Abstract
Background
Mobilized resistance to colistin is evolving rapidly and its global dissemination poses a severe threat to human health and safety. Transferable colistin resistance gene, mcr-3, first identified in Shandong, China, has already been found in several countries in multidrug-resistant human infections. Here we track the spread of mcr-3 within 13 provinces in China and provide a complete characterization of its evolution, structure and function.
Methods
A total of 6497 non-duplicate samples were collected from thirteen provinces in China, from 2016 to 2017 and then screened for the presence of mcr-3 gene by PCR amplification. mcr-3-positive isolates were analyzed for antibiotic resistance and by southern blot hybridization, transfer analysis and plasmid typing. We then examined the molecular evolution of MCR-3 through phylogenetic analysis. Furthermore, we also characterized the structure and function of MCR-3 through circular dichroism analyses, inductively coupled plasma mass spectrometry (ICP-MS), liquid chromatography mass spectrometry (LC/MS), confocal microscopy and chemical rescue tests.
Findings
49 samples (49/6497 = 0.75%) were mcr-3 positive, comprising 40 samples (40/4144 = 0.97%) from 2017 and 9 samples (9/2353 = 0.38%) from 2016. Overall, mcr-3-positive isolates were distributed in animals and humans in 8 of the 13 provinces. Three mcr-3-positive IncP-type and one mcr-1-bearing IncHI2-like plasmids were identified and characterized. MCR-3 clusters with PEA transferases from Aeromonas and other bacteria and forms a phylogenetic entity that is distinct from the MCR-1/2/P(M) family, the largest group of transferable colistin resistance determinants. Despite that the two domains of MCR-3 not being exchangeable with their counterparts in MCR-1/2, structure-guided functional mapping of MCR-3 defines a conserved PE-lipid recognizing cavity prerequisite for its enzymatic catalysis and its resultant phenotypic resistance to colistin. We therefore propose that MCR-3 uses a possible “ping-pong” mechanism to transfer the moiety of PEA from its donor PE to the 1(or 4′)-phosphate of lipid A via an adduct of MCR-3-bound PEA. Additionally, the expression of MCR-3 in E. coli prevents the colistin-triggered formation of reactive oxygen species (ROS) and interferes bacterial growth and viability.
Interpretation
Our results provide an evolutionary, structural and functional definition of MCR-3 and its epidemiology in China, paving the way for smarter policies, better surveillance and effective treatments.
Keywords: Lipid A, Polymyxin resistance, Acquired colistin resistance, MCR-P(M), MCR-3, MCR-2, MCR-1, Gut bacteria, Microbiome
Research in Context.
Evidence Before this Study
On July 18, 2018, we searched PubMed with the terms “mcr-3 and China [21 from local epidemiology]”, “MCR-3 and function [1 references]”, “MCR-3 and evolution [no references]”, for reports published between January 2000 and July 2018. We did not restrict our search by language of publication. Our search identified some group reported the sporadic cases of MCR-3 in very confined area and very limited in number in China. Evidently, comprehensive epidemiology of MCR-3 remains unclear in China. More importantly, we found no reports addressing mechanisms of MCR-3 action. Therefore, it is very necessary to elucidate its potential spread, evolution and functional aspects of MCR-3 polymyxin resistance.
Added Value of this Study
Our results represent a first multi-province study on the dissemination of MCR-3 in China. Also, we report the origin and possible evolution of MCR-3. We have integrated multiple approaches to systematically address the biochemical mechanism and physiological roles of MCR-3 action.
Implications of all the Available Evidence
Our data shows that 49(49/6497 = 0.75%) isolates from 13 provinces in China, comprising 40 samples (40/4144 = 0.97%) from 2017 and 9 samples (9/2353 = 0.38) from 2016, were mcr-3 positive. This study suggests that the threat of mcr-3 to public health should be assessed because of the potential prevalence of mcr-3. In addition, given that the resistance mechanism of MCR-3 is similar to mcr-1, we suggest that further studies are needed to clarify the evolutionary pattern of mcr-3.
Alt-text: Unlabelled Box
1. Introduction
Antimicrobial resistance (AMR) has become a global public health priority. The accelerated development of multidrug resistance (MDR) is attributed in part (if not completely) to the massive and inappropriate use of antimicrobials in agricultural and clinical settings. Human infections caused by MDR pathogens result in over 70,000 deaths in the United States each year [1, 2]. In fact, a team led by Prof. Lord Jim O'Neil has estimated that AMR could result in 10 million deaths a year worldwide by 2050 [3]. Although the accuracy of this frightening prediction is uncertain, we acknowledge the enormous burden AMR causes at multiple levels (economic, social, clinical and public health) [4]. This highlights the importance and urgency of a coordinated international action to prevent and control the worldwide spread of AMR [4, 5].
Polymyxins refer to an array of non-ribosomally-synthesized, cationic antimicrobial cyclic-peptides (CAMP) [6]. Among the five known subtypes, polymyxin B and polymyxin E (Colistin) are extensively used in agricultural production and clinical therapies [[6], [7], [8]]. Historically, the primary target of colistin is thought to be the negatively charged lipid A moiety of lipopolysaccharides (LPS) on the outer-leaflet of the bacterial outer membrane [9]. Despite its potential nephrotoxicity and neurotoxicity [6, [10], [11], [12]], colistin is still used for treatment as an ultimate line of defense against critical infections caused by MDR pathogens (esp. carbapenemase-producing Enterobacteriaceae) [7, 13, 14]. However, an acquired resistance to polymyxin has been frequently found in certain species of bacterial pathogens like Klebsiella pneumoniae (K. pneumoniae) [15] and Salmonella enterica (S. enterica) [16, 17]. The chemical mechanism underlying the colistin resistance consistently involves bacterial lipid A-centered surface remodeling, including i) The addition of 4-amino-4-deoxy-L-arabinose in S. enterica [16, 17] and Pseudomonas aeruginosa [18]; ii) The attachment of phosphoethanolamine (PEA) in Neisseria [19], Acinetobacter baumannii [20] and Campylobacter jejuni [21]; and iii) Glycine/diglycine modification in the pandemic Vibrio cholerae biotype EI Tor [[22], [23], [24], [25]]. Intrinsic resistance to polymyxin is limited to the originally-resistant population. However, the recent emergence and global discovery of plasmid-borne mobilized colistin resistance determinants (mcr-1) potentially threatens the clinical effectiveness of colistin as a last-resort antibiotic against carbapenem-resistant superbugs [26].
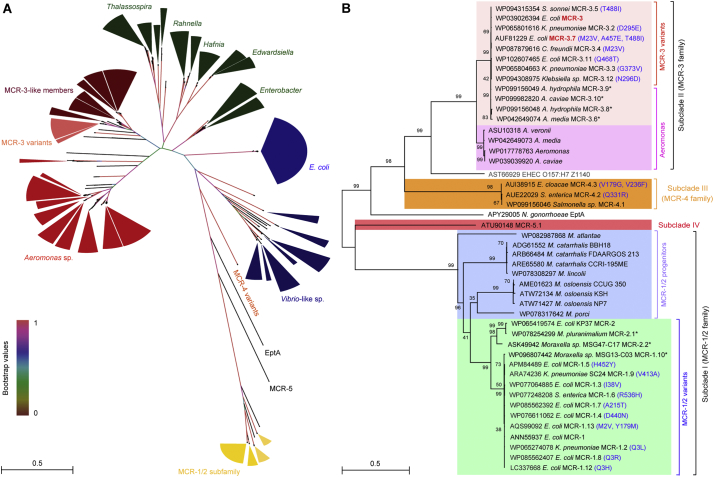
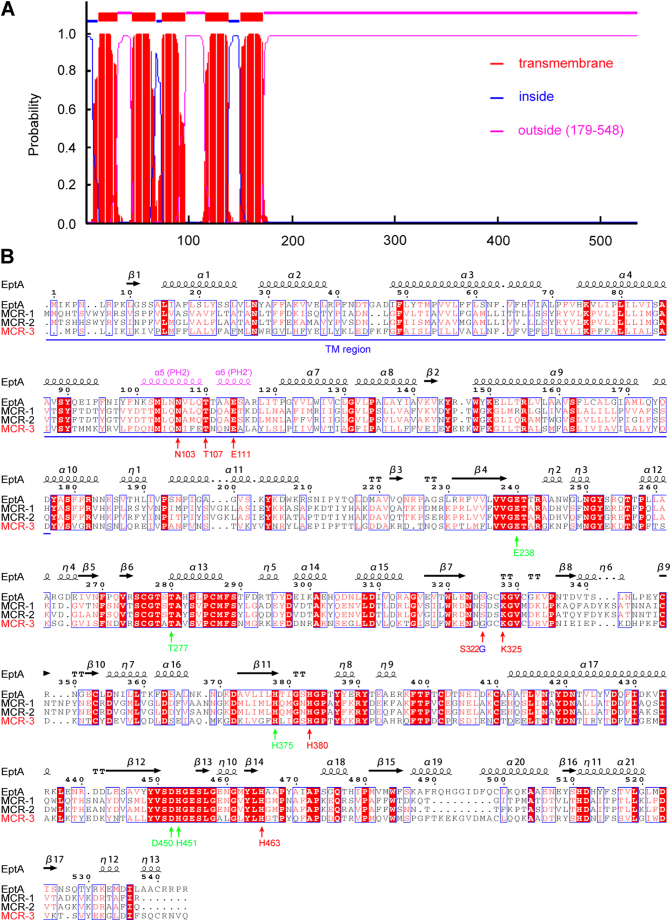
The mcr-1 gene product, MCR-1, is a PEA lipid A transferase, belonging to the “YhjW/YjdB/YijP” alkaline phosphatase super-family [26, 27]. MCR-1 catalyzes the transfer of the PEA group from its physiological donor phosphatidylethanolamine (PE) to the 1(4′)-phosphate position of lipid A glucosamine (GlcN) moieties [19, 28, 29]. Structure-guided functional studies have determined this mechanism and demonstrated that the enzymatic activity of MCR-1 renders the recipient strains resistant to polymyxin [27, [30], [31], [32], [33], [34], [35]]. Intriguingly, the determinants of transferable colistin resistance have extended beyond MCR-1, to a number of new MCR-like members [36] (namely MCR-2 [[37], [38], [39]], MCR-3 [40], MCR-4 [41], MCR-5 [42], MCR-6 [Genbank no.: ASK49942] (Indeed, it is a MCR-1/2 progenitor from Moraxella sp. MSG47-C17 [43], and exhibits high level of homology to ICR-Mo of M. osloensis [44]. Thus, it is supposed to be renamed as ICR-M), MCR-7 [45] and MCR-8 [46]), as well as over a dozen of new heterogeneous MCR-1 variants (e.g., MCR-1.2 [47] and MCR-1.6 [48]). Unlike the predominant MCR-1 which is distributed world-wide [49], both MCR-2 (81% identity to MCR-1 and originally found in Belgium [37, 38], and very recently detected in pigs/poultries [39] and human vaginal swabs [50] from China) and MCR-5 (only detected in Germany [42]) are thought to be two rare members of the MCR-like protein family. This is slowly changing with the discovery of MCR-2 and its variants in countries like China [39]. As for MCR-4, it has been detected in a pig isolate of S. enterica in Italy 2013 [41], swine isolates of E. coli from Spain and Belgium in 2015–2016 [41], and clinical isolates of carbapenemase-producing Enterobacter cloacae from Singapore in 2017 [51]. In terms of epidemiological/geographic distribution, MCR-3 seems to be second only to MCR-1. Phylogenetic analysis indicates that MCR-3 is evolutionarily distinct from MCR-1 and closely clustered with chromosomally-encoded MCR-like proteins in certain species of Aeromonas (Fig. 3) [52, 53]. To the best of our knowledge, the new mcr-3 gene has been discovered in 3 of 7 continents, namely Asia (China [40, 54], Singapore [51], Japan [55], Thailand [40] and Malaysia [40]), Europe (Denmark [56, 57], France [58] and Spain [59]) and North America (the United States [40]). Given that i) In Europe, colistin is used to treat bacterial infections of livestock (such as pigs, cows, and goats) [60]; and ii) Colistin is heavily supplemented as a growth promoter of livestock (pigs and poultries) in Asian countries (e.g., China, Japan, and Vietnam) [61], it is possible that indiscriminate antibiotic use has selected for the emergence of new colistin resistance determinants like mcr-3.
Fig. 3.
Phylogeny of MCR-3.
A. A radial representation of a phylogenetic tree of MCR-3 and its homologs.
Protein sequences of MCR-1 homologs are included in the analysis as a reference for evolutionary distance from transferable colistin resistance. Four distinct subclades are observed and are clustered as two distinct groups: MCR-3 like proteins (in light blue) with MCR-3 variants highlighted (in blue), Intrinsic determinants (in orange), Vibrio species (in pink) and MCR-1/2 proteins and their progenitors (in green). MCR-3 is highlighted in Bold font.
B. An unrooted phylogenetic tree of MCR-3 and its close homologs.
Two distinct subclades are observed: i) Subclade I with MCR-3 variants and MCR-3-like genes ii) MCR-1/2 variants and their progenitors. Paraphyletic branches within each subclade are indicated in different colors, MCR-3 variants (in light blue), MCR-3 like proteins (in blue), MCR-1/2 (in light green) and MCR-1/2 progenitors (in dark green), MCR-3 and MCR-1 is highlighted in bold font. Asterisk denotes >4 amino acid substitutions in comparison to MCR-3.
The evolutionary history of MCR-3 was inferred using the maximum likelihood method. The trees presented here have been inferred from 1000 bootstrap replicates using a LG amino acid substitution model. The percentages of replicate trees in which the associated taxa are clustered in the bootstrap test (1000 replicates) are shown next to the branches. A discrete gamma distribution was used to model evolutionary rate differences among sites with some evolutionarily invariable sites.
Protein accession numbers of individual members have been indicated in the figure.
Given that it is a putative member of PEA lipid transferase without detectable activity conferring colistin resistance, Z1140 of the E. coli O157:H7 EDL933 is used here as an internal reference for phylogeny.
In fact, the growing body of mcr-3 variants includes mcr-3.2 [D295E] [40], mcr-3.3 [G373V] [40], mcr-3.4 [M23V], mcr-3.5 [T488I] [57, 58], and mcr-3.7 [M23V, A457E, T488I] [54]. Most of them are located on IncHI2-type plasmids, like pWJ-1 (~260 kb), IncP-like plasmid (~50 kb), F46: A-: B20 IncF plasmid [58], respectively. The range of mcr-3-harboring host bacteria includes E. coli [40, 54, [57], [58], [59]], K. pneumoniae [40] and S.typhimurium [40, 56]. Of note, mcr-3-bearing bacteria in Europe are frequently prevalent in either human infections [40, 56] (even bloodstream infections [57]) or epidemic MDR lineages of E. coli (ST744 [58] and ST131 [57]). To our surprise, mcr-3 has been found to coexist with mcr-1 [56, 59] (or mcr-4 [51]) in a single cell, in MDR pathogens [56, 59] and even in blaNDM5-harboring superbugs [54]. This suggests an urgent need to introduce mcr-3 into national surveillance programs for monitoring of antibiotic resistance. Although two independent research groups recently evaluated the prevalence and clinical risk of mcr-1 carriage in human infections [62, 63], the molecular epidemiology and functional aspects of mcr-3 in China are poorly understood. In this work, we aim to close this knowledge gap and present an overview of the dissemination, comparative genomics, evolution and mechanism of MCR-3-mediated colistin resistance.
2. Methods
2.1. Ethics and Consents
Ethical approval was sought and informed consent was provided by patients, healthy volunteers and farm owners. Individual consent forms were translated into Mandarin Chinese and the study vocally explained to each subject. All documents complete with patient signatures have been retained. All participants held the right to withdraw from the study at any stage.
2.2. Epidemiological Study
We undertook a retrospective cross-sectional study to assess the prevalence of mcr-3-carrying isolates in animal, farmer, inpatient and healthy volunteer in China. A total of 6497 non-duplicate fecal samples were collected from thirteen provinces in China, from Oct. 2016 to Dec. 2017. Specifically, 1951 fecal samples of animal were collected from 32 swine farms in 13 provinces, and the 174 fecal samples of farmer who approved and provided consent forms were collected from 16 farms in 9 provinces (Tables 1 and S1). The 2281 inpatients submitting fecal specimens during the study timeframe to the hospital microbiology laboratories for diagnostic purposes were asked to participate in the study by means of an invitation included with the diagnostic test report returned to the patient. The 2091 healthy volunteers were including who came into the hospitals for routine physical examinations. Fecal samples from inpatient and healthy volunteers were recovered from 5 hospitals which contain >6000 beds in Guangdong province. All these samples were isolated from Oct. 2016 to Dec. 2017 (Tables S1 and S2).
