The invasive and recurrent nature of glioblastoma multiforme (GBM) is linked to a small subpopulation of cancer cells, which are self-renewing, resistant to standard treatment regimens, and induce formation of new tumors. Matrix stiffness is implicated in the regulation of cell proliferation, drug resistance, and reversion to a more invasive phenotype. Therefore, understanding the relationship between matrix stiffness and tumor cell behavior is vital to develop appropriate in vitro tumor models. Here, we fabricate chitosan-hyaluronic acid (CHA) polyelectrolyte complex (PEC) scaffolds with statistically significant stiffness variances to characterize the effect of scaffold stiffness on morphology, proliferation, drug resistance, and gene expression in human glioblastoma cells (U-87 MG). All scaffolds supported GBM proliferation over a 12-day culture period, yet larger spheroids were observed in scaffolds with higher stiffness. Additionally, GBM cells cultured in stiffer (8% CHA) scaffolds proved significantly more resistant to the common chemotherapeutic temozolomide. Moreover, the stiffer 8% CHA scaffolds exhibited an increase in expression of drug resistance and invasion related genes compared to 2D culture. CHA scaffolds present a tunable microenvironment for enhanced tumor cell malignancy and may provide a valuable in vitro microenvironment for studying tumor progression and screening anti-cancer therapies.
1. Introduction
Glioblastoma multiforme (GBM) is a highly invasive, primary, malignant brain tumor.[1] Clinical treatment generally utilizes tumor resection followed by chemotherapy and radiation therapy; yet median survival time is approximately 15 months.[2] Unlike other solid tumors, GBMs rarely invade the vasculature to metastasize outside of the brain.[3] Instead, high mortality is attributed to the invasion of therapy-resistant cancer cells located away from the primary tumor.[4] This behavior suggests that the local brain microenvironment plays a key role in regulation of GBM progression and recurrence. [3] Studies investigating the correlation between matrix stiffness and GBM malignancy yield disparate in vivo and in vitro results, indicating a critical lack in understanding of GBM progression. Development of novel therapeutics also remains challenging because the response of drug candidates differs among in vitro cell lines, animal tumor models, and human tumors. Therefore, to improve our understanding of tumor biology and to develop novel therapeutics for complete GBM eradication, new tools are required to model GBM tumor progression.
Malignant cancer cell behavior is highly influenced by biochemical and biomechanical cues in the surrounding microenvironment. Studies show that tumor microenvironment biomechanics play a significant role in cancer progression including tumor invasion and metastasis, and the reversion of tumor cells into cancer stem cells (CSCs).[5] The tumor extracellular matrix (ECM) is often stiffer than the ECM of surrounding stroma, as is the case for GBM and studies indicate that increasing matrix stiffness induces the epithelial-to-mesenchymal transition (EMT).[6] EMT plays a significant role in tumor formation and metastasis[7] and is often linked to a more malignant cell phenotype. [5a, 5b, 8] Therefore matrix elasticity of in vitro platforms for GBM cell culture should be tunable to encompass both healthy and cancerous tissue in order to study how differences in elasticity affect cell behavior and tumor progression of GBM.
Evidence that matrix stiffness regulates multiple characteristics of cancer cells has led to many studies of matrix stiffness and corresponding cell behavior in 2D cultures. Two-dimensional platforms, such as tissue culture polystyrene (TCPS), are effective in promoting GBM cell proliferation when used for monolayer culture, but do not adequately mimic in vivo tumor environments. More complex, spherical cancer models such as non-adherent cancer cell line-derived spheroids, or spheroids derived from primary tumor dissociation, promote cell-cell interactions and are frequently used for cancer cell culture. Importantly, non-adherent cultures lack the cell-matrix interactions present in native tumor stroma.[9] Three-dimensional culture platforms, such as hydrogels or scaffolds, promote both cell-cell and cell-matrix interactions and present a diffusion-limited environment not found in 2D cultures. This can result in enrichment of tumor-derived cells with a more malignant, adherent spheroid morphology.[10] Many three-dimensional (3D) environments for cancer cell culture are fibrillar protein-based hydrogels,[11] which do not reflect the glycosaminoglycan (GAG)-based brain ECM. Further, in animal-derived hydrogels, batch-to-batch variability and compositional complexity pose problems.[10g, 12] More recently, studies utilizing hyaluronic acid (HA)-based hydrogels have elucidated many important aspects of GBM CSC phenotype, motility, and behavior. Yet HA gels are mechanically weak, thus control over matrix stiffness and tumor sphere formation is challenging.
Here we present the fabrication and characterization of HA-based, porous scaffolds as potential in vitro platforms for modeling the GBM microenvironment. The porous chitosan-hyaluronic acid (CHA) scaffolds of varied stiffness were fabricated using a simple phase separation method, providing an alternative to hydrogels. HA, an anionic natural polymer is a major GAG component of the brain ECM.[13] Chitosan, a naturally occurring polysaccharide with a structure similar to GAGs, is cationic and biocompatible.[14] When blended together, chitosan and HA form a stable, polyelectrolyte complex (PEC) enhancing the stability of both natural polymers. Changing the total polymer concentration of CHA scaffolds by varying the chitosan content alters the scaffold microstructure and stiffness in a range encompassing the mechanical properties of normal brain tissue to malignant gliomas. Material properties of CHA scaffolds were characterized including stiffness, density, porosity, polyelectrolyte composition, and swelling behavior. U-87 MG RFP GBM cells were cultured in CHA and on TCPS to compare and evaluate cell morphology, proliferation, dose-dependent drug response, and gene expression.
2. Results and Discussion
2.1. CHA scaffold structural and chemical properties
Brain ECM, unlike fibrillar protein-rich ECM of other tissues, is predominantly composed of hyaluronic acid (HA), one of major glycosaminoglycan (GAG) components.[15] The structure and physical cues of brain ECM are implicated in the induction of highly aggressive cancer cell behavior resulting in metastatic characteristics unlike those of other solid tumors.[16] HA is highly prevalent in glioblastomas where GBM cells overexpress HA receptors, CD44 and RHAMM.[13, 17] Moreover, the presence of HA in the tumor stroma is associated with enhanced tumor growth and progression.[13] Therefore a 3D culture platform that mimics the properties of the GBM microenvironment should contain HA. Pure HA scaffolds are not suitable for this purpose because of their insufficient mechanical strength and failure to promote cell adhesion due to their anionic nature.[18] Chitosan, a naturally occurring polysaccharide with a chemical structure similar to GAGs, is cationic and biocompatible.[14] When blended together, chitosan and HA form a stable, polyelectrolyte complex (PEC) combining the positive attributes of both materials representing a rational material system to recapitulate brain ECM for in vitro GBM cell culture. Fabrication of chitosan-hyaluronic acid (CHA) PEC scaffolds can be accomplished via thermally-induced phase separation (TIPS) of an aqueous phase and polymer-rich phase followed by lyophilization resulting in a highly porous structure amenable to cell culture.
As summarized in Table 1, the hyaluronic acid content of PEC scaffolds remained constant (1 wt%) whereas the chitosan concentration was varied from 2 wt% to 8 wt%. With 1 wt % HA in all scaffolds, the overall scaffold polymer content was 1.5 wt% (“2% CHA” scaffold), 2.5 wt% (“4% CHA” scaffold) or 4.5 wt% (“8% CHA” scaffold).
Table 1.
