Abstract
Circular RNA (circRNA) is a group of endogenous noncoding RNA characterized by a covalently closed cyclic structure lacking poly-adenylated tails. Recent studies have suggested that circRNAs play a crucial role in regulating gene expression by acting as a microRNA sponge, RNA binding protein sponge and translational regulator. CircRNAs have become a research hotspot because of their close association with the development of diseases. Some circRNAs are reportedly expressed in a tissue- and development stage-specific manner. Furthermore, due to other features of circRNAs including stability, conservation and high abundance in body fluids, circRNAs are believed to be potential biomarkers for various diseases. In the present review, we provide the current understanding of biogenesis and gene regulatory mechanisms of circRNAs, summarize the recent studies on circRNAs as potential diagnostic and prognostic biomarkers, and highlight the major advantages and limitations of circRNAs as novel biomarkers based on existing knowledge.
Keywords: Circular RNA, Biomarker, Diseases, Liquid biopsy
Highlights
-
•
Circular RNAs are a novel class of endogenous noncoding RNAs characterized by their stable closed loop structure
-
•
Circular RNAs are prospective disease biomarkers due to their tissue- and development stage-specific expression pattern
-
•
Mechanism of circular RNAs' functions in disease development need to be clarified to ensure reliability as biomarkers
1. Introduction
The recent advances in molecular biology techniques enable researchers to explore the complex mediatory network of coding and noncoding transcriptome. Circular RNAs (circRNAs) are a novel class of endogenous noncoding RNAs and a field of much research interest and activity. Unlike linear RNAs, such as mRNAs, microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), with a 5′ cap and 3′ tail structure; circRNAs are characterized by a covalently closed loop structure formed by back-splicing event. As early as 1970s, circRNA molecules were first discovered in RNA viruses by scientists with electron microscope. However, back then the circularized RNAs were thought to be splicing artefacts; and were continuously considered as “junk” RNAs for about two decades until the recent developments in transcriptome sequencing and bioinformatics analysis. Currently, circRNAs have emerged as the most interesting molecules because of their high abundance, stability, and conservation in mammalian cells [1]. CircRNAs have been reported to orchestrate gene expression by acting as miRNAs sponges, interacting with RNA binding proteins (RBPs) and modulating transcription. Importantly, tissue-specific regulation of circRNAs expression has been found associated with initiation and progression of numerous diseases, including various kinds of cancers, cardiovascular diseases, and neurological diseases [[2], [3], [4]]. Here we summarize the present understanding of the formation and function of circRNAs in disease development and discuss the feasibility of circRNAs to serve as biomarkers for different human diseases.
2. Biogenesis and Classification of Circular RNAs
CircRNAs are mainly synthesized by the transcription of protein-coding genes with RNA polymerase II (Pol II); but unlike linear RNAs, they are not produced by canonical mode of RNA splicing [5]. CircRNA molecules are circularized by joining the 3′ and 5′ ends together with unique back-splicing [6]. CircRNAs are commonly named according to their parental genes or specific functions, for example cerebellar degeneration-related protein 1 antisense RNA (CDR1as) is also known as ciRS-7 (circRNA sponge for miR-7) [7]. In this way, the same circRNA may be described with distict names by different researches. Recently, along with the ongoing reseach of circRNAs, several circRNA databases have been constructed to enable orgnization of discovered and idntified circRNAs. A serial number is given to every detected back-spliced juntoin site. Databases like circBase (http://www.circbase.org/) and CircNet (http://circnet. mbc.nctu.edu.tw/) provides tissue-specific circRNA expression profiles as well as circRNA-miRNA-gene regulatory networks [8,9]. Circ2Traits (http://gyanxet-beta.com/circdb/) also allows user to search circRNAs by mutiple diseases [10].
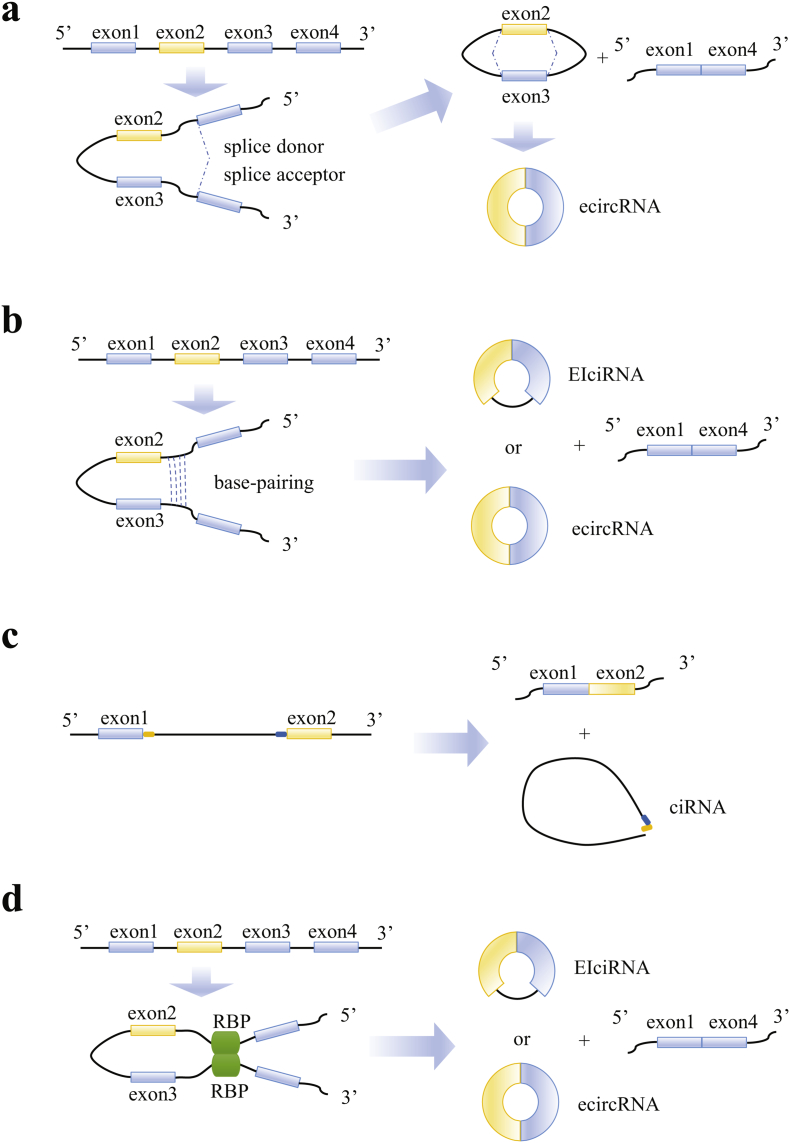
