Abstract
Background
OGN could modify tissue inflammation and immune response via local and circulating innate immune cells, which was suggestive of a reciprocal relationship between OGN and T cell infiltration in cancer. Hence, we aim to measure the OGN expression patterns and immune cells response in colorectal cancer(CRC).
Methods
This study enrolled three independent sets of patients from TCGA and the Fudan University Shanghai Cancer Center(FUSCC). The effect of OGN on T cell infiltration and the mechanism were examined in vitro and in vivo.
Findings
Tumor OGN expression levels were positively associated with CD3, CD8, and PTPRC expressions in the training and testing sets from TCGA, respectively. In validation set from FUSCC, OGN expression level also paralleled positively with CD8+ cell density in colorectal cancer tissue (p < .001). For a unit decrease in outcome quartile categories, multivariable OR in the lowest (vs highest) OGN expression was 0.17 (95% CI 0.08–0.33). Consistently, immunofluorescence validated that OGN was preferentially expressed with CD8+ cells in both normal epithelium and cancer tissue. Xenograft tumors arising from MC38 cells with OGN-over-expression displayed a significant increase in CD8+ cells recruitment. Hence, high expression of OGN was associated with a profound longer survival (P = .009). In mechanism, elevated OGN expression inhibited the activation of the transcriptional genes HIF-1α in CRC cells, then significantly impeded the expression of VEGF. As a result of this, T cell tumor infiltration was reduced.
Interpretation
OGN expression is positively associated with CD8+ cell density in colorectal cancer tissue, suggesting a possible influence of OGN expression on tumor reactive T cells in the tumor niche.
Fund
No
Keywords: Osteoglycin, Tumor-infiltrating lymphocytes, Colorectal cancer, Immune response, Survival
Highlights
-
•
OGN is positively associated with T cell density in CRC
-
•
OGN inhibited the activation of HIF-1α, then significantly impeded the expression of VEGF.
Research in Context.
Evidence Before This Study
High density of CD3+, CD8+, PTPRC+ or FOXP3 T cells were supposed to be recruited, when strong immune response was manifested in colorectal cancer with better clinical outcome. But the determinant of immune response or immune cells recruits in CRC is under unraveled and it is a crucial role in understanding their dynamics as to tumor invasion, immune-surveillance and evasion. A wide range of findings were suggestive of a reciprocal relationship between OGN and T cell infiltration.
Added Value of this study
In TCGA and FUSCC cohorts, tumor OGN expression had a positive association with CD8+ cell density in colorectal cancer, suggesting a possible mechanism of a profound longer survival in the CRC with OGN expression. In mechanism, elevated OGN expression inhibited the activation of the transcriptional genes HIF-1α in CRC cells, then significantly impeded the expression of VEGF. As a result of this, T cell tumor infiltration was reduced.
Implications of all available evidence
Our consolidated data draws a possible effect of OGN level on CD8 T cells in colorectal cancer microenvironment, and can promote further translational research on the associations of OGN with host immunity in colorectal cancer.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancers(CRC) are a heterogeneous group of neoplasms and are frequently affected by tumor-host interactions [1, 2]. T-cell-mediated adaptive immunity are involved in tumor initiation and progression, emerging as a novel strategy to treat various cancers [3]. High density of CD3+, CD8+, PTPRC+ or FOXP3+ T cells were supposed to be recruited, when strong immune response was manifested in colorectal cancer with better clinical outcome [4, 5]. Although some certain tumor molecular status, just as high-level microsatellite instability, has been identified to associate with enhanced infiltration of T cells [6], while the determinant of immune response or immune cells recruits in CRC is under unraveled. Osteoglycin (OGN) has been observed down-expression in a variety of malignances including gastric cancer [7], squamous cervical, vaginal cancer [9], colorectal adenoma [8], invasive ductal breast carcinoma [10], and laryngeal carcinoma [11]. However, it is still undetermined according to the significance of OGN in CRC. OGN has been found in local and circulating innate immune cells [12], as OGN clearly co-stained with markers of both neutrophil and monocyte/macrophage in the myocardium. What is more, great distinguished phenotypic characteristics were indicated by significantly altered phosphorylation of c-jun in the innate immune cells with OGN high expression. In addition, OGN over-expressed cells were able to result in altered cell death and autophagy by leading to mTOR pathway activation [13]. Given the crucial role of tumor autophagy activity in modifying T cells and FOXP3+ Treg cells, there was suggestive of a reciprocal relationship between OGN and T cell infiltration. Based on these previous findings, we will measure oncologic outcomes for CRC based upon OGN expression patterns, further test the effect of OGN on immune cells.
2. Patients and Methods
2.1. Antibodies and Reagents
We utilized these antibodies for research: human Osteoglycin (OGN) antibody for Western blotting(WB)from R&D Systems, Vascular endothelial growth factor A (VEGF-A) antibody, hypoxia inducible factor-1α (HIF-1α) from Abcam; human and mouse Osteoglycin (OGN) for immunohistochemistry (IHC) from Sigma-Aldrich and Santa Cruz Biotechnology; Akt, phospho-Akt (Ser473), epidermal growth factor receptor (EGFR), phospho-epidermal growth factor receptor (EGFR, Y1068), Erk1/2 antibody, CD3, CD8, PTPRC, and FOXP3 from Cell Signaling Technology; beta-actin antibody from proteintech; AKT activator: sc79 from Selleck.
2.2. Cell Culture
We originally purchased SW620, HT29 cell lines of human colon cancer and MC38 murine colon cancer cell line, which will be utilized in following experiments, from the American Type Culture Collection (Manassas, VA), and the cells were cultivated in DMEM medium according to the Defense Technical Information Center recommendation (DTIC) in addition of 10% fetal bovine serum (FBS, Gibico, Life Technology, Austria), 1% penicillin/streptomycin (PS) in a humidified 5% (v/v) atmosphere of CO2 at 37 °C.
2.3. Study Population
2.3.1. Patients
This study enrolled three independent sets of patients with colorectal cancer from the cancer genome atlas (TCGA) and the Fudan University Shanghai Cancer Center (FUSCC). The training set and testing set including 170 consecutive patients and 434 consecutive colorectal cancer patients with radical surgery were obtained from TCGA database available from Cancer Genomics Browser of University of California Santa Cruz (https://genome-cancer.ucsc.edu/). The validation set comprised 276 consecutive colorectal cancer patients after surgery at the Department of colorectal Surgery, FUSCC, Shanghai, China. All patient data were prospectively entered in the FUSCC database since 2006, such as: age at the diagnosis, race, tumor localization, diagnostic year, tumor diameter, histological grade, number of lymph nodes retrieved, post-operative multimodal treatment (adjuvant chemotherapy or radiation), details of surgical procedures, complications rate, postoperative histopathology, and follow-up information (date of last visit, tumor relapse, tumor-related or unrelated death, overall survival, OS and disease-free survival, DFS). The research protocol was reviewed and approved by the institutional review board of the FUSCC. Informed consent was obtained from all patients.