Table 1.
Prevalence of mcr-3 in samples from swine and farmers in this study.
| Provinces | Farms | No. of swine samples | mcr-3-positive rate of pigs | No. of farmer samples | mcr-3-positive rate of farmers |
|---|---|---|---|---|---|
| Guangdong | GD1 | 166 | 0.60% (1) | - | - |
| GD2 | 122 | 0.00% (0) | - | - | |
| GD3 | 208 | 3.85% (8) | 10 | 0.00% (0) | |
| GD4 | 15 | 0.00% (0) | - | - | |
| GD5 | 38 | 0.00% (0) | - | - | |
| GD6 | 97 | 0.00% (0) | 20 | 0.00% (0) | |
| GD7 | 150 | 1.33% (2) | 30 | 0.00% (0) | |
| GD8 | 50 | 0.00% (0) | 10 | 0.00% (0) | |
| GD9 | 50 | 10.00% (5) | 10 | 2.00% (2) | |
| GD10 | 31 | 0.00% (0) | 6 | 0.00% (0) | |
| GD11 | 49 | 4.08% (2) | - | - | |
| GD12 | 146 | 6.85% (10) | - | - | |
| Anhui | AH1 | 10 | 40.00% (4) | 2 | 0.00% (0) |
| AH2 | 50 | 10.00% (5) | 10 | 0.00% (0) | |
| AH3 | 49 | 0.00% (0) | 7 | 0.00% (0) | |
| Guangxi | GX1 | 33 | 0.00% (0) | - | - |
| GX2 | 47 | 0.00% (0) | - | - | |
| GX3 | 48 | 0.00% (0) | 9 | 0.00% (0) | |
| GX4 | 41 | 2.44% (1) | - | - | |
| Hunan | HN1 | 3 | 0.00% (0) | - | - |
| HN2 | 50 | 2.00% (1) | - | - | |
| HN3 | 45 | 0.00% (0) | - | - | |
| Jiangxi | JX1 | 1 | 0.00% (0) | - | - |
| JX2 | 79 | 1.27% (1) | - | - | |
| Hebei | HB | 50 | 0.00% (0) | 10 | 0.00% (0) |
| Heilongjiang | HLJ | 50 | 2.00% (1) | 9 | 0.00% (0) |
| Hubei | HuB | 51 | 1.96% (1) | 4 | 0.00% (0) |
| Jilin | JL | 50 | 0.00% (0) | 1 | 0.00% (0) |
| Liaoning | LN | 50 | 0.00% (0) | 7 | 0.00% (0) |
| Sichuan | SC | 120 | 0.83% (1) | 29 | 13.79% (4) |
| Shaanxi | SX | 1 | 0.00% (0) | - | - |
| Hainan | HaiN | 1 | 0.00% (0) | - | - |
| Total | / | 1951 | 2.20% (43) | 174 | 3.45% (6) |
2.3. Bacterial Isolation, Identification and Growth Condition
Fecal samples were collected by sterile rectal swab and sent to the lab immediately. All of the fecal samples were cultured by adding three milliliter enrichment broth and incubated at 37 °C overnight. The negative control was performed in every cultivation and identified eligible. Subsequently, the total DNA was extracted by water-boiling method and screened for the presence of mcr-3 using PCR as previously described [64]. The resulting amplicons were confirmed by sequencing. Screening of MCR-3-producing isolates from PCR-positive fecal samples was by inoculating Columbia Blood Agar (Oxoid, Hampshire, United Kingdom) with colistin (2 μg/ml) while species identification was first conducted by the API20E system (bioMérieux, Marcy l'Etoile, France) and then by MALDI-TOF MS (BrukerDaltonik GmbH, Bremen, Germany). When the species could not be well interpreted by MALDI-TOF, 16S rDNA sequencing was applied.
E. coli [DH5α and BL21 (DE3)] were used as the gene cloning host and the protein expression host, respectively (Table S3). E. coli MG1655, a colistin-susceptible strain, is used to test the ability of mcr-3 and/or its mutants to confer colistin resistance (Table S3). All bacterial cultures were maintained in the Luria-Bertani (LB) broth. Solid LB agar plates supplemented with appropriate antibiotics were used to either screen the mcr-3-containing clones or determine the level of bacterial colistin resistance.
2.4. Measurement of Bacterial Growth
To address effect exerted on bacterial growth by the eptA and/or mcr-like genes, the E. coli strain MG1655 alone [or with either the pBAD24 empty vector or plasmid-borne eptA/mcr genes] was subjected to the cultivation at 37 °C (180 rpm) in LB broth (Table S3). Totally, three different versions of mcr-like gene used here included mcr-1, mcr-2, and mcr-3, respectively. To generate bacterial growth curves, over-night cultures were inoculated (1:1000) in fresh LB broth (37 °C, 180 rpm) with varied level of arabinose (0.00%, 0.02%, and 0.20%). Spectrophotometer (Spectrum lab S32A) was used to measure the optical density (OD) at 600 nm at a regular interval of one hour for a total of 20 h.
2.5. Plasmid Conjugations and Multi-Locus Sequence Typing (MLST)
Conjugation experiments were performed to test the transferability of the mcr-3-harboring plasmid, using a Streptomycin-resistant E. coli C600 as the recipient. Briefly, overnight culture of mcr-3-positive isolates and recipient strain E. coli C600 were mixed (ratio of 1:9) in LB broth, which was subjected to overnight incubation. The mixture was then spread on LB agar plates containing sodium streptomycin (2000 μg/ml) plus colistin (4 μg/ml) to select trans-conjugants that had acquired the mcr-3-harboring plasmid. Carriage of such a plasmid in the parental strain and corresponding trans-conjugants was confirmed by PCR and Sanger sequencing. MLST was determined using BLASTn and publicly available database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (Table S1).
2.6. Sequencing, Assembly and Annotation of Plasmid Genomes
The acquired mcr-1/3-positive plasmids were used to construct a sequencing library using the TruePrepTM DNA Library Prep Kit V2 from Illumina® (Vazyme, USA). The qualified libraries were sequenced on an Illumina HiSeq X-ten platform (Illumina, USA), in which 150 bp paired-end reads were generated. Following removal of low-quality reads with software Trimmomatic [65], the high-quality clean reads were assembled into long contigs with the software of SPAdes (version 3.11.0) [66]. The assembled contigs with resistance genes were identified and compared to those closely-related plasmids (with known genome sequences deposited in NCBI database) using the program Blastall (version 2.2.22) with an e-value cutoff of 1 × 10−5. The suspected gaps between contigs were closed with Sanger quenching of PCR products. RAST [67] and GLIMMER (version 3.02) [68] were utilized for the prediction of genes on the complete plasmids. The GC content and GC skew analysis of the whole plasmid sequence were performed by homemade Python Scripts. Finally, the plasmid map was drawn with the software of Circos (version 0.69).
2.7. Sequence Data
GenBank accession numbers of new plasmids with full genome sequences are listed as follow: MH043623 for pGDZJ002–1; MH043624 for pGDZJ003–1; MH043625 for pGDZJ003–2; and MH043626 for pGDZJ004.
2.8. S1-PFGE and Southern Blotting
The plasmid and/or chromosome location of mcr-3 and mcr-1 genes was determined by S1-nuclease digestion and pulsed-field gel electrophoresis (S1-PFGE), followed with southern blotting hybridizations. The mcr-3-harboring isolates were suspended in PBS and embedded in gold agarose gel plugs (SeaKem® Gold Agarose, Lonza, USA). The plugs were digested with S1 nuclease (TaKaRa, Dalian, China) and the DNA fragments were separated by PFGE. Southern blotting hybridizations of plasmid DNA were performed with DIG-labelled mcr-3 or mcr-1 probes according to the manufacturer's instructions (Roche Diagnostics, Germany).
2.9. Antimicrobial Susceptibility Tests
We determined minimum inhibitory concentrations (MICs) of colistin, polymyxin B, tigecycline, ampicillin, amoxicillin-clavulanate, cefotaxime, ceftazdime, cefepime, gentamicin, amikacin, ertapenem, imipenem, meropenem, fosfomycin, nitrofurantoin and ciprofloxacin for all isolates using the agar dilution method in accordance with the Clinical and Laboratory Standards Institute guidelines, and we interpreted the results according to the breakpoints of the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2016) for colistin, polymyxin B, and tigecycline and those of the Clinical and Laboratory Standards Institute (CLSI, 2016) for other antimicrobials (Table S2).
2.10. Structure Modeling and Molecular Docking
The software Swiss-Model (https://swissmodel.expasy.org/interactive/qMEvX5/models/) [69] was used to obtain the modelled structure of MCR-3 using the structure of the Neisseria meningitidis EptA (PDB, 5FGN) as the template [70]. Although MCR-3 shows only 37.55% identity to EptA, its modelled structure exhibits the coverage score of 98% (6–540). It was evidently a qualified structural prediction, in which GMQE (Global Model Quality Estimation) is 0.73, and QMEAN (quality estimation on the modelled structure) is −3.68. The ready-to-dock 3D structure of phosphatidylethanolamine (PE) (ID: ZINC32837871) and head group of PE (ID: ZINC02798545) was obtained from ZINC database [71]. As we recently described with EptA/MCR-1 [72], the program DOCK6.7 was used to dock the PE ligand to designated binding pocket into MCR-3 [73].
2.11. Overlap Extension PCR and Site-directed Mutagenesis
Using appropriate primers (Table S3), overlap extension PCR was conducted, generating the hybrid derivatives of MCR-1/3 and Z1140 [30]. Site-directed mutagenesis was applied to produce all the point-mutants of mcr-3 [27, 30, 72]. The Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech Co.,Ltd) was used along with specific primers for mcr-3 (Table S3).
2.12. Expression and Identification of MCR-3 Membrane Protein
To produce MCR-3 integral membrane protein, the strain FYJ1153 (BL21 with pET21a::mcr-3) was engineered (Table S3). As we descrbed with MCR-1/2 [30, 72], the expression of MCR-3 was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial pellets were harvested and lysed through a French Press (JN-Mini, China) [at 500 p.s.i. once and 1300 p.s.i twice]. Following the routine purification process for the membrane proteins MCR-1/2, MCR-3-containing fraction solubilized in buffer B [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5% glycerol, 1% detergent dodecyl-β-D-maltoside (DDM, M/V)] were incubated overnight with pre-equilibrated Ni-NTA agarose beads at 4 °C. Then, the intramembrane protein MCR-3 was eluted from the Ni-NTA agarose beads using the elution buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 100 mM immidazole, 5% glycerol (vol/vol), 0.03% DDM (M/V)]. The acquired protein was concentrated with a 30 kDa cut-off ultra-filter (Millipore, USA), and analyzed with 12% SDS-PAGE. To further verify its identity of recombinant MCR-3, the expected protein bands were cut from the SDS-PAGE, digested with Trypsin (G-Biosciences St. Louis, MO), and subjected to the confirmation with A Waters Q-Tof API-US Quad-ToF mass spectrometer [74, 75]. Finally, data analyses were conducted using the Waters Protein Lynx Global Server 2.2.5, Mascot (Matrix Sciences) and BLAST against the NCBI nr database.
2.13. Circular Dichroism Analyses
The protein secondary structure of MCR-3 was determined using circular dichroism (CD) [72]. Prior to CD analyses, MCR-3 protein (~0.2 mg/ml) was dissolved in Tris-buffer [20 mM Tris-HCl, 300 mM NaCl, 0.03% DDM, 10% (vol/vol) glycerol, pH 8.0)]. For every experiment, ~600 μl protein sample was dropped into a quartz cylindrical cuvette. The CD spectra were recorded on a Jasco Model J-1500 spectrometer (Jasco Corp., Tokyo, Japan) through continuous wavelength scanning (in triplicate) from 200 to 260 nm at a scan rate of 50 nm/min [76] and smoothed with a Savitsky-Golay filter [77].
2.14. Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
As recently demonstrated with MCR-1 [72], ICP-MS was adopted to examine the protein-bound zinc in MCR-3. The MCR-3 protein sample was subjected to an NexION™ 300× ICP-MS instrument (PerkinElmer, USA) in which the mass-to-charge ratio (m/z) was calculated [78]. The carrier gas was helium.
2.15. Assays for Enzymatic Reaction of MCR-3
As recently showed for EptA [79] and MCR-1/2 [72, 79] and ICR-Mo [44], the reaction catalyzed by MCR-3 in vitro was conducted. In brief, the 1-acyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl) amino] dodecanoyl}-sn-glycero-3-phosphoethanolamine (Avanti Lipids, USA), acted as the substrate for MCR-3, which is abbreviated as NBD-PEA. The reaction system (50 μl in total) consisted of the following components [50 mM HEPES (pH 7.50), 100 mM NaCl, 0.03% of DDM, 0.2 mM NBD-PEA and 40 μM MCR-3] and kept for 20 h at room temperature. Subsequently, the reaction products were separated with thin layer chromatography (TLC) in a mobile phase consisting of ethyl acetate: methanol: water (7:2:1, vol/vol). The fluorescent signal on the TLC plate was visualized under Epi blue light (455–485 nm) with a gel imaging system (Bio-Rad) as Anandan et al. described [70].
2.16. Preparation and Structural Determination of Lipopolysaccharide-Lipid A
The crude lipopolysaccharide (LPS) was extracted as described by Caroff et al. [26, 80] with modifications. The resulting LPS species were freeze-dried, and then dissolved in the buffer of 30 mM Tris-HCl [pH 8.0] containing 0.2% SDS. DNase I (25 μg/ml) and RNase A (100 μg/ml) were used to remove the nucleic acid contamination. The removal of protein contaminants was proceded via 1 h of treatment with proteinase at 37 °C. To release the Kdo linkage from lipid A, the crude LPS was heated at 100 °C for 1 h in the sodium acetate buffer (10 mM, pH 4.5) supplemented with aqueous 0.2% SDS. Then, the residual SDS was cleaned through the precipitation with acidified ethanol [81].
SDS-PAGE coupled with silver staining was utilized to detect the purity of lipid A pools from different bacterial strains [82]. The qualified lipid A species were subjected to MALDI-TOF/TOF-MS (Bruker, ultrafleXtreme) in negative-ion mode with the linear detector [23, 83]. Briefly, the samples of lipid A were dissolved in 20 ul of chloroform: methanol (2:1) solution and then loaded onto the MALDI sample plate, giving MS spectrum. In general, each spectrum was collected from an average of 500 shots and 50% laser power [26, 80, 84].
2.17. Liquid Chromatography Mass Spectrometry (LC/MS)
The LC/MS system (Agilent technologies 6460 Triple Quad LC/MS) was applied in structural measurement of the alternative lipid substrate, NBD-Glycerol-3-PEA [85]. Similarly, the reaction mixture catalyzed by MCR-3 was assayed. In the trials of GC/MS, the analytical chromatographic column [Zorbax SB C18 (2.1*50 mm, 3.5 μm)] was utilized. The samples were eluted with methanol/0.1% methanoic acid (95:5) at 0.3 ml/min. Of particular note, an electrospray ionization (ESI) source was connected with mass spectrometry (MS), and the positive ion scanning appeared in the mode of neutral loss ion (m/z, 141).
2.18. Confocal Microscopy
Confocal microscopy was applied in both the determination of ROS level and the differentiation of ALIVE/DEAD status of E. coli. First, to evaluate the potential effects of MCR-3 (and/or EptA) on colistin-induced ROS production, the E. coli strains with or without the gene of mcr-3/eptA were subjected to confocal microscopy analyses. All the E. coli strains in mid-log phase [OD600 = ~0.8] (Table S3) were divided into two equal fractions: one half acted as control, the remaining fraction was challenged with 4 μg/ml colistin for 0.5 h. The oxidant-sensitive dye used for ROS detection was 2′,7′-dichlorodihydrofluorescein diacetate (DCFH2DA, Sigma). The fluorescent images were captured with a Zeiss LSM 510 Meta confocal laser scanning microscope (100× oil immersion objective), in which the fluorescence of dichloro-fluorescein (DCF, an oxidation product of DCFH2DA) was excited by 488 nm, and resultant emission was detected at 530 nm [86].