Based on the containing components of exons and introns from the parental genes, circRNAs can be divided into three categories: exonic circRNAs (ecircRNA) that only contain back-spliced exons; circular intronic RNAs (ciRNA) that come from introns; and exon-intron circRNAs (EIciRNAs) which is circularized with both exons and introns [11]. Two different models of exon cyclization have been proposed by Jeck and colleagues, which is termed “lariat-driven circularization” and “intron-pairing driven circularization” (Fig. 1) [12]. The former model is associated with “exon skipping”, which leads to a covalent splice from the 3′ end of splice donor to 5′ end of splice acceptor, resulting in an exon-containing lariat structure. The lariat is then joined by spliceosome and form an exonic circle after the introns being removed (Fig. 1a). The latter one is based on pairing of complementary motifs in the transcripts. EcircRNA and EIciRNAs are formed respectively by removing or retaining the introns. Complementary flanking Alu elements are suggested important for circRNA biogenesis, whereas other inversed repeat sequences are also sufficient to drive circRNA formation (Fig. 1b) [13]. Accumulating evidence has verified the model of intron pairing driven circularization suggesting it might occur more frequently than lariat-driven circularization [14]. Thereafter, a new type of circRNA derived from intron was discovered in human cells and a novel model of ciRNA formation due to failure in debranching was proposed (Fig. 1c) [15]. Additionally, recent studies also suggested that circRNAs can be generated by bridging of RNA molecules with RNA binding proteins (RBPs). RBPs such as Muscleblind (MBL) protein and Quaking (QKI) protein can bind to flanking introns and mediate the circularization (Fig. 1d) [1,5].
Fig. 1.
Possible models of circular RNA biogenesis. (a) Lariat-driven circularization: Exon skipping event results in covalently splices from 3′ splice donor to 5′ splice acceptor, which forms a lariat structure containing the exon 2 and 3 and a linear product of exon 1 and 4. The introns are removed by splicesome to form an ecircRNA (exonic circRNA). (b) Intron-pairing-driven circularization: Direct base-paring of the complementary sequence motifs (such as Alu elements) forms a circulation structure and a linear product. The introns are removed or retained to form an ecircRNA or an EIciRNA (exon-intron circRNA). (c) Circular intronic RNA (ciRNA): The intron lariat is generated from splicing reaction. GU-rich element near the 5′ splice site (orange box) and C-rich element near the branch point (blue box) makes it stable to escape debranching. (d) RNA binding proteins (RBPs)-driven circularization: The interaction between two RBPs can bridge two flanking introns together and form a circRNA and a linear product.
3. Putative Mechanisms of Gene Regulation by Circular RNAs
3.1. Competing Endogenous RNA or miRNA Sponges
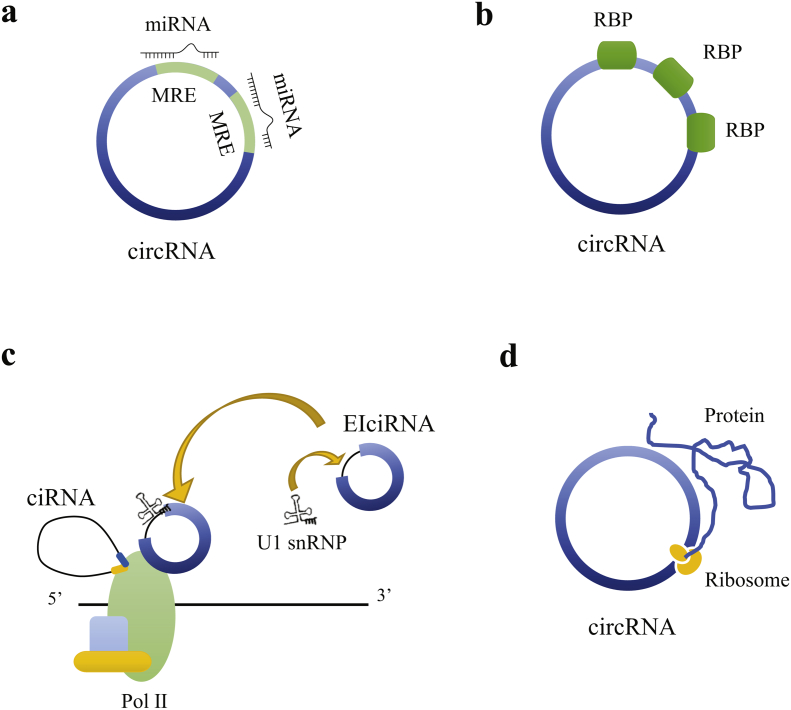
Competitive endogenous RNA hypothesis is currently the most intensively studied and well accepted mechanism on regulatory function of circRNAs on gene expression. CircRNAs contain plenty of miRNA response elements (MREs) that allow them to competitively bind to miRNAs, leading to decreasing of the functional miRNA molecules and subsequent upregulation of target miRNAs [6,16]. This phenomenon is also described as miRNA sponge since circRNAs can “absorb” miRNAs like a sponge (Fig. 2a). CDR1as is the most representative miRNA sponge circRNA. It has been reported contain more than 70 conserved binding sites for miR-7, and therefore pronouncedly reduced miR-7 level when elevated. Similarly, circ-SRY (sex-determining region Y), which is responsible for mammalian sex determination and specifically expressed in testis, has 16 binding sites for miR-138 [7]. Additionally, circ-HIPK3, HRCR and many other circRNAs have all been documented with miRNA sponge function [[17], [18], [19]]. Although sponge effect is a classical model of circRNA-mediated gene regulation, some recent studies have revealed that only few circRNAs exhibit properties of miRNA sponges and physiological ceRNA expression changes do not have impact on highly expressed miRNAs [20,21]. The interaction between circRNAs and miRNA are regarded also related to storage, sorting and localization of miRNA other than simple inhibition [22].
