Abstract
Preventive efforts for serious immunologically mediated adverse drug reactions (IM-ADRs) have been fueled by discovery of strong class I human leukocyte antigen (HLA) associations; however, the low positive predictive value of HLA for IM-ADRs has limited translation. Studies were undertaken to explain why most patients carrying an HLA risk allele do not develop IM-ADR on exposure to the risk drug. Tissue-specific approaches defined the T-cell receptor (TCR) repertoire and phenotype of the pathogenic T cells found in the skin and blister fluid of IM-ADRs. Dominant CD8+ T cell clonotypes representing >50% of total TCRαβ sequences among CD8+ CD137+ T cells were identified in tissue to identify the pathogenic activated T cells. Identification of the specific molecular and cellular signatures of the antigen-driven pathogenic T cells will facilitate more specific mechanisms to determine the small percentage of individuals carrying an HLA risk allele who are likely to develop an IM-ADR before drug exposure.
INTRODUCTION
The Scope of the Problem
ADRs account for considerable patient morbidity and mortality. In addition, they represent a major economic burden on health care and drug development. The majority of ADRs are predictable based on the known pharmacological action of the drug and its target of activity. However, serious IM-ADRs result from an “off-target” interaction of the drug with an immune receptor which is either dose-independent in the case of antibody and immunoglobulin E – mediated reactions or dose-dependent with many other reactions including T-cell–mediated reactions. Similar to emerging infectious diseases, serious IM-ADRs are currently unpredictable during drug development, affect otherwise healthy individuals, are life-threatening, and often cause long-term disability. These ADRS are also over-represented in drugs that are used to treat infections of global importance such as HIV, tuberculosis, and leprosy. In addition to their acute impact, they can significantly constrict future treatment choices for patients. Research progress and funding of these serious IM-ADRs has been challenging because, although relevant to all medical disciplines, they are not owned by any single constituency.
CLINICAL PHENOTYPES
Serious IM-ADRs comprise a number of different clinical phenotypes. In relation to T-cell–mediated reactions, these can either be diseases with multisystem involvement such as drug reaction with eosinophilia and systemic symptoms (DRESS) and abacavir hypersensitivity (1,2). They also include severe cutaneous diseases such as Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN), acute generalized exanthematous pustulosis, and generalized bullous fixed-drug eruption where the primary disease involves the skin and mucous membranes, but other organs can be involved as a sequela of the disease. Single-organ diseases such as drug-induced liver disease (DILI), drug-induced pancreatitis, and interstitial nephritis are characterized by prominent involvement of one organ. The most serious of these drug-induced diseases is SJS/TEN which in the case of TEN involves >30% body surface area and is associated with overall mortalities of 20% or higher in the elderly and immune compromised populations.
TRANSLATIONAL OPPORTUNITIES
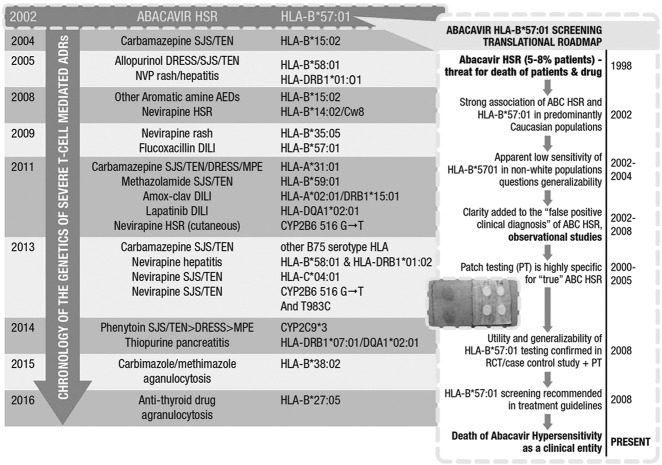
During the last 10 to 15 years, significant translational opportunities have been created from the discovery of strong associations between HLA alleles and T-cell–mediated drug hypersensitivity syndromes such as abacavir hypersensitivity, DRESS, SJS/TEN, and DILI, caused by specific drugs. The strong associations between variation in class I HLA and abacavir hypersensitivity, DRESS, and SJS/TEN have provided important clues to their immunopathogenesis. Class I major histocompatibility complex molecules (HLA-A, B, and C) are expressed on all nucleated cells and activate CD8+ T lymphocytes. The year 2002 marked the discovery by two independent groups of the association between HLA-B*57:01 and abacavir hypersensitivity. In the subsequent 6 years, important studies defined the roadmap from the discovery of this genetic association through to translation into the clinic (Figure 1). This included demonstration of the clinical utility and 