Abstract
Background
Global tuberculosis (TB) control is challenged by uncontrolled transmission of Mycobacterium tuberculosis complex (Mtbc) strains, esp. of multidrug (MDR) or extensively resistant (XDR) variants. Precise analysis of transmission networks is the basis to trace outbreak M/XDR clones and improve TB control. However, classical genotyping tools lack discriminatory power due to the high similarity of strains of particular successful lineages, e.g. Beijing or outbreak strains. This can be overcome by whole genome sequencing (WGS) approaches, but these are not yet standardized to facilitate larger investigations encompassing different laboratories or outbreak tracing across borders.
Methods
We established and improved a whole genome gene-by-gene multi locus sequence typing approach encompassing a stable set of core genome genes (cgMLST) and linked it to a web-based nomenclature server (cgMLST.org) facilitating assignment and storage of allele numbers.
Findings
We evaluated and refined a previously suggested cgMLST schema by using a reference strain set (n = 251) reflecting the global diversity of the Mtbc. A set of 2891 genes showed excellent performance with at least 97% of the genes reliably identified in strains of all Mtbc lineages and in discriminating outbreak strains. cgMLST allele numbers were automatically retrieved from and stored at cgMLST.org.
Interpretation
The refined cgMLST schema provides high resolution genome-based typing of clinical strains of all Mtbc lineages. Combined with a web-based nomenclature server, it facilitates rapid, high-resolution, and harmonized tracing of clinical Mtbc strains needed for prospective local and global surveillance.
Keywords: Whole genome sequencing, Mycobacterium tuberculosis, Core genome MLST, Molecular epidemiology, Genotyping
Research in context.
Evidence Before This Study
Whole genome sequencing (WGS) is currently widely developed for genotyping of bacterial pathogens. While an efficient tool for high resolution strain typing employing genomic data for comparison of Mycobacterium tuberculosis complex (Mtbc) strains and tuberculosis (TB) surveillance is urgently needed, workflow standardization and a unified nomenclature for genome-based strain typing have not yet been established. At present, this makes “genomic epidemiology” an interesting academic endeavour, but with limited practical use for cross border transmission surveillance. Here, the extension of multi locus sequence typing (MLST) which has previously been used for harmonization of classical sequencing data of few conserved genes, to the genome level has been recently suggested as an efficient approach for a unified and easily standardizable genomic typing method. Core genome MLST (cgMLST) schemes have already been suggested for some bacterial pathogens, e.g. Staphylococus aureus, Legionella pneumophila, or Campylobacter jejuni. Previous to the present work, there was only a provisional cgMLST scheme of 3257 genes suggested by our group for clinical Mtbc strains. This scheme had been established from a limited number of genomes, i.e. four strains from M. tuberculosis Lineage 4, one M. africanum Lineage 6 strain, and two M. bovis strains. While the scheme could successfully be used for the analysis of a TB outbreak caused by a M. tuberculosis Lineage 4 strain, the strains used to construct the scheme do not represent the global phylogenetic diversity of the Mtbc and it remained unclear whether the scheme could be used for efficient analysis of the full Mtbc species diversity. Furthermore, it has not been linked to an open web-based nomenclature system ensuring harmonized assignment of cgMLST allele types.
Added Values of This Study
Using an extensive set of strains representing the known diversity of the Mtbc, we show the limits of the previously suggested cgMLST scheme for defined subgroups of the Mtbc. This clearly demonstrates the necessity to consider the full species/complex diversity for construction of a cgMLST scheme, even for a pathogen with a relatively stable genome such as the Mtbc. Using a set of 45 strains covering the known diversity of the Mtbc, we developed an improved cgMLST scheme consisting of 2891 genes and showed its applicability to the whole Mtbc diversity using a comprehensive set of reference strains. In order to enable its use in pathogen surveillance, we define critical thresholds for likely (at most five distinct alleles) and unlikely recent transmission events (>12 distinct alleles), the evolutionary rate of allele change, and the correlation with classical genotyping data. As a necessary prerequisite for standardized use of the cgMLST scheme for unified genotyping across different laboratories, a central web-based nomenclature server has been established.
Implications of All the Available Evidence
The improved cgMLST scheme can be directly used for standardized typing and surveillance of clinical isolates of all lineages of the Mtbc. This is especially critical for setting up a surveillance tool for multi-country, cross-border analysis of strain transmission and outbreak dynamics. WGS data are transferred into a set of 2891 numeric values that can be efficiently exchanged between laboratories and public health agencies. The thresholds for likely transmission and allele mutation rates can directly be used for the detection of recent transmission and the analysis of pathogen evolution. Importantly, we also suggest the definition of a cgMLST-based complex type, which facilitates even easier communication in outbreak situations such as the recently confirmed outbreak of a multidrug resistant Mtbc strain among Somalian refugees in the European Union. Finally, the web-based cgMLST allele server provides a universally harmonized allele nomenclature and generation of comparable cgMLST genotypes on a global scale.
Alt-text: Unlabelled Box
1. Introduction
Tuberculosis (TB) remains a main global health challenge, with roughly a third of the human population latently infected, approx. 10.4 million new TB cases and 1.7 million deaths in 2016 [1]. The efforts in controlling TB are seriously challenged by the increasing numbers of multiple (MDR) or extensively resistant (XDR) Mycobacterium tuberculosis complex (Mtbc) strains [1]. Overall, approx. 490,000 new MDR-TB cases have been reported in 2016, with about 6% of them being XDR [1].
