterns than mRNAs based on the unsupervised clustering of expression profiles (only expressed genes were analyzed, Fig. 3a). Furthermore, we calculated subtype specificity score for each gene using an entropy-based metric that relies on Jensen-Shannon (JS) divergence [31] (Materials and Methods). The expression of lncRNAs was found to be more subtype-specific than mRNAs significantly, with median specificity score of 0.302 and 0.282 for lncRNAs and mRNAs, respectively (Fig. 3b). To rule out the possibility that this effect was caused by increased noise from low expressed genes, we also calculated the specificity scores of only highly expressed genes (i.e., 50th-percentile RPKM values >0 and 95th-percentile RPKM values >0.25). There were 7382 and 530 highly expressed mRNAs and lncRNAs, respectively. Again, we observed these highly expressed lncRNAs showed a higher subtype specificity than mRNAs (Fig. 3b, with median specificity score of 0.291 and 0.279 for lncRNAs and mRNAs, respectively).
Fig. 3.
Subtype specificity of lncRNAs and mRNAs. (a) Heatmaps of normalized expression levels of 14,570 mRNAs (left) and 4596 lncRNAs (right). (b) Distributions of maximal subtype specificity scores for all expressed lncRNAs and protein-coding genes (left) and highly expressed ones (right).
3.5. Functional Annotation of Amygdala lncRNAs Through Expression Correlation
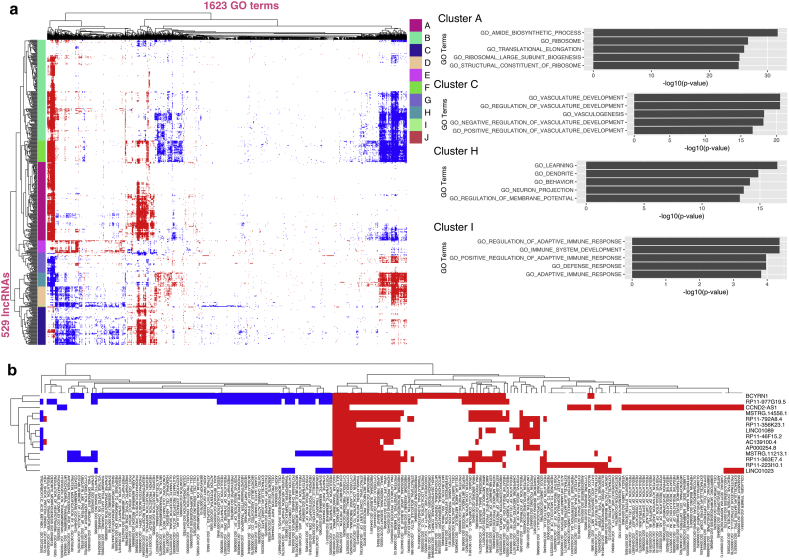
The lack of annotated features makes it a challenging task to assign functions to lncRNAs. However, “guilt-by-association” analysis helps to predict roles of mammalian lncRNAs [31, 43, 45, 46]. Because genes with similar co-expression patterns tend to have similar functional coherency [47], we conducted gene set enrichment analysis (GSEA) to associate GO terms and lncRNAs by analyzing the correlation between the expression dynamics of each lncRNA with the expression dynamics of each mRNA across all 45 SCZ and normal samples using the highly expressed 530 lncRNAs and 7382 mRNAs (Materials and Methods). As a result, 529 lncRNAs were identified to have significantly associated GO terms (Fig. 4a). We grouped lncRNAs into 10 clusters by hierarchical clustering (Supplementary Table S5). We found that several clusters were associated with protein-coding gene sets of distinct functional categories, such as ribosome and protein synthesis (cluster A), blood vessel development (cluster C), nervous system and synaptic transmission (cluster H) and immune system (cluster I) (Fig. 4b). These pathways were found dysregulated in schizophrenia.
Fig. 4.
Prediction of lncRNA functions. (a) Expression-based association matrix of 529 highly expressed lncRNAs and functional 1623 GO terms (left). Rows are lncRNAs, columns are GO terms. In heatmap, red, blue, and white dots represent positive, negative, and no correlation respectively. Top 5 most enriched GO terms in cluster A, C, H, and I are showed (right). (b) Association heatmap of dysregulated lncRNAs in all SCZs (all SCZs vs controls).
Interestingly, by focusing on differential expressed lncRNAs (all SCZ vs. controls), we observed that these lncRNAs were associated with protein synthesis, blood vessel development, nervous system pathways and immune system pathways (Fig. 4b), and these pathways were dysregulated in all of three subtypes of schizophrenia. These observations indicate that dysregulated amygdala lncRNAs are putative contributors to the pathogenesis of schizophrenia.