Fig. 2.
Putative mechanisms of gene regulation by circular RNAs. (a) CircRNA can act as microRNA (miRNA) sponge which prevents miRNA from interacting with their target messenger (m)RNAs at 3′-untranslated region (UTR). (b) CircRNA can bind to RNA binding proteins (RBPs) that regulate mRNA processing and hence alter the splicing pattern or mRNA stability. (c) CircRNA can regulate the transcription of their parental gene. CiRNA can interact with RNA polymerase II (Pol II) and modulates transcription; EIciRNA (exon-intron circRNA) can interact with U1 small nuclear ribonucleoproteins (snRNPs) and then binds to Pol II. (d) CircRNA can be translated with ribosome and encode proteins.
3.2. Interaction with RNA Binding Proteins and mRNAs
In addition to the role of circRNAs as miRNA sponge, circRNAs can also act as sponges for RBPs (Fig. 2b). For example, strong direct interaction between circMbl and MBL protein enable circMbl to function in gene regulation by competing with linear splicing [5]. CircPABPN1 suppresses PABPN1 translation by competitively binding to HuR preventing interaction between HuR with PABPN1 miRNA [23]. Circ-Foxo3 can repress cell cycle progression by binding to G1 to S phase transition-related CDK2 and p21 [24]. Besides, circRNAs are able to interact with mRNAs as well. CircRNAs that contain translation start site can act as mRNA traps and regulate protein translation of mRNA [14]. Moreover, several circRNAs have been reported capable of modulating the stability of mRNAs. CDR1as is suggested to form a duplex structure with CDR1 mRNA which in turn stabilizes it [22]. LPS-inducible mouse circRNA circRasGEF1B has been found facilitate the stabilization of mature ICAM-1 mRNAs in macrophages [25].
3.3. Regulation of Parental Gene Transcription
CircRNAs have also been reported to regulate transcription of parental gene through distinctive mechanisms (Fig. 2c). CiRNAs that are enriched in nuclei can interact with Pol II machinery and modulate host transcription activity in a cis-acting manner [15]. Nuclear EIciRNAs containing intronic sequence from their parental gene, such as circEIF3J and circPAIP2, can interact with the U1 small nuclear ribonucleoproteins (snRNPs) which then bind to Pol II on the promoter of their parental genes and thus enhance gene expression [11]. Also, circ-ITCH and the 3′-untranslated region (UTR) of ITCH gene share some identical miRNA binding sites with miR-1, miR-17 and miR-214, and thereby circ-ITCH regulates the expression level of ITCH by indirectly by interacting with its target miRNA [18].
3.4. Protein Translation
Recent studies have demonstrated the potential of circRNAs in proteins translation (Fig. 2d). The first natural circRNA found able to encode protein is the genome of hepatitis virus back to 1980’s [26]. Whereas, at that time most researchers believed circRNAs was endogenous noncoding RNAs in eukaryotic cells until solid evidences start to be uncovered. It has been confirmed that circ-ZNF609 can be translated into protein functioning in myogenesis [27]. CircMbl3 is found to be translated in a splicing-dependent but cap-independent way in fly head extracts [28]. N6-methyladenosine can promote the initiation of protein translation from circRNA in human cells, and a single N6-methyladenosine residue in circRNA is sufficient to drive the translation [29]. Additionally, study on human U251 and U373 cell line has displayed a novel protein encoded form circ-FBXW7 [30]; and a novel tumor suppressive protein encoded by circ-SHPRH has been identified in glioblastoma [31]. Moreover, computational analysis on sequencing of human transcriptomes has revealed the universal existence of circRNAs with coding potential, which provide a new direct for the functional studies of circRNAs [32].