Transmission is the major driving force of the epidemic, esp. of the MDR pandemic [[2], [3], [4], [5]]. As there is no environmental reservoir, controlling human-to-human transmission is key for successful local and global TB control and for achieving the targets of the “End TB strategy” proposed by the World Health Organisation [1]. Targeted interventions to stop transmission esp. of M/XDR strains requires in depth epidemiological knowledge that can only be provided by a combination of effective genotyping with classical epidemiology in molecular epidemiological studies [4, 6]. Classical genotyping techniques such as spoligotyping or MIRU-VNTR typing (Mycobacterial Interspersed Repetitive Units - Variable Number of Tandem Repeats) interrogating just a small fraction of the genome have been used for years to study transmission dynamics in low and high incidence settings, population structure of the pathogen, and global spread of particular strains/lineages [[3], [4], [5], [6]]. While they provide standardized, easily computable typing results with an on-line nomenclature, recent studies indicate that their discriminatory power is too low to differentiate outbreak strains esp. in high incidence settings with the resolution needed to trace recent transmission chains [4, [7], [8], [9], [10]].
Several studies have now demonstrated that whole genome sequencing (WGS) based genotyping has an improved discriminatory power compared to classical genotyping, e.g. better discrimination of outbreak strains [[7], [8], [9], [10]]. Furthermore, WGS provides a highly accurate identification of the diverse lineages of the Mtbc and potentially allows for comprehensive detection of drug resistance mediating genomic variants [6, [10], [11], [12], [13]].
While these advantages of WGS-based strain analysis are commonly accepted, there are several drawbacks limiting the routine application of WGS for transmission analysis and diagnostic questions. Most importantly, there are not yet any standardized analysis pipelines, leading to inherent problems in defining single nucleotide polymorphisms (SNPs) later included in SNP-based similarity analysis commonly used for strain comparison. Furthermore, there is not yet a common nomenclature to standardize WGS data to facilitate data exchange in an easily extendable classification scheme. The advantages and problems of WGS-based pathogen tracing have recently been demonstrated by a European wide MDR-TB outbreak, in which high resolution WGS-based investigation delineated likely transmission routes, however, analysis was done in a central laboratory to allow strain comparison [14].
These limitations can be overcome by using a whole genome gene-by-gene allele calling approach [15, 16], which extends the concept initially developed for sets limited to six or seven house-keeping genes used in multi locus sequence typing (MLST) [15], to a genome-wide level. Such an approach transfers genome wide diversity into a more easily processed and standardizable allele numbering system, ideally using a web-based nomenclature server assigning and storing allele variants such as BIGSdb [15, 17, 18]. This approach is based on a predefined scheme of genetic loci, e.g. encompassing the core genome set of genes (cgMLST). We have previously developed a preliminary cgMLST scheme for Mtbc strains that showed sufficient resolution for strains from the outbreak investigated [18]. However, this initial schema was not systematically tested with a larger collection of clinical strains representing the various Mtbc lineages present in different regions of the world. In addition, allele thresholds for defining recent transmission chains have not yet been fully established. Furthermore, no publicly accessible web-based cgMLST Mtbc nomenclature server has been set up yet.
To address these questions, we first evaluated the preliminary cgMLST scheme using a reference strain set (n = 251) reflecting the global diversity of the Mtbc. Based on the performance data, we developed an improved cgMLST schema and tested its performance in outbreak strains and the comprehensive dataset used to define SNP thresholds for recent transmission analysis [9, 19, 20]. In line with this, we developed a web-based nomenclature server that facilitates assignment and storage of allele numbers.
2. Materials and Methods
2.1. Study Design and Datasets Used
To set up a comprehensive collection of Mtbc strains, we combined 19 completed and closed Mtbc genomes listed in the NCBI GenBank archive with WGS data from strains used in studies on the global population structure of the Mtbc [21, 22], excluding datasets with <50-fold mean coverage after mapping to the H37Rv genome (GenBank ID: NC_000962.3), and with <95% genome positions covered by at least ten reads and 75% allele frequency. The resulting collection was divided into a training (45 strains) and an evaluation set (251 strains), both fully covering the Mtbc diversity. Full details for all strains are listed in Supplementary Table S1, and depicted in Fig. 1. The training set was employed to construct an improved cgMLST scheme, and the evaluation set was used to assess the performance of the cgMLST scheme, both using the Ridom SeqSphere+ software (Ridom GmbH, Münster, Germany).
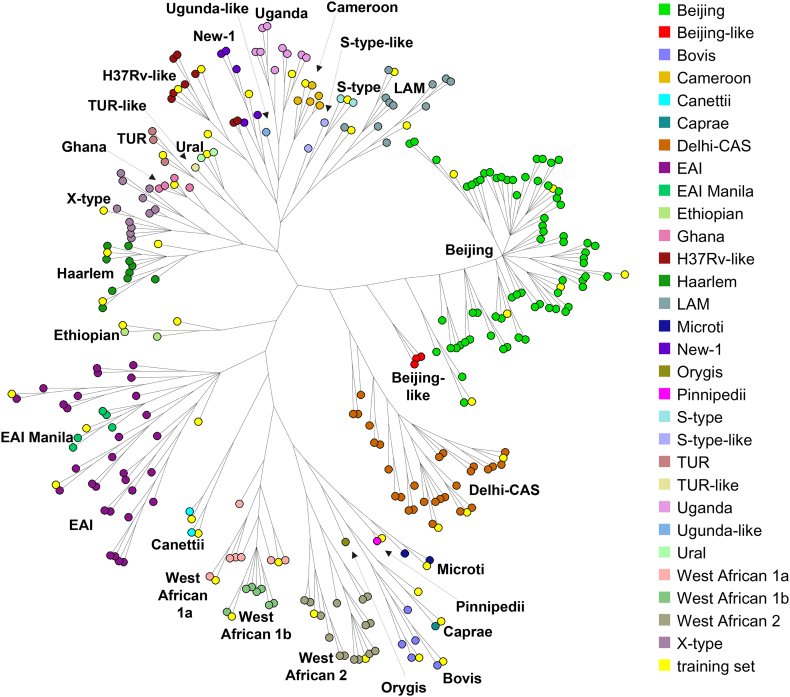
Fig. 1.