2.3.2. Tissue Microarray(TMA) Construction and Immunohistochemistry(IHC) Staining
In all, 276 unselected, non-consecutive, primary colorectal cancers were enrolled form January 2007 to November 2009 in FUSCC to construct the tissue microarray(TMA). Li et al. has previously described the construction in detail for TMA [14]. There were two independent pathologists scoring the TMA section utilizing a semiquantitative scoring system. This scoring system included staining intensity score defined as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong), and extent score defined as 0 (<5%), 1(range from 5% to 25%), 2 (range from 26% to 50%), 3 (range from 51 to 75%), 4 (>75%) according to the ratio of the positive staining areas in the whole carcinoma area. The immunoreactivity score (IRS) was generated as the results of the percentage score multiplied by the staining intensity score. OGN expression with High, Intermediate and Low pllevel was defined as detectable immunoreactions in cytoplasm and stoma with IS>6, 6 ≥ IS>2 and IS ≤2, respectively. And the densities of CD3+, CD8+, PTPRC+, and FOXP3+ cells in tumor tissue were measured as the average density per microarray cores.
2.3.3. Plasmids Construction and Viral Transduction
For protein over-expression, gene-specific overexpression vectors (pCDH-CMV-MCSEF1-Puro or CMV-MCS-3FLAG-SV40) were used. The lentiviral expression vector with OGN over-expression or the control vector were used to transfect the human or murine colon cancer cell lines (SW620, HT29, MC38).
2.3.4. Immunofluorescence
Cells reaching 80% confluent on a chamber slide or xenografts were fixed with paraformaldehyde (PFA), and we permeate the cells at room temperature for 15 min with 0.5% Triton X-100. Thereafter, sufficient and diluted primary antibodies for OGN, CD3+, CD8+, PTPRC+, and FOXP3+ were added to be incubated at 4 °C overnight. At last, the secondary antibody of the alexa-flours 488, 594 with anti-rabbit or mouse (1:200, Invitrogen) was added for an hour at room temperature. In addition, DAPI was utilized to stain nuclei when necessary. Fluorescence images were photographed with a fluorescence microscope.
2.3.5. Western Blot
Whole-cell lysates were collected in RIPA buffer for total protein extraction. Protein were separated by SDS-PAGE (10% gel) and then transferred to PVDF membranes. The primary antibodies against specific protein were incubated with PVDF membranes for overnight. HRP conjugated secondary antibodies were used at a 1:5000 dilution for 1-h incubation at room temperature. At last, specific proteins were detected using ECL (Pierce, Thermo Scientific) in a Bio-Imaging System.
2.3.6. Xenotransplant Murine Models
MC38 cells (5 ∗ 106 cells/mouse) which transfected with OGN over-expression or empty vector control were injected subcutaneously into the right flank of C57 mice (n = 5, male; 4-week C57 mouse), and mice were sacrificed after 4 weeks. Tumors were removed for pathological analysis.
2.3.7. Matrigel Plug Assay
Matrigel plug assay was performed to investigate endothelial activation and the angiogenesis properties of OGN. Liquid Matrigel was mixed at 4° with MC38 cells dissolved in PBS at a final concentration equal to 106/ml and injected subcutaneously (0.5 ml/mouse) into the flank of 5-week old C57 mice. The Matrigel plugs were harvested 7 days after implantation. The plugs were fixed in 4% formalin, embedded in paraffin and used for IHC and IF analysis.
2.4. Statistical Analysis
SPSS 22.0 (SPSS Inc., Chicago, IL, USA) was used to perform statistical analysis All quantification data are presented as mean ± s.d. Statistically significant differences were determined by one-way ANOVA test and Student's t-test. And all two-sided P values <.05 were considered statistically significant. The relationship between baseline clinicopathological parameters and OGN levels was evaluated with the chi-square test. To imbalance potential confounding, we carried out multivariable ordinal logistic regression array, in which each T cell sub-population (CD3, CD8, PTPRC, and FOXP3) were considered as an ordinal quartile outcome variable, and OGN levels as the ordinal predictor variable. The proportional odds ratio (OR) in the ordinal logistic regression model was used, which was generally satisfied (P > .05).
3. Results
3.1. Association of OGN Expression with T-cell Antigens (CD3, CD8, PTPRC, FOXP3) in the Training and Testing Sets
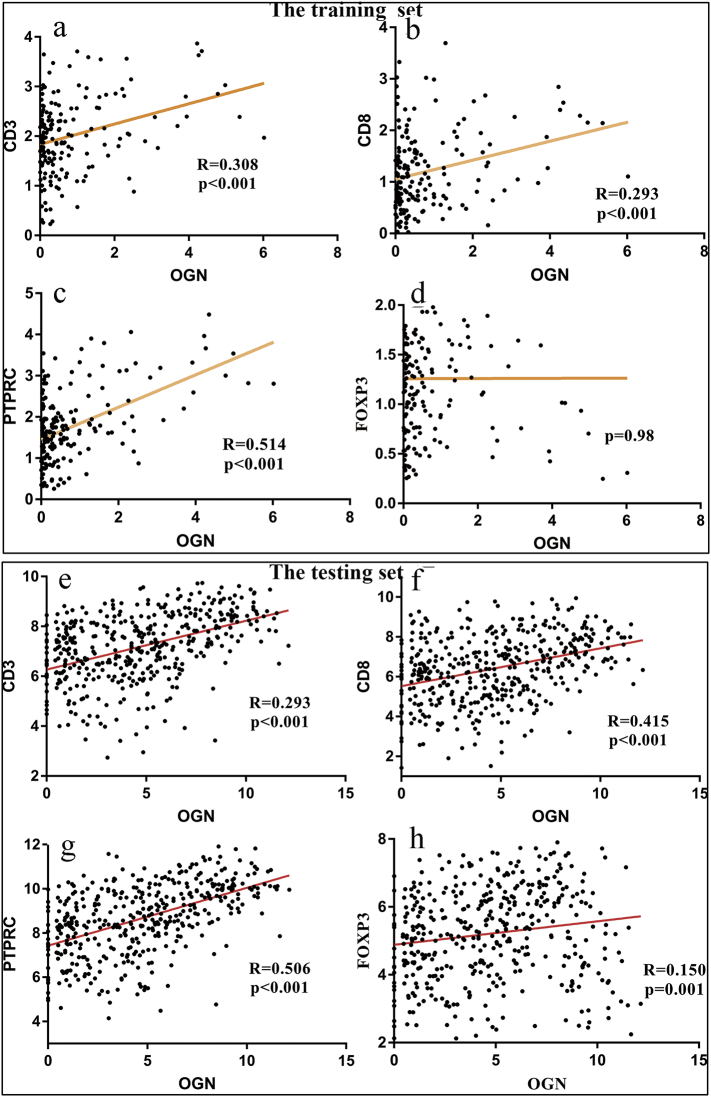
In our primary hypothesis testing, we conducted linear regression analyses to assess the association of tumor OGN mRNA level with the level of CD3+, CD8+, PTPRC+, and FOXP3+ antigen for colorectal cancers in the training and testing sets from TCGA. It turned out that tumor OGN mRNA level was positively associated with T-cell antigens CD3, CD8, PTPRC, and FOXP3 in linear logistic regression analyses (R = 0.308, 0.293, 0.514 and 0.06, respectively in the training set, all p < .01 except FOXP3+, Fig. 1a–d; R = 0.293, 0.415, 0.506 and 0.15, respectively in the testing set, all p < .01, Fig. 1e–h).
Fig. 1.
OGN mRNA are associated with T-cell antigens CD3, CD8, PTPRC, and FOXP3 in the training and testing sets.