4. Relevance of Circular RNAs as Biomarkers of Diseases
The use of biomarkers has emerged to be one of the main approaches for diagnosis and prognosis of various diseases. The features of a qualified biomarker include stability, sensitivity, specificity, accuracy and reproducibility [33]. According to the current knowledge about circRNAs, there are several remarkable characteristics of circRNA make them potential biomarkers for human diseases. [1] Stability: due to the covalently closed loop structure lacking of free 5′ and 3′ ends, circRNA molecules are highly resistant to exonuclease RNase R, which makes them much more stable compared to linear RNAs [6,34]. The average half-life of circRNAs in plasma exceed 48 h, much longer than 10 h of the average value of mRNAs [14]. [2] Universality: circRNAs are suggested to be the most universal molecules distributed in human cells [35]; and in some circumstances circRNAs are way more abundant than their linear isoforms [8]. Additionally, the abundance of circRNA is higher in brain compared to other organs [36]; and the population of circRNAs circulating in the blood is also very prevalent, attributed to their structure with high stability [37]. [3] Specificity: circRNAs are suggested to express in a tissue-specific and developmental stage-specific manner, which makes them potential biomarkers of specific diseases. In particular, lots of studies have demonstrated that circRNAs are distinctively expressed between cancerous and non-cancerous tissues [6]; and the presence and abundance of circRNAs in different types of cancer cells are distinctive as well [38]. [4] Conservatism: circRNAs are found evolutionally conserved in different species, which means some circRNA biomarkers identified in murine modes hold the potential to be translated to clinical application for human beings [12].
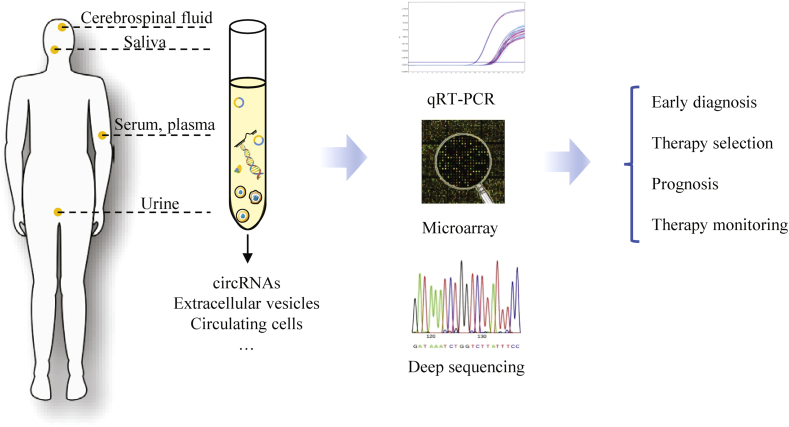
Minimal-invasive liquid biopsies are associated with easier and frequent assessments with less morbidity compared to conventional biopsies [39]. Thus, there is a growing trend in exploring the use of suitable circulating biomarkers for liquid biopsy. Owing to the general features of circRNA including stability, specificity and abundance, circRNAs are considered to be suitable biomarkers for liquid biopsies. CircRNAs are enriched in body fluid including blood, cerebrospinal fluid, saliva and urine. They can also be specifically detected in the free-floating cells inside these body fluids such as circulating blood cells and circulating tumor cells. Furthermore, circRNAs are found highly enriched in extracellular vesicles circulating in blood and other body fluids [40]. Similar to other non-coding RNAs, the technology applied to quantification circRNAs is mainly qRT-PCR, however the primers need to be designed specific for back-splicing site. Likewise, microarray technology can be adopted for high-throughput analysis of biological samples; and deep sequencing is required to verify the results. Collectively, circRNAs are thought to be a group of potential biomarkers especially for liquid biopsies (Fig. 3). CircRNA biomarkers can be detected effectively in body fluid or in circulating cells and extracellular vesicles, to allow early diagnosis, appropriate therapy selection, precise prognosis and frequent therapy monitoring for different type of diseases. Next, we will summarize recent explorations on developing circRNAs as potential biomarkers for various human diseases.
Fig. 3.
Potential application of circular RNAs as liquid biopsy biomarkers. CircRNA biomarkers can be isolated from liquid clinical samples including blood, cerebrospinal fluid, saliva and urine; as well as from the circulating cells and extracellular vesicles contained in these body fluids. Analytical results are used for early diagnosis, therapy selection, prognosis and therapy monitoring of different type of diseases.
5. Circular RNAs as Potential Biomarkers for Human Diseases
5.1. Circular RNAs as Biomarkers for Cancer
The regulatory function of non-coding RNA on cancer development has always been a hotspot in the field of cancer research. Recent studies have revealed that circRNAs may play an important role in the initiation and progression of many types of cancer, and hence may serve as biomarkers of cancers to facilitate diagnosis and prognosis. CircRNAs as biomarker for cancer are most intensively investigated among all the explanatory studies for biomarkers. A representing summary of recently reported potential biomarkers for different diseases are listed in Table 1.
Table 1.
Summary of recent studies on circular RNA as potential biomarker of different diseases.