To further determine bacterial viability, confocal microscopy-based LIVE/DEAD assays were conducted as Yang et al. described with little change [87]. The strains tested here are same as those of growth curves. In brief: it can be described as follows: i) overnight cultures were sub-cultured into 5 ml LB broth and incubated for 16 h 37 °C (180 rpm); ii) bacterial culture was concentrated via 10 min of centrifugation at 4000 rpm, gently removing the supernatant, and the dissolved pellets were further incubated for 8 h in fresh LB broth with/without arabinose (0.2%, w/v); iii) after the second round of spinning at 4000 rpm for 10 min, the resultant pellet was washed twice with 0.085% NaCl; iv) bacterial bio-film was stained with the kit of LIVE/DEAD® (v/v; BacLight TM Bacterial Viability Kit, Invitrogen) in the solution of 0.085% NaCl (of note: 3 μl of the dye mixture was added to each 1 ml of the bacterial suspension, mixed thoroughly at room temperature in the dark, and kept for 15 min); v) the images were obtained by the Confocal Laser Scanning Microscopy (CLSM, Zeiss LSM 800) with a 63× oil immersion lens. The CLSM images were analyzed using COMSTAT image analysis software for quantification of biofilm biomass. The COMSTAT data was evaluated using one-way ANOVA followed by Tukey-Kramer Multiple comparison post hoc test. Statistical significance was set at p < .05.
2.19. Chemical Rescue Tests
Accumulating evidence suggests that the hydroxyl radical death pathway could be activated as a downstream event of colistin exposure [88, 89]. The fact that MCR-3 can modify lipid A moieties of LPS on bacterial membrane, an initial target of colistin, allowed us to hypothesize that expression of MCR-3 might physiologically prevent colistin-induced ROS production. To test this hypothesis, chemical rescue experiments were conducted routinely using two ROS inhibitors: bipyridine (the ferric chelator) and L-cysteine (a ROS scavenger). To compare the bacterial survival, E. coli strains (with or without mcr-1) were subjected to the different challenges as follows: i) the treatment of colistin alone; ii) the combined treatment of colistin plus 2,2′-dipyridine [90]; iii) the stress of colistin mixed with L-cysteine [91]. Briefly, overnight cultures were sub-cultured at a ratio of 1: 500 into LB medium for ~5 h to OD600 nm of 1. For the iron chelation, 2,2′-dipyridine (500 μM, Sangon Biotech) was added simultaneously with colistin (20 μg/ml, Sigma). As for the ROS quenching experiments, L-cysteine (10 mM, Sangon Biotech) was mixed simultaneously with colistin (20 μg/ml, Sigma). Following a 0.5 h growth at 37 °C, cultures were centrifuged at 1800 g for 5 min. The pellets were re-suspended with 1× PBS (pH 7.2) and serially diluted. 5 μl of 3 selected dilutions [10–5~10–7] were dropped onto LB agar plates. After overnight incubation at 37 °C, colony forming units (CFU) were enumerated [92].
2.20. Phylogenetic Tree
The amino acid sequence of MCR-3 was used as a query to identify similar protein sequences using protein BLAST with options enabled to exclude models and uncultured environmental samples. The default parameters were modified to reduce the word size to 2 and to return 500 target sequences. Sequences of distantly related MCR proteins including those in the MCR-1/2/4/5 family were manually included to obtain a comprehensive analysis. Redundant sequences were eliminated using the Unique seq server (https://www.ncbi.nlm.nih.gov/CBBresearch/Spouge/html_ncbi/html/fasta/uniqueseq.cgi) and aligned using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/). In total, 511 unique amino acid sequences were utilized for the subsequent phylogenetic analysis.
jModeltest (via MEGA 7) was used to identify the best-fit protein substitution model and the best model was used to generate a maximum-likelihood tree with 1000 bootstrap replicates. A LG model with Gamma distribution and Invariant sites was used. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The results are presented as a radial phylogram. A subset of the sequences was re-analyzed using the about method to obtain a smaller, more detailed phylogenetic tree.
2.21. Role of the Funding Source
The funder had no role in the study design, data collection, data analysis, data interpretation, or writing of the article. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Molecular Epidemiology of mcr-3 in China
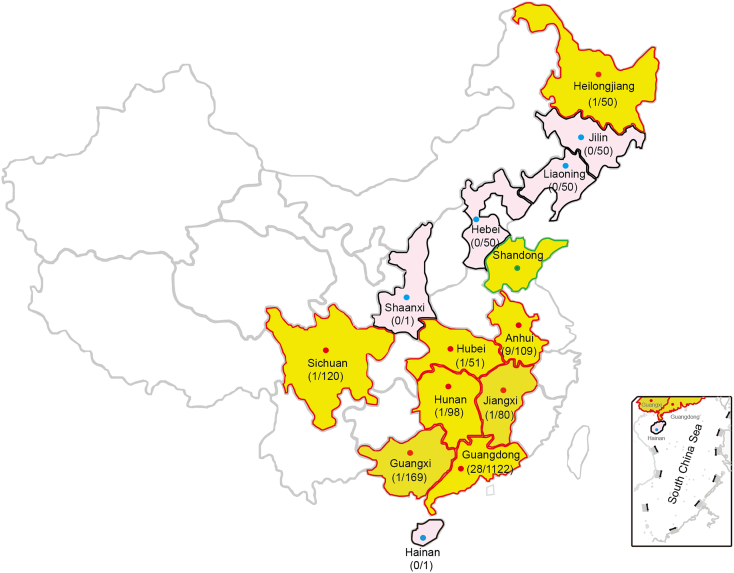
Of the 6497 unique samples collected, a total of 6497 non-duplicate samples were collected from thirteen provinces in China, from Oct 2016 to Dec 2017. Of them, 49 samples (49/6497 = 0.75%) were mcr-3 positive when verified by PCR and Sanger sequencing. Specifically, 3.45% (6/174) of samples from farmers were mcr-3 positive, followed by 2.20% (43/1951) from swine-derived samples. None of the mcr-3-positive samples were from patients or other healthy individuals. In addition, 14.02% (911/6497) samples were mcr-1-positive. In particular, mcr-3-positive samples were identified in 8 out of 13 provinces, namely Guangdong, Guangxi, Heilongjiang, Anhui, Hubei, Jiangxi, Hunan and Sichuan (Fig. 1 and Table 1). The rate of incidence of mcr-3 was as low as 0.59% in the isolates from Guangxi to as high as 8.26% from Anhui (Fig. 1). No mcr-3 was detected in the neighboring provinces of Hebei, Jilin, Liaoning, Shaanxi and Hainan (Fig. 1).
Fig. 1.
Dissemination of mcr-3 (and/or its variants) in swines of China.
Shandong Province where mcr-3 is initially detected [64] is highlighted with green boundary. Those with mcr-3-positive isolates in our study are labelled with red boundaries, whereas those without mcr-3-positive isolates are indicated with black boundaries. Of note, a mcr-3 variant, mcr-3.7, was recently detected in Sichuan Province of China [54].
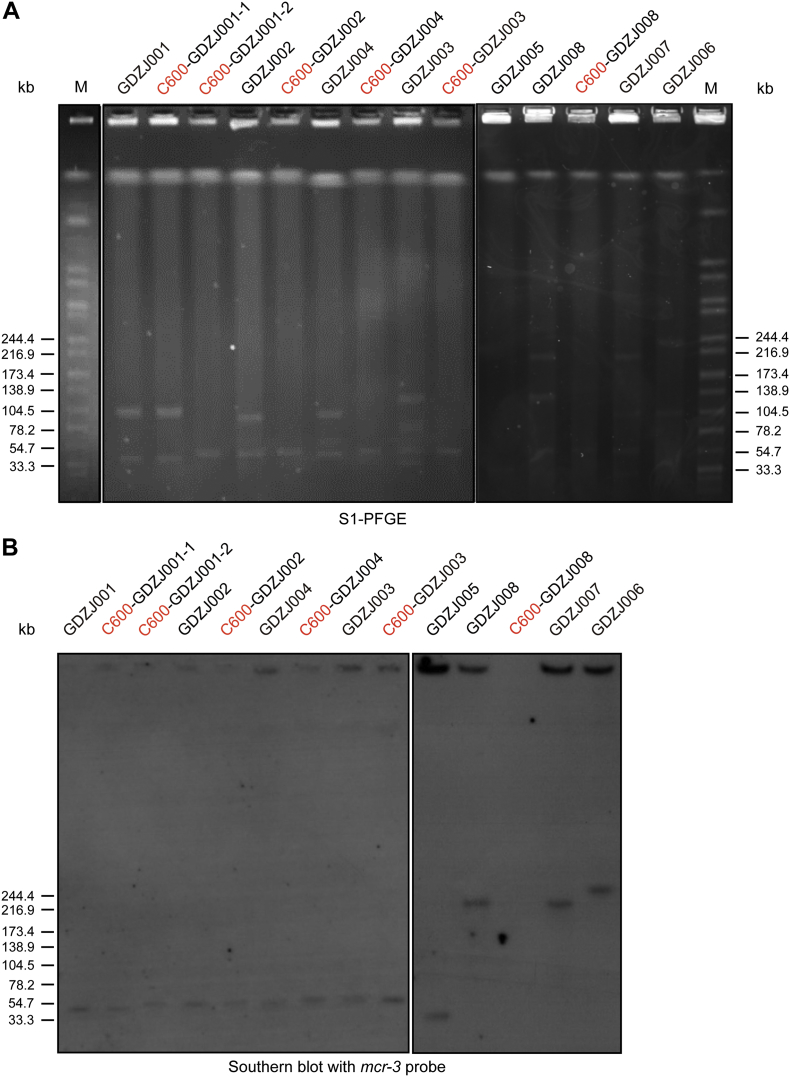
We randomly chose 8 isolates out of 49 mcr-3-positive samples for microbial analyses (Table S1). All the 8 mcr-3-carrying isolates were resistant to colistin and polymyxin B. Most of them were susceptible to cefotaxime (6 of 8 isolates), ceftazdime (7 of 8 isolates), cefepime (8 of 8 isolates), gentamicin (5 of 8 isolates), amikacin (8 of 8 isolates), ertapenem (7 of 8 isolates), imipenem (8 of 8 isolates), meropenem (8 of 8 isolates), fosfomycin (7 of 8 isolates), tigecycline (8 of 8 isolates), only some of the isolates remained susceptible to amoxicillin-clavulanate (1 of 8 isolates), nitrofurantoin (4 of 8 isolates), ciprofloxacin (4 of 8 isolates) and one isolate was susceptible to ampicillin (1 of 8 isolates) (Table S2). We further investigated the plasmids harboring mcr-3. In conjugation experiments, mcr-3-bearing plasmids were successfully transferred to E. coli C600 in 4 out of 8 mcr-3-carrying isolates, and 2 of 8 exhibited high conjugation rates (~10−3) (Table S1). Five isolates co-carried mcr-1, 2 of which were on plasmids and successfully transferred to E. coli C600. S1-PFGE and southern blotting of 8 mcr-3-positive isolates and their trans-conjugants showed that mcr-3 genes were located on plasmids with sizes of ~35-kb, ~48-kb, ~210-kb and ~240-kb, whereas mcr-3 genes of GDZJ005, GDZJ006, GDZJ007 and GDZJ008 were not successfully transferred to E. coli C600. The mcr-3 genes of GDZJ005, GDZJ006, GDZJ007 and GDZJ008 were located on plasmids, which is based on S1-PFGE and Southern blot. (Fig. S1). In addition, we identified two novel variants of mcr-3 from 3 isolates, namely mcr-3.4 and mcr-3.7 (Table S2 and Fig. S4). This brings the number of mcr-3 variants up to eleven (Table S2).
3.2. Comparative Genomics of mcr-3-Harboring Plasmids
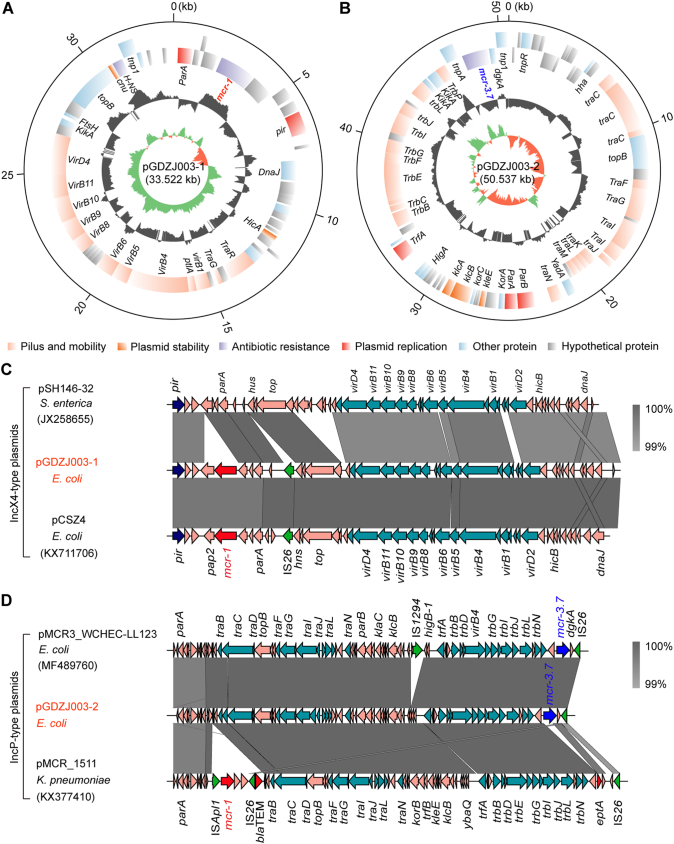
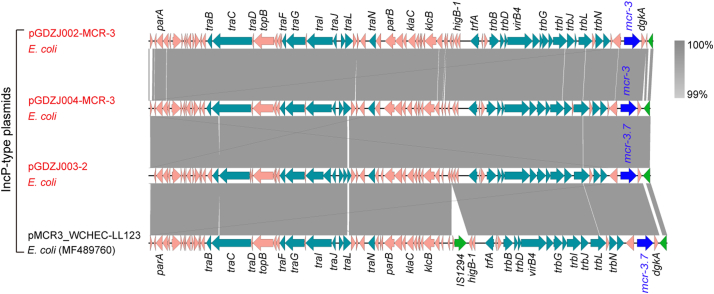
We succeeded in sequencing of MCR-3-producing plasmids pGDZJ002-1, pGDZJ003–2 and pGDZJ004 from GDZJ002, GDZJ003 and GDZJ004, respectively, and mcr-1-encoding plasmid pGDZJ003-1 from the GDZJ003. Of the two plasmids isolated from GDZJ003, whole genome sequencing shows that plasmid pGDZJ003–2 is a 50,537 kb plasmid that contains 73 ORFs including the mcr-3.7 gene. MCR-3.7 has 18 amino acid substitutions when compared to MCR-3. This IncP-type plasmid is also seen to possess the tra and trb conjugative regions, portioning modules par, replication initiator trfA, host-lethal protein encoding genes klc and their regulators kor and a toxin-antitoxin system higA-B (Fig. 2B). It shows very high sequence identity to another IncP-type mcr-3.7-bearing plasmid pMCR3_WCHEC-LL123 [54] (Accession no.: MF489760) isolated from human isolates of E. coli. However, unlike pMCR3_WCHEC-LL123, pGDZJ003–2 is missing the insertion sequence IS1294 (Fig. 2C) and does not have additional antibiotic resistance genes like blaTEM (conferring ampicillin resistance). When compared to another IncP type mcr-1-carrying plasmid, pMCR_1511 [93], pGDZJ003–2 also lacks the transposition unit of mcr-1 (ISApl1-mcr-1-hp) on pMCR_1511 (Fig. 2D). The other plasmid from GDZJ003 is pGDZJ003-1 which harbors the mcr-1 gene and is 33,522 kb in length with a GC content of 42%. This plasmid contains 43 predicted ORFs, with 17 associated with formation of the type IV pilus (Fig. 2A). Comparative analysis shows that its backbone is very closely related to two narrow-host-range IncX4-type plasmids [94] pSH146-32 (Accession no.: JX258655) and the mcr-1-harboring plasmid pCSZ4 [95] (Accession no.: KX711706) (Fig. 2C). Like most IncX4 plasmids, both pCSZ4 and pGDZJ003–1 are missing the ISApl1 insertion sequence in front of the mcr-1 gene.