(a–d) OGN mRNA level was positively associated with T-cell antigens: CD3, CD8 and PTPRC (R = 0.308, 0.293, and 0.514, respectively in training set. (e-h) OGN mRNA level was positively associated with T-cell antigens: CD3, CD8, PTPRC and Foxp3 R = 0.293, 0.415, 0.506 and 0.15, respectively. The statistical test was used for linear logistic regression analyses.
3.2. Immunohistochemical Findings in the Validation Set
In the validation set from FUSCC, we evaluated the level of OGN expression in 276 cases of colorectal cancer by immunohistochemical (IHC) analysis. OGN expression was observed in both cytoplasm and a clear membrane in tumor. At the meantime, staining both in the epithelial cancer cells and the stroma was frequently observed, while stromal staining usually was diffuse. Among the selected colorectal cancers, 130 (47%), 105 (38%), and 41 (15%) tumors presented low, intermediate, and high-level OGN expression, respectively. Clinical, pathological, and molecular features were summarized in Table 1 based on the tumor OGN expression levels in colorectal cancers. No demographic or baseline clinical data were statistically associated with any pattern of OGN expression.
Table 1.
Description of the study population among colorectal cancer patients according to tumor OGN expression level.
| Variables, n (%) | OGN |
P value | ||
|---|---|---|---|---|
| Low(n = 130) | Intermediate(n = 105) | High(n = 41) | ||
| Gender | ||||
| Male | 81(62.3) | 48(45.7) | 13(31.7) | 0.885 |
| Female | 49(37.7) | 57(54.3) | 28(68.3) | |
| Age, years | 56.73 ± 10.887 | 57.86 ± 11.741 | 56.83 ± 9.620 | 0.723 |
| T stage | 0.114 | |||
| T2 | 18(13.8) | 19(18.1) | 6(14.6) | |
| T3 | 22(16.9) | 17(16.2) | 15(36.6) | |
| T4 | 90(69.2) | 69(65.7) | 20(48.8) | |
| TNM stage | 0.584 | |||
| I | 10(7.7) | 8(7.6) | 3(7.3) | |
| II | 42(32.3) | 24(22.9) | 15(36.6) | |
| III | 52(40.0) | 61(58.1) | 19(46.3) | |
| IV | 26(20.0) | 12(11.4) | 4(9.8) | |
| N stage | 0.8 | |||
| N0 | 62(47.7) | 38(36.2) | 20(48.8) | |
| N1 | 32(24.6) | 35(33.3) | 15(36.6) | |
| N2 | 36(27.7) | 31(29.5) | 6(14.6) | |
| N3 | 0(0) | 1(1.0) | 0(0) | |
| M stage | 0.1 | |||
| M0 | 104(80.0) | 93(88.6) | 37(90.2) | |
| M1 | 26(20.0) | 12(11.4) | 4(9.8) | |
| Grade | 0.597 | |||
| Well/ moderate | 95(73.1) | 77(73.3) | 32(78.0) | |
| Poor | 28(21.5) | 21(20.0) | 7(17.1) | |
| Histological type | 0.846 | |||
| Adenocarcinoma | 122(93.8) | 101(96.2) | 38(92.7) | |
| Mucinous | 8(6.2) | 4(3.8) | 3(7.3) | |
| Lymph node examined | 0.937 | |||
| Median | 14.8 ± 6 | 15.1 ± 6 | 15 ± 5 | |
| Tumor location | 0.269 | |||
| Colon | 54(1.5) | 56(53.3) | 19(46.3) | |
| Rectum | 76(8.5) | 49(6.7) | 22(53.7) | |
| Perineural invasion | 0.31 | |||
| Negative | 109(83.8) | 83(79.0) | 39(95.1) | |
| Positive | 21(16.2) | 22(21.0) | 2(4.9) | |
| Vascular invasion | 0.235 | |||
| Negative | 87(66.9) | 68(64.8) | 33(80.5) | |
| Positive | 43(33.1) | 37(35.2) | 8(19.5) | |
| Adjuvant Chemotherapy | 0.177 | |||
| No | 25(19.2) | 17(16.2) | 7(17.1) | |
| Yes | 82(63.1) | 79(75.2) | 28(68.3) | |
| MS /MMR status | 0.697 | |||
| MSS/MMR-proficient | 87(66.9) | 61(58.1) | 28(68.3) | |
| MSI/MMR-deficient | 43(33.1) | 44(41.9) | 13(31.7) | |
| CEA status | 0.125 | |||
| Normal | 71(54.6) | 70(66.7) | 29(70.7) | |
| Elevated | 53(40.8) | 31(29.5) | 11(26.8) | |
MMR indicates mismatch repair; MS, microsatellite; MSS, microsatellite stability; MSI, microsatellite instability.
We investigated the correlation of the expression pattern of OGN and T-cell densities by immunohistochemistry assay (IHC). The correlation of OGN expression score and T-cell densities in colorectal cancers was shown in Table 2. Tumor OGN expression score was positively correlated with CD8+ cell density (p < .001, by Spearman test). Moreover, the ordinal logistic regression analysis was carried out to identify the linkage of OGN expression levels (an ordinal predictor variable) with the density of CD3+, CD8+, PTPRC+, or FOXP3+ cells (an ordinal quartile outcome variable) in FUSCC prospective cohort (Table 3). Uni- and multi-variable analyses both showed OGN expression level was positively associated with CD8+ cell density (p < .001). Exactly, when a unit decreased in the category of CD8+ cell density, the multi-variable OR in the lowest tumor OGN expression score relative to the highest score was 0.17 (95% CI 0.08–0.33). In addition, the OGN expression level did not have any significant association with CD3+, FOXP3+ or PTPRC+ cell density (all p > .05).
Table 2.
Distribution of colorectal cancers according to tumor OGN expression level and the T cells expression score.
| Variables | OGN level |
P value | ||
|---|---|---|---|---|
| Low | Intermediate | High | ||
| CD3+ cell density | 0.74 | |||
| 1 (lowest) | 34(26.2) | 21(20.0) | 6(14.6) | |
| 2 | 49(37.7) | 43(41.0) | 17(41.5) | |
| 3 | 26(20.0) | 26(24.8) | 11(26.8) | |
| 4 (highest) | 21(16.2) | 15(14.3) | 7(17.1) | |
| FOXP3+ cell density | 0.774 | |||
| 1 (lowest) | 45(34.6) | 33(31.4) | 9(22.0) | |
| 2 | 51(39.2) | 41(39.0) | 17(41.5) | |
| 3 | 13(10.0) | 14(13.3) | 7(17.1) | |
| 4 (highest) | 21(16.2) | 17(16.2) | 8(19.5) | |
| PTPRC cell density | 0.078 | |||
| 1 (lowest) | 35(26.9) | 31(29.50) | 17(41.5) | |
| 2 | 50(38.50) | 28(26.7) | 10(24.4) | |
| 3 | 24(18.5) | 34(32.4) | 9(22.0) | |
| 4 (highest) | 21(16.2) | 12(11.4) | 5(12.2) | |
| CD8+ cell density | <0.001 | |||
| 1 (lowest) | 45(34.6) | 51(39.2) | 0(0.0) | |
| 2 | 51(39.2) | 17(16.2) | 2(4.9) | |
| 3 | 16(12.3) | 19(18.1) | 26(63.4) | |
| 4 (highest) | 18(13.8) | 41(39.0) | 13(31.7) | |
P value was calculated by Pearson Chi-Square test between the tumor OGN expression score (ranging from Low to High) and the level of CD3+, CD8+, PTPRC, or FOXP3+ T cells (immunoreactivity score; as continuous variables).
