Abstract
We sought to investigate the underlying mechanism of action of the long noncoding RNA (lncRNA) LOC283070 in the development of androgen independence in prostate cancer. The interactions between LOC283070 and target proteins were investigated by RNA pull-down and RNA-binding protein immunoprecipitation (RIP) assays. Subcellular fractionation and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) were used to detect the subcellular localization of LOC283070. Western blotting was performed to detect the expression of prohibitin 2 (PHB2). Luciferase activity assays were performed to evaluate the effects of LOC283070 and PHB2 on the androgen receptor (AR) signaling pathway. A methyl thiazolyl tetrazolium (MTT) assay and a growth curve assay were used to test cell viability. Flow cytometry was performed to analyze cell cycles. A transwell assay was employed to test cell migration. We identified PHB2 as an interaction partner of LOC283070 in the pull-down and RIP experiments. Furthermore, we confirmed that the enrichment of LOC283070 with PHB2 in androgen-independent LNCaP (LNCaP-AI) cells was much greater than that in LNCaP cells. Moreover, the expression of PHB2 was not significantly different between the two cell lines, and the expression of LOC283070 in the nuclei of the LNCaP-AI cells was significantly greater than that in the LNCaP cells. In vitro data revealed that PHB2 overexpression significantly inhibited AR activity and cell proliferation and migration and induced accumulation of prostate cancer cells in G0/G1 phase. Moreover, the overexpression of LOC283070 fully abrogated the effects of PHB2 overexpression. In conclusion, we found that LOC283070 can bind to PHB2 located in the nucleus and inhibit its effect, and this is one of the mechanisms by which LOC283070 is involved in the transition of LNCaP cells into androgen-independent cells.
Keywords: androgen-dependent prostate cancer, androgen-independent prostate cancer, LOC283070, long noncoding RNA, prohibitin 2
INTRODUCTION
Prostate cancer (PCa) is the most prevalent cancer among males. The American Cancer Society reported that PCa has become the cancer with the second-highest mortality rate among cancer in men.1 Androgen ablation therapies are the mainline treatment for patients with advanced PCa. Despite the high initial response rates, these tumors eventually develop into castration-resistant prostate cancer (CRPC),2 which is the major cause of death among prostate cancer patients.3 However, the underlying molecular mechanism of the emergence of CRPC is not fully understood.
We established a CRPC cell line termed androgen-independent LNCaP (LNCaP-AI)4,5 from the androgen-dependent prostate carcinoma (ADPC) cell line LNCaP via prolonged androgen deprivation culture. Next, we determined that LOC283070, a novel long noncoding RNA (lncRNA), plays an important role in the progression of CRPC.6
The microarray expression profiles revealed that LOC283070 was significantly upregulated in the LNCaP-AI cells compared with the LNCaP cells. Further studies demonstrated that LOC283070 participated in the transformation of LNCaP cells into androgen-independent cells by modulating the expression of calcium/calmodulin-dependent protein kinase ID (CAMKID).6 CAMKID is an identified oncogene in basal-like tumors that can promote cell proliferation and increase cell cycle progression by directly targeting the cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB) pathway.7,8 Ectopic LOC283070 expression upregulated the expression of CAMKID, which further increased the proliferation and migration of LNCaP cells, even in androgen-independent circumstances, and promoted tumor growth in both normal and castrated mice. LOC283070 knockdown in LNCaP-AI cells downregulated the expression of CAMKID and decreased proliferation, cell cycle progression, and migration.