Fig. 2.
Comparative genomics of the plasmids carrying mcr-3 and/or mcr-1 genes.
A. Genomic map of the mcr-1-bearing plasmid pGDZJ003_mcr-1 that coexists with the mcr-3.7-harboring plasmid pGDZJ003_mcr-3.7 in E. coli.
B. Schematic representation for circular genome of the mcr-3.7-harboring plasmid pGDZJ003_mcr-3.7.
Circles (from inside to outside) separately denote the GC screw, GC content and the open-reading frames in both DNA strands [27]. The plasmid sequences were annotated by RAST, and the maps were generated using Circos program [27].
C. Colinear genomic analyses of three plasmids pGDZJ003_mcr-1, pSH146–32 and pCSZ4.
D. Linear genome alignment of the mcr-3-positive plasmid pGDZJ003_mcr-3.7 with the other two plasmids (pMCR3_WCHEC-LL123 and pMCR_1511).
Similarly, genome sequences for two other mcr-3-carrying plasmids, pGDZJ002-1 (50,818 kb; Fig. S2) and pGDZJ004 (50,519 kb; Fig. S3) were obtained. Both plasmids have a GC content of 47% and contain similar elements for replication, conjugation, and partitioning (Figs S2 and S3). Colinear genome analysis of pGDZJ003–2 and pMCR3_WCHEC-LL123 shows that they possess similar genetic arrangements (Fig. S4) and belong to the IncP family of plasmids. Except for little bite of SNPs and deletions of short fragments, both pGDZJ002–1 and pGDZJ004 carry the mcr-3 gene within the same context as pGDZJ003–2 and pMCR3_WCHEC-LL123, which carry the mcr-3 variant, mcr-3.7 instead. However, unlike pMCR3_WCHEC-LL123, the other 3 plasmids do not contain IS1294 (Fig. S4). Together, these plasmids are additive members of the mcr-1-carrying (pGDZJ003-1) and mcr-3-carrying family of plasmids (pGDZJ002-1, pGDZJ003–2 and pGDZJ004). mcr-like genes have been increasingly identified on plasmids of multiple incompatibility groups within a wide range of bacterial hosts. Though these plasmids were isolated from diverse hosts, their frequent sharing of similar genetic elements suggests a common evolutionary origin.
3.3. Evolution of MCR-3
We utilized phylogenetic analysis to try and understand the evolution of MCR-3, which shares a closer homology to Neisseria EptA (53.1%) than MCR-1 (44.1%) using MCR-3 as a template, >500 homologs with >95% sequence coverage and 60% identity were identified via blastp and compared with representative protein sequences from the MCR-1/2/P families. The radial phylogram shows three broad groups within the MCR-3 family and their homologs and a fourth distinct and detached group consisting of the MCR 1/2/P family (Fig. 3A). The first group primarily consists of MCR-3-like genes exclusively from the Aeromonas species. One branch from this group consists of MCR-3 and its variants (MCR-3.1, MCR-3.2, MCR-3.4, MCR-3.7, MCR-3.9, MCR-3.10) (Fig. 3B). This branch is surrounded by other MCR-3-like genes that encode putative PE-transferases from Aeromonas species (Fig. 3A), primarily found in fresh and brackish water but also implicated in human infections. The second group consists of paraphyletic branches with PE-lipid A transferases from several bacterial genera including Thalassospira, Rahnella, Hafnia, Edwardsiella, Buttiauxella and Enterobacter, some of which are normal human commensals that have been reported to be resistant to a number of antibiotics (e.g., Hafnia spp.), while others (e.g., Edwardsiella spp.) are naturally resistant to colistin. The third group consists of other putative PE-lipid A transferases from E. coli and Vibrio/Photobacterium species. The marine bacterium Photobacterium damselae is a human and fish pathogen with colistin resistance [96]. The E. coli members of this group form a very tight subclade. One of these members, E. coli O157:H7 strain EDL933 containing a putative PEA-transferase Z1140 was tested experimentally and conferred no appreciable colistin resistance on LB agar plates (<0.5 μg/ml) (Fig. S9). Swapping the transmembrane domain of MCR-3 with that from Z1140 still maintained activity (8 μg/ml) (Fig. S9). The fourth subclade is further organized into 2 sub-groups: i) MCR-1/2 and their point mutant variants; ii) MCR-P(M)and other transitional intermediates of the MCR-1/2 family from Moraxella spp. (Fig. 3B). MCR-1/MCR-2 represent a group undergoing rapid evolution with the MCR-P(M) family serving as a potential reservoir of chromosomally-encoded transitional intermediates in the evolution of the MCR-1/2 family (Fig. 3B). Surprisingly, Neisseria EptA, a chromosomal colistin resistance determinant (Fig. 3A) is an evolutionarily-distant member.
The findings indicate that MCR-3 and its variants share a common ancestor with MCR-like proteins from Aeromonas species and constitute a rapidly evolving branch of transferable colistin resistance genes that are more closely related to chromosomally encoded intrinsic determinants found in commensal and environmental bacteria. Aeromonas species might occupy an unexplored niche in the marine ecosystem that is yet to be examined for colistin resistance. A complete biochemical and physiological study of MCR-3 is missing and would provide a better understanding of its evolutionary pattern.
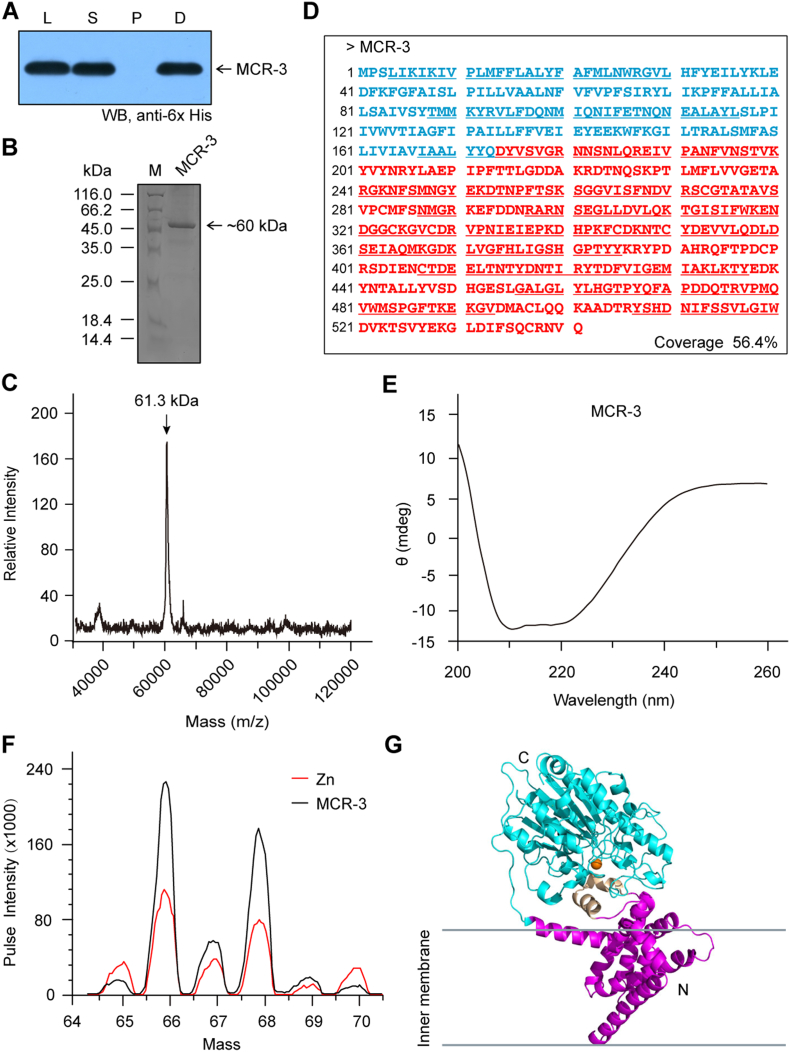
3.4. Characterization of MCR-3 Intramembrane Protein
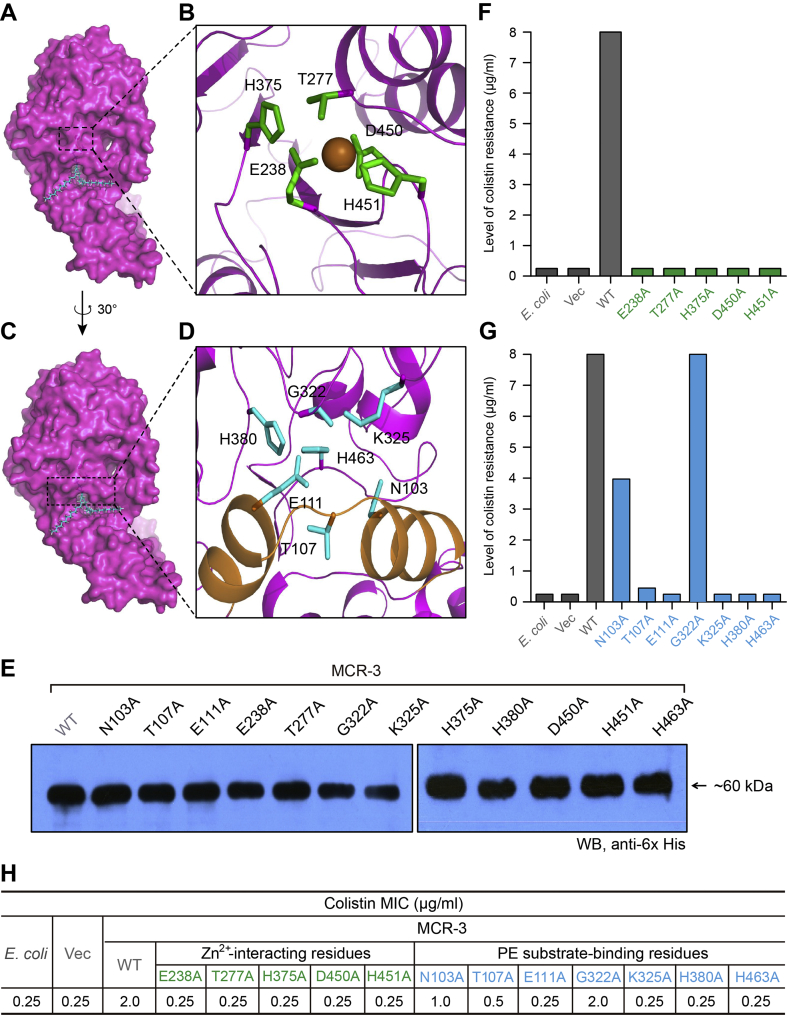
Predictions with the TMHMM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM) suggest that MCR-3 is an integral membrane protein possessing five N-terminal helices (Fig. S5A), which is almost identical to those of MCR-1 [27], MCR-2 [30] and EptA [19]. The hexa-histidine-tagged MCR-3 protein was tracked throughout the purification process (via Ni affinity chromatography) by Western blot with an anti-6× His antibody (Fig. S6A). The purified MCR-3 protein was visualized by 12% SDS-PAGE and its mass was estimated to be around 60 kDa (Fig. S6B). Its exact mass was then determined to be 61.3 kDa (Fig. S7C) by MALDI-TOF MS (Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry) and its sequence identity was verified by peptide mass fingerprinting followed by MALDI-TOF MS with a 56.4% fragment coverage (Fig. S8D). Further, the secondary structure and folding properties of MCR-3 were determined using circular dichroism (CD). The spectrum obtained had negative dips at 208 nm and 222 nm, characteristic of alpha helices (Fig. S6E) and consistent with earlier observations made for MCR-1 [27] and MCR-2 [30]. More importantly, consistent with crystallographic evidence [27, [31], [32], [33], [34]], our results indicate the presence of protein bound zinc in full-length MCR-3 (Fig. S6F). This was measured using inductively coupled plasma mass spectrometry (ICP-MS). In agreement with our recent biophysical description of MCR-1 and its derivatives [72], MCR-3 seemed to be well expressed and purified to homogeneity while maintaining its structural integrity.
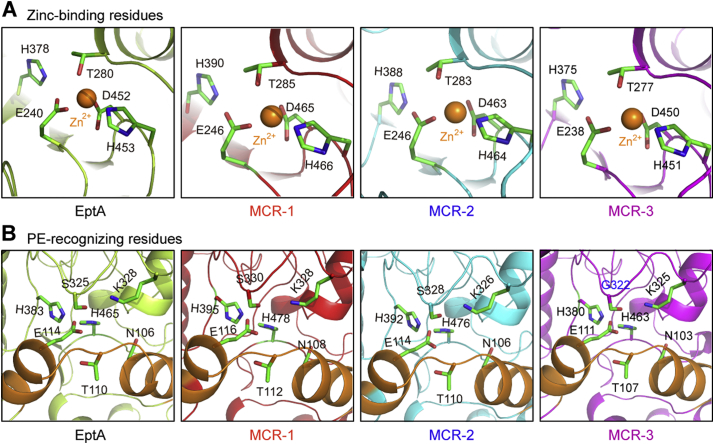
MCR-3 was modelled (Fig. S6G) against the structure of full-length EptA (PDB: 5FGN) [70] using Swiss-Model (https://swissmodel.expasy.org/interactive/qMEvX5/models/) [69] with a coverage of 98%. The model was found to retain a similar topology [70], despite possessing only 37.55% identity to EptA (Fig. S5B). As observed with EptA [70], the overall architecture of MCR-3 contains an N-terminal trans-membrane (TM) domain and a periplasm-facing catalytic domain at C-terminus (Fig. S6G). The catalytic domain has a hydrolase-fold (Fig. S6G) comprising 10 α-helices and 7 β-sheets (Figs S5B and S6G). This is connected to a TM domain containing six α-helices (Fig. S5A) by four short periplasmic loops (PH2, PH2’, PH3 and PH4), a bridge helix (BH) and a long-coiled loop (Figs S5B and S6G). Certain residues (Fig. S5B) within these structural elements are found to be deeply conserved via amino acid sequence alignments between MCR-1, MCR-2, EptA and MCR-3. These include five potential zinc-interacting residues (E238, T277, H375, D450 and H451, Fig 6A–B) and seven possible PE substrate-binding residues (N103, T107, E111, G322, K325, H380, and H463, Fig 6C-D), consistent with experimental evidence from EptA [70] and MCR-1 [72]. The roles of these residues in MCR-3 catalytic activity require further biochemical analysis.
Fig. 6.
Structure-guided functional dissection of MCR-3 colistin resistance.
A. Surface structure of MCR-3 with the cavity required for its PE lipid substrate entry.
B. An enlarged illustration for a five residues-containing, Zn2+-binding motif.
The five residues in Zn2+-binding motif of MCR-3 refers to E238, T277, H375, D450 and H451, respectively.
C. Surface architecture of MCR-3 in counter-clockwise rotation (30 °).
D. An enlarged view of the seven residues-containing motif that is involved in binding of MCR-3 to the PE lipid substrate.
The seven residues denote N103, T107, E111, G322, K325, H380 and H463, respectively.
E. Western blot analyses of the expression of MCR-3 and its 12 point-mutants in E. coli.
F. Structure-guided site-directed mutagenesis analyses for the Zn2+-binding motif of MCR-3 using the colistin susceptibility assays.
G. Functional mapping of the PE-interactive residues of MCR-3 in the context of colistin resistance.
H. The measurement of colistin MIC of the E. coli strains carrying the wild-type mcr-3 (and/or its point-mutants).
Level of colistin resistance was measured using the LBA plates. A representative result is given from no less than three independent trials.
In terms of level of colistin resistance, MCR-3 is almost identical to that of EptA, but only half of that seen with MCR-1.