Table 3.
Ordinal logistic regression analysis evaluating the linkage of tumor OGN level (predictor) with the density of T cells (outcome).
| Variables | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| Odd ratio | 95% CI | P value | Odd ratio | 95% CI | P value | |
| Model for CD3+ cell density as an ordinal outcome variable | ||||||
| Tumor OGN expression level | ||||||
| Low | 0.67 | 0.36–1.27 | 0.22 | 0.63 | 0.33–1.21 | 0.17 |
| Intermediate | 0.79 | 0.41–1.52 | 0.48 | 0.69 | 0.36–1.40 | 0.28 |
| High | 1 (referent) | 1 (referent) | ||||
| Model for FOXP3+ cell density as an ordinal outcome variable | ||||||
| Tumor OGN expression level | ||||||
| Low | 0.61 | 0.32–1.15 | 0.13 | 0.64 | 0.33–1.24 | 0.19 |
| Intermediate | 0.69 | 0.36–1.34 | 0.28 | 0.73 | 0.37–1.45 | 0.38 |
| High | 1 (referent) | 1 (referent) | ||||
| Model for PTPRC cell density as an ordinal outcome variable | ||||||
| Tumor OGN expression level | ||||||
| Low | 1.45 | 0.77–2.75 | 0.25 | 1.4 | 0.72–2.71 | 0.32 |
| Intermediate | 1.54 | 0.79–2.97 | 0.19 | 1.56 | 0.79–3.08 | 0.20 |
| High | 1 (referent) | 1 (referent) | ||||
| Model for CD8+ cell density as an ordinal outcome variable | ||||||
| Tumor OGN expression level | ||||||
| Low | 0.17 | 0.09–0.33 | <0.001 | 0.17 | 0.08–0.33 | <0.001 |
| Intermediate | 0.5 | 0.26–0.98 | 0.04 | 0.47 | 0.24–0.94 | 0.03 |
| High | 1 (referent) | 1 (referent) | ||||
CI indicates confidence interval.
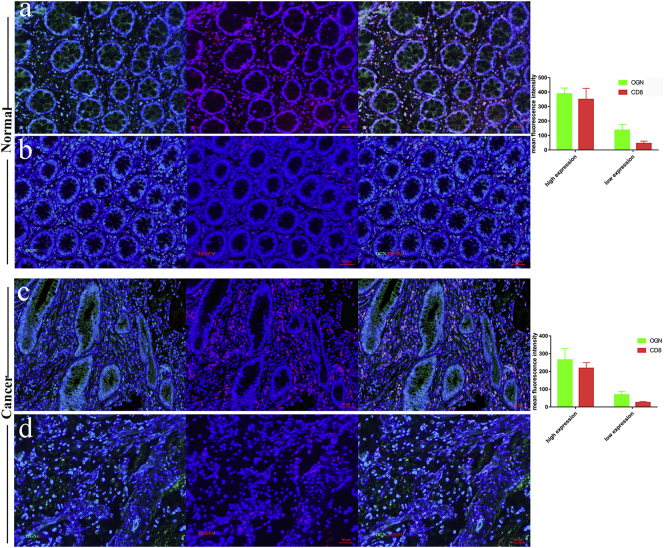
To validate the result that OGN was preferentially expressed with CD8+ cells, multi-color immunofluorescence was performed using monoclonal antibodies recognizing OGN, CD8+ and DAPI in normal epithelium and cancer tissues. Both normal epithelial and cancer tissue OGN expression levels were correlated with CD8+ cells as above depicted. The number of CD8+ cells was significantly higher in almost both normal epithelial and tumors, when OGN expression elevated (Fig. 2).
Fig. 2.
OGN was preferentially expressed with CD8+ TIL in both normal epithelium and cancer tissue.
Representative image of normal epithelium tissue with (a) high OGN expression (green) and high density of CD8+ infiltration (red). In the same way, (b) the low OGN expression accompanied by low density of CD8+ infiltration. Representative image of colorectal cancer tissue in (c) high OGN expression (green) and high density of CD8+ infiltration (red). In the same way, (d) the low OGN expression accompanied by low density of CD8+ infiltration. The multivariable ordinal logistic regression array was performed to evaluate the correlation.
3.3. OGN Recruited More CD8+ Cells, and Inhibited VEGF As Well as Angiogenesis In Vivo: Xenograft Models
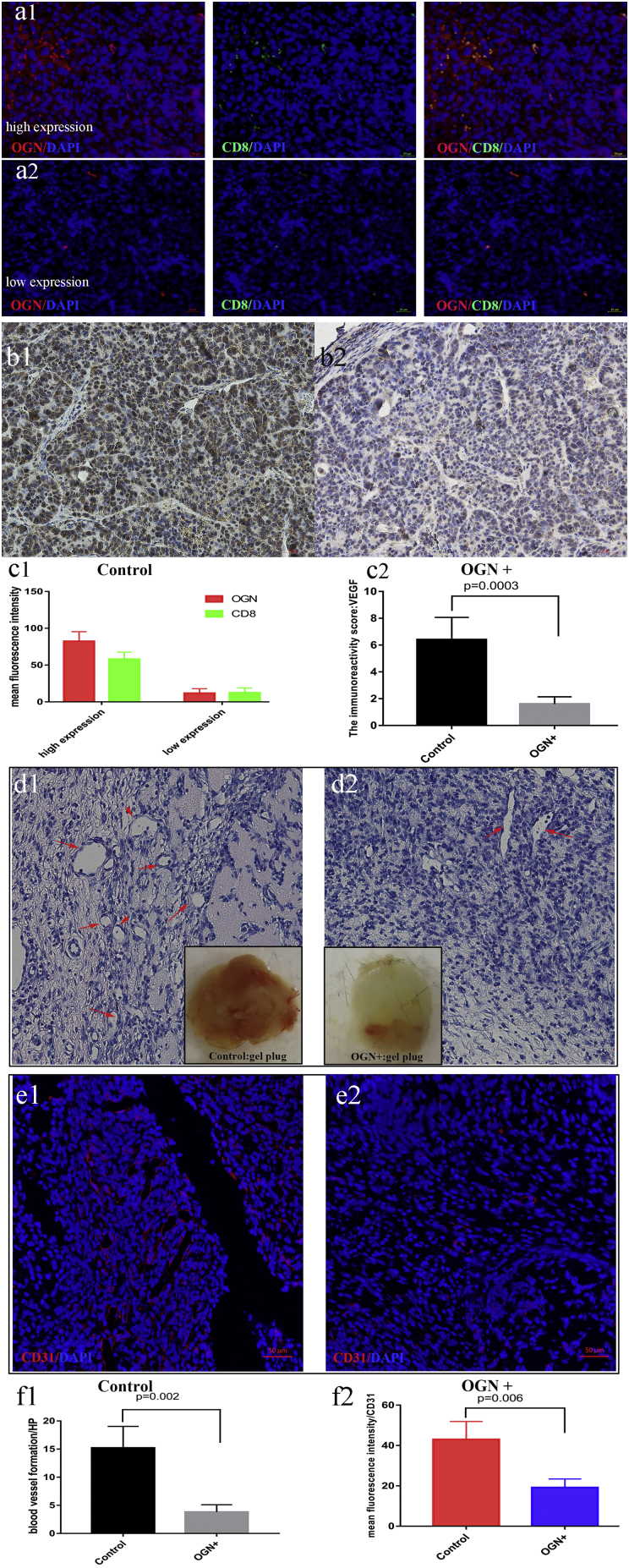
To test directly whether OGN mediated T cell recruitment in vivo, murine colon cancer cells MC38, with or without OGN over-expression (MC38-OGN+ and MC38-Control), were implanted into the right flank of C57 mice. Xenograft tumor sections were stained for above T cell antigens to assess the constituents of the tumor microenvironment. Tumors arising from OGN-over-expression cells displayed a significant increase in CD8+ cells recruitment (Fig. 3a1-a2 and c1), compared with the control group (20 ± 5 cells /HP in OGN + group vs. 5 ± 2 cells /HP in Control group, p < .05).
