Abstract
Background:
Depression is a common medical condition with a high prevalence leading to emotional abnormality. Despite some drawbacks, depression currently diagnosed using a combination of patient interviews and self-report questionnaires. Recently, there is emerging emphasis to establish biomarkers to diagnosis and clinical management of depression. This case–control study was designed to develop microRNA (miRNA)-based serum biomarker for depression.
Materials and Methods:
In this study, 39 patients with depression and 36 healthy controls were enrolled. Serum miRNAs gene expression was measured using real-time polymerase chain reaction (PCR) analysis; finally, the data represent as the 2–ΔCt followed by further statistical analysis.
Results:
The serum level of miR-16 was significantly (P < 0.001) down-regulated (mean: 0.9123 and standard deviation [SD]: 0.06) in compared to normal individuals (mean: 1.6848 and SD: 0.09). The concentration of miR-135a was also catastrophically decreased (P < 0.001) in the patients (mean: 1.160 and SD: 0.07) in compared to control (mean: 1.819 and SD: 0.09). The relative miR-1202 expression levels were significantly lower (P < 0.001) in the patients (mean: 0.1755 and SD: 0.01) than in the healthy individuals (mean: 0.2939 and SD: 0.01). The receiver operating characteristic curve analysis indicated the obvious separation between patient and healthy control, with an AUC of 0.75 (95% confidence interval [CI] = 0.642–0.858, P < 0.001), 0.72 (95% CI = 0.607–0.834, P < 0.001), and 0.74 (95% CI = 0.630–0.861, P < 0.001) for miR-16, miR-135a, and miR-1202, respectively. The data suggest that these miRNAs have a potential to be used as a biomarker of depression with sensitivity 77.8% and specificity of 61.5% for miR-16, 94.4% and 41.0% for miR-135a as well as 86.1% and 61.5% for miR-1202, respectively (P < 0.001).
Conclusion:
Our findings showed that these miRNA can be used as a biomarker of depression diagnosis. MiR-135a and miR-1202 exhibited better sensitivity and specificity, respectively.
Keywords: Depression, microRNAs, sensitivity, serum biomarker, specificity
INTRODUCTION
Depression is a common disorder recognized by the World Health Organization as one of the major causes of psychiatric disability worldwide causing to a poor quality of life.[1] It also has been predicted that depression will become the second disorder affecting individuals by the year 2020.[2] Emerging data have been exhibited that up to 32% of patients who visited primary health-care centers of European countries showed signs and symptoms of depression and other related disorders such as anxiety and stress-related disorders.[3,4] Despite a few years of unremitting studies, the etiology of depression has not been well elucidated. However, to date, large bodies of studies have suggested that the etiopathogenesis of depression appears to be influenced by a wide variety of factors, such as neural and structural plasticity, neurotransmitter systems, and epigenetic and genetic susceptibility.[5,6,7,8,9] Serotonin or 5-hydroxytryptamine (5-HT), a monoamine neurotransmitter, is produced from tryptophan in the neurons of raphe nuclei (RN) that modulates a variety of cognitive, emotional, and physiological functions.[10] It is well documented that serotonin is an important contributor to feelings of happiness.[11] The link between the serotonergic system of brain and depression is generally well accepted. It also has been indicated that serotonin synthesis, secretion, reuptake, and deactivation were observed to be deregulated in depression.[12] Moreover, most of the antidepressant strategies target the function of 5-HT system-related proteins, leading to increased 5-HT levels in the brain.[13]
In a recent decade of research, the emergence of small noncoding RNAs as a posttranscriptional regulator of gene expression has gained lots of attention in almost all of the disease pathophysiologies.[14] MicroRNAs (miRNAs) are emerging class of highly conserved, noncoding RNAs altering gene expression by ribosomal RNA modifications, alternative splicing as well as inducing messenger-RNA degradation.[15] It has been shown that miRNAs are expressed highly in the central nervous system to make an extremely powerful mechanism to dynamically regulate the protein content of neuronal compartments.[16] Thus, miRNAs appear to play the vital role in brain functions such as neurogenesis, neuronal metabolism, proliferation, and apoptosis.[17] Furthermore, miRNAs are present in the peripheral blood in a partially stable form. Therefore, they seem to be the potential blood-based biomarkers.[18] Over a decade of studies, circulating miRNAs, as the potential diagnostic biomarkers, have been well demonstrated in a wide variety of psychiatric diseases.[19] For instance, recently, it has been observed that circulating miRNAs have altered expression in the blood of patients with major depressive disorder; however, their diagnostic value remains undefined.[20,21]
To date, there is no generally accepted experimental biomarker for depression. This is because of the brain tissue that is not simply accessible. It has been shown that serum or plasma miRNAs also are constantly stable and tissue specific. Recently, it has been observed that serum and brain may share a common miRNA expression pattern. In this regard, several lines of examination used easy and noninvasive samples such as circulating blood samples, mainly including serum and plasma to develop the diagnostic approach. The reported miRNAs biomarkers for depression and other behavioral disorder have several drawbacks. First, miRNAs are often impaired during the majority of diseases such cancer, inflammation, infection, and other human disorders and thus not specific for depression. Second, most reported miRNA diagnostic biomarkers for depression could aid in diagnosis, but their predictive and diagnostic values remain to be determined.