Summary of CHA scaffold naming convention and polymer content. The scaffold naming is based on chitosan content in solution. For all scaffolds the HA content was 1 wt%.
| CHA scaffold | Chitosan solution polymer content | HA solution polymer content | Combined CHA solution polymer content |
|---|---|---|---|
| 2% CHA | 2 wt% | 1 wt% | 1.5 wt% CHA |
| 4% CHA | 4 wt% | 1 wt% | 2.5 wt% CHA |
| 8% CHA | 8 wt% | 1 wt% | 4.5 wt% CHA |
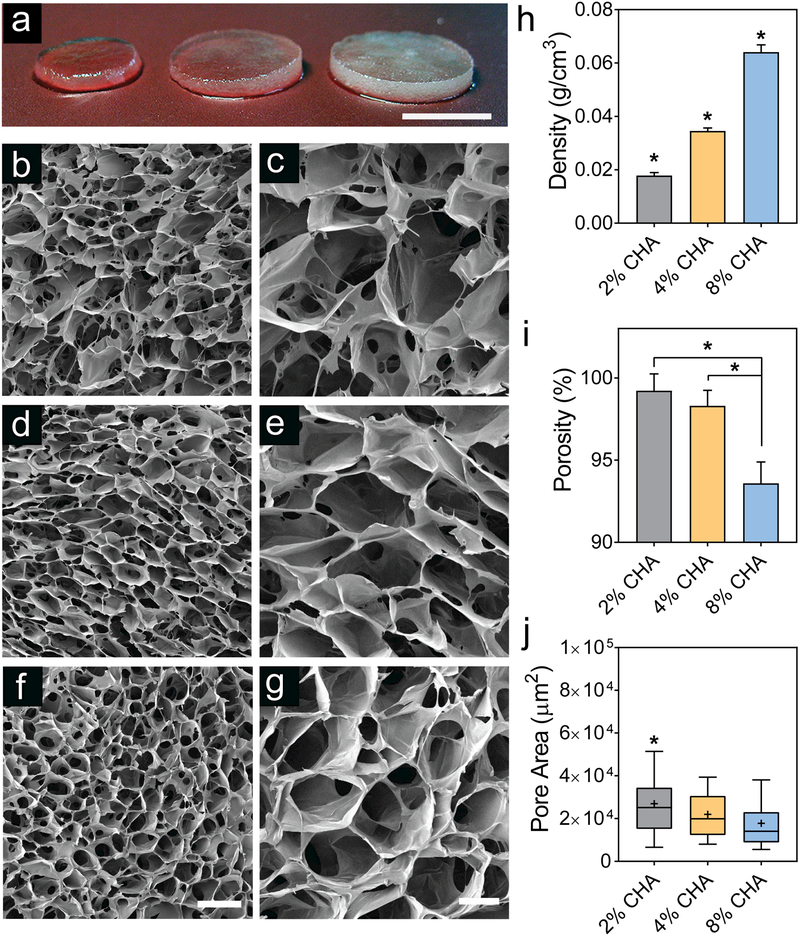
Figure 1a displays hydrated CHA scaffolds where 2% CHA appears most translucent due to low polymer content. Figure 1b-g depicts the scaffold microstructure as observed using scanning electron microscopy (SEM). CHA scaffolds of all compositions are highly porous with interconnected pores. The formation of a porous structure is attributed to the phase separation of the aqueous solvent and PEC, followed by solvent crystal nucleation and growth during the freezing step. The open porous structure remains following sublimation during lyophilization. The 2% CHA scaffold (Figure 1b, c) displays a more bimodal pore distribution with small pores embedded in larger pores, forming interconnections between adjacent pores. As the polymer content increased to 8% CHA (Figure 1f, g), the presence of these small interconnections decreased due to the increase in solution viscosity. The viscosity of the higher polymer content solution hinders dendritic ice crystal formation thereby decreasing the interconnections between adjacent walls of the pore structure.[19] Figure 1h summarizes the apparent density of CHA scaffolds, which increased with increasing polymer concentration and is significantly different among all scaffold compositions (p ≤ 0.05). By visual examination, hydrated 8% CHA scaffolds were more opaque than 4% and 2% CHA scaffolds (Figure 1a). This trend of increasing density with increasing polymer concentration was expected as more polymer is present per scaffold volume.
Figure 1.
Physical properties of CHA scaffolds. (a) Macroscale photograph of D-PBS-hydrated CHA scaffolds (left to right: 2% CHA, 4% CHA, 8% CHA). Scanning electron micrographs of (b-c)_2% CHA, (d-e) 4% CHA, and (f-g) 8% CHA scaffolds depicting a highly porous microstructure. Scale bars represent (a) 4 mm, (b, d, f) 300 μm, and (c, e, g) 100 μm. (h) Apparent density of CHA scaffolds presented as mean ± SD (n = 8). (i) Total scaffold porosity presented as mean ± SD (n = 12). (j) Pore area of CHA scaffolds (n ≥ 90) where “+” denotes mean pore size. P-values were calculated using one-way ANOVA where (*) indicates a significant difference from all other conditions (p ≤ 0.05).
Liquid displacement was utilized to measure bulk scaffold porosity, which was greater than 90% for all scaffold compositions (Figure 1i) thereby confirming the presence of interconnected pores. Importantly, an open, interconnected pore structure is critical for maintenance of healthy cells in vitro by allowing for diffusion of nutrients, oxygen, and waste. The porosity of 8% CHA scaffolds (94%) is significantly lower than that of 2% CHA (99%) and 4% CHA (98%) scaffolds (p ≤ 0.05), which corresponds to higher polymer content, thicker pore walls, and decreased interconnections between pores.
The pore diameter associated with the different scaffold compositions was estimated and compared by measuring the area of pore cross-sections from SEM micrographs. Figure 1j presents the mean (denoted by “+”), median, interquartile range, and 10/90 margin of the pore size distribution for each scaffold composition. The median pore size, in terms of cross-sectional area, was 2.51 × 104 μm2, 1.99 × 104 μm2, and 1.41 × 104 μm2 for the 2%, 4%, and 8% CHA scaffolds, respectively, with the 2% CHA scaffold containing significantly larger pores (p ≤ 0.05). The CHA scaffold pores were not perfectly spherical thereby precluding direct measure of pore cross-sectional diameter. For reference, if the average pore areas reported above represented circular pore cross-sections, the corresponding pore diameters would be approximately 179 μm (2% CHA), 159 μm (4% CHA) and 134 μm (8% CHA). These values fall in the range of relevant pore sizes for porous scaffolds useful for in vitro cell culture.[20] Overall, there was a trend of decreasing pore size associated with increasing polymer content.