Studies have demonstrated that lncRNAs can be involved in gene expression regulation in a variety of manners.9 (1) LncRNAs serve as scaffolds that assemble chromatin remodeling machinery at the site of regulation. HOX transcript antisense RNA (HOTAIR) was the first noncoding gene that was discovered to perform this function;10 the upregulation of HOTAIR in a subset of primary breast tumors led to a genome-wide retargeting of the polycomb repressive complex 2 (PRC2) and histone 3 lysine 27 trimethylation (H3K27me3) patterns, resulting in gene expression changes to promote tumor metastasis.11 (2) LncRNAs may serve as recruiters that regulate gene expression in cis and trans fashions. (3) LncRNAs serve as molecular sponges because they harbor binding sites for microRNAs and titrate them away from their mRNA targets. (4) LncRNAs act as precursors of small noncoding RNAs (ncRNAs). (5) LncRNAs can interact with proteins, nucleic acid molecules, and even RNA-RNA-protein and RNA-DNA-protein complexes through their secondary structures. Moreover, lncRNAs can play a variety of roles simultaneously. For example, lncRNAs activated by transforming growth factor-beta (LncRNA-ATB) not only competitively bind microRNA-20012 as a sponge, but also bind to interleukin (IL)-11 mRNA.13,14 These observations indicate the flexibility of the mechanisms of action of lncRNAs.
Our previous study revealed that LOC283070 participates in the progression of CRPC via promoting proliferation and migration that is partially mediated by increasing CAMKID protein expression.6 LncRNAs function through a variety of mechanisms, and understanding this functional versatility is critical for the clinical exploitation of lncRNAs. Therefore, in this article, we further explored the molecular mechanisms of action of LOC283070.
MATERIALS AND METHODS
Cell cultures
The androgen-dependent human prostate cancer cell line LNCaP was obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences and cultured in RPMI-1640 (Life Technologies, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) at 37°C in 5% CO2. The LNCaP-AI cells were cultured in RPMI-1640 supplemented with 10% charcoal-treated (stripped) fetal bovine serum.
Construction of the expression and reporter vectors
The prohibitin 2 (PHB2) coding sequence was amplified by qRT-PCR and then cloned into the pcDNA3.1(+) vector (Invitrogen, Shanghai, China). The resulting vector was named pcDNA3.1(+) - PHB2. The following primers were used: F: 5’-CCCAAGCTTATGGCCCAGAACTTGAAGGACTTGG-3’ and R: 5’-CTCTAGATCATTTCTTACCCTTGATGAGGCTGTCAC-3’.
The androgen response element (ARE) sequences (F: 5′-TGGAGGAACATATTGTATTTATT-3′ and R: 5′-AATAAATACAATATGTTCCTCCA-3′) were synthesized and cloned into pGL4.23[luc2/minP] (Promega, Madison, WI, USA) to construct pGL4-ARE.
The vectors were transfected into prostate cancer cells using X-tremeGENE HP DNA transfection reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Luciferase assay
After the transfections of PHB2 and LOC283070 for 24 h, pGL4-ARE was co-transfected with pGL4.74 [hRluc/TK] in the LNCaP cells. The pGL4.23[luc2/minP] was also co-transfected with pGL4.74[hRluc/TK] as a control. Firefly and Renilla luciferase activities were measured at 48 h posttransfection using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Western blotting
Equal amounts of protein were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, blotted onto polyvinylidene fluoride membranes, and incubated overnight at 4°C in Tris-buffered saline with Tween 20 containing 5% skim milk. Next, the membranes were incubated with rabbit anti-human PHB2 primary antibody (1:5000; ab71970, Abcam, Cambridge, UK) overnight at 4°C. Mouse anti-β-actin antibody (1:5000; Santa Cruz, Dallas, TX, USA) was used as a control. The immunoblots were detected using an electrochemiluminescence kit (Santa Cruz) and visualized after X-ray film exposure.
Methyl thiazolyl tetrazolium assay
For all cell viability studies, the cells were plated in 96-well plates after transfection with the pcDNA3.1(+) and pcDNA3.1(+)-PHB2 vectors, the pcDNA3.1(+)-PHB2 and pcDNA3.1(+)-LOC283070 vectors, or the pcDNA3.1(+) vector. MTT (tetrazolium salt, 10 μl, 5 mg ml−1) cell proliferation assays were then performed on various days according to the manufacturer's instructions. The assays were performed in triplicate.