Overview of the 251 isolates included in the evaluation and 45 isolates in the training set of isolates, shown in a neighbour joining tree built with BioNumerics from 41,774 SNP positions in logarithmic scaling. The 45 strains included in the training set are marked in yellow.
Using the newly constructed cgMLST scheme, we re-analyzed 390 Mtbc sequences previously published as part of a study calibrating whole genome sequencing as an epidemiological tool [20]. In line with the original publication, we used these data to estimate thresholds for the maximum number of allele differences suggestive of recent transmission.
To evaluate the performance of the cgMLST scheme for outbreak scenarios, we performed WGS for isolates from seven clusters defined by traditional genotyping methods (IS6110 DNA fingerprint and spoligotype patterns) in a longitudinal prospective molecular epidemiological surveillance study undertaken in Hamburg starting from January 1st, 1997 [9, 18, 23].
2.2. Classical Genotyping
Extraction of genomic DNA from mycobacterial strains, DNA fingerprinting using IS6110 as a probe, spoligotyping, and 24-loci MIRU-VNTR genotyping were performed using standardized protocols as described previously [[24], [25], [26]].
2.3. Whole Genome Sequencing
Isolated genomic DNA of individual strains was sequenced using Illumina sequencing platforms and Nextera or Nextera XT library preparation kits as instructed by the manufacturer (Illumina, San Diego, CA, USA). All isolates were sequenced with a minimum coverage of 50-fold. The raw reads were submitted to the EMBL ENA sequence read archive (Supplementary Table S1).
2.4. SNP Based Analysis Pipeline
Sequence reads were mapped to the M. tuberculosis H37Rv genome (GenBank ID NC_000962.3) using the exact alignment program SARUMAN [27]. SNPs were extracted from mapped reads using a minimum coverage of ten reads and a minimum allele frequency of 75% as thresholds for detection. All positions for which a SNP was detected in at least one isolate were combined, and positions within resistance associated genes or repetitive regions discarded. Likewise, positions were excluded if two SNPs appeared in a window of 12 bp. All remaining positions with reliable base calls (10-fold coverage, 75% allelic frequency) in 95% of all isolates were combined into a concatenated alignment. The BioNumerics software (version 7.5, Applied Maths) was used to calculate neighbour joining or minimum spanning trees from aligned SNP positions.
2.5. cgMLST Based Analysis Pipeline
We used the functionality implemented in the Ridom SeqSphere+ software with default settings to perform core genome MLST analysis. Using a set of 45 datasets covering the known diversity of the MTBC, we defined a cgMLST scheme using the cgMLST Target Definer tool of the Ridom SeqSphere+ software with default settings. The finished genome of the M. tuberculosis strain H37Rv (GenBank ID NC_00962.3) served as seed genome. Subsequently, query genomes from the training set were compared with the seed genome to establish a list of core genome genes. Query genomes are indicated in Supplementary Table S1. Here, default settings include the removal of the shorter of two genes overlapping by more than four bases and of genes with an internal stop codon in >80% of all query genomes from the scheme. Finally, additional repetitive genes described previously, e.g. all members of the PPE / PE-PGRS gene families were manually excluded from the scheme. cgMLST based neighbour joining and minimum spanning trees were both calculated and drawn with the SeqSphere+ software. A cgMLST server for M. tuberculosis complex strains was set up (http://www.cgmlst.org/mtbc) to allow standardized typing based on a cgMLST allele nomenclature.
To estimate allele distance thresholds for likely transmission, we exported from SeqSphere+ the cgMLST allelic profiles for further analysis. As previously described, we plotted the number of alleles distinguishing paired isolates sampled from individuals (across different anatomical foci at a single point in time, and from a single anatomical focus over time), between individuals in household clusters, and from 24-locus MIRU-VNTR defined community clusters [20].
2.6. Role of the Funding Source
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author (SN) had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
The cgMLST scheme comprising 3257 genes we previously developed as an ad hoc tool for classification of Mtbc strains was established from a limited number of genomes, i.e. four strains from Lineage 4, one from Lineage 6, and two M. bovis [18], not representing the global phylogenetic diversity of the Mtbc. To validate its applicability and representativeness for clinical strains of all Mtbc lineages, we evaluated its performance in an extended collection of 251 strains reflecting the global Mtbc population diversity (Supplementary Table S1).
Due to the high clonality of the Mtbc, we set the level of good targets to be achieved to at least 97% of the cgMLST genes present in each of the 251 validation genomes. We employed SeqSphere+ to determine the number of genes fulfilling the quality criteria for allele calling in each of the test strains (good cgMLST targets). While the cgMLST performed well overall, with 98.1% of genes fulfilling the criteria on average (Table 1), the scheme performed remarkably worse in individual strains of virtually all lineages with good cgMLST target percentage going down to 94.4% in M. canettii strains.
Table 1.
Evaluation of the cgMLST scheme suggested previously (Kohl et al. 2014).
| Number of strains | Perc. mean1 | Perc. min1 | Perc. max1 | |
|---|---|---|---|---|
| Lineage 1 | 35 | 97.7 | 96.4 | 98.1 |
| Lineage 2 | 71 | 97.8 | 96.9 | 98.2 |
| Lineage 3 | 28 | 98.1 | 97.4 | 98.4 |
| Lineage 4 | 74 | 99.0 | 96.9 | 99.7 |
| Lineage 5 | 14 | 97.2 | 96.6 | 97.4 |
| Lineage 6 | 15 | 97.7 | 97.3 | 98.0 |
| Lineage 7 | 2 | 97.7 | 97.6 | 97.7 |
| Canettii | 2 | 94.4 | 94.2 | 94.6 |
| Animal⁎ | 10 | 97.1 | 96.6 | 97.5 |
| total | 251 | 98.1 | 94.2 | 99.7 |
Percentage of cgMLST genes fulfilling quality criteria, given as mean, minimum, and maximum value.