2.1.1. FTIR analysis of the CHA polyelectrolyte complex
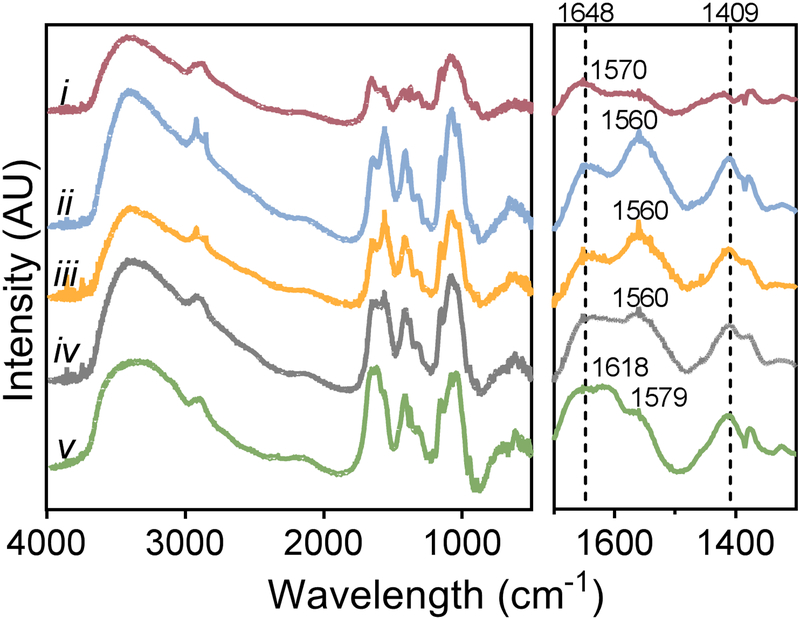
The CHA scaffolds were analyzed using FTIR to confirm the formation of a CHA polyelectrolyte complex after blending individual polymer solutions (Figure 2). In the range of 2800–3500 cm−1, two broad absorption bands characteristic of polysaccharide structures are visible for all five spectra. These bands represent the O-H stretching vibration (3000–3700 cm−1) and the C-H stretching vibration (2800–3000 cm−1).[21] In the fingerprint regions of the spectra (Figure 2, right panel), amide and carbonyl vibrations result in several distinctive bands between 1500 cm−1 and 1700 cm−1. The chitosan spectrum (Figure 2i) displays two characteristic bands: a prominent amide I band (C=O vibration) at 1653 cm−1 and the overlapped band of amid II and the N-H bending vibration of the deacetylated amine groups at 1570 cm−1. [10a] The carbonyl stretching vibration region of the HA spectra (Figure 2v) (1500–1800 cm−1) reveals three absorption bands where the highest intensity peak at 1618 cm−1 is derived from the asymmetric stretching vibration of the carbonyl group (COO-).[21] Additionally, the carbonyl band overlaps an amide I shoulder (-C=O- stretching of carboxylic acid groups) at 1648 cm−1 and an amide II band at 1579 cm−1.
Figure 2.
Chemical analysis confirming CHA polyelectrolyte complex (PEC) in scaffolds. Fourier Transform Infrared Spectroscopy (FTIR) analysis of (i) pure chitosan, (ii) 8 wt%, (iii) 4 wt%, and (iv) 2 wt% CHA PEC scaffolds, and (v) pure hyaluronic acid.
In the spectra representing CHA blends, Figure 2(ii-iv), the amide II band intensifies and shifts to lower wavelengths (~1560 cm−1) when compared to the pure chitosan or pure HA spectra. The amide I band (~1648 cm−1), prominently displayed in the pure chitosan, is more pronounced in CHA blends, but presents as a small shoulder to the carbonyl stretching vibration band in pure HA. Additionally, a prominent peak in HA at 1409 cm−1 is mirrored in all CHA blends with a similar high intensity, but the peak intensity is lower in the pure chitosan. These peak shifts and changes in band intensities suggest the formation of a PEC between the positive amino groups in chitosan and the negative carboxyl groups in HA.[10a]
2.1.2. Mechanical properties of CHA scaffolds
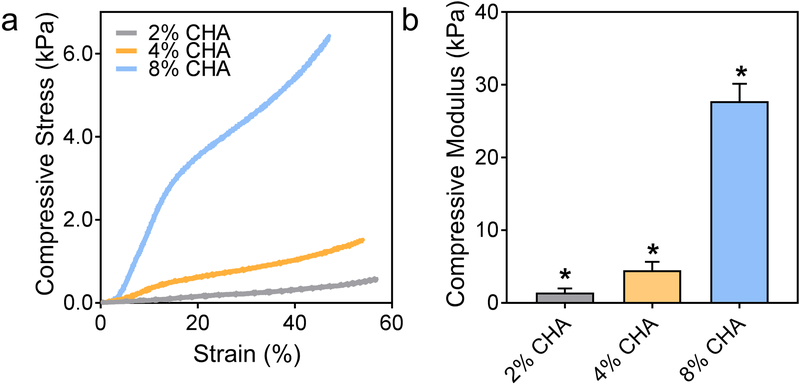
CHA scaffold stiffness was determined via mechanical testing under compression of hydrated scaffolds. Figure 3a displays representative compressive stress-strain curves for hydrated CHA scaffolds of varying polymer composition. As the polymer concentration and density of the scaffolds increase, the characteristic regimes for compressing polymer foams are more pronounced and are best depicted by the 8% CHA scaffold. Here, the scaffold initially experiences linear elastic deformation followed by a slight plateau where the stress is less affected by each successive increase in strain, and finally densification, where the pore walls collapse, and the polymer is compressed as a bulk solid.[22] The differing densities among the scaffolds are apparent during the densification phase where stress loading on the scaffold increases with polymer content. Figure 3b shows that the compressive Young’s modulus of fully hydrated CHA scaffolds increases with increasing polymer content from approximately 1.41 kPa for 2% CHA to 27.7 kPa for 8% CHA. The compressive moduli of all scaffolds are significantly different (p ≤ 0.05), and the moduli range is biologically relevant. Native brain tissue has a compressive modulus in the range of ~0.2–1 kPa[23] and glioma tissue is thought to be stiffer although conclusive values have not been determined due to the challenges associated with procuring test samples of relevant size.[23b, 24] Netti et al reported a compressive modulus of 26 kPa for tumors generated from human GBM cells using a mouse xenograft model.[25]
Figure 3.
Increasing compressive stiffness with increasing polymer content in hydrated CHA scaffolds. (a) Representative compressive stress-strain curves of hydrated CHA scaffolds and (b) mean compressive moduli where the moduli of all scaffolds are significantly different (n = 8). (*) indicates significant difference from all conditions (p ≤ 0.05).
2.2. Proliferation and morphology response of GBM to varied CHA scaffold stiffness
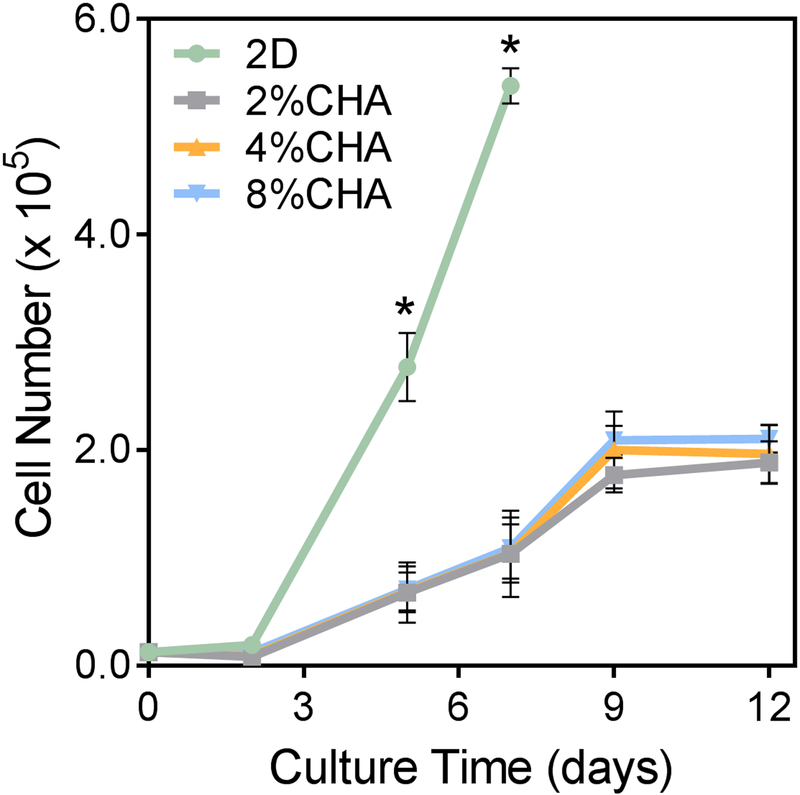
GBM cells were cultured in vitro for 12 days to evaluate cell proliferation in response to the CHA scaffold microenvironment at varying polymer contents. The growth kinetics of U-87 MG RFP cells cultured on 2D TCPS and CHA scaffolds were quantified by measuring cell metabolic activity as a function of fluorescence intensity (Figure 4). The proliferation rate increased significantly (p≤ 0.05) for 2D monolayer cells cultured on TCPS, reaching confluence by day 7. The initiation of substantial cell proliferation did not appear until day 3 for all scaffold conditions. Increasing cell number with culture time was observed for all scaffolds, but no statistical difference in cell number among CHA conditions was observed. Diffusion limitations and the high surface area in 3D systems have been cited as reasons for differences in 2D and 3D culture proliferation rates.[26] Additionally, the growth rates in 3D culture in the presence of increased cell-cell and cell-matrix interactions may be more indicative of native cell behavior.[20a, 20c]
Figure 4.