3.6. lncRNA-mRNAs Co-Expression Network
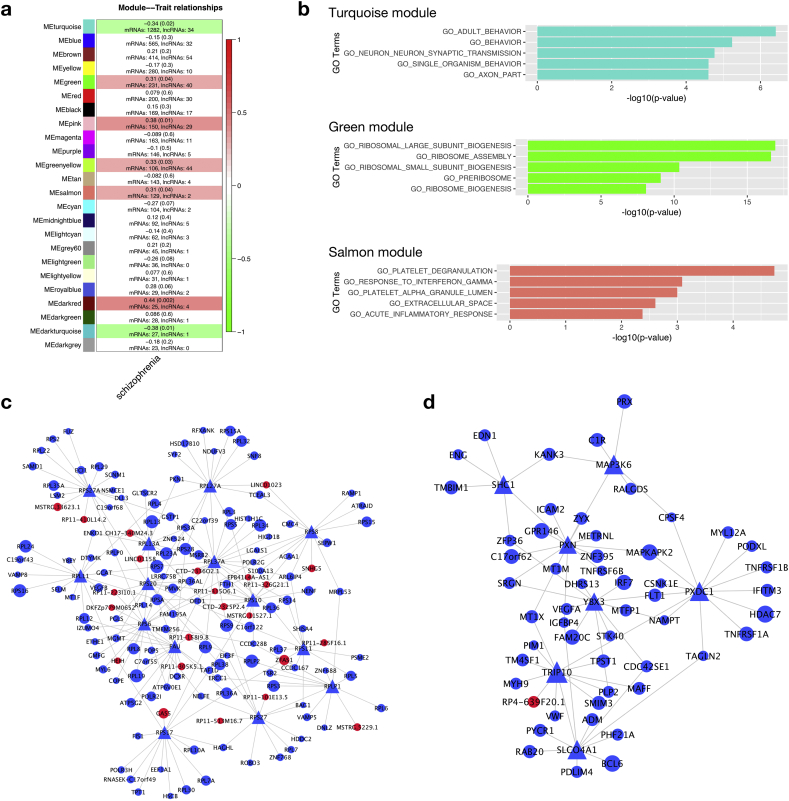
We built co-expression networks over the highly expressed 7912 genes (including 530 lncRNAs, and 7382 mRNAs) using WGCNA [37], followed by network modules testing (total 45 samples were used). We identified 23 co-expressed modules (Supplementary Table S6), 7 of which were highly correlated with the schizophrenia trait (Fig. 5a): turquoise, green, pink, greenyellow, salmon, darkred and darkturquoise (correlation p-value < 0.05). To identify functions of each module, we performed a hypergeometric test on mRNAs to detect enriched GO terms in each module. We found that turquoise module was enriched with neuron related pathways, the green module enriched with ribosomal pathways, and the salmon module enriched with immune response related pathways (Fig. S2). Our previous GSEA analysis showed these pathways were dysregulated in schizophrenia. The numbers of genes (including both mRNAs and lncRNAs) of these modules varied. We then ran the Fisher exact test to determine the enrichment level of associated GO terms for lncRNAs in each module. The associated functions of lncRNAs (GO terms associated with lncRNA were determined in the Section “Functional annotation of amygdala lncRNAs through expression correlation”) are consistent with their coding counterparts in each module: lncRNAs in the turquoise module are primarily associated with neuronal and synaptic pathways, lncRNAs in the green module are primarily associated with ribosomal pathways, and lncRNAs in the salmon module are primarily associated with immunity pathways (Fig. 5b).
Fig. 5.
Co-expression network of lncRNAs and mRNAs. (a) Module-trait relationships. Each row represents a module, the column is the trait (schizophrenia or normal). Each cell contains corresponding correlation coefficient and p-value in the bracket. Numbers of mRNAs and lncRNAs in each module are also shown. (b) Enriched lncRNA-correlated GO terms in turquoise, green and salmon module. (c, d) Graphic view of lncRNA-mRNAs co-expression network of (c) green and (d) salmon module. Node size represents corresponding intramodular connectivity degrees, blue and red nodes represent mRNAs and lncRNAs respectively, hub genes are displayed in triangles. Connection lines between nodes indicate direct interactions determined by ARACNE. Only hub genes and genes directly connected to hub showed.