100% negative predictive value (NPV) of HLA-B*57:01 as a genetic screening test to prevent abacavir hypersensitivity in the first and largest clinical trial to study the utility of a specific genetic marker to prevent a toxicity, as well as the generalizability of HLA-B*57:01 across populations of different ethnicity (3,4). Observational studies also highlighted that HLA-B*57:01 screening reduced not only true immunologically mediated abacavir hypersensitivity but also false positive clinical diagnosis (5,6). Translation of laboratory testing was fueled by the development of molecular assays for HLA-B*57:01 with a rapid turn-around-time and a paired HLA-B*57:01 allele–specific quality assurance program (7). By 2008, HLA-B*57:01 screening before abacavir prescription was part of guideline-based routine HIV practice in the developed world. The discovery of the association between HLA-B*57:01 and abacavir was of fundamental clinical importance but also led to further discovery of its immunopathogenesis and a new model of drug hypersensitivity known as the altered peptide repertoire model (8,9). Structural and peptide elution studies showed that abacavir binds noncovalently to the peptide binding cleft. This leads to presentation of peptide pools only bound to HLA-B*57:01 in the presence of abacavir that are recognized as foreign by the immune system leading to a T-cell hypersensitivity response. Based on this model, the crystal structure of HLA-B*57:01 in complex with abacavir and peptide was solved independently by two groups in 2012 (8,9).
Fig. 1.
Translational roadmap of HLA-B*57:01 as a marker to predict and prevent abacavir hypersensitivity in relation to the discovery timeline of other HLA-associated serious immunologically mediated drug reactions. Adapted with permission (13). Abbreviations: HSR, hypersensitivity reaction; ABC, abacavir; PT, patch testing; RCT, randomized controlled trial; SJS/TEN, Stevens-Johnson syndrome/toxic epidermal necrolysis; DRESS, drug reaction with eosinophilia and systemic symptoms; NVP, nevirapine; AED, anti-epileptic drug; DILI, drug-induced liver disease; MPE, maculopapular eruption; ADR, adverse drug reaction.
Subsequent associations of importance were defined between HLA-B*15:02 and carbamazepine SJS/TEN and HLA-B*58:01 and allopurinol SJS/TEN and DRESS (10,11). HLA-B*15:02 screening before carbamazepine prescription has been introduced into routine clinical practice in many Southeast Asian countries such as Taiwan, Singapore, and Hong Kong where significant reductions in carbamazepine-associated SJS/TEN have been noted (12).
TRANSLATIONAL GAPS
From the PREDICT-1 (Prospective Randomized Evaluation of DNA Screening in a Clinical Trial) study, a randomized double-blind controlled study that examined the utility of HLA-B*57:01 as a screening test to predict and prevent abacavir hypersensitivity, it could be determined that the NPV of HLA-B*57:01 testing to prevent immunologically confirmed, patch test (PT) – positive abacavir hypersensitivity was 100% and the positive predictive value (PPV) was 55% (3). Although similar findings of 100% NPV have been determined for HLA-B*15:02 and HLA-B*58:01 for SJS/TEN associated with carbamazepine and SJS/TEN/DRESS associated with HLA-B*58:01, the PPV for the at-risk allele for drug hypersensitivity syndromes outside of abacavir hypersensitivity has been universally <10% and typically <5% (1,13). This means that dependent on the prevalence of disease, the number needed to test to prevent one case of a severe hypersensitivity in a population varies from 300 to >10,000. Although the evidence indicates that the presence of an HLA risk allele appears necessary for the development of most HLA-class I –restricted drug hypersensitivities, it is not sufficient. The mechanisms surrounding both this “positive predictive gap” and the specific clinical phenotype and tissue distribution of a given reaction remain unexplained. This has come to light more recently in cohorts of family members carrying the same HLA class I risk allele in the case of nevirapine SJS/TEN where mismatch in nevirapine SJS/TEN versus tolerance has occurred between mother and child pairs (unpublished). Clues to the mechanistic basis for this gap have come from: 1) Identification