Growth kinetics of U-87 MG RFP cells cultured on 2D and CHA scaffolds using the alamarBlue® metabolic assay (n = 4). (*) Proliferation in the 2D culture condition was significantly higher than all 3D conditions (p ≤ 0.05).
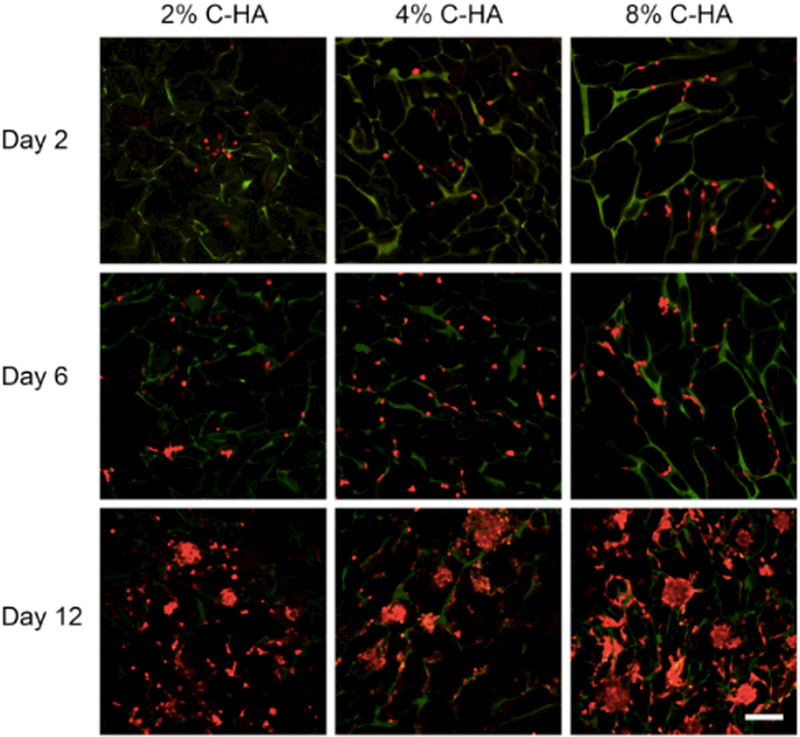
The influence of CHA scaffold stiffness on cell morphology was evaluated using confocal imaging of the red fluorescent protein-expressing U-87 MG cells (U-87 MG RFP) at day 2, 6, and 12 (Figure 5). At day 2, single cells attached to the pore walls were observed on all scaffolds. On day 6, a higher density of single cells and some small cell aggregates formed within 8% CHA scaffolds. By day 12, tumor spheroids were visible on all scaffolds. Cell aggregates on 2% CHA scaffolds were smaller than the spheroids present on 4% and 8% CHA scaffolds. The highest density of tumor spheroids was seen on the stiffest CHA scaffold (8%). Differences in spheroid size and density were not reflected in the metabolic assay with all CHA scaffolds exhibiting similar growth curves. This may be due to diffusion limitations such that the metabolic activity measured is limited to cells on the outer surface of spheroids. Tumor sphere morphology is linked to a more clinically-relevant GBM behavior and often yields tumors with enhanced malignant potential[26], indicating that the 4% and 8% CHA scaffolds may be well suited for use as in vitro GBM tumor models.
Figure 5.
Fluorescence imaging of U87-MG RFP cells (red) progressing from single cells to spheroids after 12 days when cultured on 2%, 4% and 8% CHA scaffolds. CHA scaffolds are visible (green) due to autofluorescence. The scale bar represents 300 μm.
2.3. Drug response of GBM cells in CHA scaffolds with various stiffness
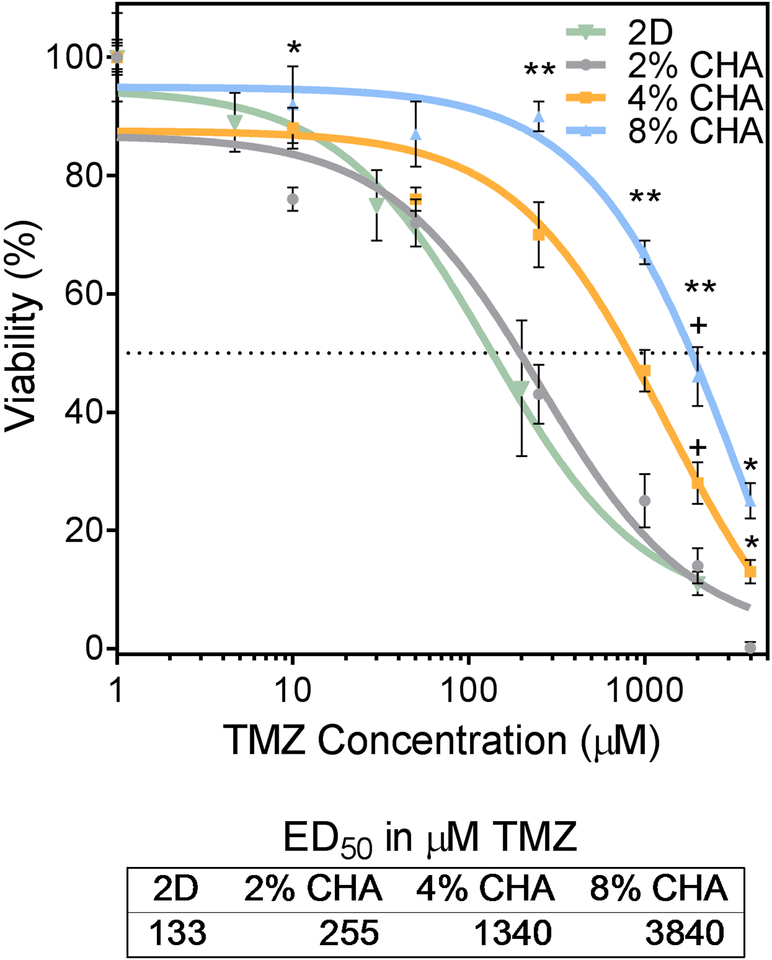
GBM cells cultured on 2D TCPS and CHA scaffolds for 12 days were treated with a common cancer chemotherapeutic, temozolomide (TMZ), and the drug response was assessed. Cells were exposed to doses of TMZ ranging from 0–4000 μM for 72 hours before viability was evaluated with alamarBlue® (Figure 6). GBM cells in CHA scaffolds displayed increased drug resistance relative to those on 2D for TMZ doses greater than 40 μM. The TMZ ED50 for cells cultured in 2D (133 μM) is consistent with similar studies reporting 2D culture of U-87 cells.[27] The highest cell viability and thus greatest resistance to TMZ-induced cell death, was associated with cells cultured in 8% CHA scaffolds. Increased drug resistance of cells cultured in CHA scaffolds may be the result of diffusion gradients, where the TMZ must penetrate the porous scaffold to reach the cells. This is in contrast to 2D culture conditions where all cells are equally exposed. Cells cultured in 8% CHA exhibited a significant increase in resistance to TMZ-induced cell death, as compared with other 3D CHA cultures (p≤ 0.05). This may reflect diffusion gradients within individual tumor spheres, where TMZ was unable to reach cells localized at the core.