Due to the role of serotonin system roles in depression, recent studies have shown that miR-16 strongly regulates serotonin transporter (SERT) gene expression, which is associated with depression.[22,23] Similarly, it has been shown that there is a strong relationship between miR-135a and both the 5-HT transporter and 5-HT1A receptor transcripts.[24] In addition, emerging data have shown that miR-1202 which is specific to primates brain, regulates serotonergic neurotransmission.[25] Previous studies have developed these miRNAs as a single biomarker which needs to be confirmed in different population either alone or in combination together. Therefore, the current study was designed to investigate these three serotonergic miRNAs expression in the serum of patients with depression in compared with healthy controls in the Ilam city, Iran, from January to December 2014.
MATERIALS AND METHODS
Participants
All 39 new cases, diagnosed according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders-Fourth Edition (DSM-IV) based on the Structured Clinical Interview for DSM-IV under no treatment at the time of blood collection (in the Ilam city of Iran in 2014).
The inclusion criteria for this case–control study were as follows: age 20–40 years, medication-free for at least 2 months before sampling, a minimum total score of 14 on the Hamilton Rating Scale for Depression-17 item, according to the American Psychiatric Association task force for the handbook of psychiatric measures.[26] Patients also with any current or previous diagnosis of mental disorder, multiple sclerosis, schizophrenia, or other psychotic disorder, the heavy drinker and or heavy smoker, pregnancy, and obese patients were excluded from this study. Thirty-six age- and sex-matched healthy volunteers (in term of depression) were recruited as controls.
Demographic criteria and clinical status of the individuals were obtained through the administration of a semi-structured interview. The current study was performed based on the ethical standards Declaration of Helsinki and all individuals provided written informed consent which was approved by the Local Ethics Committees (ir.medilam.rec1395.110).
Sample collection
Five milliliters of venous blood samples were collected from the participants. The blood samples were centrifuged at 1500 g for 15 min at room temperature, and the supernatant was transferred to RNase/DNase-free tubes, and finally, the samples were stored immediately at −80°C until the time of the experiment.
Real-time quantitative reverse transcription-polymerase chain reaction detection of microRNAs expression
Total RNA was extracted using TRIzol reagent (Life Technologies, Grand Island, NY) and purified using a miRNeasy Serum/Plasma kit (Qiagen, Frederick, MD, USA) based on the manufacturer's protocol. The purity of RNA was assayed by the absorbance ratio at 260/280 nm using NanoDrop 2000 spectrophotometer (Thermo Scientific, Worcester, MA). SYBR green-based stem-loop reverse transcription-polymerase chain reaction (RT-PCR) analysis to detect the level of mature miRNAs in all samples was performed as described previously.[27] In brief, extracted RNA (2 μg) was reverse transcribed using AccuPower RT-PCR Premix in the presence of each specific stem-loop primers which were as follows: The sequences of the stem-loop RT primer used were; miR-16: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC GCC AA-3, miR-1202: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC TCC CC-3′, miR-135a: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACT CAC AT-3′, U6 snRNA; 5′GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA AA ATA-3′; Quantitative real-time PCR was performed using CFX96 real-time system (Bio Rad-USA) for 40 cycles (15 s at 95°C, 15 s at 60°C, and 30 s at 72°C). The following primers were used for SYBR Green-based real-time PCR, miR-16 forward: 5′-CAC GCA CGT AGC AGC ACG TA-3, miR-1202 forward: 5′-ACT CTG GTG CCA GCT GCA G-3′, miR-135a forward: 5′-GCG CCG GTA TGG CTT TTT ATT CC-3′, U6 forward: 5′-GCG CGT CGT GAA GCG TTC-3′ and universal reverse has-miR primer is 5′-GTG CAG GGT CCG AGG T-3′. Comparative quantification analysis (2-ΔCt method) was used to determine the relative expression and normalized to the housekeeping gene, U6, which has been validated as the housekeeping gene for miRNAs studies.