of memory T cell responses in HLA-B*57:01 abacavir–naïve subjects; 2) PT-positive abacavir hypersensitivity occurring as early as 1.5 days of first dosing; 3) Long-lasting immune responses despite lack of drug re-exposure in the peripheral blood detected in patients with HLA-B*57:01+ abacavir hypersensitivity and HLA-B*15:02+ carbamazepine SJS/TEN. In addition, further clues have come from the identification of a public TCR clonotype that has been found in the blister fluid and peripheral blood of unrelated Taiwanese patients with HLA-B*15:02+ carbamazepine SJS/TEN. This forms the basis of the double major histocompatibility complex restriction-heterologous immunity model which postulates that T-cell–mediated drug hypersensitivity represents a cross-reaction between a memory T-cell immune response that has occurred much earlier in life to a prevalent and persistent viral pathogen with a neo-epitope created when the drug is present (14). The finding of a dominant and public clonotype in HLA-B*15:02-restricted carbamazepine SJS/TEN in the blister fluid at the site of immune damage to the skin supports the scientific premise for taking tissue-specific approaches to study these serious immunologically mediated ADRs. Because SJS/TEN is a rare disease involving 1 to 5 patients/million/annum, it is imperative that opportunities are not missed for tissue direct approaches to study immunopathogenesis.
METHODS
We accrued paired samples from peripheral blood mononuclear cells (PBMCs), blister fluid, and skin from patients with acute serious cutaneous ADRs such as SJS/TEN and DRESS (Vanderbilt IRB#131836 and 150754). The first part of this study which will be reported here has focused on known HLA-class I–restricted SJS/TEN. Detailed analyses have been performed on a 70-year-old African American woman who presented with pharyngitis, conjunctivitis, and a blistering skin eruption 2 weeks following initiation of allopurinol. She was found to carry HLA-B*58:01 on high resolution HLA typing (Illumina Miseq: Illumina, Inc., San Diego, CA). Blister fluid was collected into ethylenediaminetetraacetic acid tubes and blister cell extraction yielded 52 × 106 cells that were cryopreserved in liquid nitrogen in aliquots of 10 million cells per cryovial. Multiparameter mass cytometry (CyTOF) was used to characterize the cellular composition of the blister fluid. A validated 24-parameter mass cytometry panel was developed that targeted 22 leukocyte cell surface markers (CD3, CD45, CD4, CD8a, TCRγδ, CD62L, CD45RO, CCR7, CD69, CD25, CD27, CD19, CD20, CD14, CD16. CD56, CD57, CCR6, CCR4, CCR5, CXCR3, and CXCR4). To visualize and compare cell phenotypes between total blister fluid cells and autologous PBMCs collected during acute allopurinol SJS/TEN, unsupervised nonlinear multidimensional reduction analysis were performed using ViSNE software (Cytobank, Santa Clara, CA) which was followed by manual gating to determine cell frequencies.
Single Cell Studies
Blister fluid cells and PBMCs were thawed, washed, and stained immediately with T-cell – identifying markers (CD3, CD4, and CD8), markers of T-cell activation (CD137 and CD69), markers of T-cell memory (CCR7 and CD45RO), markers of T-cell homing or residence (CLA and CD103), natural killer (NK) cell (CD56) and live/dead for exclusion. Single, live gated CD3+CD8+CD137+ cells were sorted into individual wells of a 96-well polymerase chain reaction plate containing barcoded oligo deoxythymidine primer engineered with an 8-nucleotide unique molecular identifier. A nonactivated control population was sorted (CD3+CD8+CD137−) for comparison. Full-length amplified cDNA libraries were created (15). Paired single-cell TCRαβ sequencing (scTCRseq) was performed using previously published primers to amplify the TCRαβ CDR3 and surrounding hypervariable regions. Barcoded amplicons were pooled and sequenced (Illumina Nextseq: Illumina Inc., San Diego, CA). Whole transcriptome analysis single-cell RNAseq (scRNAseq) was performed using the same amplified cDNA library as used for scTCRseq. Amplified cDNA was fragmented by tagmentation using the Nextera XT system and pooled and sequenced (Illumina Nextseq) with well- and plate-specific unique molecular identifiers to allow quantitation of transcripts.