Figure 6.
Dose dependent response of U87-MG RFP cells after 12 days of culture and 72 hrs after TMZ exposure. CHA scaffolds containing 3D tumor spheroids showed increased ED50 relative to 2D culture signifying increased drug resistance. Viability was determined using the alamarBlue® assay (n = 4). (*) indicates significant difference from 2% CHA condition (p ≤ 0.5). (**) indicates all scaffolds are significantly different from each other (p ≤ 0.5). (+) indicates significant difference from 2D condition (p ≤ 0.5).
2.4. Gene expression of GBM cells in CHA scaffolds with various stiffness
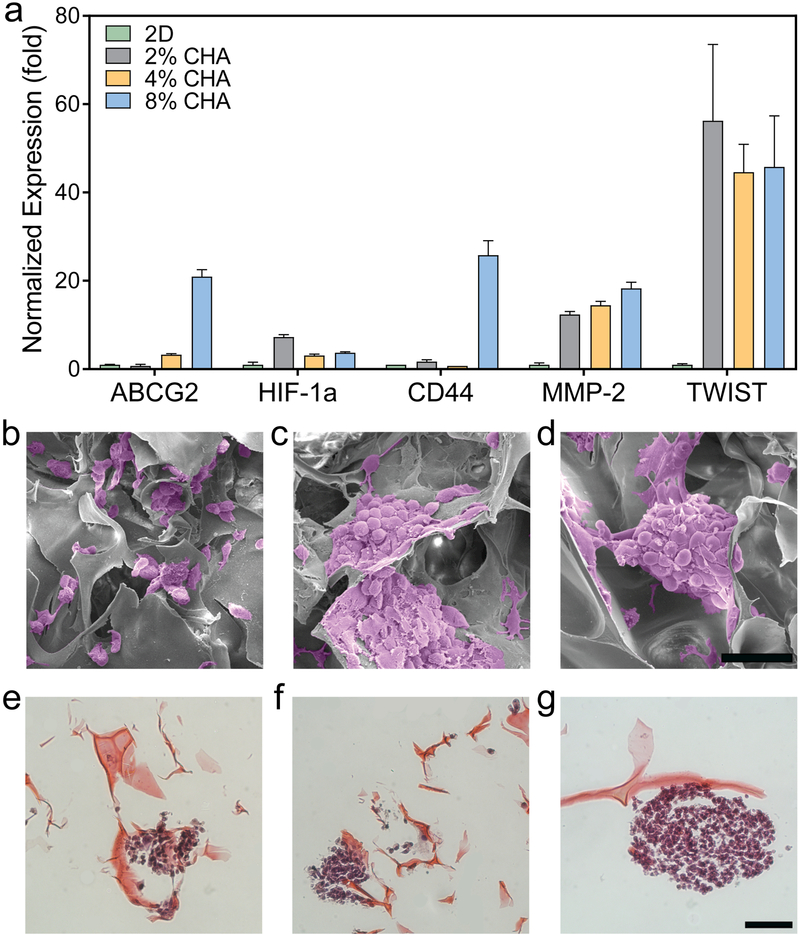
To better understand the influence of CHA scaffolds on GBM cell characteristics and behavior, the expression profile of a subset of relevant genes was assessed. Markers of chemoresistance (ABCG2), hypoxia (HIF-1α), and invasion (CD44, MMP-2, and TWIST1) were analyzed using quantitative real-time PCR (qRT-PCR) (Figure 7a) where, in general, upregulation of gene expression was observed for cells cultured on stiffer CHA scaffolds. Here, expression of ABCG2, a member of the ATP-binding cassette (ABC) transporter superfamily, was upregulated in cells cultured on 4% CHA (3.3 ± 0.23 fold) and 8% CHA (21.0 ± 1.6 fold) scaffolds relative to 2% CHA cultures. In glioma, post-treatment tumor recurrence has been linked to a subpopulation of cells that express chemotherapeutic drug resistance. A potential mechanism of this resistance is increased drug efflux, leading to increased cell survival.[28] ABCG2 functions as an efflux pump for chemotherapeutic drugs like TMZ and its increased expression has been implicated in cell self-renewal and poor patient prognosis.[29]
Figure 7.
Gene expression and morphology of cells cultured on different substrates. (a) Relative expression displaying increased drug resistance and invasion specific genes in 3D scaffolds by U87-MG RFP cells after 12 days of culture (n = 3). Error bars represent standard error of mean. Scanning electron micrographs of GBM cells in (b) 2% CHA, (c) 4% CHA, and (d) 8% CHA where tumor spheroid size increases with increasing stiffness. Histological staining (H&E) of tumor spheroids on (e) 2% CHA, (f) 4% CHA, and (g) 8% CHA scaffolds. Scale bars represent 100 μm (b-d) and 50 μm (e-g).
Cellular response to hypoxia, or insufficient tissue oxygenation, is mediated by the hypoxia-inducible factors (HIF) transcription factor family and contributes to aggressive tumor phenotypes, drug resistance, and poor prognosis.[30] The HIF-1α protein is overexpressed in a variety of common solid tumors.[31] As expected, in a 3D microenvironment containing diffusion gradients, expression of HIF-1α was upregulated and was 3-fold in all CHA scaffolds than in 2D cultures (Figure 7a). Expression of genes regulated by HIFs is implicated in influencing many of the cancer “hallmarks” such as unchecked proliferation, apoptosis, invasion, metastasis, and appear to contribute to increased resistance to chemotherapy and radiation.[30–31]
Cell invasion in GBM, a key factor in tumor recurrence and poor prognosis, is driven by cell motility, ECM degradation, and the epithelial to mesenchymal transition (EMT). Cell motility in the extracellular matrix is promoted by CD44, a principal cell surface hyaluronic acid receptor that plays a key role in cell adhesion. [32] When cultured for 12 days in 8% CHA scaffolds, GBM cells displayed an increase in CD44 expression (25.8 ± 3.3 fold) relative to those on the 2D control (Figure 7a). The CD44 signaling cascade in glioma is an important driver of tumor invasion[33] and upregulation of CD44 here was likely a response to the HA content of the scaffold indicating increased cell motility.
Matrix metalloproteinase (MMP) expression is upregulated when invasion occurs via ECM degradation and specifically, expression of MMP-2 is considered a marker of GBM cell invasiveness.[34] MMP-2 expression was upregulated (>12 fold) for all CHA scaffolds relative to the 2D control indicating increased cell-matrix interactions. TWIST1 expression is also frequently upregulated in gliomas with elevated expression of MMP-2 and cell invasion genes. TWIST1 is considered an essential therapeutic target in GBM because the TWIST transcription factors act to repress cell-matrix adhesion, a hallmark of a more invasive cancer cell phenotype.[35] Relative to 2D monolayer culture, TWIST1 expression by GBM cells cultured in all CHA scaffolds was upregulated (> 45 fold) (Figure 7a). TWIST1 expression in 2% CHA scaffolds was slightly higher than those in 4% and 8% CHA scaffolds. This may be attributed to increased cell-matrix interactions in 2% CHA scaffolds due to the presence of single cells and small cell clusters. Importantly, upregulation of markers implicated in increased chemoresistance, invasion, and EMT provides further evidence that considering the biochemical and biomechanical properties 8% CHA scaffolds may be an excellent in vitro model for study of GBM pathogenesis and drug interactions.