3.5. Enzymology of MCR-3 Catalysis
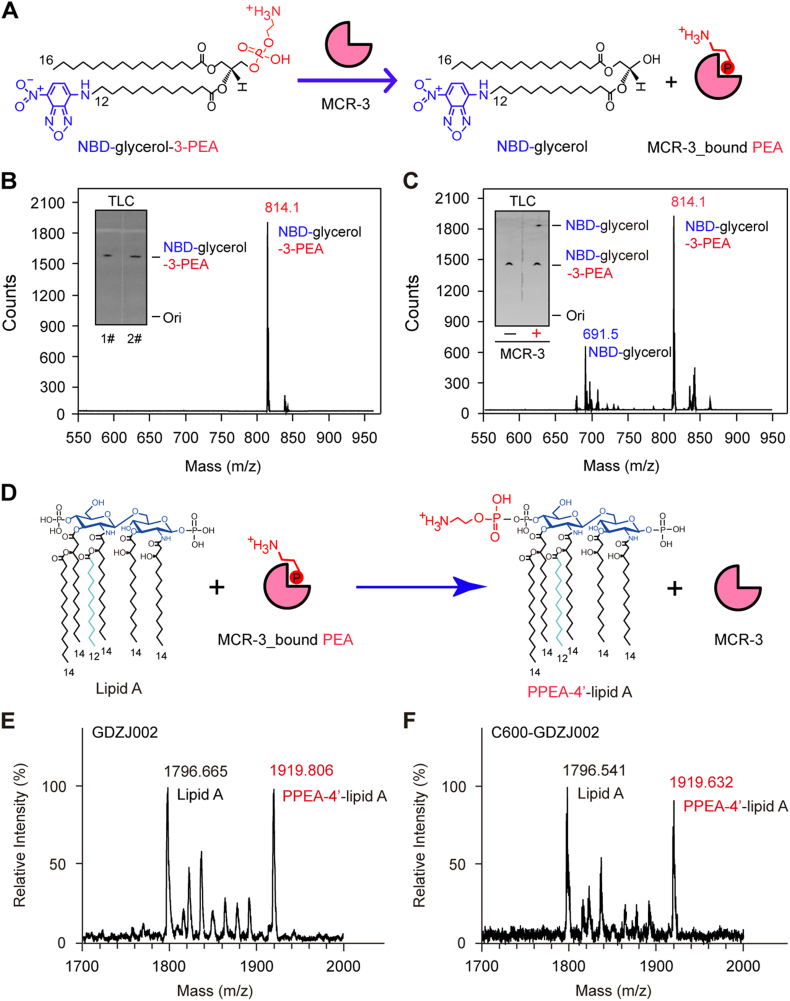
In an in vitro reaction with a fluorescent substrate NBD-glycerol-3-PEA, (Fig 5A-B), MCR-3 catalyzes the production of NBD-glycerol (Fig. 5C) from the substrate. NBD-glycerol-3-PEA (Fig. 5B) is observed as a faster migrating product when separated on a TLC. Presence of both the substrate and the product were further confirmed by liquid chromatography mass spectrometry (LC/MS). This clearly demonstrated that MCR-3 can enzymatically catalyze the removal of the PEA moiety from the lipid substrate in vitro like other well studied PEA transferases such as MCR-1 and MCR-2. Through MALDI-TOF MS, this PEA group is shown to modify the Lipid A of a clinically isolated, mcr-3-carrying E. coli strain GDZJ002 and its E. coli conjugant C600-GDZJ002. The observance of a PPEA-1(4′)-lipid A peak [m/z 1919.806 (Fig. 5E) or 1919.632 (Fig. 5F)] in addition to the unmodified lipid A peak [m/z 1796.665 (Fig. 5E) and 1796.541 (Fig. 5F)] demonstrates that the PEA moiety is enzymatically cleaved from the PE lipid substrate and is transferred to lipid A.
Fig. 5.
Enzymology of MCR-3.
A. Scheme for the removal of PEA from an alternative substrate NBD-glycerol-3-PEA by MCR-3 to give the product of NBD-glycerol and an intermediate of MCR-3-bound PEA.
PEA is an abbreviation of phosphoethanolamine. NBD was indicated in blue, and PEA was labelled in red.
B. LC/MS determination of the alternative substrate of MCR-3, NBD-glycerol-3-PEA.
C. LC/MS-based detection of the NBD-glycerol product from the hydrolysis reaction of the NBD-glycerol-3-PEA substrate by MCR-3.
The inside gels denote the TLC assays of the NBD-glycerol-3-PEA substrate (in Panel B) and the MCR-3-catalyzed hydrolysis product, NBD-glycerol (in Panel C). NBD-glycerol-3-PEA is eluted at m/z of 814.1, whereas the resultant product NBD-glycerol is present at m/z of 691.5.
Abbreviation: TLC, thin layer chromatography.
D. Schematic illustration for generation of PPEA-4′-lipid A product from lipid A by transferring of PEA from MCR-3-bound PEA
MALDI-TOF MS evidence for the chemical modification of the lipid A moieties of lipopolysaccharide (LPS) in GDZJ002, a clinical strain of mcr-3-positive E. coli strain (E) and its conjugant C600-GDZJ002 (F).
The peak of the bis-phosphorylated hexa-acylated lipid A varies at m/z (1796.541~1796.665), whereas resultant derivative with PEA modification (PPEA-4′-lipid A) is at m/z (1919.632~1919.806).
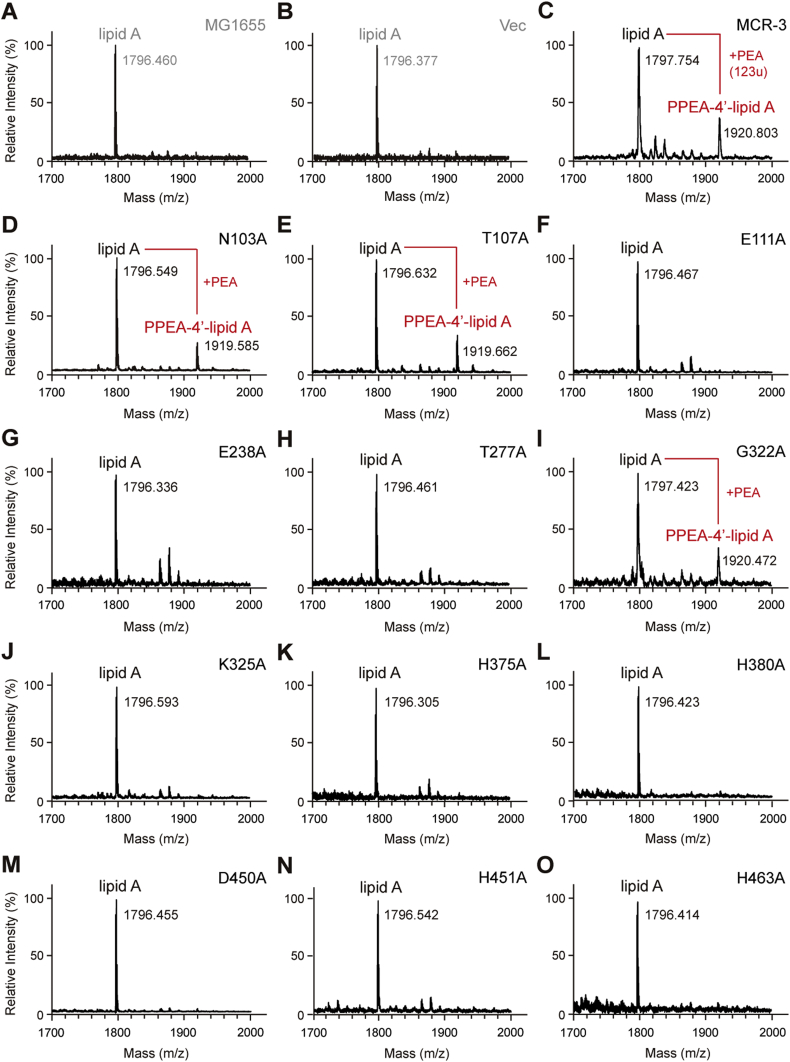
To unequivocally demonstrate that MCR-3 is responsible for the modification of lipid A in vivo, we engineered an E. coli MG1655 strain carring an arabinose-inducible plasmid pBAD24-borne mcr-3. Through MALDI-TOF MS, lipid A isolates from MG1655 or MG1655 with an empty pBAD24 vector showed the presence of a single peak [m/z 1796.46 (Fig. 7A) or 1796.377 (Fig. 7B)] corresponding to bis-phosphorylated hexa-acylated lipid A, similar to the unmodified peak observed in the clinical strains above (Fig 5E–F). Minor strain to strain variations were observed in the mass of unmodified lipid A. The expression of mcr-3 lead to the production of an additional peak at m/z 1920.803 (Fig. 7C) corresponding to a single addition of a PEA group (mass difference of 123) to lipid A (Fig. 5D). The position of this addition may be at the 1 or 4′ position.
Fig. 7.
Metabolic evidence that the PE lipid substrate-interactive cavity of MCR-3 participates in chemical modification of the lipid A moieties of lipopolysaccharides in E. coli.
MALDI-TOF MS spectrum of the LPS-lipid A species isolated from the two negative controls, the E. coli strain MG1655 alone (A) and with the empty vector pBAD24 (B).
C. Expression of MCR-3 in E. coli leads to the appearance of an additional peak of PPEA-4′-lipid A, the modified form of lipid A.
The substitution of N103A (D) and T107A (E) in MCR-3 cannot completely impair the enzymatic activity in the structural modification of lipid A moieties.
MALDI-TOF MS analyses confirm that the three point-mutants of MCR-3 [namely E111A (F), E238A (G) and T277A (H)] are nonfunctional in the transfer of PEA to lipid A species.
I. The point mutation of MCR-3 (G322A) does not influence its enzymatic activity in the addition of PPEA to the 4′-phosphate group of lipid A moieties.
The six point-mutants of MCR-3 whose enzymatic activities are fully inactivated include K325A (J), H375A (K), H380A (L), D450A (M), H451 (N) and H463A (O), respectively.
The MS peak of lipid A species in E. coli is shown at m/z of 1796.305–1797.630, whereas its modified form occurs at m/z of 1919.585–1920.803, upon the presence of functional (and/or partial active) versions of mcr-3 in E. coli.
Given the similarity between the Thr280-PEA adduct seen in EptA [70, 97] and its counterparts in reactions involving alkaline phosphatase-type phosphate transferases [98] along with the in vivo and in vitro evidence presented before, it can be hypothesized that in the first half of the reaction, MCR-3 binds to the PE lipid substrate and releases diacyl glycerol, resulting in a MCR-3 bound PEA adduct (Fig. 5A). In the second half-reaction, PEA is transferred from this adduct to the 1(4′)-phosphate position of lipid A GlcN moieties, generating PPEA-4′- lipid A (Fig. 5D). Thus, MCR-3 might utilize a “ping-pong” mechanism of catalysis like that observed in EptA [70].
3.6. Mechanistic Insights into MCR-3 Polymyxin Resistance
To understand the relationship between the two domains of MCR-3 observed in the modelled structure and the evolution of its function from MCR-like enzymes such as EptA and MCR-1, we utilized domain swapping analysis to engineer four hybrid proteins as denoted below: i) TM1-MCR-3, MCR-3 catalytic domain and TM region of MCR-1; ii) TM3-MCR-1, MCR-1 catalytic domain with TM from MCR-3; iii) TM-MCR-3, MCR-3 catalytic domain with TM of EptA; iv) TM3-EptA, catalytic domain of EptA fused to TM3 of MCR-3 (Fig. 4A). The chimeric proteins expressed as well as the full-length proteins, as evaluated by western blot (Fig. 4B). Further, functional assays for resistance to colistin indicated that in E. coli MG1655, all four hybrids were susceptible to colistin in MIC assays [0.25 μg/ml] (Fig. 4C), indistinguishable from the negative controls (strain with either no plasmid or empty vector). In contrast, expression of the full-length proteins, MCR-1, EptA and MCR-3, allowed the E. coli strain to tolerate 4 μg/ml, 2 μg/ml and 2 μg/ml of colistin, respectively (Fig. 4C). The incompatibility of the TM and PEA transferase domains of MCR-3 and EptA or MCR-1/2 (Fig. 4A) hints at an evolutionary distance between them (Fig. 3).
Fig. 4.
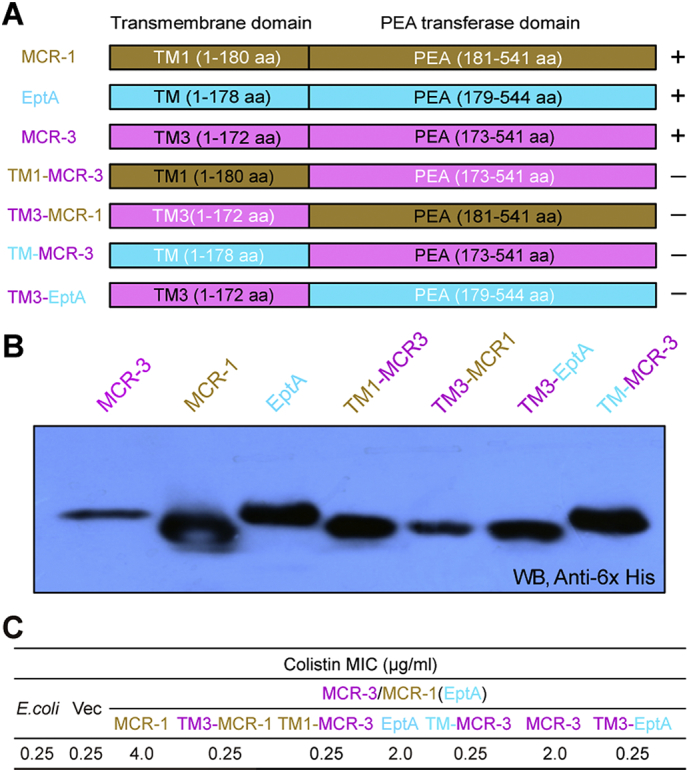
Domain-swapping analyses of MCR-3 and its two paralogues (MCR-1 plus EptA).
A. Scheme for domain-swapped constructs between MCR-3 and MCR-1 (or EptA).
B. Western blot analyses for the expression of mcr-3 and its mosaic versions in E. coli.
C. Measurement of colistin MIC of E. coli strains carrying mcr-3 and its hybrid derivatives.
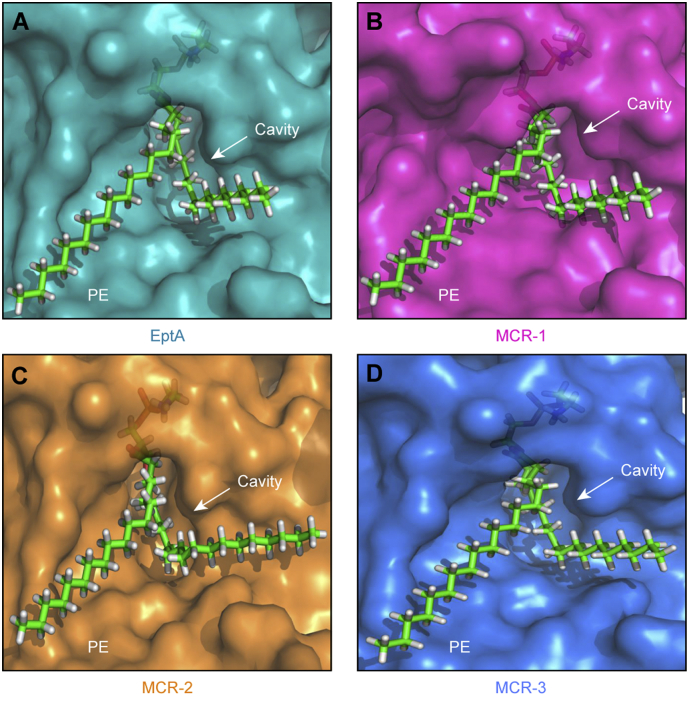
To obtain detailed experimental evidence regarding the specific residues responsible for PE substrate binding, we utilized molecular docking (MD) to analyze the modelled structure of MCR-3 with its physiological substrate, PE (Fig. 6A and C). By comparing similarly docked models of MCR-1, MCR-2 and the X-ray structure of EptA [70], we propose a similar cavity (Figs S7A–D) that accommodates the substrate in all three enzymes. This is also observed through multiple sequence alignments of the amino acid sequences of these four proteins (Fig. S5B). Using, MSA and MD data, the substrate binding cavity in MCR-3 has been defined by 12 residues of which five (E238, T277, H375, D450 and H451) seem to be involved in the interaction with zinc (Fig. 6D and S8A) and seven (N103, T107, E111, G322, K325, H380 and H463) are implicated in the recognition of the substrate, PE (Fig. 6B and S8B). 11 of these 12 residues are highly conserved between MCR-3, MCR-1, MCR-2 and EptA (Figs S8 and S5B) except for a glycine (G322) in MCR-3 instead of a conserved serine in EptA, MCR-1 and MCR-2 (S325, S330 and S328, respectively).