| Disease | circRNA | Features and potential application |
|---|---|---|
| Gastric cancer | hsa_circ_002059 | Downregulated in gastric cancer (GC) tissues compared with noncancerous tissues; Potential diagnostic biomarker of GC |
| hsa_circ_0000190 | Downregulated in GC tissues and plasma of GC patients; Potential noninvasive diagnostic biomarker of GC | |
| circPVT1 | Upregulated in GC tissues; Acting as sponge of miR-125 family; Potential independent prognostic marker for overall survival and disease-free survival of GC patients | |
| hsa_circ_0001895 | Downregulated in GC cell lines, GC tissues and gastric precancerous lesions; Potential prognostic biomarker for clinical prediction of GC | |
| hsa_circ_101308, hsa_circ_104423, hsa_circ_104916, hsa_circ_100269 | Hsa_circ_101308 upregulated, other three downregulated in GC tissues; four-circRNA-based classifier to predict early recurrence of stage III GC after radical surgery | |
| hsa_circ_0001017, hsa_circ_0061276 | Downregulated in GC tissues and plasma of GC patients; Potential diagnostic biomarker for GC | |
| Hepatocellular carcinoma | hsa_circ_0001649 | Downregulated in hepatocellular carcinoma (HCC) tissue; Potential diagnostic biomarker for HCC and metastasis of HCC |
| Cdr1as | Comparable between HCC tissue and noncancerous tissue; Potential independence biomarker of hepatic microvascular invasion in HCC | |
| circZKSCAN1 | Downregulated in HCC tissues; Potential diagnostic biomarker for HCC | |
| circ-ITCH | Downregulated in HCC tissues; Potential prognostic biomarker of HCC, correlated with favorable survival of HCC | |
| Hepatitis B-induced hepatocellular carcinoma | circ_100,338 | Upregulated in HCC tissues; Directly target miR-143-3p; Potential diagnostic biomarker for hepatitis B-induced HCC |
| Lung cancer | has_circ_0013958 | Upregulated in lung cancer (LC) tissues, LC cell and patient plasma; Promote cell proliferation and invasion in vitro; Potential non-invasive biomarker for the early detection and screening of LC |
| hsa_circ_0000064 | Upregulated in LC tissues and LC cell A549 and H1229; Knockdown of hsa_circ_0000064 attenuates cell proliferation, block cell cycle progression, promote cell apoptosis; Potential biomarker and therapeutic target of LC. | |
| circRNA_102231 | Upregulated in lung adenocarcinoma (LAC) tissues; Potential biomarker and therapeutic target of LAC | |
| circRNA_100876 | Upregulated in Non-small cell lung cancer (NSCLC) tissues; Potential prognostic biomarker and therapeutic target for NSCLC |
|
| hsa_circ_0014130 | Upregulated in NSCLC tissues; Potential biomarker related to carcinogenesis of NSCLC | |
| circFARSA | Upregulated in cancerous tissues, plasma; Promote cell migration and invasion; Potential noninvasive biomarker for NSCLC | |
| Colorectal cancer | hsa_circRNA_10380, hsa_circRNA_104700 | Downregulated in colorectal cancer (CRC) tissues; Potential diagnostic biomarkers of CRC |
| circRNA0003906 | Downregulated in CRC tissues and cell lines; Potential diagnostic biomarker and treatment target of CRC | |
| circHIPK3 | Upregulated in CRC tissues and cell lines; Inhibit miR-7 and elevate proto-oncogenes (FAK, IGF1R, EGFR, YY1); Potential prognostic biomarker and therapeutic target of CRC | |
| Breast cancer | circGFRA1 | Upregulated in triple negative breast cancer (BC) cell and tissue; Potential diagnostic biomarker and therapeutic target of triple negative BC |
| hsa_circ_0001785 | Upregulated in plasma of BC patients; Potential circulating biomarkers for diagnosis and progress of BC | |
| Pancreatic cancer | circ-LDLRAD3 | Upregulated in cancer cell lines, cancer tissues and plasma from patients with pancreatic cancer; Potential diagnostic biomarker for pancreatic cancer |
| Coronary artery disease | hsa_circ_0124644 | Upregulated in peripheral blood of patients; Potential diagnostic biomarker of coronary artery disease |
| Intracranial aneurysms | hsa_circ_0021001 | Downregulated in peripheral blood of patient with intracranial aneurysm; Potential diagnostic marker for intracranial aneurysms |
| Hypertension | hsa-circ-0005870 | Increased in plasma of hypertensive patients; Targeting hsa-miR-6807-3p, hsa-miR-5095, hsa-miR-1273 g-3p, hsa-miR-5096, hsa-miR-619-5p; Potential diagnosis target for hypertension |
| Heart failure | MICRA | Potential prognostic biomarker for risk stratification of heart failure after myocardial infarction |
| Eclampsia | circ_101222 | Increased in blood corpuscles from pregnant women with pre-eclaspsia; Potential biomarker (together with endoglin) for early diagnosis for eclampsia in pregnancies |
| Diabetes | hsa_circ_0054633 | Increased in peripheral blood of type 2 diabetes patients; Potential diagnostic biomarker for pre-diabetes and type-2 diabetes |
| Rheumatoid arthritis | circRNA_104871, circRNA_003524, circRNA_101873, circRNA_103047 |
Upregulated in peripheral blood mononuclear cells (PBMCs); Potential diagnostic biomarkers of rheumatoid arthritis |
| Tuberculosis | hsa_circ_0000414, hsa_circ_0000681, hsa_circ_0002113, hsa_circ_0002362, hsa_circ_0002908, hsa_circ_0008797, hsa_circ_0063179 |
(7-signuture) Upregulated in (peripheral blood mononuclear cells) PBMCs from TB patients; Potential diagnose biomarker of TB infection |
| hsa_circRNA_001937 | Downregulated in PBMCs from active pulmonary tuberculosis patients; Potential diagnostic biomarker of TB infection |
Gastric cancer (GC) is one of the major cancer types characterized by its notable heterogeneity; and therefore tumor markers are essential for precise diagnosis and proper management of GC. Standard biomarkers for GC include serum carcinoembryonic antigen (CEA) and cancer antigen 19–9 (CA19–9), however their sensitivity and specificity are low since they can be detected in patients with other types of cancer. Therefore, researchers have been searching for novel biomarkers for GC with diagnostic and prognostic significance. Hsa_circ_002059 was downregulated in GC tissues and suggested to be a prognostic marker since its level in plasma was associated with tumor grade and distal metastasis [41]. Hsa_circ_0000190 was found lower in plasma samples from patients with GC; and the expression levels was correlated with tumor diameter, lymphatic metastasis and distal metastasis and thereby a potential non-invasive diagnostic biomarker superior to traditional CEA and CA19–9 [42]. CircPVT1 was found upregulated in GC tissues and suggested as an independent prognostic marker for overall survival and disease-free survival of patients with GC [43]. Likewise, hsa_circ_0001895, hsa_circ_101308, hsa_circ_104423, hsa_circ_104916, hsa_circ_100269, hsa_circ_0001017 and hsa_circ_0061276 were also thought to be potential biomarker for GC [[44], [45], [46]].