Further characterization of cell and tumor spheroid morphology was completed with SEM and histological staining. Scanning electron micrographs (Figure 7b-d) show the presence of tumor spheroids within the CHA scaffolds. This confirms the morphology observed using fluorescence microscopy and indicates that formation of GBM cell tumor spheroids is not limited to the scaffold surface. GBM cells present within 2% CHA scaffolds (Figure 7b) appear to form multicellular aggregates surrounded by disperse single cells that do not resemble the spheroids observed in 4% and 8% CHA scaffolds (Figure 7c and 7d, respectively). To quantify, the area of at least 6 tumor spheroids were measured using ImageJ. Although the spheroids are not perfectly spherical, if the area of each spheroid corresponds to the area of a perfect sphere, then the diameter of tumor spheroids in the 2% CHA, 4% CHA, and 8% CHA would be 73 μm, 111 μm, and 110 μm, respectively, which confirms spheroids in the 2% CHA are smaller than those in the 4% and 8% CHA scaffolds. The SEM and confocal microscopy (Figure 5) results were further confirmed by histological staining (Figure 7e-g), again showing tumor spheroids with increasing diameter as polymer content and scaffold stiffness increased. Overall, the formation of cell aggregates and tumor spheroids within CHA scaffolds indicates that the 3D scaffolds provide a more biomimetic microenvironment than 2D TCPS likely by allowing cells to form more cell-cell and cell-ECM contacts. The dramatic difference in GBM cell morphology between monolayer culture and CHA scaffold culture may indicate a more primitive cancer cell phenotype in the latter.
3. Conclusions
Chitosan-hyaluronic acid PEC scaffolds were fabricated with varying chitosan content leading to significant differences in scaffold total density, porosity, microstructure, and compressive modulus. Overall, an increase in polymer content led to a higher apparent density, lower total porosity, smaller pore area, and higher compressive modulus. The scaffolds characterized in this work exhibited >90% total porosity, pores on the order of 100–200 μm in diameter and compressive moduli in a range that encompasses native brain tissue to tumor tissue. When scaffolds were utilized for culture of U-87 MG RFP cells, cells formed small, irregularly-shaped aggregates on 2% CHA scaffolds and tumor spheroids in 4% and 8% CHA scaffolds. Differences in cell morphology and proliferation among the CHA scaffold conditions examined suggest an effect of matrix stiffness on GBM cells in vitro. The 8% CHA scaffold formed larger tumor spheres with increased drug resistance, and elevated expression of drug resistance, hypoxia, and invasion-related genes. CHA scaffolds represent a useful tool for in vitro culture of GBM cells in a spheroid morphology allowing modeling of the relationship between GBM cells and the surrounding tumor microenvironment. The ability to tune matrix stiffness allows for extended investigation of cancer cell behavior during tumorigenesis providing a tool to target and study therapy effectiveness at different stages of cancer progression.
4. Experimental Section
Materials:
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Chitosan (from shrimp shells, practical grade, > 75% deacetylated) and hyaluronic acid (hyaluronic acid sodium salt, from Streptococcus equi) were used as received.
Preparation of chitosan-hyaluronic acid scaffolds:
Chitosan and hyaluronic acid (CHA) blend scaffolds were prepared by first dissolving chitosan (2, 4, or 8 wt%) and hyaluronic acid (1 wt%) separately in 1 wt% acetic acid solution. The solutions were individually mixed for three minutes using a Thinky mixer (ARM-300, Thinky USA, Laguna Hills, CA) at 2000 rpm. Mixing was repeated at least three times to ensure each solution was completely dissolved. The solutions were aged overnight at room temperature to ensure complete dissolution. After aging, the chitosan and hyaluronic acid solutions were mixed again individually for 3 minutes at 2000 rpm. The solutions were combined and mixed three times with the Thinky prior to casting into 24-well tissue culture plates. The plates were refrigerated at 4ºC for 12 hours, frozen at −20°C for 24 hours, and lyophilized for 36 hours with a Labconco Freezone 6 Plus freeze drier (Labconco, Kansas City, MO). The scaffolds were sectioned to obtain 2 mm thick discs, neutralized in 15 vol% ammonium hydroxide for 1 hour under vacuum, washed four times with DI water, and allowed to soak in D-PBS overnight to ensure the removal of any residual base. Neutralized scaffolds were sterilized with 70% ethanol for 1 hour under vacuum, transferred to sterile D-PBS, and placed on an orbital shaker overnight prior to in vitro cell culture experiments. Scaffold naming convention and corresponding polymer content (2% CHA, 4% CHA, 8% CHA) are summarized in Table 1.
Scanning electron microscopy:
Scanning electron microscopy (SEM) was utilized to visualize scaffolds before and after cell culture. Prior to cell culture, dry scaffolds were mounted directly on stubs using carbon tape. After cell culture, samples were fixed with 10% formalin at 4ºC overnight, dehydrated through an ethanol series (0%, 30%, 50% 70%, 90%, 100% ethanol), critical point dried with a Denton DCP-1 critical point dryer (Denton Vacuum, Moorestown, NJ), sectioned, and mounted on stubs using carbon tape. All samples were sputter coated with Au/Pd for 90 s before imaging with a FEI Sirion XL30 Field Emission SEM (FEI, Hillsboro, OR).
Fourier Transform Infrared Spectroscopy:
The interaction between chitosan and hyaluronic acid in forming polyelectrolyte complexes was characterized using Fourier transform infrared spectroscopy (FTIR) and compared to spectra of pure chitosan and pure hyaluronic acid samples. Scaffolds of each blend were sectioned, ground into a powder with KBr and pressed into pellets. Spectra of 64 scans at 2 cm−1 resolution were obtained using a Nicolet 5DX spectrometer.
Scaffold density:
The apparent bulk density of scaffolds was calculated after measuring individual scaffold mass and volume (n = 8). Scaffold diameters were measured using a micrometer to calculate scaffold volume and mass measured to the ten-thousandth of a gram.
Scaffold porosity:
The porosity of CHA scaffolds was measured using a modified liquid displacement method.[36] Isopropanol was utilized as the displacement liquid as it is a nonsolvent and easily penetrates the porous structure with minimal volume change. Briefly, the dry scaffold volume (Vi) and weight (Wi) were recorded. The scaffold was fully immersed in 5 mL of isopropanol of known density (ρi) for 15 minutes during which time the scaffold stopped releasing air bubbles and sank to the bottom of the container. The impregnated scaffold was removed from the isopropanol and weighed (Wf). Using Equation 1, the porosity was calculated as a ratio of volume of solvent within the scaffold pores to the volume of the dry scaffold (n = 12).
| (1) |
Scaffold pore size:
The individual pore area was measured from three representative SEM micrographs using ImageJ software. Three separate SEM micrographs were used per scaffold condition to determine median pore area. To ensure consistency in selection of measured pores, two intersecting lines were overlaid on each micrograph. One line was placed from the top left corner to the bottom right corner of the image. A second line was placed extending from the top right corner to the bottom left corner resulting in an “X”. Pores intersecting the lines were outlined using the freehand selection function in ImageJ and the area measured. At least 90 pores were measured per scaffold condition.