Data and Statistical Analyses
The bulk and single-cell analysis pipelines were used to integrate and visualize bulk and single-cell TCR and RNAseq data. Nonparametric, iterative subsampling via the grouping of lymphocyte interactions by paratope hotspots (GLIPH) algorithm and a Bayesian multinomial models were used to score TCRs for HLA specificity, clonal expansion, and significantly enriched TCR sequence motifs. The comparative populations for both scTCR and scRNAseq were activated (CD137+) and nonactivated (CD137−) CD8+ T cells in the our HLA-B*58:01+ allopurinol SJS/TEN patient. For the scRNAseq data deconvoluted reads in FASTQ format were trimmed and aligned to the human transcriptome (hg38) using Bowtie2 and quantified using the RSEM (RNA-Seq by Expectation-Maximization), General Public License v3 software package. Raw count matrices were imported into single-cell differential expression (SCDE) for differential expression and normalized count matrices were delivered into single-cell consensus clustering (SC3), Seurat, and Monocle 2.0 for consensus clustering, differential expression, marker identification and pseudotemporal ordering. Differential expression among populations of interest was assessed using a Bayesian posterior likelihood model (SCDE), AUROC classification curve (SC3), nonparametric Kruskal-Wallis test between consensus clusters (SC3), branched expression modelling (Monocle 2.0: Massachusetts Institute of Technology, Boston, MA), and a modified likelihood ratio test optimized for single-cell differential expression (Seurat: GNU Public License (GPL 3.0), New York University, New York).
RESULTS
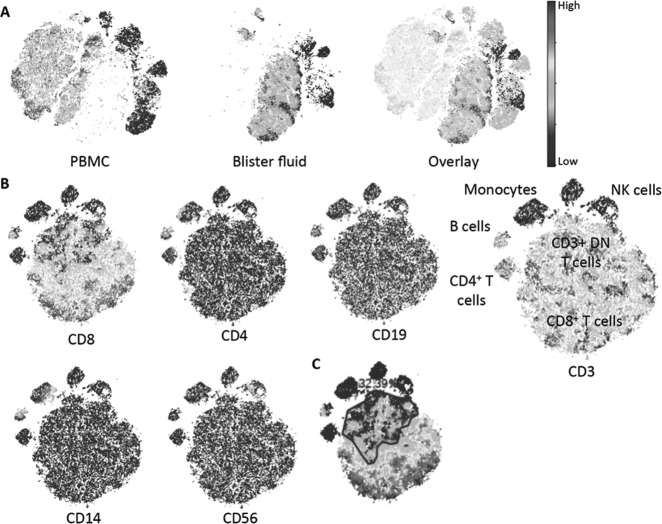
CyTOF analysis of blister fluid cells from the HLA-B*58:01+ allopurinol SJS/TEN patient shows that most cells are T cells (CD3+) with the majority of these being CD8+ T cells. Although present in the blister fluid, other populations of cells such as CD4+ T cells, B cells, monocytes, and NK cells were the minority. Furthermore, 32.5% of total blister fluid cells were CD3+CD4−CD8−. Strikingly, blister fluid cells form populations distinct from that seen in the peripheral blood during acute SJS/TEN (Figure 2). Bulk CDR3 sequencing was performed on total genomic DNA on the ImmunoSEQ platform (Adaptive Biotechnologies).
Fig. 2.
CyTOF analysis of blister fluid from an HLA-B*58:01+ patient with allopurinol SJS/TEN. (A) VisNE analysis of total blister fluid and autologous PBMC with overlay showing distinction between cell populations represented in blister fluid and PBMCs. (B) ViSNE analysis of blister fluid cells alone. (C) Gating for frequency of CD3+ double-negative T cells colored for CD8 expression. Abbreviations: SJS/TEN, Stevens-Johnson syndrome/toxic epidermal necrolysis; PBMC, peripheral blood mononuclear cell; NK, natural killer.