The catalytic role of these sites was experimentally confirmed by generating 12 alanine point mutants of MCR-3 and demonstrating their enzymatic effectiveness using colistin resistance assays and MS analysis of the target lipid A. Empty vector (0.25 μg/ml) and plasmid-free E. coli (0.25 μg/ml) were susceptible to colistin and showed a single dominant MS peak corresponding to hexa-acylated lipid A species (m/z, 1797.75) while full-length MCR-3 (2 μg/ml) was resistant to colistin and its expression results in the appearance of an additional spectral peak at m/z 1920.803 corresponding to the addition of a PEA moiety (123 mass units) to the wildtype lipid A. All twelve alanine mutants express well in E. coli, as verified by western blot (Fig. 6E). Mutants with alanine substitutions in the zinc-binding residues cannot support growth in the presence of colistin (Fig. 6E) and cannot modify lipid A (unmodified peak at m/z 1796.305-1796.632) (Fig. 6A–O) indicating their importance in enzymatic activity. Of the PE-binding residues, four are completely non-functional with respect to colistin resistance (0.25 μg/ml) and modification of lipid A (Fig. 6A-O). Two others retain partial activity [N103A (1 μg/ml) and T107A (0.5 μg/ml)] while one mutant [G322A (2 μg/ml)] shows no phenotypic defect as judged by colistin resistance trials (Fig. 6G). These three mutants also [N103A (Fig. 6D), T107A (Fig. 6E) and G322A (Fig. 6I)] show a PPEA-1(4′)-lipid A product peak (m/z, 1919.585–1920.803), indicating their functionality. Similar results were observed with all 12 mutants during colistin MIC measurements (Fig. 6H). These in vivoin vivo and in vitro data give us a mechanistic insight into the importance of cavity-forming residues in the ability of MCR-3 in providing resistance to colistin.
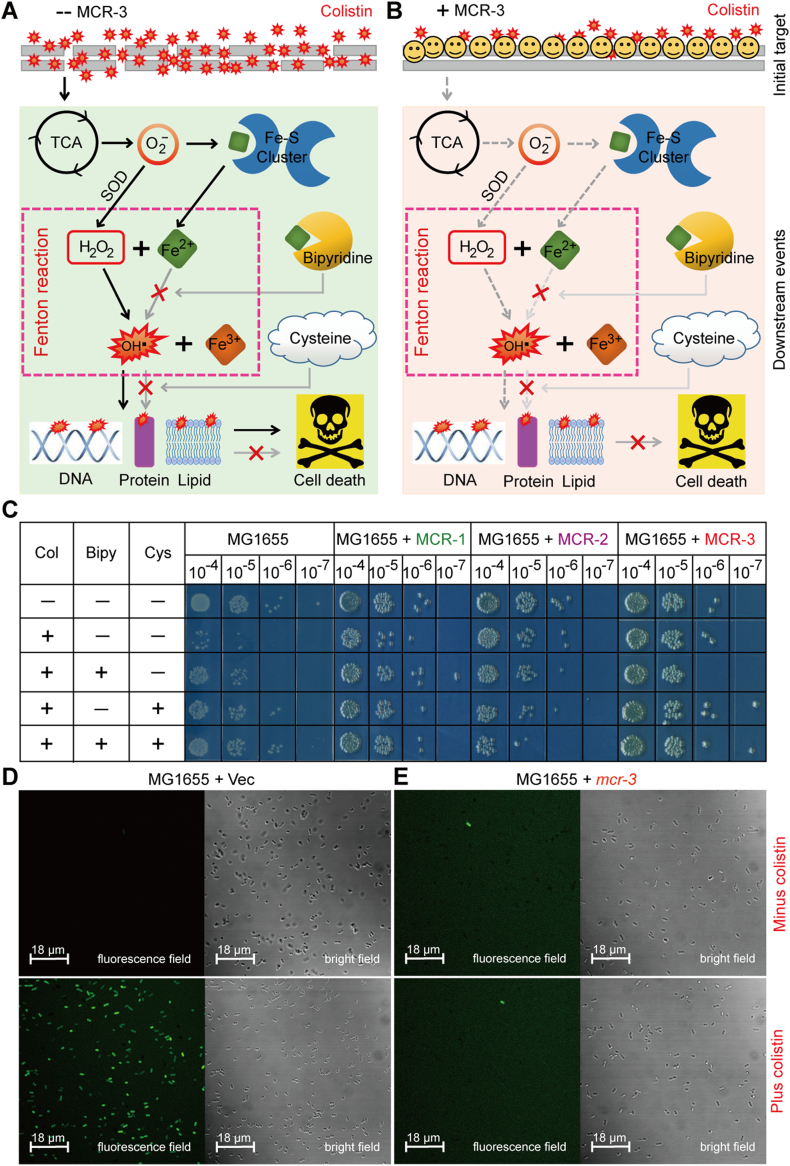
3.7. MCR-3 Quenches Hydroxyl Radical Death Pathway
Colistin is known to activate a downstream hydroxyl radical mediated cellular death pathway [89, 99]. Recently, the intracellular level of ROS was found to be prevented by the presence of MCR-1/2 and its progenitor ICR-Mo [44]. Thereby, we utilized two methods [chemical rescue assay (Fig. 8B) and confocal microscopy (Fig. 8C)] to evaluate the effect of MCR-3, another lipid A modifier on colistin-induced ROS formation. We show the involvement of the Fenton reaction in the production of free hydroxyl radicals (Fig. 8A and B) through chemical rescue experiments, where the presence of the ferric chelator bipyridine significantly bypasses the effect of colistin treatment on mcr-3-negative E. coli (Fig. 8C). This is also true in the presence of the ROS scavenger, L-cysteine (Fig. 8C). Surprisingly, E. coli is prevented from entering the hydroxy radical death pathway during the expression of mcr-3 (or mcr-1/mcr-2) in the presence of colistin, independent of bipyridine and L-cysteine (Fig. 8C). To monitor the intracellular production of H2O2 species in E. coli with/without MCR-3 (or MCR-1/MCR-2), an oxidant-susceptible dye DCFH2_DA (2′,7′-dichlorodihydrofluorescein diacetate) was utilized and cells were monitored in a fluorescent field (Fig. 8D). As expected, treatment with colistin induced ROS production in E. coli MG1655 with an empty vector (Fig. 8D), which was significantly alleviated upon the expression of MCR-3 (Figs 8E and 9). MCR-3 expression, thus, seems to prevent the entry of colistin into the cells thereby quenching/alleviating ROS production in vivo, and consequently bypassing antibiotic killing by colistin (Fig. 8B).
Fig. 8.
A working model for MCR-3-mediated impairment of the hydroxyl radical death pathway in E. coli.
A. Scheme for membrane disruption by the cationic antimicrobial polypeptide colistin and the resultant ROS production in E. coli.
B. Functional expression of MCR-3 prevents the penetration of colistin into bacterial membrane and thereafter attenuates the production of ROS in E. coli.. It was modified appropriately from Wei et al. [44] with permission.
C. Chemical rescue experiments reveal that Fenton reaction involves colistin-triggered hydroxyl radical killing pathway in E. coli.
The LPS-lipid A moiety denotes the first target for colistin treatment. Bipyridine is a well-known ferric chelator, and L-cysteine is the ROS scavenger.
D. Exposure to colistin boosts the accumulation of hydrogen peroxides in the negative control, the strain MG1655 of E. coli with the empty vector pBAD24.
E. The expression of plasmid-borne MCR-3 attenuates the production of hydrogen peroxides in E. coli, regardless of the presence of colistin.
An oxidant-sensitive dye, DCFH2-DA, was used to detect the level of intracellular ROS, which was oxidized by hydrogen peroxides into the fluorescent product of DCF. The fluorescence intensity of DCF was measured with a Zeiss LSM 510 Meta confocal laser scanning microscope (100× oil immersion objective). Hydrogen peroxides appear in green.
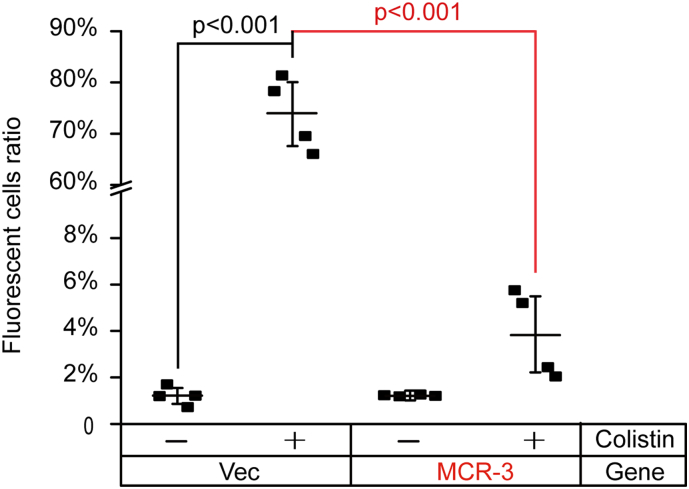
F. Quantitative comparison of colistin-promoted ROS levels in E. coli with or without expression of mcr-3.
Ratio of fluorescent cells was calculated through counting the number of cells with/without fluorescence. In each group, no <500 cells were counted from 4 individual photographs. The data was evaluated through one-way analysis of variance (ANOVA) as well as Tukey–Kramer multiple comparisons post hoc test.
Fig. 9.
Measurement of the quenched ROS levels in mcr-3-bearing E. coli even with the treatment of colistin.
Ratio of fluorescent cells was determined through counting the number of cells with/without fluorescence (seen in Fig. 8). In total, 500 cells from 4 individual photographs were counted for each group. The data was presented after one-way analysis of variance (ANOVA) along with Tukey–Kramer multiple comparisons post hoc test.
3.8. Effects of Intrinsic/Transferable Colistin Resistance Determinants on Metabolic Fitness of Gut Bacteria
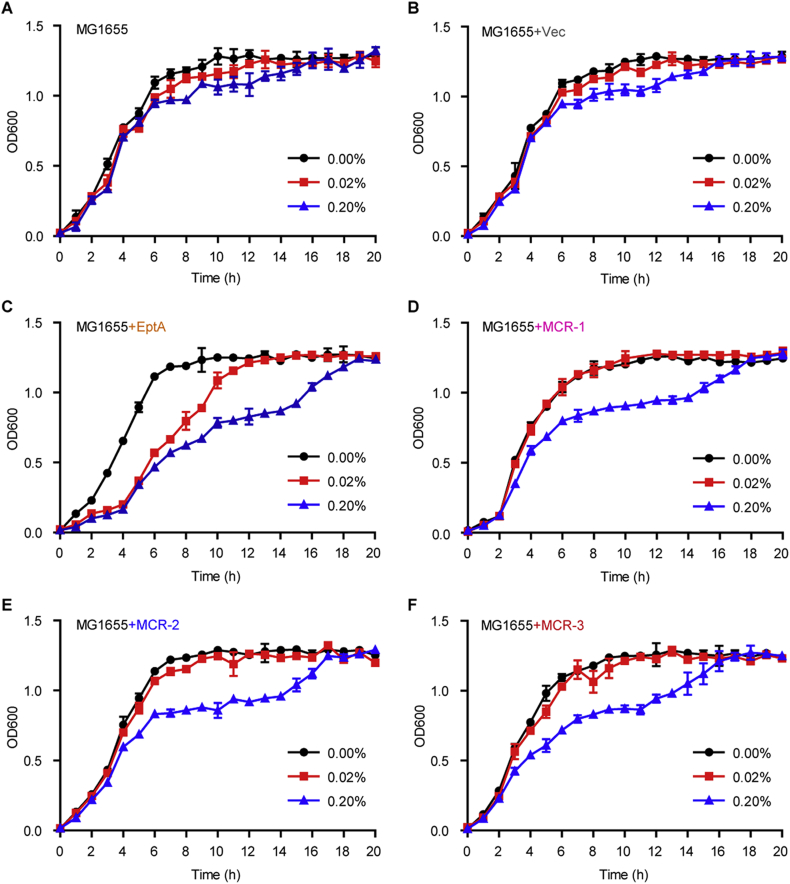
To systematically compare the metabolic fitness of gut bacteria in response of expression of intrinsic and/or transferable colistin resistance determinants, we engineered an array of derivatives of E. coli MG1655 [with the pBAD24 empty vector alone or harboring eptA/mcr-like gene, in Table S3]. The five strains used here separately included the negative control FYJ796 (MG1655/pBAD24) [72], the neisserial intrinsic colistin resistance gene eptA-expressing strain FYJ832 (MG1655/pBAD24::eptA) [79], and three mcr-like gene-bearing strains (FYJ795 [MG1655/pBAD24::mcr-1] [72], FYJ855 [MG1655/pBAD24::mcr-2] [79] & FYJ1125 [MG1655/pBAD24::mcr-3]). As expected, the following four points are recorded: i) the addition of arabinose (an inducer of the arabinose promoter of pBAD24) failed to exert significant effect on growth curves of MG1655 alone (Fig. 10A) or with the empty vector pBAD24 (Fig. 10B); ii) the expression of eptA obviously interferes the growth of E. coli MG1655 even on the condition of induction with as low as 0.02% arabinose (Fig. 10C); iii) in general agreement with the observation of mcr-1 inTOP10 strain [87], the expression of MCR-1 (only induced by 0.2% arabinose) can greatly cause growth retardation of the recipient strain MG1655 (Fig. 10D); iv) the suppressed growth of gut bacterium MG1655 harboring either mcr-2 (Fig. 10E) or mcr-3 (Fig. 10F) is indistinguishable from that seen with mcr-1 (Fig. 10D). Despite that its ability is comparable with that of eptA [72], BUT weaker than those of mcr-1/2 in rendering the recipient strain MG1655 resistant to polymyxin [79], the cellular effect of mcr-3 on bacterial growth is surprisingly seen to be similar to those of mcr-1/2 (Fig. 10D & E), but somewhat appreciably-less than that of eptA, an intrinsic determinant of colistin resistance (Fig. 10C).
Fig. 10.
Comparison of growth curves of E. coli with or without carrying mcr-like genes.
Growth curves of two negative control strains, the E. coli MG1655 alone (A) or carrying the empty vector pBAD24 (B).
C. Arabinose-induced expression of the neisserial EptA inhibits the growth of E. coli MG1655.
D. Expression of MCR-1 exerts effect on the growth of E. coli MG1655.
E. Expression of MCR-2 interferes the growth of E. coli MG1655.
F. The presence of MCR-3 inhibits the growth of E. coli MG1655.
The expression of eptA and/or mcr-1 (mcr-2 & mcr-3) was triggered through the addition of different levels of arabinose (0.00%, 0.02%, and 0.20%, w/v) into LB media. The means of three independent replicates were shown and the error bars represent the standard deviation (SD).