Mechanical testing:
The stress versus strain behavior of D-PBS-hydrated scaffolds under compression was measured at room temperature using a Shimadzu universal tester (AGS-X Series, Kyoto, Japan). Scaffolds of 2 mm in height and 17 mm in diameter were compressed at a rate of 0.4 mm/min until at least 40% strain was attained (n = 8). The compressive modulus was calculated as the slope of the linear region of the stress-strain curve (0–10% strain).
Cell culture:
The human glioblastoma cell line, U-87 MG, was purchased from American Type Culture Collection (Manassas, VA). Cells were transfected with pRFP-N2 using Lipofectamine 2000 reagent according to manufacturer’s instructions. Forty-eight hours after transfection, the cells were washed with D-PBS and cultured in fresh media containing G418 (500 μg/mL) for selection of the stably transfected population. Two weeks after selection, U-87 MG RFP cells were sorted by fluorescence activated cell sorting (FACS; Vantage SE). Cells were maintained in Minimum Essential Medium (MEM) (Thermo Fisher Scientific, Grand Island, NY) containing 10% FBS and 1% antibiotic-antimycotic at 37°C and 5% CO2 in a humidified environment. U-87 MG RFP cells (37,500 cells at 1.25 × 106 cells/mL) were seeded onto scaffolds and maintained for 2 hours in an incubator for cell attachment. Following the attachment period, fully supplemented media were added, and cells cultured for 12 days with regular medium changes. Cells were imaged on days 2, 6 and 12 with a Leica SP8X confocal microscopy (Leica, Buffalo Grove, IL).
Proliferation analysis:
Growth kinetics of U-87 MG RFP cells cultured on CHA scaffolds and 2D tissue culture polystyrene (TCPS) were determined using the alamarBlue® metabolic assay and following the manufacturer’s protocol (Life Technologies). Briefly, cells (37,500) were cultured on TCPS or CHA scaffolds in a 24-well plate for 3, 6, 9, or 12 days. An alamarBlue® working solution (10% Resazurin and 90% fully supplemented medium) was then added to each well and incubated at 37°C for a predetermined time (2 hrs for 2D and 4 hrs for CHA scaffolds). Then aliquots of the alamarBlue® solution were transferred to a black-bottom 96-well plate and the fluorescence intensity was measured on a microplate reader. The cell number was determined using standard curves. Cells were rinsed with D-PBS to remove residual alamarBlue® solution and the fully supplemented medium was added to each well for further culture.
Drug response analysis:
For 3, 6, or 12 days U-87 MG RFP cells were grown in 2D and CHA scaffolds. The scaffolds were then treated with temozolomide (TMZ) for 24 hrs. After 24 hrs, the medium containing the drug was replaced with fresh medium. Cell viability was examined 24, 48, or 72 hrs after treatment using the alamarBlue® assay as described previously. Viability was reported as a percent of viable cells relative to an untreated control.
PCR:
Cells were detached from TCPS or the CHA scaffolds using TripLE and RNA was extracted using the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. An iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) was used for reverse transcription (RT) to prepare cDNA following the manufacturer’s instructions. DNA transcripts were probed using SsAdvanced Universal SYBR Green Supermix (Bio-Rad) with the primers listed in Table 2. Thermocycling was performed in 10 μL solution containing 5 μL SYBR Superrmix, 300 nM primers (Integrated DNA Technologies, Coralville, IA), and cDNA at concentration 4 ng/μL. The thermocycle was performed on BioRad CFX96 System at 95°C for 2 min, 40 cycles at 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s. All qRT-PCR data was analyzed with the CFX Manager software (Bio-Rad) with expression levels normalized to GAPDH and standard error of mean calculated.
Table 2.
Primers used for PCR to evaluate chemoresistance (ABCG2), hypoxia (HIF-1α), and invasion (CD44, MMP-2, and TWIST1). Expression levels were normalized to GAPDH.
| Target | Forward (5’−3’) | Reverse (5’−3’) |
|---|---|---|
| GAPDH | GGT GTG AAC CAT GAG AAG TAT GA | GAG TCC TTC CAC GAT ACC AAA G |
| ABCG2 | GTC GGT GTG CGA GTC AGG GC | CTT GCC TCC GCC TGT GGG TC |
| CD44 | AAC GCT TCA GCC TAC TGC AAA | TCT TCC AAG CCT TCA TGT GAT G |
| MMP-2 | GAG TTG GCA GTG CAA TAC CT | GCC GTC CTT CTC AAA GTT GT |
| TWIST1 | AAT CGA GGT GGA CTG GGA AC | CTT ACG AGG AGC TGC AGA CG |
Statistical analysis:
Results are presented as mean values ± standard deviation unless otherwise specified. Box plot whiskers in Figure 1 represent the 10 and 90 percentile with the “+” denoting the mean. The drug response profiles were fitted using a nonlinear three parameter dose response curve in GraphPad Prism 7 (Prism version 7.04, Graph Pad Software, San Diego, CA). Statistical significance was determined using one-way or two-way analysis of variance (ANOVA) followed by post-hoc Tukey’s test for multiple comparisons conducted using GraphPad. Values were considered statistically significant at p ≤ 0.05.
Porous scaffolds composed of chitosan and hyaluronic acid (CHA) can support the growth and proliferation of glioblastoma cells. CHA scaffolds of varying stiffness regulate cell morphology changes from small cell aggregates to tumor spheroids on scaffolds with stiffnesses ranging from normal brain tissue to tumor tissue. Cells on scaffolds of higher stiffnesses have increased drug resistance and increased expression of drug resistance, hypoxia, and invasion-related genes.
Acknowledgements
This work is supported in part by NIH grant NIH/NCI R01CA172455 and Kyocera Professor Endowment to M.Z. A.E. acknowledges support from the National Science Foundation Graduate Research Fellowship Program. S.K.L.L. acknowledges support from the Ruth L. Kirschstein NIH Training grant T32CA138312. We acknowledge the use of resources at the Molecular Analysis Facility and the Biology Imaging Facility at the University of Washington. We also acknowledge the support from the NIH to the UW W. M. Keck Microscopy Center (510 OD016240).