Single-Cell TCR
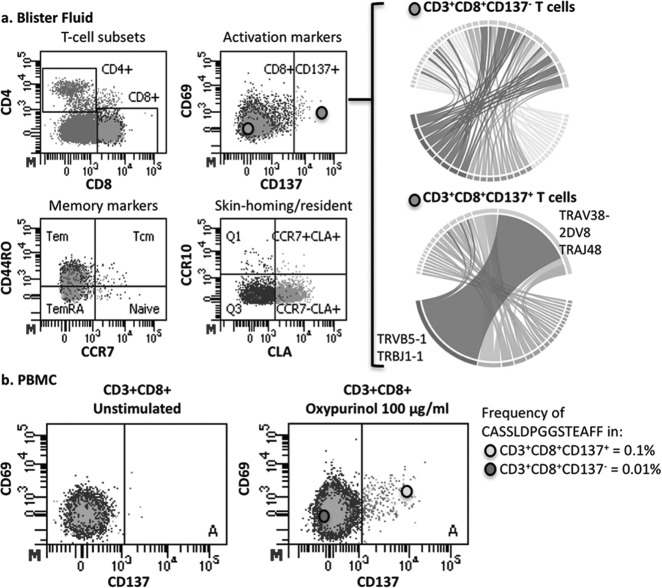
A dominant CD8+ T cell clonotype comprised of TRBV5-1/J1-1 and TRAV38-2/DV8/J48 bearing the TCRβ CDR3 sequence CASSLDPGGSTEAFF was identified in the blister fluid of the HLA-B*58:01+ allopurinol SJS/TEN patient among the activated CD8+CD137+ T cells that was not seen among the CD8+CD137− blister fluid T cells. This clonotype was also not seen among PBMCs from unrelated donors. PBMCs from the same patient with allopurinol SJS/STEN had been subjected to overnight stimulation with oxypurinol (the primary metabolite of allopurinol that has been shown to activate T cells). When stained for the same markers (CD3, CD4, CD8, CD69 and CD137) and bulk sorted for TCRβ CDR3 sequencing, these stimulated cells showed that even with oxypurinol stimulation the TCRB CDR3 sequence that was dominant in the blister fluid was rare in the peripheral blood and found in 0.1% of CD8+CD137+ T cells and 0.01% of CD8+CD137- T cells (Figure 3).
Fig. 3.
(A) Single cell sorting and TCR repertoire of activated CD8+ T cells shows dominant TCRA/B clonotype (TCRA CDR3: CAYRSGAFGNEKLTF; TCRB CDR3: CASSLDPGGSTEAFF) HLA-B*58:01+ allopurinol SJS/TEN blister fluid. Single cell TCR sequencing of CD8+CD137+ T cells from blister fluid. Circus plots represents CDR3 amino acids. (B) TCRβ CDR3 sequencing of bulk sorted PBMC showing that the dominant TCR clonotype in CD8+CD137+ T cells that was prevalent in blister fluid is rare in the peripheral blood. Abbreviations: TCR, T-cell receptor; SJS/TEN, Stevens-Johnson syndrome/toxic epidermal necrolysis; PBMC, peripheral blood mononuclear cell.
scRNAseq
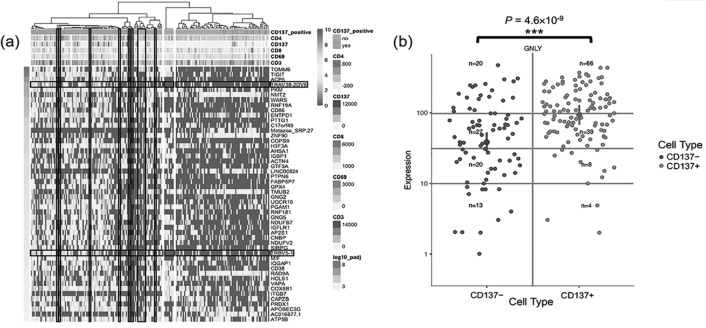
Using the same analytic strategy as for the scTCRseq data to distinguish populations of CD8+ T cells based on CD137+ and CD137-, distinct transcriptional signatures were identified that were unique to the CD8+CD137+ T-cell population in the blister fluid from the HLA-B*58:01+ patient with allopurinol SJS/TEN (Figure 4A). This included an enrichment of extracellular exosome genes false discovery rate (FDR = 7.8 × 10-8) and identification of other activation markers relevant to CD8+ T cells such as CD38, TIGIT, TNFRSF1B and KLRC1. The scRNAseq analysis (Figure 4) independent to the scTCRseq analysis identified increased expression of the same TCR identified by the scTCRseq primer-based assay (TRBV5-1 and TRAV38-2/DV8) and again this was represented in the CD8+CD137+ T cells. In addition, the activated CD8+CD137+ T cells showed significantly higher expression of granulysin, (p = 4.6 × 10-9), which is known to be the main cytotoxic peptide that mediates keratinocyte cell death (Figure 4B).