Statistical analysis
All statistical analyses were performed the SPSS version 19 software (SPSS, Inc, Chicago, IL, USA). Data of quantitative RT (qRT)-PCR were analyzed using 2–ΔCt method, as described previously.[28] All data were presented as mean ± standard deviation (SD) The normal distribution of data was checked by Kolmogorov–Smirnov analysis. Differences in miRNA concentrations between two groups were compared using Mann–Whitney test.[29] Receiver operating characteristic (ROC) curves were generated to assess the diagnostic accuracy of each miRNA. A difference was considered statistically significant at P < 0.05.
RESULTS
Clinical characteristics of patients and controls
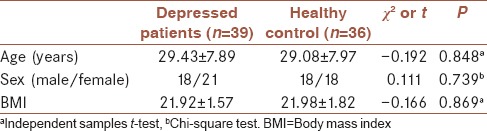
Clinical characteristics of the study participant are summarized in Table 1. In brief, 39 patients with depression who had never been treated with antidepressant drugs such as amitriptyline, sertraline, and tranylcypromine, 36 controls also participated in the present study. Based on the demographic data, patients and control groups were compared statistically. Two groups of study have been well paired in terms of age, gender, and BMI. There was no significant different between two groups (P < 0.05).
Table 1.
Basic demographic characterization of patients and healthy controls
MiR-16, miR-135a, and miR-1202 were decreased in serum of patient with depression
As shown in Figure 1, the results of present study showed that the serum concentration of miR-16 was significantly (P < 0.001) down-regulated in the group of patients with depression (mean: 0.9123 and SD: 0.06) compared to normal individuals (mean: 1.6848 and SD: 0.09).
Figure 1.
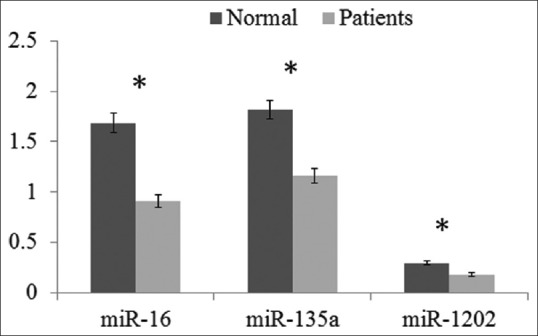
Serum level of miR-16, miR-135a, and miR-1202 relative expression by quantitative real-time polymerase chain reaction of patients with depression (n = 39) compared to healthy control (n = 36). The data represent as the 2–ΔCt. Statistical analyses were done using Mann–Whitney test. *P < 0.001 is statistically significant
The serum concentration of miR-135a was also catastrophically decreased (P < 0.001) in the group of depressed patients (mean: 1.160 and SD: 0.07) in comparison to healthy individuals (mean: 1.819 and SD: 0.09). As shown in Figure 1, the relative miR-1202 expression levels were significantly lower (P < 0.001) in the patients with depression (mean: 0.1755 and SD: 0.01) than in the healthy individuals (mean: 0.2939 and SD: 0.01).
Receiver operating characteristic analysis of miR-16, miR-135a, and miR-1202
To investigate the association between the three significantly differently expressed miRNAs and depression, the ROC analysis was performed to estimate the sensitivity and specificity of the diagnostic ability of miR-16, miR-135a, and miR-1202. As shown in Figure 2, the ROC curves exhibited the obvious separation between patient and healthy groups, with an AUC of 0.75 (95% confidence interval (CI) = 0.642–0.858, P < 0.001), 0.72 (95% CI = 0.607–0.834, P < 0.001), and 0.74 (95% CI = 0.630–0.861, P < 0.001) for miR-16, miR-135a, and miR-1202, respectively. The specificity of miR-16, miR-135a, and miR-1202 were 61.5%, 41.0%, and 61.5%, respectively. The sensitivity of miR-16, miR-135a, and miR-1202 were 77.8%, 94.4%, and 86.1%, respectively. MiR-16, miR-135a, and miR-1202 displayed a high sensitivity and specificity for the diagnosis of depression.
Figure 2.