Including M. caprae, M. bovis, M. orygis, M. microti, and M. pinnipedii.
As this was below our set threshold, we used the genomic data of 45 strains (19 fully finished genomes and 26 isolates from the reference collection) representing the different lineages of the Mtbc (Fig. 1 and Supplementary Table S1) to construct a more representative cgMLST scheme. Employing the core genome MLST definer tool of SeqSphere+ software, a refined cgMLST scheme consisting of 2891 genes was established (for the defined genes it is referred to the Show Targets functionality of the cgMLST.org server), containing 2.86 million bases.
Testing this refined cgMLST scheme in the evaluation set of 251 isolates resulted in an improved performance, considering both, the mean percentage and minimum percentage of good cgMLST targets with at least 97% of genes successfully retrieved across strains of all lineages (Table 2), including M. caprae, M. bovis, M. orygis, M. microti, and M. pinnipedii strains.
Table 2.
Evaluation of the improved cgMLST scheme constructed from the 45 isolates representing the phylogenetic diversity of the Mtbc.
| Number of strains | Perc. mean1 | Perc. min1 | Perc. max1 | |
|---|---|---|---|---|
| Lineage 1 | 35 | 98.4 | 97.4 | 98.7 |
| Lineage 2 | 71 | 98.6 | 98.0 | 98.8 |
| Lineage 3 | 28 | 98.9 | 98.5 | 99.1 |
| Lineage 4 | 74 | 99.5 | 98.0 | 99.8 |
| Lineage 5 | 14 | 98.2 | 98.0 | 98.4 |
| Lineage 6 | 15 | 98.3 | 98.1 | 98.5 |
| Lineage 7 | 2 | 98.3 | 98.2 | 98.3 |
| Canettii | 2 | 97.8 | 97.7 | 97.9 |
| Animal⁎ | 10 | 97.8 | 97.4 | 98.3 |
| total | 251 | 98.8 | 97.4 | 99.8 |
Percentage of cgMLST genes fulfilling quality criteria, given as mean, minimum, and maximum value.
Including M. caprae, M. bovis, M. orygis, M. microti, and M. pinnipedii.
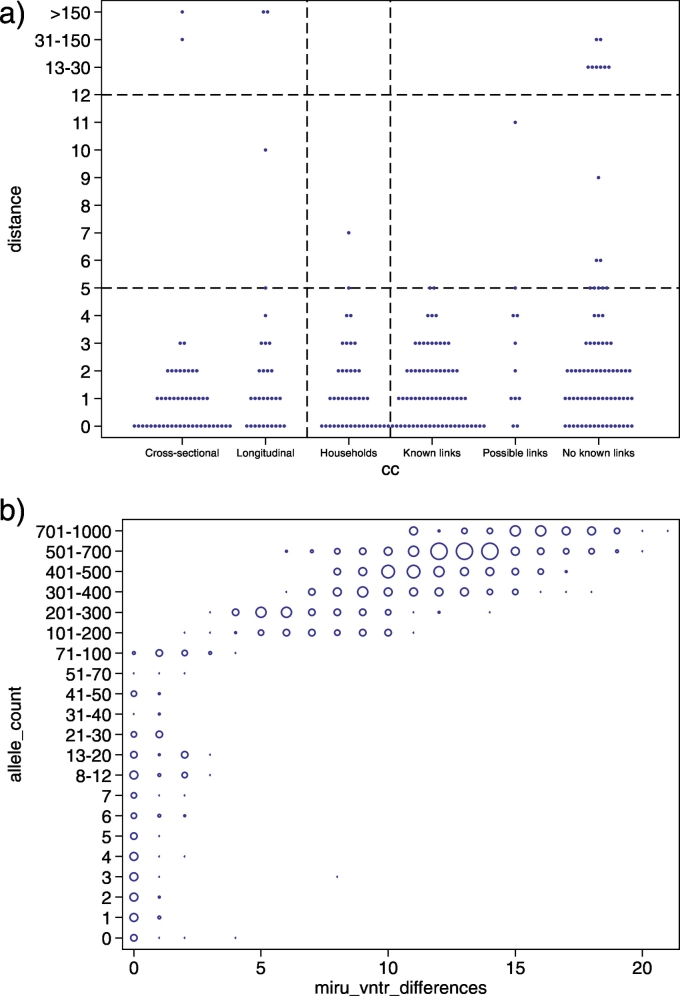
Using the newly constructed cgMLST scheme, we then re-analyzed WGS sequences of 390 Mtbc strains previously published as part of a study calibrating WGS as an epidemiological tool [20]. The dataset comprises cross-sectional isolates, longitudinal isolates from the same patient, household outbreak isolates, and community isolates and is thus well suited to define thresholds for the maximum number of allele differences suggestive of recent transmission. The cgMLST diversity among longitudinal isolates was similar to the diversity seen among isolates from epidemiologically linked cases and was highly similar to the original SNP-based analysis of the dataset (Fig. 2a). Accordingly, the same thresholds of 5 and 12 alleles were determined for the allele-based approach (Fig. 2a). In particular, the allelic difference associated with epidemiologically linked cases was five or fewer in 37/38 instances (number of patients minus number of outbreaks).
Fig. 2.
a: Within host and between host observed allelic diversity across paired isolates. From left to right, paired isolates from within host (pulmonary and non-pulmonary sampled within 6 months of each other); paired pulmonary isolates sampled >6 months apart from within individual hosts; pairwise distances between patients within households; and pairwise distances between patients across 11 different community clusters, stratified by type of epidemiological link. 22 of the 38 links within the 25 household clusters also occur within community clusters (ie, known linkage) but are shown with household isolates and not with community isolates. Top horizontal dashed line indicates the threshold above which direct transmission can be judged to be unlikely; bottom horizontal dashed line indicates the threshold below which transmission should be investigated. b: All against all comparison of the number of allele differences and MIRU-VNTR locus differences for genomes for which full 24-locus MIRU-VNTR profiles were available. Zero MIRU-VNTR differences implies identical MIRU-VNTR profiles. The size of the blue circles indicates the number of comparisons with X MIRU-VNTR locus differences and Y allele differences.