Fig. 4.
(A) Single-cell RNAseq from HLA-B*58:01+ allopurinol SJS/TEN blister fluid showing the integration of 9 flow cytometric markers measured for each cell. CD137+CD8+ T cells are enriched in immune activation and extracellular exosome markers and showed selective upregulation of TRBV 5-1 and TRAV38-2DV8 not seen in CD8+CD137- cells. (B) Monocle 2.0 reveals differential expression of granulysin (GNLY) in the cells falling along the pseudo-time trajectory of the CD137+ population (CD137+ [green]; CD137-[blue]). Abbreviations: SSJ/TEN, Stevens-Johnson syndrome; TRBV, T-cell receptor beta variable; GNLY, granulysin.
DISCUSSION
Serious IM-ADRs are associated with significant short- and long-term morbidity and mortality, increased health care costs, and uncertainty in drug development. More recently, strong associations between class I HLA alleles and T-cell–mediated reactions have contributed to our knowledge of their mechanisms. For some drugs, such as HLA-B*57:01 and abacavir hypersensitivity across most of the developed world and HLA-B*15:02 and carbamazepine SJS/TEN in Southeast Asia, the NPV of these HLA alleles is 100% providing a viable screening strategy to predict and prevent these serious immunologically mediated ADRs. For most T-cell –mediated drug reactions, the risk HLA allele has either not been defined, or prevalence of disease and PPV are in an unfavorable ratio for screening for the HLA allele before drug prescription to be feasible in clinical practice. The mechanistic basis for why serious T-cell–mediated ADRs are associated with long-lasting immunity, why they can affect one organ preferentially, and why the HLA risk allele appears to be necessary but not sufficient for the development of a serious immunologically mediated ADR has remained elusive.
In a single individual with HLA-B*58:01+ allopurinol-associated SJS/TEN using technologies with single-cell resolution, we have been able to define cellular and molecular signatures that are unique to this reaction as well as those that more generally define the clinical phenotype of the patient. We have defined on mass cytometry analysis that responses seen in the peripheral blood and blister fluid during a severe systemic cutaneous disease such as SJS/TEN differ significantly from each other quantitatively and qualitatively. These studies support that pathogenic CD8+ T cells are enriched at the site of tissue damage and that these antigen-driven T-cells that are causing the immune response and damage in these patients are at a tissue level. Definition of a dominant T-cell clonotype in the blister fluid of a patient with HLA-B*58:01+ SJS/TEN at the primary site of keratinocyte death that is rarely found in the peripheral blood of oxypurinol stimulated cells is in keeping with previous studies showing a dominant clonotype in the blister fluid of HLA-B*15:02-associated carbamazepine SJS/TEN which cannot be found in an unbiased way in the peripheral blood. Furthermore, we can show the expression of the same TCR in activated T-cell populations on RNAseq and definition of the specific phenotype of these CD8+CD137+ is in keeping with what we could predict based on what is known about their functional activity.
Currently it remains unknown as to whether these antigen-driven cells that are present at the site of acute tissue damage maintain long-lasting residency in the skin following serious cutaneous ADRs such as SJS/TEN. Future studies will help to define the presence of CD8+ resident T-cells in the skin following SJS/TEN and whether these share an αβ TCR clonotype with the CD8+ T cells of an effector memory phenotype present during the acute reaction. In the future it will be important to build on these models for drugs of known HLA restriction and confirm the model with appropriate drug-exposed single HLA antigen presenting cells in a TCR reporter system.
This work has significant translational outputs in its capacity to explain why not all patients carrying an HLA risk allele when exposed to the causal drug will develop a serious immunologically mediated ADR. It is likely, given the short latency period of many of these reactions such as SJS/TEN and abacavir hypersensitivity, that pre-existing memory T-cell responses that are formed early in life are important drivers of cross-reactive T-cell responses that arise when a drug is introduced many years later. The capacity to define these signatures, which are dominant at a tissue level during the acute disease, particularly when these are associated with public TCR clonotypes, will translate into measures to screen patients for risk. This will help shape and define mechanisms not only for drug hypersensitivity but other immunologically driven diseases.