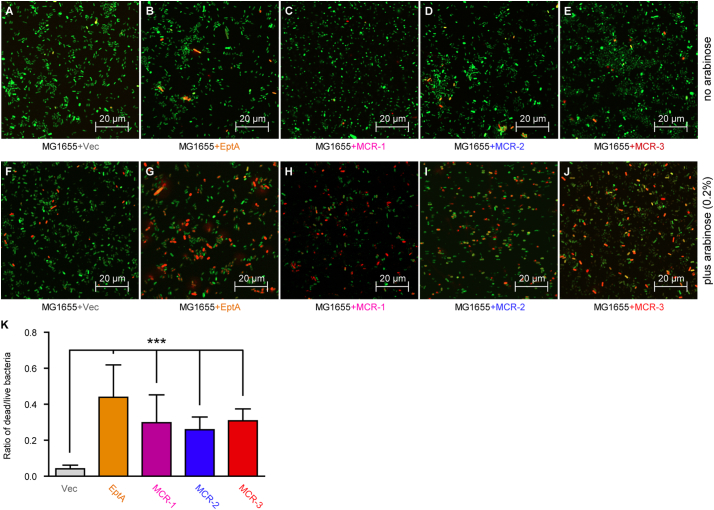
To further address the unexpected scenarios aforementioned amongst MCR-1/2/3 and EptA, we conducted confocal microscopy-based investigation on these gut bacteria using the “LIVE/DEAD” assays of bacterial viability (Fig. 11). As predicted, low percentage of dead cells was visualized for the log-phase culture of MG1655 carrying pBAD24 (Fig. 11A and F), whereas appreciably-increased level of killed cells was observed upon the induced expression of EptA with 0.2% arabinose (Fig. 11B and G). As for every one of mcr-like genes (Fig. 11C–E) in MG1655, activation of its expression apparently produced metabolic pressure featuring with the decrement of bacterial viability (Fig. 11H-J). In general agreement with scenarios seen in the experiments of growth curves (Fig. 10), the side effects exerted by MCR-1/2/3 on bacterial viability are consistently less profound than that of EptA (Fig. 11K), BUT clearly stronger than that of the negative control strain with the empty vector alone (Fig. 11K). In summary, cellular effects of the three resistance proteins MCR-1/2/3 on bacterial metabolic fitness (at least bacterial growth and viability, if not all) are relatively comparable, despite they are in quite differentiation of gene sequence (Fig. S5) as well as its resultant phenotypic resistance level (Fig. 4, Fig. 6).
Fig. 11.
Alteration in bacterial viability of E. coli by the expression of eptA and/or mcr-like genes.
Visualization of bacterial viability of the negative control, the empty vector pBAD24-carrying E. coli without the addition of arabinose (A) or with the induction of 0.2% arabinose (F).
Arabinose-induced expression of eptA promotes cell death of E. coli (G) when compared with that on the condition lacking arabinose (B).
Confocal microscopy-aided detection of LIVE/DEAD biomass of the E. coli MG1655 on the growth condition without (C) and with (H) the supplementation of 0.2% arabinose.
Visualization of the LIVE/DEAD biomass of the E. coli MG1655 expressing mcr-2 (I) with that of MG1655 without mcr-2 (D).
Comparative analysis of the LIVE/DEAD biomass ratio of the mcr-3-harboring E. coli MG1655 with (E) or without (J) the induction of 0.2% arabinose.
K. Measurement of the relative ratio of LIVE/DEAD E. coli strains carrying different colistin resistance determinants.
The bacterial cells were incubated with/without 0.2%(w/v) L-arabinose to trigger the expression of eptA (mcr-1/mcr-2/mcr-3) and stained with LIVE/DEAD kit, giving the images with Confocal laser scanning microscopy. Green and red separately denotes live and dead cell. ***, p < .001.
4. Discussion
More than 11 genetic variants of MCR-3 have been identified since its original discovery in the Shandong province of China. This study has now identified mcr-3 in bacterial isolates (Fig. 1) from 8 of the 13 provinces examined in China. This suggests a reasonable level of control in the spread of mcr-3 within China. This might be indicative of a broader reduction in mcr partially caused by the ban of colistin use in animal feedstock in China and by improved animal husbandry practices, though further studies would be necessary. A previous study reported a significantly higher prevalence of mcr-3 (>10%) from animals in 9 provinces in China, while the prevalence of mcr-3 is lower in this study [39]. The reasons could attribute to the different sampling time or/and the conditions of antibiotic resistance could be various in different farms.
All four randomly-selected mcr-3-positive clinical strains for comparative genomics of mcr-3-harboring plasmids also harbored the mcr-1 gene. In our study, all the three mcr-3-positive plasmids were identified through whole genome sequencing to be IncP type conjugable plasmids, while mcr-1 was on a non-conjugable IncX4-type plasmid. In fact, these 3 plasmids lack the IS1294 insertion element found on other mcr-3-bearing plasmids like pMCR3_ECHEC-LL123 or the ISApl1 insertion element found on mcr-1 plasmids like pMCR_1511. These insertion elements have been proposed to be involved in the initial mobilization of these genes through transposition. It has been shown that mcr-1 genes lacking insertion elements on either side must have lost them by abortive transposition and without at least a single downstream insertion element incapable of mobilization. Phylogenetically, MCR-3 is very distinct from MCR-1 and its variants (Fig 3A–B) and clusters within a larger family of MCR-3 like putative PE-lipid A transferases exclusively from the Aeromonas species. Interestingly, the neighboring sub-clades consist of putative PE-lipid A transferases from human commensal and naturally colistin-resistant bacteria from Hafnia and Edwardsiella species. Another neighboring subclade predominantly consists of PE-lipid A transferases from Vibrio species where a chromosomal system that attaches glycine to LPS-lipid A GlcN moieties and provides ‘intrinsic’ resistance to colistin has been described in Vibrio cholerae.
Taken together, we can be reasonably confident that MCR-3 and its variants might have a parallel evolutionary path to MCR-1 from a yet to be identified chromosomal progenitor. Despite that the co-occurrence of mcr-1 and mcr-3 on certain plasmids [59] raises the possibility of mcr-1/3 evolved through gene duplication events, the poor homology between MCR-1 and MCR-3 argues the aforementioned hypothesis. The divergence of these two groups is probably a distant event and hence, the current source of genetic diversity for each of these species might be different. Recently, Moraxella species have been proposed as reservoirs for MCR-1/2 species [43, 100]. A similar reservoir might exist in the Aeromonas (Fig. 3B), though little is known about the functionality of their MCR-like genes. Unique selection pressures in the environment might have triggered a recent mobilization of some of these intrinsic genes. Z1140 might be an excellent example of a protein caught in the act of evolving either through gain or loss of function over time (Fig. S9). This idea is further strengthened by the biochemical and physiological characterization of MCR-3 which shows that the two distinct protein domains of MCR-1 and MCR-2 are functionally exchangeable, while those in MCR-3 and MCR-1/EptA are not (Fig. 4). However, we show that MCR-3 and MCR-1/MCR-2/EptA share a common PE substrate binding cavity, which is essential for its activity and for conferring resistance to colistin (Fig. S7). This indicates an evolutionarily conserved mechanism for both intrinsic and transferable polymyxin resistance wherein they share the same catalytic scheme involving a non-sequential or ‘ping-pong’ reaction mechanism. Further, MCR-3 displays similar abilities in both modifying lipid A structure (Fig. 5, Fig. 7) thereby reducing its affinity to colistin. Though polymyxins like colistin have been shown to activate a downstream hydroxyl radical mediated cellular death pathway [89, 99], MCR-3 also quenches ROS production in vivo (Fig. 8) similar to MCR-1/MCR-2/EptA.
These data together suggest that despite a vast amount of evolutionary diversity in the MCR family, a mechanistic and functional unification is observed. By understanding these aspects of a phylogenetically-distinct member like mcr-3, we might be able to track and arrest the origins and spread of colistin resistance. This might also indicate the selection pressures that have resulted in this rapid discovery of mcr-3 variants across the globe suggesting better policies for antibiotic use in agriculture and medicine. It seems very true that further comprehensive in vivo and clinical studies are necessary to understand the threat of mcr-3 gene to clinical and public health.
The following are the supplementary data related to this article.
Fig. S1.
Genetic analyses for mcr-3-carrying plasmids isolated from clinical strains
A. S1-PFGE analyses for clinical E. coli strains and their trans-conjugants
B. Southern blotting for clinical E. coli strains and their trans-conjugants using mcr-3 probe
Fig. S2.
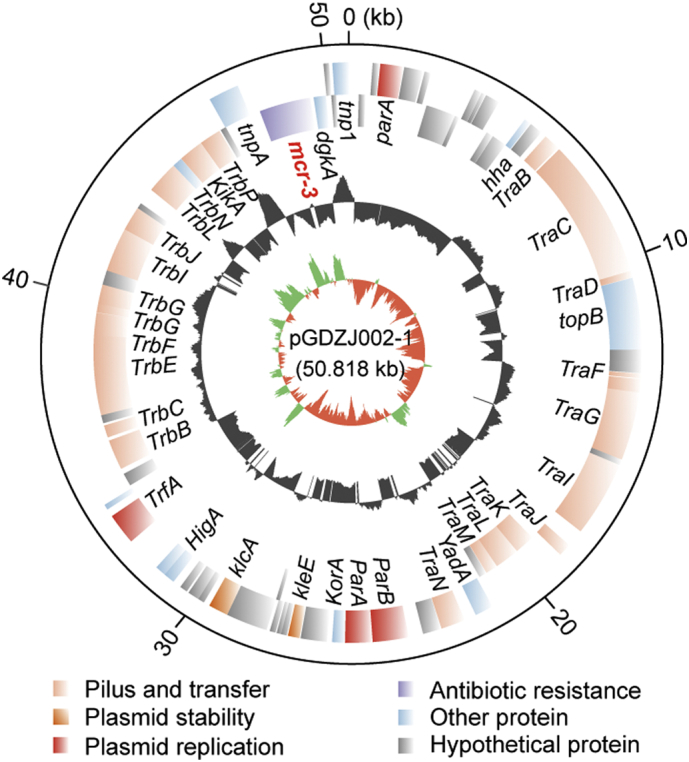
Physical map of the mcr-3-carrying plasmid pGDZJ002-1
Circles (from inside to outside) represent the GC screw, GC content and ORF, respectively [27]. The plasmid genomes are annotated by RAST. The plasmid maps are generated with Circos program [27].
Fig. S3.
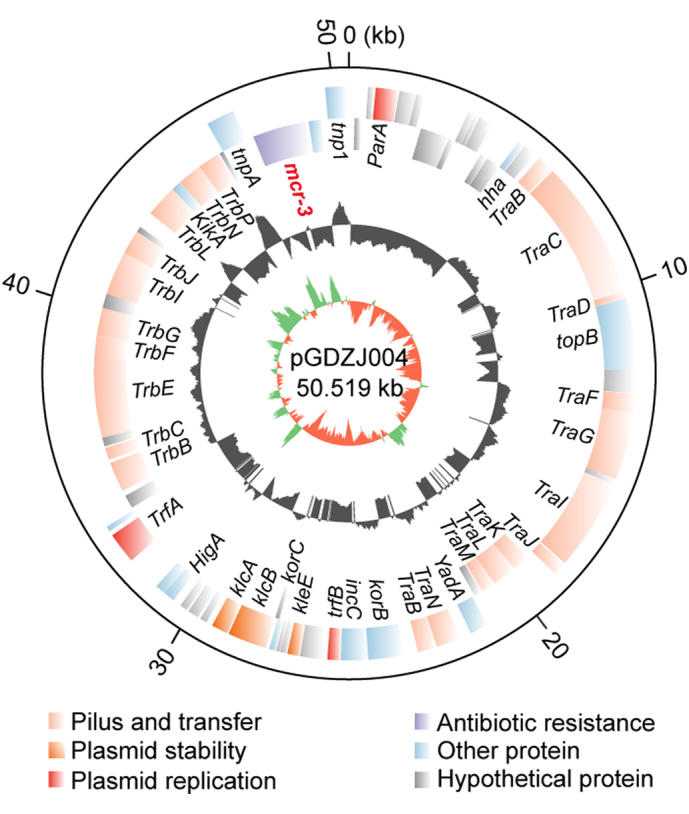
Genomic map of the mcr-3-harboring plasmid pGDZJ004
Fig. S4.
Colinear genome analyses of four mcr-3-bearing plasmids.
Boxed arrows represent the position and transcriptional direction of the ORFs. Regions of >99% identity are given by grey shading. The antibiotic resistance gene mcr-3 and its variant mcr-3.7 are colored in blue.
Fig. S5.
Bioinformatic analyses of MCR-3.
A. Transmembrane prediction of MCR-3
B. Sequence alignment of MCR-3 with EptA and MCR-1/2 homologs.
Fig. S6.
Characterization of MCR-3 protein.
A. Western blot detection suggests sub-cellular localization of MCR-3 in the membrane-associated fraction from E. coli lysates Designations: L, Lysate of E. coli following sonication; S, Supernatant of bacterial crude extracts undergoing ultra-centrifugation; P, Pellet of bacterial crude extracts undergoing ultra-centrifugation; D, Detergent-solubilized fraction. B. SDS-PAGE (15%) profile of the purified MCR-3 C. Use of MALDI-TOF to determine the molecular mass of MCR-3 The mass of MCR-3 is unveiled to be 61.3 kDa. D. MS identification of the recombinant MCR-3 integral membrane protein. The transmembrane region of MCR-3 is in light blue, and the enzymatic domain is in red. The polypeptides that have been identified by MS and match MCR-3 are underlined with coverage of 56.4%. E. Circular dichroism (CD)-based illustration of the protein secondary structure of MCR-3. F. Use of ICP/MS to determine the existence of MCR-3 in the zinc-bound form G. Overall architecture of the modelled MCR-3 structure. Abbreviations: C, C-terminus; N, N-terminus.
Fig. S7.
Parallels in PE-recognizing cavities of MCR-like enzymes.
A. Enlarged view of surface structure of EptA having a cavity for the entry of PE lipid substrate B. Structural snapshot of the PE lipid substrate-interactive cavity in MCR-1. C. A similar PE-recognizing cavity is present in MCR-2. D. A conservative PE-binding cavity is also shared by MCR-3. The enlarged surface structures of PE-binding cavities are generated through molecular docking together with structural modelling. The PE molecules are denoted with green sticks, and cavity is indicated with an arrow. The photographs are generated using PyMol. Designations: PE: phosphatidylethanolamine.
Fig. S8.
Structural similarity of PE-recognizing cavities of MCR-like enzymes.
A. A putative Zn2+-centering, five-residues forming motif is conserved amongst PE lipid substrate-recognizing cavities of four MCR-like paralogues EptA, MCR-1, MCR-2 and MCR-3. B. Comparative analyses of the conserved PE-recognizable residues of four MCR-like enzymes EptA, MCR-1, MCR-2 and MCR-3 With an exception that the conserved serine (S325 in EptA, S330 in MCR-1 and S328 in MCR-2) is replaced with glycine on the residue 322 (G322) in MCR-3, all the other six residues are identical in these cavities. The photographs are produced with PyMol.
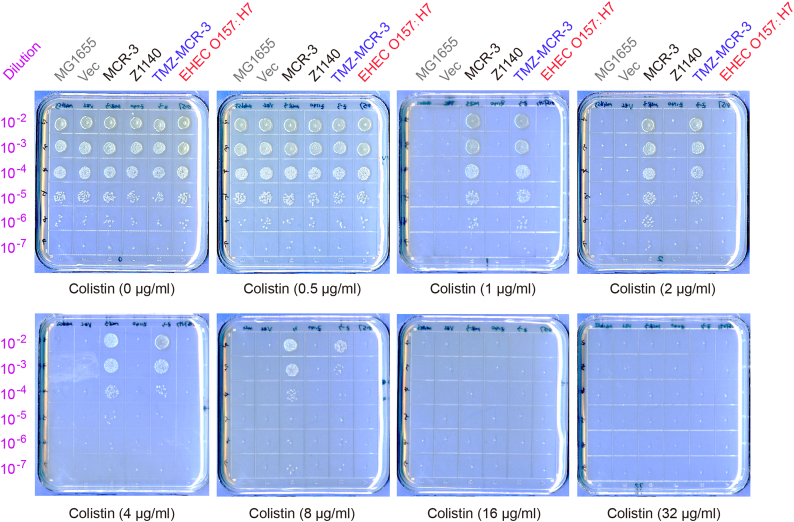
Fig. S9.
Both the E. coli O157: H7 strain EDL933 and the E. coli MG1655 strain carrying Z1140 (a putative PEA lipid A transferase from Strain EDL933) are susceptible to colistin, whereas the expression of mcr-3 (and its hybrid version of TMZ-MCR-3) confers the recipient strain MG1655 resistant to colistin
The mid-log phase cultures (in serial dilution) were spotted on LBA plates containing varied levels of colistin (0, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0 and 32.0 11 μg/ml) and kept overnight at 37°C. Plasmid-borne expression of mcr-3 (TMZ-MCR-3) is induced with the addition of 0.2% arabinose. Designations: Vec, the empty vector pBAD24; TMZ-MCR-3, a hybrid version of MCR-3 in which the TM region is replaced with the counterpart in Z1140 of the E. coli O157:H7 strain EDL933. A representative result of three independent experiments is given.