Fig. 3.
OGN over-expression recruited more CD8+ cells in xenograft models.
(a1) Representative image from MC38 OGN-over-expression (green) displayed a significant increase in CD8+ cells recruitment (red), while the control group (a2) recruited much fewer CD8+ cells. The Quantification was shown in c1. (b2) VEGF expression was reduced in the OGN over-expression group in xenograft compared to the control group (b1). The Quantification was shown in c2. The gross images and the H&E staining of Matrigel plug showing the number of blood vessel formation was markedly reduced in OGN over-expression group (d2) compared to the control group (d1). Immunofluorescent staining of CD31 (endothelial cell marker) in OGN group (e2) was much weaker than the control group (e1). The Quantification was determined by Student's t-test in f1-f2.
Furthermore, we explored the VEGF expression and angiogenesis responding to OGN in vivo. So the Matrigel plug analysis was performed to evaluate the angiogenesis properties. We also observed that VEGF expression was reduced in the OGN over-expression group in xenograft (Fig. 3b1-b2 and c2). In the gel plug assay, the red color of plugs was much lighter in the presence of OGN over expression compared with the control group, suggesting the formation of fewer blood vessels. The H&E staining also demonstrated the number of blood vessel formation was markedly reduced by OGN (Fig. 3d1-d2 and f1). At last, the CD31 microvascular density (an endothelial cell marker) was significantly reduced displayed by Immunofluorescence (Fig. 3e1-e2, and f2).
3.4. CD8+ Cells Infiltration and OGN Expression Predicted Prognostic Roles in Survival of CRC
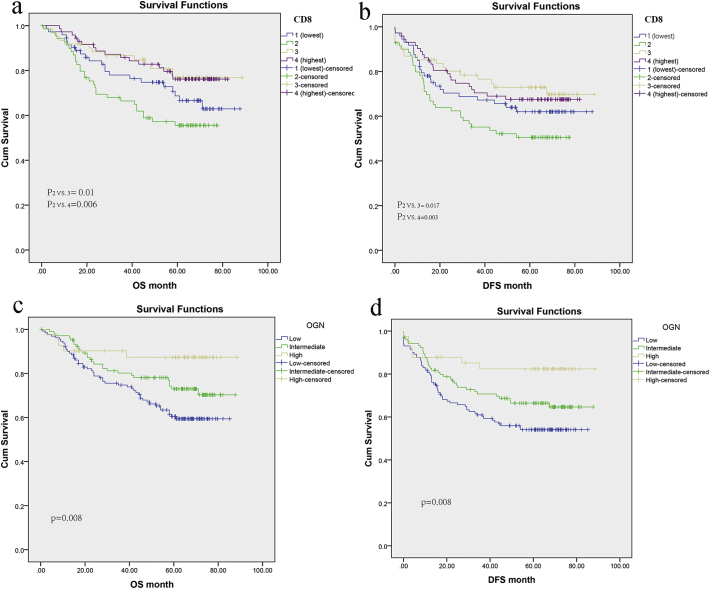
We compared overall and disease-free survivals in patients with different levels of infiltrating CD8+ cells. Fig. 4a showed Kaplan–Meier plots of overall survival in patients with different levels of infiltrating CD8+ cells. An increased infiltration of CD8+ cells (high score) was closely associated with an improved prognosis (p < .05). A similar, but even stronger association was seen in disease-free survivals (p < .05) (Fig. 4b).
Fig. 4.
Kaplan–Meier plots according to CD8+ cells infiltration and OGN expression in FUSCC.
Kaplan Meier curves of overall survival (a) and Disease-free survival (b) showing CD8 score closely associated with an improved prognosis. Kaplan Meier curves for overall survival (c) and Disease-free survival (d) displaying high OGN expression associated with an improved prognosis.
As exploratory analyses, Cox proportional hazards regression analysis was performed to evaluate the prognostic role of OGN expression in colorectal cancers. Initially, the univariate Cox regression model for overall survival indicated OGN levels had an association with prognosis of CRC, with much less HR in the high OGN level group (P = .009, Table 4). Consistently, multivariate analysis also revealed that OGN expression was an independent prognostic factor for overall survival in CRC (HR 0.34, 95%CI:0.13–0.91), even when age, gender, tumor localization, MMR status, TNM classification, venous/ perineural invasion and adjuvant chemotherapy were adjusted. Moreover, the benefit of improved OS by OGN can translate into prolonged disease-free survival (DFS) evidenced by Cox proportional hazards model (Table 4), and even after adjusting for other confounding, high OGN expression (HR 0.45, 95%CI:0.2–0.99) was also independently associated with prolonged DFS. Hence, Kaplan-Meier analysis demonstrated high OGN level was markedly linked with longer time post-surgical resection, with prolonged cancer specific survival as 75.7 months in the High OGN expression group versus 61.6 months in the Low OGN expression group. Moreover, so was the disease-free survival (Fig. 4c, d).
Table 4.
Tumor OGN expression and colorectal cancer patient survival.