Following [48], we defined 5% of nodes with highest intra-modular connectivity as hub genes (Supplementary Table S6). Connectivity reflects how frequently a node interacts with other nodes in a co-expression module. Hub genes, which had the highest connectivity, were usually considered as key regulators in gene co-expression networks since they intended to interact with more genes than others [[49], [50], [51]]. The turquoise module was enriched by genes from neuron pathways and many of their members were observed as hub genes. We did find lncRNA “ENSG00000225465.8” (RFPL1S) as a hub gene in turquoise (Fig. S3), which suggested that this lncRNA might play as a key regulator in this functional module. Interestingly, the gene pathways positively correlated with “ENSG00000225465.8” included axon, synapse, dendrite as well as other similar pathways (Supplementary Table S7). There were 20 lncRNAs directly connected to the hub genes in the turquoise module. We found most of them were positively associated with synapse pathways (Supplementary Table S7). In contrast, in the green and salmon modules, no lncRNA was identified as hub genes, but there were several lncRNAs directly connected with the hub genes (Fig. 5c, d). In the green module, there were 23 lncRNAs connected to the hub genes directly. GSEA showed that all of these lncRNAs were associated with ribosome pathways (Supplementary Table S7). In the salmon module, we identified one lncRNA connected to the hub genes directly: “ENSG00000235501.5” (RP4-639F20.1), which was associated with immune pathways (Supplementary Table S7) as indicated by GSEA. To take all these results together, by gene co-expression network, we identified seven co-expression modules correlated with SCZ trait. We found three of these seven modules were themed by neuron and synapse, ribosome and immunity respectively. In particular, we noticed that lncRNAs existed in all of the 3 modules, acting as the hub gene in the turquoise module while connecting to the hub genes directly in the other two modules. GSEA analysis of lncRNA-mRNA correlations showed that the lncRNAs were significantly associated with several pathways in each module (Supplementary Table S7). All these results suggested the potential involvement of lncRNAs in schizophrenia-related functional pathways.
4. Discussion
There is growing evidence that non-coding RNAs, particularly miRNAs and lncRNAs are involved in the pathogenesis of schizophrenia. For instance, the lncRNA MIAT (Gomafu), is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing [12]. However, they found MIAT was down-regulated in schizophrenia samples, whereas we didn't find significance difference of this lncRNA in schizophrenia and normal tissues. The tissue used in their study was brain cortex of mice, but here we used human amygdala tissue. The difference of tissue may account for the difference of expression change.
The GENCODE v24 annotation dataset contains 15,941 lncRNAs and we found about 24.5% of them were expressed in the amygdala under our study. Although GENCODE was presumably the most compressive gene annotation [52], we still identified 5924 novel transcripts not previously annotated. Our library preparation was based on the polyA tailed method, so the transcripts we found were all polyA tailed. We would expect more transcripts which were not polyA tailed. Among them, we defined a stringent set of 839 novel lncRNAs, which include lincRNAs (long intergenic non-coding RNAs), intronic overlapping lncRNAs, and exonic antisense overlapping lncRNAs. Our novel lncRNAs document the first novel long noncoding transcripts in amygdala tissue. To further validate these novel lncRNAs, we investigated if they can be found in other amygdala RNA-Seq data sets such as GTEx [40]. Due to the high heterogeneity of lncRNAs, we only recovered 321 novel lncRNAs completely or partially in GTEx amygdala data set. Nevertheless, read coverage was shown to be saturated for these 839 novel lncRNAs. With high numbers of reads support, these novel transcripts are expected to be bona fide. Based on this finding, we would expect that many more lncRNAs remain to be identified, and the next generation sequencing data from other tissue types will uncover these in the near future.
Consistent with previous RNA-Seq studies, our results showed that lncRNAs expressed about 10-fold lower than coding genes in bulk tissues. The recent single-cell RNA-Seq (scRNASeq) technology makes it possible to profile gene expression in each individual cell. It has been revealed using scRNASeq that many lncRNAs are abundantly expressed in individual cells and are cell type-specific [53]. We expect the better characterization of lncRNAs in the amygdala with the accumulation of single-cell transcriptomics data of this tissue.
A well-known feature of lncRNAs is their tissue specificity. Here we also observed high specificity of lncRNAs among different subtypes of schizophrenia even in a single tissue – i.e., amygdala. The interpretation of this is that lncRNAs may vary much more across different patients when compared with mRNAs even in a single tissue. To our knowledge, this is the first study to show that lncRNAs demonstrate substantially more variation between disease subtypes in schizophrenia than do mRNAs.