No evidence was found that the distribution of allele variants below this threshold differed from that of longitudinal Isolates (37/38 [97.3%] vs 27 [90%] of 30; rank-sum p = .97; Fig. 2a). Overall, with exclusion of differences of >100 allele variants, 111 (97.4%) of 114 paired isolates from within individuals and household outbreaks differed by five or fewer alleles, 109 (95.6%) by four or fewer alleles, and 106 (93.0%) by three or fewer alleles.
The rate of allelic microevolution was calculated using the first and last sequenced isolates of the longitudinally sampled patients and from the household outbreaks. A rate of change in DNA sequences of 0.55 alleles per genome per year (95% CI 0.26–0.84) was inferred by maximum likelihood.
We also compared the correlation between MIRU-VNTR and allele diversity (Fig. 2b). Strikingly, apart from two exceptions, a threshold of five distinct alleles related to a maximum distance of three distinct MIRU-VNTR loci, and 12 distinct alleles with four distinct MIRU-VNTR loci (Fig. 2b).
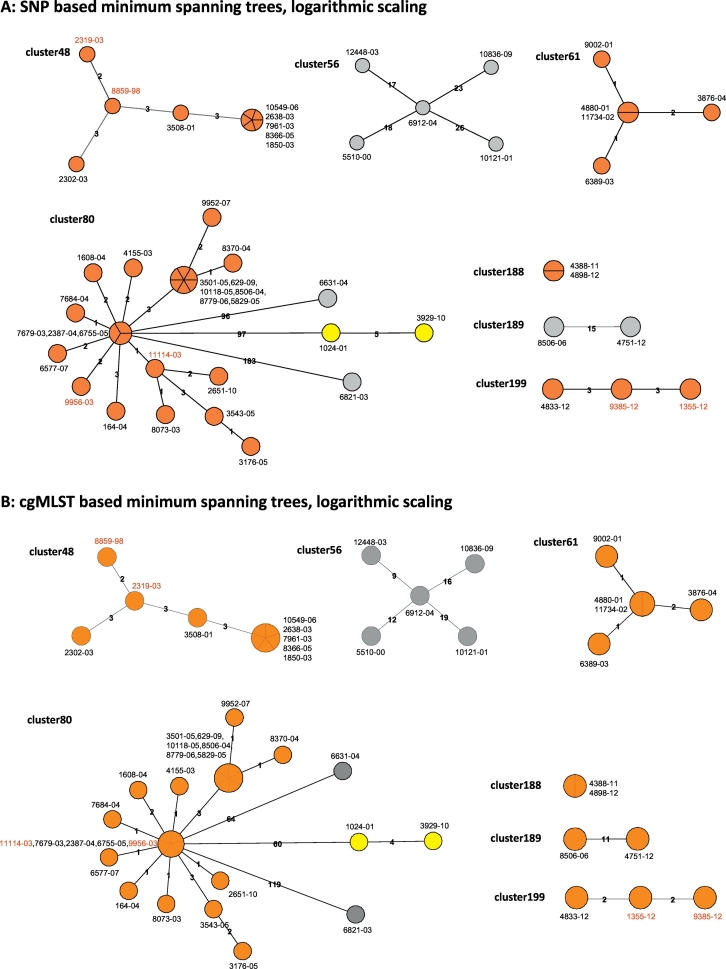
To test the performance of the refined cgMLST schema for discriminating clinical isolates for transmission analysis, we selected seven clusters comprising 52 isolates from a longitudinal molecular epidemiological study carried out in Hamburg, Germany (see Materials and Methods). All strains in a particular cluster displayed identical spoligotyping patterns and comprised up to 26 strains (Fig. 3). For all strains, at least 98.6% of the cgMLST scheme targets fulfilled quality thresholds for detection (Table 3).
Fig. 3.
Comparison of the SNP and cgMLST approach employed for seven clusters defined by traditional genotyping methods for the Hamburg surveillance study. Minimum spanning trees built from either identified SNP positions (A) or from the cgMLST approach (B), shown in logarithmic scale. Colors indicate genomic clusters defined by a maximum distance of 12 SNP positions or allele variants, with orange and yellow denoting identified genomic clusters, and isolates marked in grey ungrouped. Isolates labeled in red exhibit differences between both approaches in their position in the derived tree.
Table 3.
Evaluation of the improved cgMLST scheme with 52 isolates clustered by traditional genotyping.
| Cluster ID | Number of strains | Subgroup | Genotype (Coll 2014) | Discerning SNPs | Discerning alleles | Perc. Min1 | Perc. Max1 |
|---|---|---|---|---|---|---|---|
| Cluster 48 | 9 | LAM | 4.3.4.2 | 8 | 8 | 99.4 | 99.5 |
| Cluster 56 | 5 | Beijing | 2.2.1 | 57 | 41 | 98.6 | 98.7 |
| Cluster 61 | 5 | Euro-American | 4.1.2 | 4 | 4 | 99.7 | 99.7 |
| Cluster 80 | 26 | Haarlem | 4.1.2.1 | 322 | 206 | 99.7 | 99.7 |
| Cluster 188 | 2 | Beijing | 2.2.1 | 0 | 0 | 98.6 | 98.7 |
| Cluster 189 | 2 | LAM | 4.3.3 | 15 | 11 | 99.9 | 99.9 |
| Cluster 199 | 3 | Euro-American | 4.8 | 5 | 3 | 99.5 | 99.6 |
| Total: | 52 | – | – | 411 | 273 | 98.6 | 98.9 |
Percentage of cgMLST genes fulfilling quality criteria, given as minimum and maximum value.