| OGN expression score | Total N. | Event N. | Overall survival |
Event N. | Disease-free survival |
||
|---|---|---|---|---|---|---|---|
| Uni-HR(95% CI) | Multi-HR(95% CI) | Uni-HR(95% CI) | Multi-HR(95% CI) | ||||
| Low | 130 | 49 | [1] (reference) | [1] (reference) | 58 | [1] (reference) | [1] (reference) |
| Intermediate | 105 | 27 | 0.63(0.39–1.01) | 0.64 (0.39–1.05) | 35 | 0.69(0.45–1.0) | 0.69(0.45–1.07) |
| High | 41 | 5 | 0.29(0.12–0.74) | 0.34 (0.13–0.91) | 7 | 0.33(0.15–0.73) | 0.45(0.2–0.99) |
| P value | 0.009 | 0.0032 | 0.006 | 0.048 | |||
3.5. OGN Expression Is Negatively Correlated With VEGF Levels in Colorectal Cancer Tissues
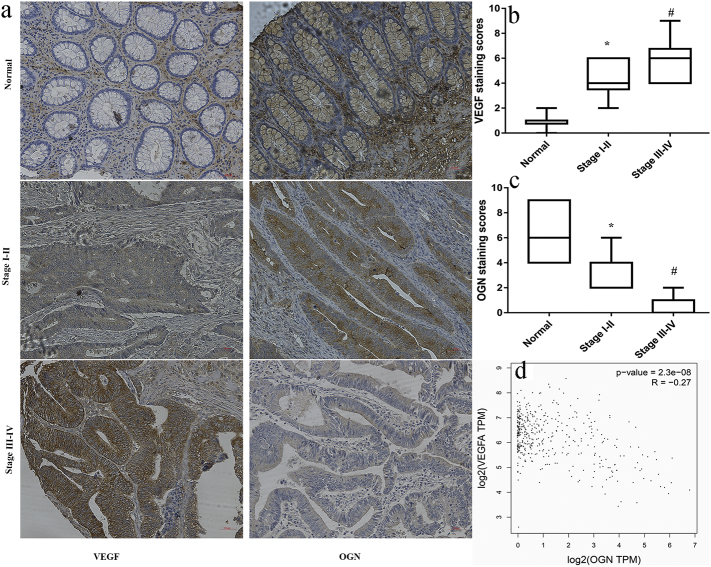
To illustrate the mechanism for OGN recruiting T-cell infiltration, we performed pairwise gene correlation analysis in sets of TCGA, finding OGN negatively correlated with VEGF closely (P = 2.3e−8, R = − 0.27, Fig. 5d) in mRNA levels besides T-cell antigens (CD3, CD8, PTPRC, FOXP3). In light of the vital role of VEGF in tumor growth and carcinogenesis, we investigated the correlation of the expression of VEGF and OGN by immunohistochemistry assay in stage I–II, stage III-IV CRC tissues and adjacent normal tissues, separately. In comparison to the normal tissues, the expression of OGN was markedly reduced in cancer tissues (Fig. 5a). What is more, the staining of OGN in stage III–IV CRC tissues were notably weaker than that of stage I–II tissues (Fig. 5c). Conversely, the expression of VEGF was found dramatically elevated in cancer tissues relative to the normal tissue (Fig. 5b). Moreover, the levels of VEGF in cancer tissues were significantly increased with the stage advanced, which was just contrast to the OGN expression. Together, in mRNA and protein levels, OGN was found to be negatively correlated with VEGF levels in colorectal tissues. It was indicated that VEGF reduction by OGN elevation might affected T cell homing through endothelial barriers.
Fig. 5.
OGN is negatively correlated with VEGF in colorectal cancer tissues.
a, Immunohistochemistry analyses of VEGF and OGN expression in varied staged colorectal tissues. b, the levels of VEGF in cancer tissues were significantly increased with the stage advanced. #P < .05 vs stage I–II or normal tissues. *P < .05 vs normal tissues. c, Conversely, the expression of OGN was found dramatically reduced with the stage advanced. #P < .05 vs stage I–II or normal tissues. *P < .05 vs normal tissues. The Quantification was determined by one-way ANOVA. d, Scatter plots showing the negative linear correlation between the mRNA expression of VEGF and that of OGN in CRC tissues of TCGA.
3.6. OGN Down-regulated HIF-1α to Inhibit VEGF
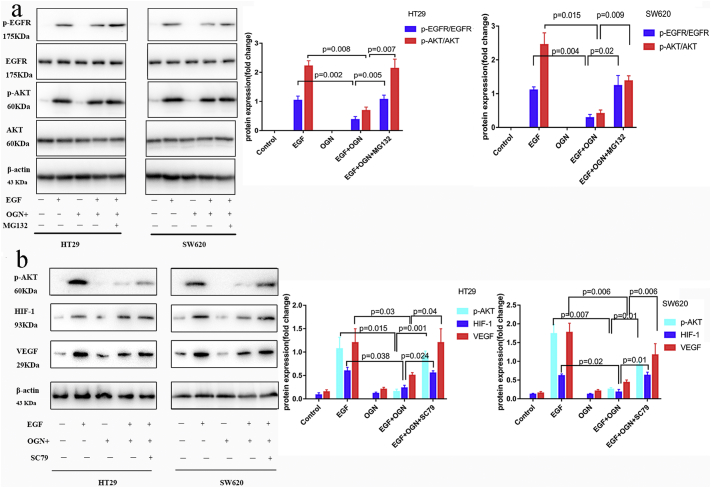
OGN, one of the Small leucine-rich proteoglycans (SLRPs), can bind several growth factors including epidermal growth factor receptors (EGFR) and insulin growth factor receptors (IGFR). Decorin, another small leucine-rich proteoglycan, was found directly interacting with EGFR to suppress EGFR tyrosine kinase. In order to further clarify the mechanism in which OGN moderated the levels of VEGF, we explored whether EGFR and concerning signaling may be involved in OGN-induced VEGF suppression. Firstly, we assessed whether the EGFR activation was regulated by OGN over-expression. We found HT29 and SW620 cells with OGN over-expression exhibited a phosphorylation reduction in EGFR protein activation level compared to Control cells exposed to extracellular EGF (100 ng/ml). After 15 min of EGF exposure, the increase in p-EGFR/EGFR levels were reduced remarkably in OGN-overexpression cells compared to the control cells (Fig. 6a). Then among the three major downstream pathways induced by EGFR, such as: PI3K/Akt, Stat3 and ERK1/2, only a decline in Akt phosphorylation was observed, while there was not any significant difference in Stat3 and ERK1/2 (Fig. 6a). It was demonstrated that OGN abolished Akt activity, what is more, the above observed Akt phosphorylation was reversed when the EGFR degradation blocked after proteosome inhibitor MG132 added.
Fig. 6.
OGN decreases VEGF expression through EGFR/HIF-1 pathway.
a, HT29, SW620 cells with OGN over-expression were treated with EGF (100 ng/ml) and co-cultured with or without MG132 (1 μM) for 24 h for the indicated intervals. Western blotting demonstrated OGN could abolish Akt activity through reducing EGFR protein activation. b, cells with OGN over-expression were challenged with EGF (100 ng/ml) and pretreated with SC79 (constitutive Akt activator) for 24 h. Western blotting presented a significant reduction in expression of HIF-1α and VEGF. Data were representative of three independent experiments with one-way ANOVA test.