In an effort to understand potential roles of lncRNAs in schizophrenia pathophysiology, we first identified differentially expressed lncRNAs across SCZ patients and healthy controls. Many mRNAs are known to be dysregulated in schizophrenia, whereas dysregulation of lncRNAs hasn't been analyzed in any considerable depth before, especially in amygdala tissue specimens. By a rigid statistical significance of FDR < 0.05, we observed 45, 0 and 171 lncRNAs that were differentially expressed in disorganized SCZs, paranoid SCZs and undifferentiated SCZs, respectively. We also observed 110 differentially expressed lncRNAs when comparing all SCZs cases vs. controls. We generated a co-expression network using all highly expressed lncRNAs and protein-coding genes. Seven modules that were correlated with SCZ were identified, and three of them were themed by the gene pathways identified to be dysregulated in SCZ. We pinpointed one lncRNA in the turquoise module (themed by neuron and synapse) as a hub gene. While no lncRNAs in the green (themed by ribosome) and salmon modules (themed by immunity) were identified as hub genes, we observed that lncRNA connected directly with hub genes in all of the three modules, which implies that lncRNAs may involve as regulatory factors in these modules.
We subsequently annotated all expressed lncRNAs through their correlation with protein-coding genes. In the context of co-expression of protein-coding genes and GSEA functional predictions, our data suggest that lncRNAs may play diverse roles in amygdala tissue. The main pathways dysregulated in schizophrenia involve immune response, ribosome, blood vessel development and synaptic transmission. Our results show that lncRNAs are involved in all of these pathways. This is an intriguing possibility and suggests that lncRNAs may be involved in the pathogenic dysregulation of schizophrenia. These potential functional roles of lncRNAs in schizophrenia etiology as determined by bioinformatics analysis are worthy further experimental validation in the future.
Due to the difficulty to collect human post-mortem samples, we were not able to obtain a very large size of samples, which put a power limit to the statistical tests. We alleviated this small sample size issue by focusing on pathway-level analysis and illustrated potential roles of lncRNAs in the pathogenic pathways of schizophrenia. Pathway analysis was proved to be more robust than single-gene analysis, and could mitigate the small sample size issue [35, 36]. Nevertheless, caution should be used that some subtle but important signals may still remain undetectable given this sample size. The current promising results call for recruiting a large cohort of samples for more comprehensive analyses in the future. In addition, it is noted that in the recent DSM-5 diagnostic criteria, DSM-IV subtypes have been omitted, and improved to a new scale, the Clinician-Rated Dimensions of Psychosis Symptom Severity (C-RDPSS), for characterizing patients [54]. In this study, the subtype information was collected following previous clinical practice but not C-RDPSS. We expect better results be obtained when using the new severity-based characterization of patients in future studies with the implementation of DSM-5.
Taken together, our study provides evidence that multiple novel lncRNAs reside within the amygdala region in schizophrenia patients whose function is differentially involved in the regulation of numerous gene expression networks in different subtypes of schizophrenia. These findings suggest that lncRNAs may have multiple roles in schizophrenia that is highly differential between different subtypes of schizophrenia, which is to be explored further in future genetic and mechanistic studies.
The following are the supplementary data related to this article.
Clinical information of 45 Postmortem brain samples.
BED format of 839 novel lncRNAs. The matching type to GTEx transcriptome was also included.
Differentially expressed genes in each subtype of schizophrenia.
Dysregulated pathways in each subtype of schizophrenia.
Clustering of lncRNAs based on associated GO terms.
Summary of co-expression modules.
Pathway associated with the hub lncRNAs and the lncRNAs connected to hub genes in the turquoise, green and salmon modules.
Supplementary material
Funding Sources
This study is funded by The Children′s Hospital of Philadelphia Endowed Chair in Genomic Research to Dr. Hakonarson, an Institutional Development Award to the Center for Applied Genomics from The Children’s Hospital of Philadelphia; MH096891-03S1 (NIMH); RC2 MH089924-02 (NIMH); R01 MH097284-03 (NIMH); and U01-HG008684 (NIH).
Declaration of Interests
The authors declare that they have no competing interests.
Author Contributions
Z.W., and H.H. conceived and supervised the project. T.T. designed and implemented the methods, and conducted the experiments. X.C., Y.L., R.E.G., and P.M.S. contributed to data acquisition. T.T., Z.W., and H.H. wrote the manuscript. All authors approved the manuscript.
Acknowledgments
We thank Joel Kleinman, Daniel R Weinberger and Thomas M Hyde for their assistance in recruiting and sequencing subjects for participation in the study.
Contributor Information
Zhi Wei, Email: zhiwei@njit.edu.