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the third leading cause of cancer-related mortality worldwide, owing to its high rate of metastasis and recurrence. The prognosis of HCC is poor unless can be diagnosed at an early stage. Alpha-fetoprotein (AFP) is the most commonly used biomarker for HCC diagnosis; however the feasibility of single application of AFP is questioned for low sensitivity [47]. According to the latest researches, circRNAs has shown some utility as biomarkers for HCC. Hsa_circ_0001649 was found downregulated with tumor status, and correlated with the occurrence of the tumor embolus as well as the size of the tumor [48]. High expression of CDR1as was suggested correlated with hepatic microvascular invasion and AFP level in HCC tissues [49]. CircRNA circZKSCAN1 and circ-ITCH were found differentially expressed in cancerous tissue from adjacent noncancerous liver tissue and could serve as diagnostic biomarkers of HCC [50,51]. Additionally, circSNX27, which was correlated with low cumulative survival rate and metastatic progression in hepatitis B-infected HCC patients, was suggested as a potential biomarker for diagnosis of hepatitis B-induced HCC [52].
Lung cancer (LC) is also the leading cause of global cancer-related death due to large cigarette-smoking population, among which non-small cell lung cancer (NSCLC) is the most common (85%–90%) type. Unfortunately, no ideal blood marker for LC is currently available, since the utility of the carcinoma antigens is limited by the poor sensitivity and specificity [53]. Recent studies on circRNAs may show a promising direction for identification of LC biomarker candidates. A constant upregulation of has_circ_0013958 has been reported in LC tissues, LC cell and plasma from patients. This circRNA promoted cancer cell proliferation and invasion in vitro, and was regarded as a potential non-invasive biomarker for the early detection and screening of LC [54]. Similarly, hsa_circ_0000064 and hsa_circRNA_103809 were proposed as potential prognostic biomarkers for patients with LC [55,56]. Moreover, circRNA_100876, hsa_circ_0014130 and circFARSA were identified as potential biomarkers particular for NSCLC detection [[57], [58], [59]].
Besides, potential utility of circRNAs as biomarkers for other types of malignancy have been reported based on related studies, including colorectal cancer [[60], [61], [62]], breast cancer [63,64], pancreatic cancer [65] and so on. These candidates all show differential expression between normal and cancerous tissues and clinical relevance to cancer development, and some may also exhibit different abundance in plasma or serum from patients, which confer them potentials of application as circulating biomarkers.
5.2. Circular RNAs as Biomarkers for Cardiovascular Disease
Cardiovascular diseases (CVDs) and eventually heart attack are leading killers of both man and woman all around the world. Early diagnosis and proper therapeutic intervention are critical for improving patients' survival rates. Blood markers including cyPA (cyclophilin A), Rho-kinase, FADD (fas-associated death domain–containing protein) and others are suggested to be biomarkers for CVDs related to vascular inflammation, such as atherosclerosis and pulmonary arterial hypertension [66,67]. Atrial ANP (natriuretic peptide), BNP (brain natriuretic peptide), cardiac troponin and myocardial band isoenzyme are recognized as blood biomarkers for heart failure [68]. Nevertheless, limitations such as untimely detection and varieties in the patients' baseline level still exist [69]. Although lots of circRNAs have been investigated and suggested to exert essential function in pathological process of various CVDs [70], not much potential circRNA biomarker candidates for CVDs have been identified and reported. The expression of circRNAs was examined in peripheral blood of patient with coronary artery disease (CAD) and results suggested that hsa_circ_0124644 could be used as a diagnostic biomarker of CAD [71]. Another circulating circRNA hsa_circ_0021001 was found decreased in peripheral blood of patients suffering intracranial aneurysms (IA) and was considered to be a potential diagnostic marker for IA [72]. Bioinformatics analysis on plasma circRNA expression profiles in hypertensive patients identified hsa-circ-0005870 as a novel biomarker for diagnosis of hypertension [73]. Moreover, circRNA MICRA showed prognostic value as a novel biomarker for risk stratification of heart failure after myocardial infarction [74].
5.3. Circular RNAs as Biomarkers for Neurological Disease
Biopsies of neuronal and brain tissues may be difficult to conduct, whereas plasma and cerebrospinal fluid is much more accessible. Also, since the majority of circRNAs is found most abundant in brain, it seems promising to find fluctuated circRNAs in cerebrospinal fluid of patients which can be utilized as biomarkers for neurological diseases. Furthermore, small molecules including free circRNAs and circRNA-contained exosomes can be transported across blood-brain barrier, therefore may be also detectable in peripheral blood [75]. Total circRNA expression in Drosophila was reported considerably increased during central nervous system aging, and circRNAs was regarded as a class of potential aging biomarkers [76]. Accumulation of circRNA during aging was later confirmed in mice, suggesting this phenomenon might occur in other mammal species including human as well [77]. Nevertheless, few potential circRNA candidates of biomarkers for neurological diseases have been documented to date. One study reported that circ_101222 level in blood corpuscles of pregnant women with pre-eclampsia was higher than those in healthy women, and circ_101222 combined with plasma protein endoglin have the potential to predict pre-eclampsia early in pregnancies [78]. More studies need to be conducted to explore the feasibility of circRNA as biomarkers of neurological diseases.