Footnotes
Potential Conflicts of Interest: Dr. Phillips is co-director of IIID Pty., Ltd., that holds a patent for HLA-B*57:01 testing for abacavir hypersensitivity; and has received grant funding from the National Institutes of Health: 1P50GM115305-01, 1RO1AI103348-01, 1P30AI110527-01A1, 1R13AR71267-01, and the National Health & Medical Research Council of Australia.
DISCUSSION
Mackowiak, Baltimore: Thank you, Dr. Phillips. Very interesting, important work. Most of the patients that I deal with are not on a single medication, and I would suspect that the ones that you have enrolled in your study are on multiple medications, certainly your HIV patients. So, the problem of drug-drug interaction must complicate the interpretation of your results. I’d be interested in your comments on how you deal with these patients who are on multiple drugs when you are trying to figure out the importance of a single drug versus drug-drug interactions.
Phillips, Nashville: So, a very important part of our program, and it’s not being rolled out to the clinic currently but is actually the development of ex vivo assays where we can look at drug causality. For our studies we have strict clinical definitions and causality assessments, so these patients that we actually study in detail would have had to have already met these criteria. There are actually causality scores. But above and beyond that, we have a pipeline of immunophenotypes that we are using on all patients that we study with severe immunologically immediate drug reactions and that includes drug and known HLA pairs. And so, we try and get these as quickly as possible into the disease as we know immune memory can wane with time. So, we’ve had cases that we’ve published with patients being on seven different antibiotics and we’ve been able to ascertain which antibiotic causes, for instance, SJS/TEN using these approaches. I think a very important part of the development process, although I didn’t speak to this in detail today, is actually developing immunophenotyping and drug causality assessment where we can do just that, and it would have great clinical relevance. But it would also be very relevant to the rigor of the research studies as well.
Molitoris, Indianapolis: You have tremendous capacity to look at the large volume three-dimensional microscopic analysis and do flow essentially on a piece of tissue. You biopsy to get your anatomic relationships of different cell types to different cell types, and different cell types to injury. I just wondered if you thought about going in that direction?
Phillips, Nashville: Yes. It’s kind of a dream to be able to do this. Our program is being directed to a tissue level just over the last couple of years for this reason. But we deal mainly with cutaneous diseases at this time. But expanding with other approaches where we can actually deal not only with T-cells from fresh skin, where we actually get a very high yield from fresh skin from SJS/TEN but also moving towards analysis of other tissue types and archived samples to use multidimensional approaches. I think in the future that the technologies, to, for instance, get RNA from archived samples, is going to move forward very quickly.
Schuster, New York: Beautiful work. Maybe you covered this when you talked about why the mouse model is not possible and I missed it. What accounts for the tropism to the skin in saliva of these rare T-cell subtypes?
Phillips, Nashville: No, I mean, I think that’s a million-dollar question. We’re trying to study drug responses in human models at a tissue level where we don’t know what the endotope is, what the specific antigen presenting cells are and what’s driving the activity in the skin at this point. We are looking at tissue homing and in some of the panels that you saw on the screen, we are looking at CLA and other markers of tissue homing. But we don’t understand for any of the immunologically mediated diseases what actually causes homing to that particular organ or why diseases amongst those different immunologically mediated diseases are so different, because they are. Even if you look at DRESS, there’s several different sub-phenotypes of DRESS, and I think these integrated all mixed approaches are going to help us to decipher that as well.
Billings, Baton Rouge: If someone has an autoimmune disease like rheumatoid arthritis or temporal arteritis or something along those lines, or also has had an immune mediated reaction to drugs such as a rash from penicillin, is there any way to test that person to see if they are more prone or less prone to have further immunological difficulties — which may be catastrophic with other drug interactions?
Phillips, Nashville: At this point in time in terms of the functional assays I have described before a patient actually receives the drug, there is no way to actually ascertain their risk other than genetic screening to learn if there is a genetic marker. So a patient has to actually have been exposed to the drug to actually detect an immune response in the peripheral blood or tissue at this point. But I think moving towards the future, one of the strategies that we are trying to move towards, is being able to define, where these cross-reactive memory T-cells are residing and their specific signature. This would open the possibility that we could actually find prior to exposure who might be at risk for one of these severe reactions. The gap right now is that we don’t have good free drug screening technologies other than HLA screening which has a low positive predictive value.