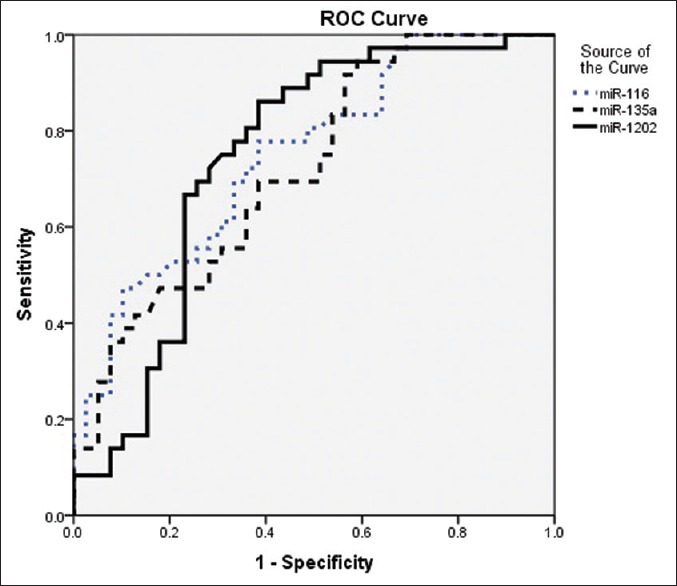
The combined receiver operating characteristic curve of the miR-16, miR-135a, and miR-1202. The plots were constructed using SPSS software. MiR-16 yields an area under the receiver operating characteristic curve of 0.75 (sensitivity of 77.8% with a specificity of 61.5%). MiR-135a also yields an area under the receiver operating characteristic curve of 0.72 (sensitivity of 94.4% with a specificity of 41.0%). MiR-1202 also yields an area under the receiver operating characteristic curve of 0.74 (sensitivity of 86.1% with a specificity of 61.5%)
DISCUSSION
It is of interest to note that in recent decades of research, it has become clear that alteration in miRNA expression are likely to play an important role in the etiopathogenesis of behavioral disorders, and have the potential to act as serum biomarkers.
The results of the current study showed that serum level of miR-16 was reduced in patients with depression. This finding is in line with Baudry et al. how have shown that SERT is a target of miR-16 which is expressed at higher level in nerve cells. In mice model, treatment with the SSRI fluoxetine increases miR-16 levels in serotonergic RN. This study showed that miR-16 posttranscriptionally regulates SERT gene that strongly involved in depression.[22,23] In this line, our study also confirmed that in depressed patients miR-16 is down-regulated.
In some previous studies, it has been indicated that miR-135a widely associates with both the 5-HT transporter and 5-HT1A receptor expression. MiR-135a levels also were up-regulated after using antidepressant administration. In addition, using genetically modified animal model showed that alteration in miR-135 levels largely affects 5-HT levels and depression-like behaviors.[24] Furthermore, previously, it has been shown that miR-135a expression appears to be antidepressant.[24,30] In parallel with the previous finding, here, we observed that the down-regulation of miR-135a expression levels is exhibited in the serum of depressed patients in compared to healthy individuals.
Using microarray analysis and qRT-PCR, Lopez et al., have shown that GRM4 which is expressed throughout the brain, and it modulates serotonergic neurotransmission, negatively associate with the expression of miR-1202.[25] In line with such observation, our findings showed that miR-1202 exhibited a similar pattern with miR-16 and miR-135a and it decreased in serum of patients with depression.
In the present study, we showed that serum miRNAs can be used as biomarkers for depression. Interestingly, aberrant pattern of these miRNAs is consistent with previous studies.[31]
Here, a model including 3 miRNAs to diagnose depression was developed. Indeed, the miRNA panel can improve the diagnostic sensitivity and specificity for depression in comparison to a single miRNA. Taken together, our observation indicated that the serum miRNA signature is an independent predictive, diagnostic, and prognostic factor for depression. However, large body of studies have been devoted to developing circulating biomarkers for depression to improve diagnosis and individualized treatment.
CONCLUSION
The present study was designed to evaluate the serum concentration of some miRNAs which have been shown to involve in the serotoninergic system, in the patients with depression. The results showed that the serum levels of 3 miRNAs (miR-16, miR-135a, and miR-1202) were significantly decreased in the serum of depressed patients in comparison with normal individuals. The current study suggests that these miRNAs could be used as a potential biomarker of depression with high sensitivity and specificity.
Financial support and sponsorship
This study was supported by Ilam University of Medical Sciences.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors appreciate Ilam University of Medical Sciences for financial support (grant no 969001/11).