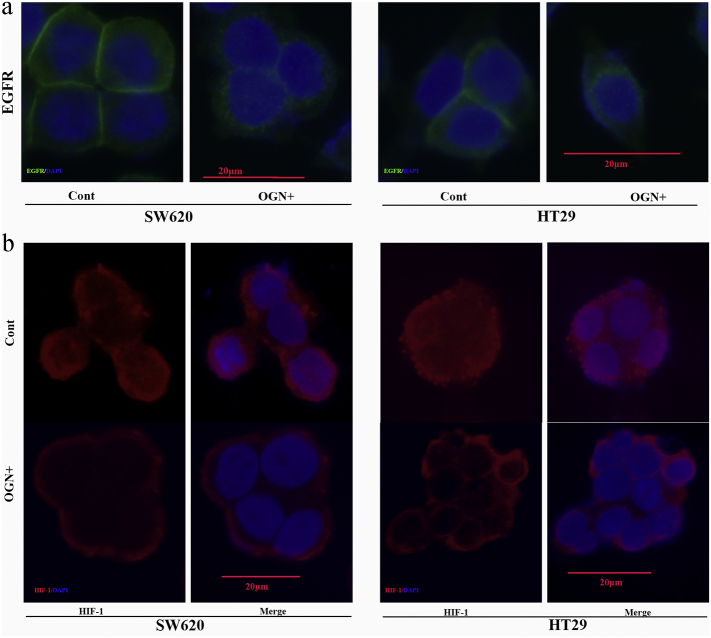
Consequently, we have observed OGN over-expressed HT29/SW620 cells presented a significant reduction in expression of HIF-1α and VEGF (the protein product of a major HIF-1α target gene) (Fig. 6b). In addition, re-expression of p-Akt leaded to resistance to the decrease in HIF-1α protein levels and VEGF previously observed after OGN over-expression (Fig. 6b), hence a critical role of Akt inhibition was indicated in the observed OGN-mediated decrease in HIF-1α and VEGF. In addition, to address the mechanism for activation reduction, immunofluorescent staining firstly demonstrated a reduction of EGFR in membranous, with a concomitant increase in cytoplasmic, after OGN over-expression (Fig. 7a). Meanwhile, it was followed that HIF-1α translocation to nuclei was blocked after OGN over-expressed (Fig. 7b). The above results not only demonstrated the fact that OGN could reduce HIF-1α activity via inhibition of the Akt pathway but also shed light on a relationship between OGN and VEGF.
Fig. 7.
OGN mediates the sublocation of EGFR and HIF-1α.
a, immunofluorescent staining of cancer cells with anti-EGFR antibody, showing OGN increased the EGFR internalization. b, immunofluorescent staining of cancer cells with anti-HIF-1α antibody, showing OGN blocked nuclear translocation of HIF-1α.
4. Discussion
Using three independent sets of colorectal cancers, we found that OGN expression is positively associated with T cell density in colorectal cancer tissue. The association persisted after balancing for potential variants, including the tumor statuses of location, microsatellite instability, TNM stages, venous/ perineural invasion, which have affected the density of tumor infiltrating T cells in colorectal cancer [15]. At last, xenograft tumors arising from OGN-over-expression cells recruited more CD8+ T cells. Maybe that is an explanation of OGN as an independent predictor for improved survival. Results from this study implied a possible role of OGN in regulating host immunity in colorectal cancer microenvironment.
Levels of OGN expression were reported to inversely associated with tumor progression, just as thyroid tumor and hepato-carcinomas. Consistently, OGN also presented as a tumor suppressor in this cohort of CRC patients, in which elevated protein expression was linked with less mortality (HR 0.45, 95%CI:0.2–0.99). To explain this, a specific chromosomal organization of osteoglycin has been implied. For instance, there was a p53 DNA-binding sequence in the OGN gene identified and confirmed, hence the OGN gene expression was assumed to account for the known tumor suppressor p53 [16]. Coincidentally, p53 is frequently mutated to inactivation in several tumors, including colorectal cancers, breast cancers, lung cancers, ovary and prostate cancers, in turn leading to the inactivation of the OGN gene [8].
Besides the malignant behavior of cancer cells modified by OGN, there was also another distinct extrinsic mechanism via nontumor cells of the microenvironment: tumor infiltrating T cells. In fact, OGN has been observed on local and circulating innate immune cells [12]. When OGN specific expression in local and circulatory immune cells was analyzed using immunofluorescence and flow cytometry, OGN was found to clearly co-stain with both neutrophil and monocyte/macrophage-markers in the myocardium. What is more, great distinguished phenotypic characteristics were indicated by significantly altered phosphorylation of c-jun in the innate immune cells with OGN expression. In the end, the change of OGN altered cytokine production by both circulating and local immune cells upon damage exposure, then leading to modify tissue inflammation and immune response. Similarly, in FUSCC colorectal cancer prospective cohort, we found OGN expression score was positively correlated with CD8+ cell density (p < .001) in immunohistochemistry assay. At last, as the possible tumor suppressing mechanism via the modified tumor T cells infiltrating, high OGN expression was independently associated with improved survival in the multivariate Cox proportional hazards model. To accurately explain the underlined mechanism of OGN in the modification of immune cells, interaction between OGN and TLRs may be responsible for the apparent altered phenotypic appearance of immune cells. Firstly, a potential interaction between the leucine-rich surface of OGN and the extracellular LRR domain of TLR4 has been predicted in silico structure modeling [12, 17, 18]. In the second, the interaction of OGN and TLR4 was observed in co-immunoprecipitation assay for both peripheral leukocytes as well as macrophage, and this observation was in line with co-expression of OGN with TLR4 in immunofluorescence. So, modification of TLR4 activation can change pro-inflammatory and Th1-type cytokine secretion [19, 20]. In addition, the maturation of APCs for releasing cytokines, and priming of T-helper (Th) cells are all based on the TLR signaling [21]. Both of these events are supposed to be crucial for robust T cell responses in the cancer immunity.
Autophagy is another mechanism responsible for mediating cellular organelles and function by OGN expression. As OGN promoter containing three conserved AP-1-binding sites [22], it was validated that OGN acted as a target gene downstream to LIP/MAPK/AP-1 [23]. So specific knockdown of OGN mRNA could attenuated cell death and autophagy triggered by ER stress in melanoma cells. And in contrast to control cells, clones transfected with OGN were significantly more sensitive to ER stress-triggered cell death and autophagy following ER inducer. Besides, OGN-overexpressed cells by mTOR pathway activation was able to result in altered cell death and autophagy [13]. Both of these were suggestive of a reciprocal relationship between OGN and autophagy. In turn, autophagic activity within tumor cells may release more extracellular immune mediators and cross antigen-present tumor-associated antigens to T cells, therefore, immune response to tumor cells was mediated [[24], [25], [26]]. Given the crucial role of tumor autophagy activity in modifying CD8+ cytotoxic T cells and FOXP3+ Treg cells, so a partial component of the mechanism underling OGN associated with T cell density can be explained.
The colorectal cancer tissue with better survival was frequently accompanied with higher densities of CD3+, CD8+ and CD45RO+ cells [4, 15, 27]. Hence, it was feasible to determine immune response by anti-tumor T-cell to colorectal cancers measuring the densities of these T cells assessed by IHC. In similar, colorectal cancer with more CD8+ cell infiltration has been generally supposed to the prediction of improved outcome [28]. One possible reason of favorable oncological outcome can attribute to increased infiltration of T cells associated with MSI-high colorectal cancer, which is a favorable tumor molecular subtype [15]. A variety of evidence suggested that role of T cells appeared to depend on tumor location and progression stage, and the microenvironment dependent function probably reflected the tailored immune milieu and contexts [29, 30]. It was for reducing tumor-promoting inflammation that T cells suppressed tumor progression in colorectal cancer microenvironment. As reported by Ladoire et al. [31] that the association of T cells with favorable outcome reflects their capacity of repressing tumor-promoting inflammatory responses to gut microbiota in colorectal cancer.
Some additional limitations are important to consider in interpreting the results of the present study. The cross-sectional nature of this study is the first drawback, which cannot distinguish the causal relationship. Exactly, the possibility of reverse causation that T cells infiltration might alter the expression of OGN. Lack of data on molecular pathology, just as KRAS, PIK3CA or BRAF mutation, was another limitation. However, distributions of tumor OGN expression levels would unlikely substantially differ according to molecular pathology. Strengths of our current study include the use database of a large number of colorectal cancer cases form a prospective cohort studies. In addition, the balanced covariates in our colorectal cancer specimens increased the generalizability of our findings.