References
- [1].a) Holland EC, Proc Natl Acad Sci U S A 2000, 97, 6242; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ortensi B, Osti D, Pellegatta S, Pisati F, Brescia P, Fornasari L, Levi D, Gaetani P, Colombo P, Ferri A, Nicolis S, Finocchiaro G, Pelicci G, Stem Cells 2012, 30, 817. [DOI] [PubMed] [Google Scholar]
- [2].a) McNamara MG, Sahebjam S, Mason WP, Cancers (Basel) 2013, 5, 1103; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Umesh V, Rape AD, Ulrich TA, Kumar S, Plos One 2014, 9, e101771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Willis AL, Sabeh F, Li XY, Weiss SJ, J Microsc 2013, 251, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Petrecca K, Guiot MC, Panet-Raymond V, Souhami L, J Neurooncol 2013, 111, 19. [DOI] [PubMed] [Google Scholar]
- [5].a) Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA, Cell 2008, 133, 704; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yin Z, Chen X, Chen JL, Shen WL, Nguyen TMH, Gao L, Ouyang HW, Biomaterials 2010, 31, 2163; [DOI] [PubMed] [Google Scholar]; c) Discher DE, Janmey P, Wang YL, Science 2005, 310, 1139; [DOI] [PubMed] [Google Scholar]; d) Pelham RJ Jr., Wang YL, Biol Bull 1998, 194, 348. [DOI] [PubMed] [Google Scholar]
- [6].Pogoda K, Chin L, Georges PC, Byfield F, Bucki R, Kim R, Weaver M, Wells RG, Marcinkiewicz C, Janmey PA, New Journal of Physics 2014, 16, 075002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J, Nat Cell Biol 2015, 17, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kievit FM, Florczyk SJ, Leung MC, Wang K, Wu JD, Silber JR, Ellenbogen RG, Lee JS, Zhang M, Biomaterials 2014, 35, 9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Weiswald LB, Bellet D, Dangles-Marie V, Neoplasia 2015, 17, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Calvet CY, Andre FM, Mir LM, Plos One 2014, 9, e89644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Florczyk SJ, Wang K, Jana S, Wood DL, Sytsma SK, Sham JG, Kievit FM, Zhang M, Biomaterials 2013, 34, 10143; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hutmacher DW, Nat Mater 2010, 9, 90; [DOI] [PubMed] [Google Scholar]; c) Kievit FM, Florczyk SJ, Leung MC, Veiseh O, Park JO, Disis ML, Zhang M, Biomaterials 2010, 31, 5903; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xu X, Farach-Carson MC, Jia X, Biotechnol Adv 2014, 32, 1256; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Rape AD, Zibinsky M, Murthy N, Kumar S, Nat Commun 2015, 6, 8129; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Martinez-Ramos C, Lebourg M, Journal of Biomedical Materials Research, Part B 2014, 103B, 1249; [DOI] [PubMed] [Google Scholar]; g) Rao SS, Bentil S, DeJesus J, Larison J, Hissong A, Dupaix R, Sarkar A, Winter JO, Plos One 2012, 7, e35852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Hegedus B, Marga F, Jakab K, Sharpe-Timms KL, Forgacs G, Biophys J 2006, 91, 2708; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS, Weitz DA, Biophys J 2005, 89, 635; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yang YL, Motte S, Kaufman LJ, Biomaterials 2010, 31, 5678. [DOI] [PubMed] [Google Scholar]
- [12].Gordon VD, Valentine MT, Gardel ML, Andor-Ardo D, Dennison S, Bogdanov AA, Weitz DA, Deisboeck TS, Exp Cell Res 2003, 289, 58. [DOI] [PubMed] [Google Scholar]
- [13].Toole BP, Nature Reviews Cancer 2004, 4, 528. [DOI] [PubMed] [Google Scholar]
- [14].Rinaudo M, Polymer International 2008, 57, 397. [Google Scholar]
- [15].Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG, Int J Biochem Cell B 2004, 36, 1046. [DOI] [PubMed] [Google Scholar]
- [16].Kleihues P, Sobin LH, Cancer 2000, 88, 2887. [DOI] [PubMed] [Google Scholar]
- [17].a) Sironen RK, Tammi M, Tammi R, Auvinen PK, Anttila M, Kosma VM, Exp Cell Res 2011, 317, 383; [DOI] [PubMed] [Google Scholar]; b) Toole BP, Clin Cancer Res 2009, 15, 7462; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Veiseh M, Turley EA, Integr Biol (Camb) 2011, 3, 304; [DOI] [PubMed] [Google Scholar]; d) Kim Y, Kumar S, Mol Cancer Res 2014, 12, 1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang X, He J, Wang Y, Cui F-Z, Interface Focus 2012, 2, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Florczyk SJ, Kim DJ, Wood DL, Zhang M, Journal of Biomedical Materials Research Part A 2011, 98, 614; [DOI] [PubMed] [Google Scholar]; b) Shapiro L, Cohen S, Biomaterials 1997, 18, 583. [DOI] [PubMed] [Google Scholar]
- [20].a) Florczyk SJ, Wang K, Jana S, Wood DL, Sytsma SK, Sham JG, Kievit FM, Zhang MQ, Biomaterials 2013, 34, 10143; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Florczyk SJ, Kievit FM, Wang K, Erickson AE, Ellenbogen RG, Zhang MQ, J Mater Chem B 2016, 4, 6326; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang K, Kievit FM, Erickson AE, Silber JR, Ellenbogen RG, Zhang MQ, Adv Healthc Mater 2016, 5, 3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Coimbra P, Alves P, Valente TA, Santos R, Correia IJ, Ferreira P, Int J Biol Macromol 2011, 49, 573. [DOI] [PubMed] [Google Scholar]
- [22].Frydrych M, Wan CY, Stengler R, O’Kelly KU, Chen BQ, Journal of materials chemistry 2011, 21, 9103. [Google Scholar]
- [23].a) Pettikiriarachchi JTS, Parish CL, Shoichet MS, Forsythe JS, Nisbet DR, Australian Journal of Chemistry 2010, 63, 1143; [Google Scholar]; b) Rao SS, Dejesus J, Short AR, Otero JJ, Sarkar A, Winter JO, ACS Appl Mater Interfaces 2013, 5, 9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rape A, Ananthanarayanan B, Kumar S, Adv Drug Deliv Rev 2014, 79–80, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Netti PA, Berk DA, Swartz MA, Grodzinsky AJ, Jain RK, Cancer Res 2000, 60, 2497. [PubMed] [Google Scholar]
- [26].Hutmacher DW, Loessner D, Rizzi S, Kaplan DL, Mooney DJ, Clements JA, Trends Biotechnol 2010, 28, 125. [DOI] [PubMed] [Google Scholar]
- [27].Lee SY, Genes Dis 2016, 3, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bleau A-M, Huse JT, Holland EC, Cell Cycle 2009, 8, 2937. [PubMed] [Google Scholar]
- [29].Emery IF, Gopalan A, Wood S, Chow K.-h., Battelli C, George J, Blaszyk H, Florman J, Yun K, Journal of Neuro-Oncology 2017, 133, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bertout JA, Patel SA, Simon MC, Nature Reviews Cancer 2008, 8, 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wigerup C, Pahlman S, Bexell D, Pharmacol Therapeut 2016, 164, 152. [DOI] [PubMed] [Google Scholar]
- [32].Radotra B, McCormick D, Crockard A, Neuropathol Appl Neurobiol 1994, 20, 399. [DOI] [PubMed] [Google Scholar]
- [33].a) Pietras A, Katz Amanda M., Ekström Elin J., Wee B, Halliday John J., Pitter Kenneth L., Werbeck Jillian L., Amankulor Nduka M., Huse Jason T., Holland Eric C., Cell Stem Cell 2014, 14, 357; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mooney KL, Choy W, Sidhu S, Pelargos P, Bui TT, Voth B, Barnette N, Yang I, J Clin Neurosci 2016, 34, 1. [DOI] [PubMed] [Google Scholar]
- [34].Ramachandran RK, Sørensen MD, Aaberg-Jessen C, Hermansen SK, Kristensen BW, Plos One 2017, 12, e0172234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].a) Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, Maxwell J-P, Glackin CA, Wakimoto H, González-Herrero I, Sánchez-García I, Silber JR, Horner PJ, Rostomily RC, Molecular Cancer 2010, 9, 194; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Iser IC, Pereira MB, Lenz G, Wink MR, Medicinal Research Reviews 2017, 37, 271; [DOI] [PubMed] [Google Scholar]; c) Medici D, Hay ED, Olsen BR, Mol Biol Cell 2008, 19, 4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].a) Gil ES, Kluge JA, Rockwood DN, Rajkhowa R, Wang L, Wang X, Kaplan DL, Journal of Biomedical Materials Research Part A 2011, 99, 16; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kirdponpattara S, Khamkeaw A, Sanchavanakit N, Pavasant P, Phisalaphong M, Carbohydr Polym 2015, 132, 146. [DOI] [PubMed] [Google Scholar]