REFERENCES
- 1.Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Geneva: World Health Organization, Department of Mental Health and Substance Dependence; 2001. Mental Health: A Call for Action by World Health Ministers. [Google Scholar]
- 3.Parikh SV, Lin E, Lesage AD. Mental health treatment in Ontario: Selected comparisons between the primary care and specialty sectors. Can J Psychiatry. 1997;42:929–34. doi: 10.1177/070674379704200903. [DOI] [PubMed] [Google Scholar]
- 4.Vázquez-Barquero JL, García J, Simón JA, Iglesias C, Montejo J, Herrán A, et al. Mental health in primary care. An epidemiological study of morbidity and use of health resources. Br J Psychiatry. 1997;170:529–35. doi: 10.1192/bjp.170.6.529. [DOI] [PubMed] [Google Scholar]
- 5.Weil-Malherbe H. Handbook of Neurochemistry. Boston, MA: Springer; 1972. The biochemistry of affective disorders; pp. 371–416. [Google Scholar]
- 6.Lee S, Jeong J, Kwak Y, Park SK. Depression research: Where are we now? Mol Brain. 2010;3:8. doi: 10.1186/1756-6606-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebmeier KP, Donaghey C, Steele JD. Recent developments and current controversies in depression. Lancet. 2006;367:153–67. doi: 10.1016/S0140-6736(06)67964-6. [DOI] [PubMed] [Google Scholar]
- 8.Leistedt SJ, Linkowski P. Brain, networks, depression, and more. Eur Neuropsychopharmacol. 2013;23:55–62. doi: 10.1016/j.euroneuro.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Ota KT, Duman RS. Environmental and pharmacological modulations of cellular plasticity: Role in the pathophysiology and treatment of depression. Neurobiol Dis. 2013;57:28–37. doi: 10.1016/j.nbd.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surtees PG, Wainwright NW, Willis-Owen SA, Luben R, Day NE, Flint J, et al. Social adversity, the serotonin transporter (5-HTTLPR) polymorphism and major depressive disorder. Biol Psychiatry. 2006;59:224–9. doi: 10.1016/j.biopsych.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 11.De Neve JE, Christakis NA, Fowler JH, Frey BS. Genes, economics, and happiness. J Neurosci Psychol Econ. 2012;5:193–211. doi: 10.1037/a0030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeroff CB. Recent advances in the neurobiology of depression. Psychopharmacol Bull. 2002;36( Suppl 2):6–23. [PubMed] [Google Scholar]
- 13.Artigas F, Perez V, Alvarez E. Pindolol induces a rapid improvement of depressed patients treated with serotonin reuptake inhibitors. Arch Gen Psychiatry. 1994;51:248–51. doi: 10.1001/archpsyc.1994.03950030084009. [DOI] [PubMed] [Google Scholar]
- 14.de Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: A review. Anal Chim Acta. 2011;699:134–52. doi: 10.1016/j.aca.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer disease and other human CNS disorders. Curr Genomics. 2009;10:154–68. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocerha J, Kauppinen S, Wahlestedt C. MicroRNAs in CNS disorders. Neuromolecular Med. 2009;11:162–72. doi: 10.1007/s12017-009-8066-1. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 19.Jin XF, Wu N, Wang L, Li J. Circulating microRNAs: A novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell Mol Neurobiol. 2013;33:601–13. doi: 10.1007/s10571-013-9940-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Tréziny C, Verrier L, et al. Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry. 2012;2:e185. doi: 10.1038/tp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23:602–11. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. MiR-16 targets the serotonin transporter: A new facet for adaptive responses to antidepressants. Science. 2010;329:1537–41. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 23.Bai M, Zhu X, Zhang Y, Zhang S, Zhang L, Xue L, et al. Abnormal hippocampal BDNF and miR-16 expression is associated with depression-like behaviors induced by stress during early life. PLoS One. 2012;7:e46921. doi: 10.1371/journal.pone.0046921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83:344–60. doi: 10.1016/j.neuron.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 25.Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, et al. MiR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med. 2014;20:764–8. doi: 10.1038/nm.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rush A, First M, Blacker D. Task Force for the Handbook of Psychiatric Measures. Handbook of Psychiatric Measures. Arlington, VA, US, American Psychiatric Association. 2008 [Google Scholar]
- 27.Gheysarzadeh A, Yazdanparast R. STAT5 reactivation by catechin modulates H2O 2-induced apoptosis through miR-182/FOXO1 pathway in SK-N-MC cells. Cell Biochem Biophys. 2015;71:649–56. doi: 10.1007/s12013-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ, et al. MiR-1254 and miR-574-5p: Serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–8. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 30.Heyer MP, Kenny PJ. MicroRNA-mediated repression combats depression. Neuron. 2014;83:253–4. doi: 10.1016/j.neuron.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: MicroRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17:359–76. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]