Hakon Hakonarson, Email: hakonarson@email.chop.edu.
References
- 1.van Os J., Kapur S. Schizophrenia. Lancet. 2009;374(9690):635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 2.Mudge J., Miller N.A., Khrebtukova I. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One. 2008;3(11) doi: 10.1371/journal.pone.0003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J.Q., Wang X., Beveridge N.J. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0036351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith R.M., Webb A., Papp A.C. Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics. 2013;14:571. doi: 10.1186/1471-2164-14-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P., Cheng J., Dike S. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 7.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin M., Pedrosa E., Shah A. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sleutels F., Zwart R., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 10.Nagano T., Mitchell J.A., Sanz L.A. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 11.Pandey R.R., Mondal T., Mohammad F. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 12.Barry G., Briggs J.A., Vanichkina D.P. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 13.Willem M., Garratt A.N., Novak B. Control of peripheral nerve myelination by the ß-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 14.Yan X., Hu Z., Feng Y. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fillman S.G., Cloonan N., Catts V.S. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2013;18(2):206–214. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- 16.Arion D., Unger T., Lewis D.A., Levitt P., Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62(7):711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi K.H., Elashoff M., Higgs B.W. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry. 2008;8:87. doi: 10.1186/1471-244X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saetre P., Emilsson L., Axelsson E., Kreuger J., Lindholm E., Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang Y., Kim J., Shin J.Y. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry. 2013;3:e321. doi: 10.1038/tp.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang X., Liu Y., Hahn C.G., Gur R.E., Sleiman P.M.A., Hakonarson H. RNA-seq analysis of amygdala tissue reveals characteristic expression profiles in schizophrenia. Transl Psychiatry. 2017;7(8) doi: 10.1038/tp.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., Sun J., Chen J. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics. 2012;13(Suppl 8):S2. doi: 10.1186/1471-2164-13-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glatt S.J., Everall I.P., Kremen W.S. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102(43):15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Chang X., Hahn C.G., Gur R.E., Sleiman P.A.M., Hakonarson H. Non-coding RNA dysregulation in the amygdala region of schizophrenia patients contributes to the pathogenesis of the disease. Transl Psychiatry. 2018;8(1):44. doi: 10.1038/s41398-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diagnostic and statistical manual of mental disorders. Fourth Edition. Text Revision (DSM-IV-TR); 2000. [DOI] [PubMed] [Google Scholar]
- 25.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L., Nie J., Sicotte H. Measure transcript integrity using RNA-seq data. BMC Bioinform. 2016;17:58. doi: 10.1186/s12859-016-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrow J., Frankish A., Gonzalez J.M. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22 doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabili M.N., Trapnell C., Goff L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Shishkin A.A., Zhu X. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016;17:19. doi: 10.1186/s13059-016-0880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 35.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 37.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P., Markiel A., Ozier O. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margolin A.A., Nemenman I., Basso K. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium G.T. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M., Amit I., Garber M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merelo V., Durand D., Lescallette A.R. Associating schizophrenia, long non-coding RNAs and neurostructural dynamics. Front Mol Neurosci. 2015;8:57. doi: 10.3389/fnmol.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinger M., Amaral P., Mercer T. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauli A., Valen E., Lin M.F. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22 doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H.K., Hsu A.K., Sajdak J., Qin J., Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Han L., Yuan Y., Li J., Hei N., Liang H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat Commun. 2014;5:3231. doi: 10.1038/ncomms4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabasi A.L. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 51.Liang H., Li W.H. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23(8):375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Zhao S., Zhang B. A comprehensive evaluation of ensembl, RefSeq, and UCSC annotations in the context of RNA-seq read mapping and gene quantification. BMC Genomics. 2015;16:97. doi: 10.1186/s12864-015-1308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S.J., Nowakowski T.J., Pollen A.A. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mattila T., Koeter M., Wohlfarth T. Impact of DSM-5 changes on the diagnosis and acute treatment of schizophrenia. Schizophr Bull. 2015;41(3):637–643. doi: 10.1093/schbul/sbu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information of 45 Postmortem brain samples.
BED format of 839 novel lncRNAs. The matching type to GTEx transcriptome was also included.
Differentially expressed genes in each subtype of schizophrenia.
Dysregulated pathways in each subtype of schizophrenia.
Clustering of lncRNAs based on associated GO terms.
Summary of co-expression modules.
Pathway associated with the hub lncRNAs and the lncRNAs connected to hub genes in the turquoise, green and salmon modules.
Supplementary material