5.4. Circular RNAs as Biomarkers for other Diseases
Some circRNAs have shown some utilities as biomarker for other types of diseases. Altered expression profile of circRNA in blood of patients with type 2 diabetes mellitus was confirmed, and among which hsa_circ_0054633 was proposed to be a potential diagnostic biomarker for pre-diabetes and type-2 diabetes [79]. CircRNA expression profile in peripheral blood mononuclear cells indicated circRNAs including circRNA_104871, circRNA_003524, circRNA_101873 and circRNA_ 103047 might serve as potential diagnostic biomarkers of rheumatoid arthritis [80]. Additionally, several circRNAs differentially expressed in peripheral blood mononuclear cells are suggested to be potential diagnostic biomarkers of active pulmonary tuberculosis [81,82].
6. Conclusion and Perspectives
In summary, circRNAs regulate gene expression during the development of various diseases by acting as miRNA sponge, RBP sponge, mRNA trap, and transcriptional modulator or even through direct encoding proteins. The fact that circRNAs are abundantly and prevalently present in human body, and are oftentimes expressed tissue- and development stage-specifically in diseases, endows circRNAs great potential to be employed as diagnostic and prognostic biomarkers for diseases. Notably, due to their excellent stability and high abundance in body fluids and in circulating extracellular vesicles, circRNAs are regarded as promising candidates of biomarkers for liquid biopsy of clinical samples such as whole blood, serum, saliva, urine and cerebrospinal fluid. Currently, clinically-used molecular biomarkers are usually proteins with low organ specificity. For example, as mentioned above, CEA and CA19–9 are standard protein biomarkers for gastric cancer, whereas they are used in diagnosis of patients with colon and pancreas tumor as well. Moreover, compared to other novel RNA biomarkers like miRNAs and lincRNAs, circRNAs have advantages such as longer half-life in blood and better cross-species conservation, which all make them favorable biomarker candidates.
On the other hand, several problems need to be addressed in exploring of circRNA as biomarkers. Firstly, some circRNAs are found differentially expressed in tissues but without difference in serum abundance. As for most intensively studied cancer biomarkers, many circRNA candidates are up- or downregulated in cancerous tissues compared to adjunct noncancerous tissues, but their plasma or serum level in patients and healthy controls are not distinctive [33,47]. These circRNAs are not suitable for noninvasive disease diagnosis or prediction. Secondary, the reliability and sensitivity of using circRNAs as biomarkers need further validation. CircRNAs have just been studied for several years, and there are still many complex functions and mechanisms of circRNAs remains elusive. Moreover, apart from several potential circRNA candidates for cancer prognosis, few circRNA has been found able to predict severity of other kinds of diseases. Finally, detecting circRNAs in blood cells or in exosomes is more expensive and time-consuming than existing protein tests, which may limit the wide application of circRNAs as biomarkers.
Collectively, although there are still functional and mechanical studies need to be conducted and problems to be overcome, circRNAs are favorable potential biomarkers for human diseases because of their stability, specificity and abundance in body fluids. It is doubtless that more and more promising circRNAs biomarkers will be identified and studied for potential clinical use thanks to the recent advance in high-throughput techniques.
7. Key Outstanding Questions
-
•
Since many circRNAs are multi-functional, are circRNA biomarkers reliable enough to predict or monitor disease development?
-
•
How do we standardize a circRNA biomarker? Can we set up normal baseline for distinguishing patient from healthy?
8. Search Strategy and Selection Criteria
Data from this review were identified by searches of PubMed and references from relevant articles using the following keywords, alone or in combination: “biomarker”, “prediction”, “clinical relevance”, “diagnosis”, “prognosis”, “disease”, “cancer”, “cardiovascular disease”, and “neurological disease/disorder”. Only articles published in English were included. Abstracts and reports from meetings were excluded.
Author Contribution
Z.Z. performed the literature search and drafted the manuscript. T.Y. draw the figs. J.X. edited and revised the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China (81722008, 91639101 and 81570362 to J.X.), Innovation Program of Shanghai Municipal Education Commission (2017-01-07-00-09-E00042 to J.X.), the grant from Science and Technology Commission of Shanghai Municipality (17010500100 to J.X.), and the development fund for Shanghai talents (to J.X.).
Competing Interests
The authors have declared that no competing interest exists.