Collectively, our work identifies OGN expression in CRC was positively associated with CD8+ cell density as a novel mechanism, through which high expression of OGN improved survival. Our consolidated data draws a possible effect of OGN level on CD8 T cells in colorectal cancer microenvironment, and can promote further translational research on the associations of OGN with host immunity in colorectal cancer.
Funding
Not applicable.
Availability of Data and Materials
Not applicable.
Authors' Contributions
Conceived and designed the experiments: H.X. and C.S.J. Analyzed the data: H.X., L.Y.Q., C.S.J., L.Q.G. Contributed reagents/materials/analysis tools: H.X. P.J.J and M.Y.L. Wrote the paper: H.X. and L.Y.Q.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by China Postdoctoral Science Foundation (2018M632024).
References
- 1.Di Caro G., Marchesi F., Laghi L., Grizzi F. Immune cells: Plastic players along colorectal cancer progression. J Cell Mol Med. 2013;17:1088–1095. doi: 10.1111/jcmm.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colussi D., Brandi G., Bazzoli F., Ricciardiello L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 4.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 5.Mei Z., Liu Y., Liu C., Cui A., Liang Z., Wang G. Tumour-infiltrating inflammation and prognosis in colorectal cancer: Systematic review and meta-analysis. Br J Cancer. 2014;110:1595–1605. doi: 10.1038/bjc.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogino S., Nosho K., Irahara N., Meyerhardt J.A., Baba Y., Shima K. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.Y., Eom E.M., Kim D.S., Ha-Lee Y.M., Lee D.H. Analysis of gene expression profiles of gastric normal and cancer tissues by SAGE. Genomics. 2003;82:78–85. doi: 10.1016/s0888-7543(03)00098-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Ma Y., Lu B., Xu E., Huang Q., Lai M. Differential expression of mimecan and thioredoxin domain-containing protein 5 in colorectal adenoma and cancer: A proteomic study. Exp Biol Med (Maywood) 2007;232:1152–1159. doi: 10.3181/0701-RM-8. [DOI] [PubMed] [Google Scholar]
- 9.Lomnytska M.I., Becker S., Hellman K., Hellstrom A.C., Souchelnytskyi S., Mints M. Diagnostic protein marker patterns in squamous cervical cancer. Proteomics Clin Appl. 2010;4:17–31. doi: 10.1002/prca.200900086. [DOI] [PubMed] [Google Scholar]
- 10.Rower C., Ziems B., Radtke A., Schmitt O., Reimer T., Koy C. Toponostics of invasive ductal breast carcinoma: combination of spatial protein expression imaging and quantitative proteome signature analysis. Int J Clin Exp Pathol. 2011;4:454–467. [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., Zhang Z., Wang C., Miao L., Zhang J., Wang J. Quantitative proteomics approach to screening of potential diagnostic and therapeutic targets for laryngeal carcinoma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rienks M., Papageorgiou A., Wouters K., Verhesen W., Leeuwen R.V., Carai P. A novel 72-kDa leukocyte-derived osteoglycin enhances the activation of toll-like receptor 4 and exacerbates cardiac inflammation during viral myocarditis. Cell Mol Life Sci. 2017;74:1511–1525. doi: 10.1007/s00018-016-2423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei Y., Du Z., Hu C., Greenwald N.F., Abedalthagafi M., Agar N.Y.R. Osteoglycin promotes meningioma development through downregulation of NF2 and activation of mTOR signaling. Cell Commun Signal. 2017;15:34. doi: 10.1186/s12964-017-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D., Peng Z., Tang H., Wei P., Kong X., Yan D. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011;17:3558–3568. doi: 10.1158/1078-0432.CCR-10-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nosho K., Baba Y., Tanaka N., Shima K., Hayashi M., Meyerhardt J.A. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasheva E.S., Maki C.G., Conrad A.H., Conrad G.W. Transcriptional activation of bovine mimecan by p53 through an intronic DNA-binding site. Biochim Biophys Acta. 2001;1517:333–338. doi: 10.1016/s0167-4781(00)00288-8. [DOI] [PubMed] [Google Scholar]
- 17.Kemball C.C., Alirezaei M., Whitton J.L. Type B coxsackieviruses and their interactions with the innate and adaptive immune systems. Future Microbiol. 2010;5:1329–1347. doi: 10.2217/fmb.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairweather D., Yusung S., Frisancho S., Barrett M., Gatewood S., Steele R. IL-12 receptor beta 1 and Toll-like receptor 4 increase IL-1 beta- and IL-18-associated myocarditis and coxsackievirus replication. J Immunol. 2003;170:4731–4737. doi: 10.4049/jimmunol.170.9.4731. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Liu B., Dai J., Srivastava P.K., Zammit D.J., Lefrancois L. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vabulas R.M., Braedel S., Hilf N., Singh-Jasuja H., Herter S., Ahmad-Nejad P. The endoplasmic reticulum-resident heat shock protein Gp96 activates dendritic cells via the Toll-like receptor 2/4 pathway. J Biol Chem. 2002;277:20847–20853. doi: 10.1074/jbc.M200425200. [DOI] [PubMed] [Google Scholar]
- 21.Pawaria S., Binder R.J. CD91-dependent programming of T-helper cell responses following heat shock protein immunization. Nat Commun. 2011;2:521. doi: 10.1038/ncomms1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tasheva E.S., Conrad G.W. The UV responsive elements in the human mimecan promoter: a functional characterization. Mol Vis. 2003;9:1–9. [PubMed] [Google Scholar]
- 23.Wassermann-Dozorets R., Rubinstein M. C/EBPbeta LIP augments cell death by inducing osteoglycin. Cell Death Dis. 2017;8:e2733. doi: 10.1038/cddis.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Wang L.X., Yang G., Hao F., Urba W.J., Hu H.M. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–6895. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Wang L.X., Pang P., Cui Z., Aung S., Haley D. Tumor-derived autophagosome vaccine: mechanism of cross-presentation and therapeutic efficacy. Clin Cancer Res. 2011;17:7047–7057. doi: 10.1158/1078-0432.CCR-11-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mlecnik B., Bindea G., Angell H.K., Sasso M.S., Obenauf A.C., Fredriksen T. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Sci Transl Med. 2014;6:228ra37. doi: 10.1126/scitranslmed.3007240. [DOI] [PubMed] [Google Scholar]
- 28.Palmqvist Richard, Wikberg Maria L., Ling Agnes, Edin S. The association of immune cell infiltration and prognosis in colorectal cancer. Current Colorectal Cancer Rep. 2013;9:372–379. [Google Scholar]
- 29.Sasaki A., Tanaka F., Mimori K., Inoue H., Kai S., Shibata K. Prognostic value of tumor-infiltrating FOXP3+ regulatory T cells in patients with hepatocellular carcinoma. Eur J Surg Oncol. 2008;34:173–179. doi: 10.1016/j.ejso.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Liston A., Gray D.H. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol. 2014;14:154–165. doi: 10.1038/nri3605. [DOI] [PubMed] [Google Scholar]
- 31.Ladoire S., Martin F., Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60:909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.