For each cluster, both, a SNP and cgMLST based analysis was carried out, including the definition of genomic clusters (GCs) by a maximum distance of 12 SNPs/ 12 distinct alleles to the nearest group member (Fig. 3). The SNP-based cluster analysis detected six GCs in the seven clusters, with one GC for clusters 48, 61, 188, and 199 each, and two GCs for cluster 80. All of these were confirmed by the cgMLST approach, with one additional GC suggested for cluster 189.
As expected, the number of distinct alleles between isolates using the cgMLST approach was smaller than the number of distinct SNP positions. For the seven clusters, this resulted in one additional GC suggested for cluster 189, and a lower discriminatory power for cluster 80, where two isolates (11114–03, 9956–03) separated by the SNP method fell into the central outbreak node in the cgMLST analysis. In addition, in two of the smaller clusters (48 and 199), the position of isolates was switched in the derived tree topology.
To allow for standardized cgMLST typing of Mtbc strains, we set up an allele nomenclature server (http://www.cgmlst.org/mtbc) for the suggested cgMLST scheme. SeqSphere+ users have the option to use either an own local or the global stable cgMLST.org allele nomenclature. If using the latter, the user may optionally submit anonymized, i.e. without storing submitter information, or non-anonymized strain allelic profiles for complex type assignment with or without additional epidemiological metadata (place, time, etc.). Thus, if a new gene sequence is detected, a new allele number is assigned, and the data are stored in the database. This facilitates the generation of standardized, comparable cgMLST allele number data from different researchers or laboratories. Furthermore, the cgMLST nomenclature server groups closely related cgMLST profiles in complex types (CT; currently 13,433) with a threshold of 12 distinct alleles. In addition to the cgMLST gene set, the server also hosts an set of 755 additional accessory genome genes. The web interface of the server allows searching strains via sample ID, cgMLST.org ID, cgMLST CT, collection year, country of isolation, PubMed ID, NCBI accession, or submission year. In addition, the scheme and the allelic sequence definitions can be downloaded. Currently, the nomenclature server hosts cgMLST allelic profiles of 35,418 Mtbc strains. At the moment, the server can be queried using the SeqSphere+ software. The implementation of a WWW interface for uploading own data to search for closely related isolates and an application programing interface (API) for other tools are planned.
4. Discussion
WGS has evolved over the last years as a prime tool for TB molecular epidemiological studies with higher discriminatory power compared to classical genotyping such as MIRU-VNTR typing esp. in high incidence settings [[7], [8], [9], [10], 14, 28]. WGS has recently been shown to facilitate European wide outbreak analysis of multidrug resistant strains [14, 29], however, in these studies, WGS data analysis was done by one core lab to enable a joint comparison of sequence data from different sources. In fact, results from different SNP-based analysis pipelines are not easily comparable due to the use of highly customized in-house pipelines [3, 18]. These limitations can be largely overcome by using a whole genome gene-by-gene allele calling approach [15, 16], extending at genome wide level the concept initially developed for sets limited to six or seven house-keeping genes, the standard MLST approach [15].
Here, we demonstrate that an improved cgMLST scheme employing 2891 loci showed excellent performance in a reference collection of 251 strains reflecting the global diversity of the Mtbc. With allele retrieval rates of >97% good targets, the cgMLST scheme proved to be universally applicable for Mtbc strains from all lineages including M. canettii. This is of particular importance for its application as a tool for standardized typing of clinical Mtbc strains for longitudinal studies as well as cross border surveillance, e.g. of MDR outbreak clones. As the scheme was also validated for the primarily animal pathogenic species M. caprae, M. bovis, M. orygis, M. microti, and M. pinnipedii, it can also be used for surveillance of these pathogens.
The performance of the cgMLST scheme in a larger set of strains previously used to establish SNP thresholds for SNP-based transmission analysis [20] and several well characterized outbreaks, was similar to SNP-based strain classification. Indeed, the thresholds of max. five variants differences in epidemiological linked cases appears to apply for SNPs and cgMLST alleles both. These data are essential to use cgMLST data for inclusion/exclusion of strains in recent transmission inference.
We also estimated that the rate of genetic change of alleles and SNPs is similar at around 0.5 allele changes per year and genome.
When we compared SNP and allele-based phylogenies for seven clusters comprising 52 isolates from a longitudinal molecular epidemiological study in Hamburg, Germany [18, 23, 30], a slight reduction in discriminatory power was seen (Table 3). However, overall tree phylogenies were not affected, the genome cluster defined by the SNP-based cluster analysis were all confirmed by cgMLST, and only one additional genome cluster was suggested.
The resolution level retained by the cgMLST scheme is thus still sufficient to analyze individual outbreaks, up to the identification of likely transmission chains. Importantly, the analysis of particular outbreaks can be done subsequently with the accessory genome set of 755 genes allowing for improved resolution.
Using cgMLST typing has several advantages compared to SNP-based approaches. Genetic relationships based on allelic profiles can be calculated at drastically reduced computational costs compared to SNP data, expandable strain databases can be created as the full allele profiles for all strains are readily available, and allele profiles can be easily exchanged between laboratories.
The latter is particular true as we have established a freely accessible web-based nomenclature server, which facilitates assignment and storage of allele numbers, thus opening the way towards standardized cgMLST typing based on an allele database. This represents the first opportunity for establishment of a globally harmonized typing scheme for Mtbc isolates based on NGS data that can be used for unified global surveillance efforts and guide the computational expensive SNP-bases analysis to suspected outbreaks.