References
- 1.Conn S.J., Pillman K.A., Toubia J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Zhong Y., Du Y., Yang X. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M., Ding W., Sun T. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018;285(2):220–232. doi: 10.1111/febs.14191. [DOI] [PubMed] [Google Scholar]
- 4.Floris G., Zhang L., Follesa P., Sun T. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54(7):5156–5165. doi: 10.1007/s12035-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashwal-Fluss R., Meyer M., Pamudurti N.R. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Memczak S., Jens M., Elefsinioti A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Hansen T.B., Jensen T.I., Clausen B.H. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y.C., Li J.R., Sun C.H. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., Huang C., Bao C. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 12.Jeck W.R., Sorrentino J.A., Wang K. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Zhang X.O., Chen T. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F., Zhang L., Li W. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Q., Bao C., Guo W. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Long B., Liu F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 20.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 21.Militello G., Weirick T., John D., Doring C., Dimmeler S., Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 22.Hansen T.B., Wiklund E.D., Bramsen J.B. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdelmohsen K., Panda A.C., Munk R. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng W.L., Marinov G.K., Liau E.S., Lam Y.L., Lim Y.Y., Ea C.K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13(9):861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 27.Legnini I., Di Timoteo G., Rossi F. Circ-ZNF609 is a circular RNA that can be translated and functions in Myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [e9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pamudurti N.R., Bartok O., Jens M. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. [e7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y., Fan X., Mao M. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018; 110(3). [DOI] [PMC free article] [PubMed]
- 31.Begum S., Yiu A., Stebbing J., Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057. doi: 10.1038/s41388-018-0230-3. [DOI] [PubMed] [Google Scholar]
- 32.Abe N., Matsumoto K., Nishihara M. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry N.L., Hayes D.F. Cancer biomarkers. Mol Oncol. 2012;6(2):140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44(3):1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rybak-Wolf A., Stottmeister C., Glazar P. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Bettegowda C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J Mol Diagn. 2017;19(1):24–34. doi: 10.1016/j.jmoldx.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Li Y., Zheng Q., Bao C. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P., Chen S., Chen H. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Chen S., Li T., Zhao Q., Xiao B., Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Li Y., Zheng Q. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 44.Shao Y., Chen L., Lu R. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4) doi: 10.1177/1010428317699125. [1010428317699125] [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Li J., Yu J. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8(14):22936–22943. doi: 10.18632/oncotarget.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T., Shao Y., Fu L. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl) 2018;96(1):85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 47.Daniele B., Bencivenga A., Megna A.S., Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S108–S112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 48.Qin M., Liu G., Huo X. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 49.Xu L., Zhang M., Zheng X., Yi P., Lan C., Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 50.Yao Z., Luo J., Hu K. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo W., Zhang J., Zhang D. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8(29):48169–48177. doi: 10.18632/oncotarget.18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X.Y., Huang Z.L., Xu Y.H. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Ding M., Xia M. A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung Cancer patients. EBioMedicine. 2015;2(10):1377–1385. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu X., Wang X., Wei S. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 55.Luo Y.H., Zhu X.Z., Huang K.W. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Liu W., Ma W., Yuan Y., Zhang Y., Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi: 10.1016/j.bbrc.2018.04.172. [DOI] [PubMed] [Google Scholar]
- 57.Yao J.T., Zhao S.H., Liu Q.P. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 58.Zhang S., Zeng X., Ding T. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hang D., Zhou J. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer; Cancer Med: 2018. Qin N, et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang P., Zuo Z., Shang W. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39(3) doi: 10.1177/1010428317694546. [1010428317694546] [DOI] [PubMed] [Google Scholar]
- 61.Zhuo F., Lin H., Chen Z., Huang Z., Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng K., Chen X., Xu M. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.He R., Liu P., Xie X. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin W.B., Yan M.G., Fang X., Guo J.J., Xiong W., Zhang R.P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2017 doi: 10.1016/j.cca.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Yang F., Liu D.Y., Guo J.T. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. doi: 10.3748/wjg.v23.i47.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satoh K., Fukumoto Y., Sugimura K. Plasma cyclophilin a is a novel biomarker for coronary artery disease. Circ J. 2013;77(2):447–455. doi: 10.1253/circj.cj-12-0805. [DOI] [PubMed] [Google Scholar]
- 67.Nihei T., Takahashi J., Hao K. Prognostic impacts of rho-kinase activity in circulating leucocytes in patients with vasospastic angina. Eur Heart J. 2018;39(11):952–959. doi: 10.1093/eurheartj/ehx657. [DOI] [PubMed] [Google Scholar]
- 68.Heil B., Tang W.H. Biomarkers: their potential in the diagnosis and treatment of heart failure. Cleve Clin J Med. 2015;82(12 Suppl 2):S28–S35. doi: 10.3949/ccjm.82.s2.05. [DOI] [PubMed] [Google Scholar]
- 69.Enroth S., Johansson A., Enroth S.B., Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan X., Weng X., Zhao Y., Chen W., Gan T., Xu D. Circular RNAs in cardiovascular disease: an overview. Biomed Res Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Z., Li X., Gao C. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teng L., Chen Y., Chen H. Circular RNA hsa_circ_0021001 in peripheral blood: a potential novel biomarker in the screening of intracranial aneurysm. Oncotarget. 2017;8(63):107125–107133. doi: 10.18632/oncotarget.22349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu N., Jin L., Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens. 2017;39(5):454–459. doi: 10.1080/10641963.2016.1273944. [DOI] [PubMed] [Google Scholar]
- 74.Salgado-Somoza A., Zhang L., Vausort M., Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grapp M., Wrede A., Schweizer M. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat Commun. 2013;4:2123. doi: 10.1038/ncomms3123. [DOI] [PubMed] [Google Scholar]
- 76.Westholm J.O., Miura P., Olson S. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gruner H., Cortes-Lopez M., Cooper D.A., Bauer M., Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y.G., Yang H.L., Long Y., Li W.L. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123(13):2113–2118. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ouyang Q., Wu J., Jiang Z. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. 2017;42(2):651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 81.Qian Z., Liu H., Li M. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Z.K., Yao F.Y., Xu J.Q. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem. 2018;45(3):1230–1240. doi: 10.1159/000487454. [DOI] [PubMed] [Google Scholar]