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Acknowledgments
Acknowledgements
This work was supported by National Key R&D Program of China (2017YFD0500202, YF), National Key Basic Research Program of China (2016YFC1200100, YF) and the National Natural Science Foundation of China (31570027 & 81772142, YF; 81722030, GT). Dr. Feng is a recipient of the “Young 1000 Talents” Award. We would like to acknowledge the contribution of Dr. Yohei Doi at Division of Infectious Diseases, University of Pittsburgh School of Medicine, for the critical review of the manuscript.
Author Contributions
YF and GT designed and supervised this project; YF, YX, LZ, JS, SS, JL and ZT performed experiments; YF, YX, LZ, MH, DP, JS, SS, JL, XL and ZT analyzed the data and prepared figures; YF, SS and GT drafted this manuscript.
Conflict of Interest
We declare that no interest conflict is present.
Funding
National Key R&D Program of China, National Key Basic Research Program of China, and National Natural Science Foundation of China.
Contributor Information
Guo-Bao Tian, Email: tiangb@mail.sysu.edu.cn.
Youjun Feng, Email: fengyj@zju.edu.cn.
References
- 1.Centers for Disease Control and Prevention, Office of Infectious Disease . 2013. Antibiotic resistance threats in the United States, 2013.http://www.cdc.gov/drugresistance/threat-report-2013 Available at: [Google Scholar]
- 2.Laxminarayan R., Amabile-Cuevas C.F., Cars O. UN High-Level Meeting on antimicrobials—what do we need? Lancet. 2016;388(10041):218–220. doi: 10.1016/S0140-6736(16)31079-0. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill L.J. Review on Antimicrobial Resistance; London: 2014. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. [Google Scholar]
- 4.de Kraker M.E., Stewardson A.J., Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13(11) doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kraker M.E., Davey P.G., Grundmann H. group Bs. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011;8(10):e1001104. doi: 10.1371/journal.pmed.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velkov T., Thompson P.E., Nation R.L., Li J. Structure--activity relationships of polymyxin antibiotics. J Med Chem. 2010;53(5):1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Nation R.L., Turnidge J.D. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 8.Rhouma M., Beaudry F., Theriault W., Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front Microbiol. 2016;7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon R.A., Chopra I. Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J Antimicrob Chemother. 1986;18(5):557–563. doi: 10.1093/jac/18.5.557. [DOI] [PubMed] [Google Scholar]
- 10.Brown J.M., Dorman D.C., Roy L.P. Acute renal failure due to overdosage of colistin. Med J Aust. 1970;2(20):923–924. doi: 10.5694/j.1326-5377.1970.tb63262.x. [DOI] [PubMed] [Google Scholar]
- 11.Koch-Weser J., Sidel V.W., Federman E.B., Kanarek P., Finer D.C., Eaton A.E. Adverse effects of sodium colistimethate. Manifestations and specific reaction rates during 317 courses of therapy. Ann Intern Med. 1970;72(6):857–868. doi: 10.7326/0003-4819-72-6-857. [DOI] [PubMed] [Google Scholar]
- 12.Nation R.L., Li J. Colistin in the 21st century. Curr Opin Infect Dis. 2009;22(6):535–543. doi: 10.1097/QCO.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron S., Hadjadj L., Rolain J.M., Olaitan A.O. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48(6):583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Falagas M.E., Kasiakou S.K. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. 2005;40(9):1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 15.Cannatelli A., D'Andrea M.M., Giani T. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother. 2013;57(11):5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z., Ribeiro A.A., Lin S., Cotter R.J., Miller S.I., Raetz C.R. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J Biol Chem. 2001;276(46):43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- 17.Helander I.M., Kilpelainen I., Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol. 1994;11(3):481–487. doi: 10.1111/j.1365-2958.1994.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 18.Miller A.K., Brannon M.K., Stevens L. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother. 2011;55(12):5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox A.D., Wright J.C., Li J., Hood D.W., Moxon E.R., Richards J.C. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J Bacteriol. 2003;185(11):3270–3277. doi: 10.1128/JB.185.11.3270-3277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beceiro A., Llobet E., Aranda J. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55(7):3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen T.W., Trent M.S. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci U S A. 2010;107(11):5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilecen K., Fong J.C., Cheng A., Jones C.J., Zamorano-Sanchez D., Yildiz F.H. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect Immun. 2015;83(3):1199–1209. doi: 10.1128/IAI.02700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hankins J.V., Madsen J.A., Giles D.K., Brodbelt J.S., Trent M.S. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proc Natl Acad Sci U S A. 2012;109(22):8722–8727. doi: 10.1073/pnas.1201313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson J.C., Herrera C.M., Trent M.S. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J Biol Chem. 2017;292(51):21205–21215. doi: 10.1074/jbc.RA117.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bengoechea J.A. Vibrio cholerae amino acids go on the defense. J Biol Chem. 2017;292(51):21216–21217. doi: 10.1074/jbc.H117.000868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y.Y., Wang Y., Walsh T.R. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 27.Gao R., Hu Y., Li Z. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12(11) doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds C.M., Kalb S.R., Cotter R.J., Raetz C.R. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem. 2005;280(22):21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 29.Lee H., Hsu F.F., Turk J., Groisman E.A. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186(13):4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J., Xu Y., Gao R. Deciphering MCR-2 colistin resistance. mBio. 2017;8(3) doi: 10.1128/mBio.00625-17. (pii: e00625–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojanoski V., Sankaran B., Prasad B.V., Poirel L., Nordmann P., Palzkill T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 2016;14(1):81. doi: 10.1186/s12915-016-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma G., Zhu Y., Yu Z., Ahmad A., Zhang H. High resolution crystal structure of the catalytic domain of MCR-1. Sci Rep. 2016;6:39540. doi: 10.1038/srep39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu M., Guo J., Cheng Q. Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci Rep. 2016;6:38793. doi: 10.1038/srep38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinchliffe P., Yang Q.E., Portal E. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci Rep. 2017;7:39392. doi: 10.1038/srep39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coates K., Walsh T.R., Spencer J., Hinchliffe P. 1.12 A resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2. Acta Crystallogr F Struct Biol Commun. 2017;73(Pt 8):443–449. doi: 10.1107/S2053230X17009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kluytmans J. Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveill. 2017,;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier B.B., Lammens C., Ruhal R. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27) doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Graells C., De Keersmaecker S.C.J., Vanneste K. Detection of plasmid-mediated colistin resistance, mcr-1 and mcr-2 genes, in Salmonella spp. Isolated from food at retail in Belgium from 2012 to 2015. Foodborne Pathog Dis. 2017;15(2):114–117. doi: 10.1089/fpd.2017.2329. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Chen L., Wang J. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci Rep. 2018;8(1):3705. doi: 10.1038/s41598-018-22084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi Z., Berglund B., Sun Q. Prevalence of the mcr-1 colistin resistance gene in extended-spectrum beta-lactamase-producing Escherichia coli from human faecal samples collected in 2012 in rural villages in Shandong Province, China. Int J Antimicrob Agents. 2017;49(4):493–497. doi: 10.1016/j.ijantimicag.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Carattoli A., Villa L., Feudi C. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30589. (pii: 30589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borowiak M., Fischer J., Hammerl J.A., Hendriksen R.S., Szabo I., Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 43.Abuoun M., Stubberfield E.J., Duggett N.A. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72(10):2745–2749. doi: 10.1093/jac/dkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei W., Srinivas S., Lin J. Defining ICR-Mo, an intrinsic colistin resistance determinant from Moraxella osloensis. PLoS Genet. 2018;14(5) doi: 10.1371/journal.pgen.1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y.Q., Li Y.X., Lei C.W., Zhang A.Y., Wang H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(7):1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Wang Y., Zhou Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Pilato V., Arena F., Tascini C. MCR-1.2: a new MCR variant encoded by a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae of sequence type 512. Antimicrob Agents Chemother. 2016;60(9):5612–5615. doi: 10.1128/AAC.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X., Hu Y., Luo M. MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica Serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother. 2017;65(5) doi: 10.1128/AAC.02632-16. (pii: e02632-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Zhang H., Sun J., Liu Y.H., Feng Y. The MCR-1 colistin resistance: a new challenge to global public health. Chin Sci Bull. 2017;62(10):1018–1029. [Google Scholar]
- 50.Zhang J., Chen L., Wang J. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Res Notes. 2018;11(1):143. doi: 10.1186/s13104-018-3255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teo J.W.P., Kalisvar M., Venkatachalam I., Tek N.O., Lin R.T.P., Octavia S. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol. 2017;56(3) doi: 10.1128/JCM.01562-17. (pii: e01562-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X., Zhai W., Li J. Presence of mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother. 2017;62(2) doi: 10.1128/AAC.02106-17. (pii: e01562-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ling Z., Yin W., Li H. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother. 2017;61(11) doi: 10.1128/AAC.01272-17. (pii: e01272-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L., Feng Y., Zhang X., McNally A., Zong Z. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother. 2017;61(12) doi: 10.1128/AAC.01757-17. (pii: e01757-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda A., Sato T., Shinagawa M. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents. 2017;51(1):163–164. doi: 10.1016/j.ijantimicag.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Litrup E., Kiil K., Hammerum A.M., Roer L., Nielsen E.M., Torpdahl M. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009–17. Euro Surveill. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roer L., Hansen F., Stegger M., Sonksen U.W., Hasman H., Hammerum A.M. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill. 2017;22(31) [PubMed] [Google Scholar]
- 58.Haenni M., Beyrouthy R., Lupo A., Chatre P., Madec J.Y., Bonnet R. Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J Antimicrob Chemother. 2017;73(2):533–536. doi: 10.1093/jac/dkx418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernandez M, Iglesias MR, Rodriguez-Lazaro D, et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015Euro Surveill. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30586. (pii: 30586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Catry B., Cavaleri M., Baptiste K. Use of colistin-containing products within the European Union and European economic area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents. 2015;46(3):297–306. doi: 10.1016/j.ijantimicag.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 61.Kempf I., Jouy E., Chauvin C. Colistin use and colistin resistance in bacteria from animals. Int J Antimicrob Agents. 2016;48(6):598–606. doi: 10.1016/j.ijantimicag.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 62.Quan J., Li X., Chen Y. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y., Tian G.B., Zhang R. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 64.Yin W., Li H., Shen Y. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8(3) doi: 10.1128/mBio.00543-17. (e00543–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bankevich A., Nurk S., Antipov D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aziz R.K., Bartels D., Best A.A. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27(23):4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biasini M., Bienert S., Waterhouse A. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue) doi: 10.1093/nar/gku340. (W252–8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anandan A., Evans G.L., Condic-Jurkic K. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc Natl Acad Sci U S A. 2017;114(9):2218–2223. doi: 10.1073/pnas.1612927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52(7):1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Lin J., Cui T., Srinivas S., Feng Y. Mechanistic insights into transferable polymyxin resistance among gut bacteria. J Biol Chem. 2018;293(12):4350–4365. doi: 10.1074/jbc.RA117.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen W.J., Balius T.E., Mukherjee S. DOCK 6: impact of new features and current docking performance. J Comput Chem. 2015;36(15):1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y., Kumar R., Ravcheev D.A., Zhang H. Paracoccus denitrificans possesses two BioR homologs having a role in regulation of biotin metabolism. Microbiol Open. 2015;4(4):644–659. doi: 10.1002/mbo3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Y., Cronan J.E. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol. 2011;80(1):195–218. doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alves A., Marques A., Martins E., Silva T., Reis R. Cosmetic potential of marine fish skin collagen. Cosmetics. 2017;4(4):39. [Google Scholar]
- 77.Powl A.M., O'Reilly A.O., Miles A.J., Wallace B.A. Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc Natl Acad Sci U S A. 2010;107(32):14064–14069. doi: 10.1073/pnas.1001793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Loeschner K., Harrington C.F., Kearney J.L., Langton D.J., Larsen E.H. Feasibility of asymmetric flow field-flow fractionation coupled to ICP-MS for the characterization of wear metal particles and metalloproteins in biofluids from hip replacement patients. Anal Bioanal Chem. 2015;407(16):4541–4554. doi: 10.1007/s00216-015-8631-4. [DOI] [PubMed] [Google Scholar]
- 79.Xu Y., Wei W., Lei S., Lin J., Srinivas S., Feng Y. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio. 2018;9(2) doi: 10.1128/mBio.02317-17. (e02317-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y.Y., Chandler C.E., Leung L.M. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother. 2017;61(6) doi: 10.1128/AAC.00580-17. (pii: e00580–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caroff M., Tacken A., Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the "isolated lipid A" fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175(2):273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 82.Tsai C.M., Frasch C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 83.Kanistanon D., Hajjar A.M., Pelletier M.R. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog. 2008;4(2) doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.El Hamidi A., Tirsoaga A., Novikov A., Hussein A., Caroff M. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res. 2005;46(8):1773–1778. doi: 10.1194/jlr.D500014-JLR200. [DOI] [PubMed] [Google Scholar]
- 85.Gros M., Rodriguez-Mozaz S., Barcelo D. Rapid analysis of multiclass antibiotic residues and some of their metabolites in hospital, urban wastewater and river water by ultra-high-performance liquid chromatography coupled to quadrupole-linear ion trap tandem mass spectrometry. J Chromatogr A. 2013;1292:173–188. doi: 10.1016/j.chroma.2012.12.072. [DOI] [PubMed] [Google Scholar]
- 86.Chen Z., Zhou Q., Zou D. Chloro-benzoquinones cause oxidative DNA damage through iron-mediated ROS production in Escherichia coli. Chemosphere. 2015;135:379–386. doi: 10.1016/j.chemosphere.2015.04.076. [DOI] [PubMed] [Google Scholar]
- 87.Yang Q., Li M., Spiller O.B. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun. 2017;8(1):2054. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Z., Zhu Y., Qin W., Yin J., Qiu J. Oxidative stress induced by polymyxin E is involved in rapid killing of Paenibacillus polymyxa. Biomed Res Int. 2017;2017:5437139. doi: 10.1155/2017/5437139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sampson T.R., Liu X., Schroeder M.R., Kraft C.S., Burd E.M., Weiss D.S. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother. 2012;56(11):5642–5649. doi: 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 91.Spagnuolo G., D'Anto V., Cosentino C., Schmalz G., Schweikl H., Rengo S. Effect of N-acetyl-L-cysteine on ROS production and cell death caused by HEMA in human primary gingival fibroblasts. Biomaterials. 2006;27(9):1803–1809. doi: 10.1016/j.biomaterials.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 92.Chen C.Y., Nace G.W., Irwin P.L. A 6 x 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods. 2003;55(2):475–479. doi: 10.1016/s0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 93.Zhao F.F., Feng Y., Lu X.J., McNally A., Zong Z.Y. IncP plasmid carrying colistin resistance gene mcr-1in Klebsiella pneumoniae from hospital sewage. Antimicrob Agents Chemother. 2017;61(2) doi: 10.1128/AAC.02229-16. (pii: e02229-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gokulan K., Khare S., Rooney A.W., Han J., Lynne A.M., Foley S.L. Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, onSalmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun J., Fang L.X., Wu Z. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep. 2017;7(1):424. doi: 10.1038/s41598-017-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lagana P., Caruso G., Minutoli E., Zaccone R., Santi D. Susceptibility to antibiotics of Vibriospp. and Photobacterium damselassp. piscicida strains isolated from Italian aquaculture farms. New Microbiol. 2011;34(1):53–63. [PubMed] [Google Scholar]
- 97.Wanty C., Anandan A., Piek S. The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J Mol Biol. 2013;425(18):3389–3402. doi: 10.1016/j.jmb.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 98.Stec B., Holtz K.M., Kantrowitz E.R. A revised mechanism for the alkaline phosphatase reaction involving three metal ions. J Mol Biol. 2000;299(5):1303–1311. doi: 10.1006/jmbi.2000.3799. [DOI] [PubMed] [Google Scholar]
- 99.Dong T.G., Dong S., Catalano C., Moore R., Liang X., Mekalanos J.J. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci U S A. 2015;112(7):2181–2186. doi: 10.1073/pnas.1425007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kieffer N., Nordmann P., Poirel L. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother. 2017;61(6) doi: 10.1128/AAC.00129-17. (pii: e00129-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4