Billings, Baton Rouge: For instance, I had a patient who had temporal arteritis and then was diagnosed subsequently as having tuberculosis which he had had for probably 50 years, and then he had an old Ghon complex and was placed on INH and became severely neutropenic and I suspect that there was something autoimmune that was resulting in his own autoimmune disorder and then translated into his drug reaction. But there was no way to really test to that prior to INH exposure.
Phillips, Nashville: We’re starting to understand that there are common risks. There are genetic variances amongst patients that may actually be shared between different diseases. And the problem is the positive predictive value is 2% to 3% and not 100%. So, similar to pleiotropic effects, you might see in families with autoimmune diseases where one family members gets one disease and another a different autoimmune disease based on common HLA risk factors we are starting to notice this with immunologically mediated severe drug reactions. We have from our cohort of South African patients that have nevirapine and SJS defined a family member with the risk gene with SJS and another family member carrying the same HLA risk gene who is tolerant. It’s very similar to other immune diseases and that way that we don’t have common mechanisms. I think for chronic diseases there may be ways of doing pre-clinical screening, risk assessment over time based on serological assessment but to see who’s destined to get a disease for drug reactions, I think this is a bit more tricky.
REFERENCES
- 1.White KD, Chung WH, Hung SI, Mallal S, et al. Evolving models of the immunopathogenesis of T cell-mediated drug allergy: the role of host, pathogens, and drug response. J Allergy Clin Immunol. 2015;136((2)):219–34. doi: 10.1016/j.jaci.2015.05.050. quiz 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redwood AJ, Pavlos RK, White KD, Phillips EJ. HLAs: Key regulators of T-cell--mediated drug hypersensitivity. HLA. 2018 Jan;91((1)):3–16. doi: 10.1111/tan.13183. PubMed PMID: 29171940; PubMed Central PMCID: PMC5743596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallal S, Phillips E, Carosi G, Molina JM, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358((6)):568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 4.Saag M, Balu R, Phillips E, Brachman P, et al. High sensitivity of human leukocyte antigen-b*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis. 2008;46((7)):1111–8. doi: 10.1086/529382. [DOI] [PubMed] [Google Scholar]
- 5.Rauch A, Nolan D, Martin A, McKinnon E, et al. Prospective genetic screening decreases the incidence of abacavir hypersensitivity reactions in the Western Australian HIV cohort study. Clin Infect Dis. 2006;43((1)):99–102. doi: 10.1086/504874. [DOI] [PubMed] [Google Scholar]
- 6.Phillips EJ. Genetic screening to prevent abacavir hypersensitivity reaction: are we there yet? Clin Infect Dis. 2006;43((1)):103–5. doi: 10.1086/504878. [DOI] [PubMed] [Google Scholar]
- 7.Hammond E, Almeida CA, Mamotte C, et al. External quality assessment of HLA-B*5701 reporting: an international multicentre survey. Antivir Ther. 2007;12((7)):1027–32. [PubMed] [Google Scholar]
- 8.Ostrov DA, Grant BJ, Pompeu YA, Sidney J, et al. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc Natl Acad Sci U S A. 2012;109((25)):9959–64. doi: 10.1073/pnas.1207934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Illing PT, Vivian JP, Dudek NL, Kostenko L, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486((7404)):554–8. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 10.Chung WH, Hung SI, Hong HS, Hsih MS, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428((6982)):486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 11.Hung SI, Chung WH, Liou LB, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A. 2005;102((11)):4134–9. doi: 10.1073/pnas.0409500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P, Lin JJ, Lu CS, Ong CT, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med. 2011;364((12)):1126–33. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]
- 13.Garon SL, Pavlos RK, White KD, Brown NJ, et al. Pharmacogenomics of off-target adverse drug reactions. Br J Clin Pharmacol. 2017;83((9)):1896–911. doi: 10.1111/bcp.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlos R, White KD, Wanjalla C, Mallal SA, et al. Severe delayed drug reactions: role of genetics and viral infections. Immunol Allergy Clin North Am. 2017;37((4)):785–815. doi: 10.1016/j.iac.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picelli S, Faridani OR, Bjorklund AK, Winberg G, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9((1)):171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]