In conclusion, the established typing ontology and nomenclature system is likely to bring Mtbc surveillance to the ultimate level of information extraction and exchange, thus allowing for most efficient pathogen surveillance which has been identified as a key priority in the fight against the major health challenges of the EU by the ECDC (www.ecdc.europa.eu) and worldwide.
The following are the supplementary data related to this article.
Supplementary Fig. S1.
Screenshot of the Mycobacterium tuberculosis/bovis/africanum/canettii cgMLST site on the cgMLST.org server.
List of all samples used in the study, indicating the analyses done, ENA SRA accession numbers, phylogenetic classification, and quality metrics of the respective datasets.
Acknowledgments
Acknowledgements
We thank Julia Zallet, Vanessa Mohr, Carina Hahn, Tanja Ubben, Anja Lüdemann, Tanja Struve-Sonnenschein, and Ilse Radzio, Research Center Borstel, for excellent technical assistance.
Funding Sources
The work was partially funded by the European Union PathoNgenTrace project (FP7-278864-2) and the German Center for Infection Research (DZIF). The funders had no role in study design, data collection, data analysis, interpretation, writing of the report, or the decision to submit the paper. The corresponding author had full access to all data of the study and the final responsibility for the decision to submit the manuscript.
Declaration of Interests
The following authors have competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article. DH and JR are shareholder and JR is employee of Ridom GmbH (Münster, Germany). The other authors have no competing interests.
Author Contributions
TAK, DH, TW, RD, and SN contributed to the study design. TAK, TW, KMW, RD, SN collected the data and performed the laboratory testing. TAK, DH, JR, TW, RD analyzed the data, which was interpreted by all authors. TAK, DH, TW, and SN drafted the report with input from all other authors. All authors agreed to the final version of the manuscript.
References
- 1.World Health Organisation . World Health Organization; Geneva: 2017. Global tuberculosis report 2016. [Google Scholar]
- 2.Cox H.S., Sibilia C., Feuerriegel S., Kalon S., Polonsky J., Khamraev A.K., Rüsch-Gerdes S., Mills C., Niemann S. Emergence of extensive drug resistance during treatment for multidrug-resistant tuberculosis. N Engl J Med. 2008;359:2398–2400. doi: 10.1056/NEJMc0805644. [DOI] [PubMed] [Google Scholar]
- 3.Merker M., Blin C., Mona S., Duforet-Frebourg N., Lecher S., Willery E., Blum M.G.B., Rüsch-Gerdes S., Mokrousov I., Aleksic E., Allix-Béguec C., Antierens A., Augustynowicz-Kopeć E., Ballif M., Barletta F., Beck H.P., Barry C.E., Bonnet M., Borroni E., Campos-Herrero I., Cirillo D., Cox H., Crowe S., Crudu V., Diel R., Drobniewski F., Fauville-Dufaux M., Gagneux S., Ghebremichael S., Hanekom M., Hoffner S., Jiao W., Kalon S., Kohl T.A., Kontsevaya I., Lillebæk T., Maeda S., Nikolayevskyy V., Rasmussen M., Rastogi N., Samper S., Sanchez-Padilla E., Savic B., Shamputa I.C., Shen A., Sng L.-H., Stakenas P., Toit K., Varaine F., Vukovic D., Wahl C., Warren R., Supply P., Niemann S., Wirth T. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemann S., Merker M., Kohl T.A., Supply P. Impact of genetic diversity on the biology of Mycobacterium tuberculosis complex strains. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.TBTB2-0022-2016. [DOI] [PubMed] [Google Scholar]
- 5.Casali N., Nikolayevskyy V., Balabanova Y., Harris S.R., Ignatyeva O., Kontsevaya I., Corander J., Bryant J., Parkhill J., Nejentsev S., Horstmann R.D., Brown T., Drobniewski F. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niemann S., Supply P. Diversity and evolution of Mycobacterium tuberculosis: moving to whole-genome-based approaches. Cold Spring Harb Perspect Med. 2014;4:a021188. doi: 10.1101/cshperspect.a021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorn-Mortensen K., Soborg B., Koch A., Ladefoged K., Merker M., Lillebaek T., Andersen A.B., Niemann S., Kohl T.A. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci Rep. 2016:6. doi: 10.1038/srep33180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee R.S., Radomski N., Proulx J.-F., Levade I., Shapiro B.J., McIntosh F., Soualhine H., Menzies D., Behr M.A. Population genomics of Mycobacterium tuberculosis in the Inuit. Proc Natl Acad Sci. 2015;112:13609–13614. doi: 10.1073/pnas.1507071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roetzer A., Diel R., Kohl T.A., Rückert C., Nübel U., Blom J., Wirth T., Jaenicke S., Schuback S., Rüsch-Gerdes S., Supply P., Kalinowski J., Niemann S. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker T.M., Kohl T.A., Omar S.V., Hedge J., Del Ojo Elias C., Bradley P., Iqbal Z., Feuerriegel S., Niehaus K.E., Wilson D.J., Clifton D.A., Kapatai G., Ip C.L.C., Bowden R., Drobniewski F.A., Allix-Béguec C., Gaudin C., Parkhill J., Diel R., Supply P., Crook D.W., Smith E.G., Walker A.S., Ismail N., Niemann S., Peto T.E.A. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coll F., McNerney R., Guerra-Assunção J.A., Glynn J.R., Perdigão J., Viveiros M., Portugal I., Pain A., Martin N., Clark T.G. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homolka S., Projahn M., Feuerriegel S., Ubben T., Diel R., Nübel U., Niemann S. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankhurst L.J., del Ojo Elias C., Votintseva A.A., Walker T.M., Cole K., Davies J., Fermont J.M., Gascoyne-Binzi D.M., Kohl T.A., Kong C., Lemaitre N., Niemann S., Paul J., Rogers T.R., Roycroft E., Smith E.G., Supply P., Tang P., Wilcox M.H., Wordsworth S., Wyllie D., Xu L., Crook D.W. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med. 2016;4:49–58. doi: 10.1016/S2213-2600(15)00466-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker T.M., Merker M., Knoblauch A.M., Helbling P., Schoch O.D., van der Werf M.J., Kranzer K., Fiebig L., Kröger S., Haas W., Hoffmann H., Indra A., Egli A., Cirillo D.M., Robert J., Rogers T.R., Groenheit R., Mengshoel A.T., Mathys V., Haanperä M., van Soolingen D., Niemann S., Böttger E.C., Keller P.M., Avsar K., Bauer C., Bernasconi E., Borroni E., Brusin S., Coscollá Dévis M., Crook D.W., Dedicoat M., Fitzgibbon M., Gagneux S., Geiger F., Guthmann J.-P., Hendrickx D., Hoffmann-Thiel S., van Ingen J., Jackson S., Jaton K., Lange C., Mazza Stalder J., O'Donnell J., Opota O., Peto T.E.A., Preiswerk B., Roycroft E., Sato M., Schacher R., Schulthess B., Smith E.G., Soini H., Sougakoff W., Tagliani E., Utpatel C., Veziris N., Wagner-Wiening C., Witschi M. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis. 2018;18:431–440. doi: 10.1016/S1473-3099(18)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiden M.C.J., van Rensburg M.J.J., Bray J.E., Earle S.G., Ford S.A., Jolley K.A., McCarthy N.D. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellmann A., Harmsen D., Cummings C.A., Zentz E.B., Leopold S.R., Rico A., Prior K., Szczepanowski R., Ji Y., Zhang W., McLaughlin S.F., Henkhaus J.K., Leopold B., Bielaszewska M., Prager R., Brzoska P.M., Moore R.L., Guenther S., Rothberg J.M., Karch H. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley K.A., Maiden M.C. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl T.A., Diel R., Harmsen D., Rothganger J., Walter K.M., Merker M., Weniger T., Niemann S. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol. 2014;52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker T.M., Merker M., Kohl T.A., Crook D.W., Niemann S., Peto T.E.A. Whole genome sequencing for M/XDR tuberculosis surveillance and for resistance testing. Clin Microbiol Infect. 2017;23:161–166. doi: 10.1016/j.cmi.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Walker T.M., Ip C.L., Harrell R.H., Evans J.T., Kapatai G., Dedicoat M.J., Eyre D.W., Wilson D.J., Hawkey P.M., Crook D.W., Parkhill J., Harris D., Walker A.S., Bowden R., Monk P., Smith E.G., Peto T.E. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comas I., Coscolla M., Luo T., Borrell S., Holt K.E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., Yeboah-Manu D., Bothamley G., Mei J., Wei L., Bentley S., Harris S.R., Niemann S., Diel R., Aseffa A., Gao Q., Young D., Gagneux S. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth T., Hildebrand F., Allix-Béguec C., Wölbeling F., Kubica T., Kremer K., van Soolingen D., Rüsch-Gerdes S., Locht C., Brisse S., Meyer A., Supply P., Niemann S. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diel R., Meywald-Walter K., Gottschalk R., Rüsch-Gerdes S., Niemann S. Ongoing outbreak of tuberculosis in a low-incidence community: a molecular-epidemiological evaluation. Int J Tuberc Lung Dis Off J Int Union Tuberc Lung Dis. 2004;8:855–861. [PubMed] [Google Scholar]
- 24.Kamerbeek J., Schouls L., Kolk A., van Agterveld M., van Soolingen D., Kuijper S., Bunschoten A., Molhuizen H., Shaw R., Goyal M., van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E., Savine E., de Haas P., van Deutekom H., Roring S., Bifani P., Kurepina N., Kreiswirth B., Sola C., Rastogi N., Vatin V., Gutierrez M.C., Fauville M., Niemann S., Skuce R., Kremer K., Locht C., van Soolingen D. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Embden J.D., Cave M.D., Crawford J.T., Dale J.W., Eisenach K.D., Gicquel B., Hermans P., Martin C., McAdam R., Shinnick T.M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blom J., Jakobi T., Doppmeier D., Jaenicke S., Kalinowski J., Stoye J., Goesmann A. Exact and complete short-read alignment to microbial genomes using graphics processing unit programming. Bioinforma Oxf Engl. 2011;27:1351–1358. doi: 10.1093/bioinformatics/btr151. [DOI] [PubMed] [Google Scholar]
- 28.Malm S., Linguissi L.S.G., Tekwu E.M., Vouvoungui J.C., Kohl T.A., Beckert P., Sidibe A., Rüsch-Gerdes S., Madzou-Laboum I.K., Kwedi S., Penlap Beng V., Frank M., Ntoumi F., Niemann S. New Mycobacterium tuberculosis complex sublineage, Brazzaville, Congo. Emerg Infect Dis. 2017;23:423–429. doi: 10.3201/eid2303.160679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiebig L., Kohl T.A., Popovici O., Mühlenfeld M., Indra A., Homorodean D., Chiotan D., Richter E., Rüsch-Gerdes S., Schmidgruber B., Beckert P., Hauer B., Niemann S., Allerberger F., Haas W. A joint cross-border investigation of a cluster of multidrug-resistant tuberculosis in Austria, Romania and Germany in 2014 using classic, genotyping and whole genome sequencing methods: lessons learnt. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.2.30439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diel R., Schneider S., Meywald-Walter K., Ruf C.-M., Rüsch-Gerdes S., Niemann S. Epidemiology of tuberculosis in Hamburg, Germany: long-term population-based analysis applying classical and molecular epidemiological techniques. J Clin Microbiol. 2002;40:532–539. doi: 10.1128/JCM.40.2.532-539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all samples used in the study, indicating the analyses done, ENA SRA accession numbers, phylogenetic classification, and quality metrics of the respective datasets.