Cell growth curve
Twenty-four hours after transfection, the cells were inoculated in 6-well plates at a density of 1 × 105 per well. The cell numbers were counted every day for 1 week using an hemocytometer. Cell growth curves were drawn with the cell numbers on the y-axis and the time on the x-axis to assess the cell proliferation abilities. The assays were performed in triplicate.
Flow cytometry
For the cell cycle assays, LNCaP and LNCaP-AI cells were transfected for 24 h. Then, the cells were cultured for 72 h and collected and fixed in 75% ethanol at 4°C overnight. After washing twice with phosphate-buffered saline (PBS), the cells were incubated with 30 μg ml−1 propidium iodide (PI), 0.2 mg ml−1 RNase A, and 0.2% Triton X-100 (all from Sigma-Aldrich, St. Louis, MO, USA) at 37°C for 30 min. The cells were then analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's manual. The assays were performed in triplicate.
Transwell assays
Cells (5 × 104) in 100 μl of serum-free medium were added to the upper chamber of a Transwell plate after transfection, and 600 μl of medium containing 10% fetal bovine serum was added to the lower chamber. After 48 h of incubation at 37°C, nonmigrated cells on the top of the membrane were scraped and removed with cotton swabs, and the migrated cells were fixed, stained with 0.1% crystal violet, and counted under an inverted microscope (ECLIPSE TE2000-U, Nikon, Tokyo, Japan). The migration assays were repeated three times with duplicate wells.
RNA pull-down
pcDNA3.1(+)-LOC283070, pcDNA3.1(+)-antisense-LOC283070, and pcDNA3.1(+) were linearized with the corresponding restriction enzymes that were used to clone the vectors at the 3’ end to prepare the template DNAs for in vitro transcription. All biotin-labeled RNA transcripts were produced in vitro using the MEGAscript T7 Kit (Ambion, Waltham, MA, USA) with biotin-16-UTP (Ambion) and purified with MEGAclear Kits (Ambion) according to the protocol provided by the manufacturer. Three micrograms of biotinylated RNA was heated at 90°C for 5 min, left at room temperature for 30 min, cooled to 4°C, mixed with 1 mg of protein extracted from the LNCaP or LNCaP-AI cells, and incubated at room temperature for 2 h. Then, 60 μl of streptavidin agarose beads (Invitrogen) was added to each binding reaction, and a further incubation at room temperature for 2 h was performed. Finally, the beads were washed five times and boiled in 1× loading buffer for 5 min. The associated proteins were resolved by SDS-PAGE, and the specific bands were excised and analyzed by mass spectrometry.
RNA-binding protein immunoprecipitation
The RIP assays were performed using EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kits (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. Briefly, LNCaP or LNCaP-AI cells were rinsed with ice-cold PBS and lysed with RIP lysis buffer. The PHB2 antibody (ab71970, Abcam) was used for the RIP. Furthermore, the co-precipitated RNAs were detected by qRT-PCR. The reaction conditions were as follows: denaturation at 95°C for 3 min; 40 PCR cycles at 95°C for 10 s and 60°C for 30 s. The total RNAs (input controls) and IgG were simultaneously assayed to demonstrate that the detected signals were the result of RNAs that specifically bound to PHB2. The gene-specific primers that were used to detect LOC283070 were as follows: 5’- AGGCGGTCTGAGGAAGATAAGG -3’ (sense) and 5’- TCCCACTGACTCTGGAGGCAT-3’ (antisense).
Subcellular fractionation
The separation of the nuclear and cytosolic fractions was performed using a PARIS Kit (Invitrogen) according to the manufacturer's instructions. The RNAs in the nuclei and cytoplasm were isolated and extracted from the LNCaP and LNCaP-AI cells, and the expression levels of LOC283070 in the nuclei and cytoplasm of both cells were examined by qRT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were detected as fractionation indicators.
Statistical analysis
All data were analyzed using the SPSS 17.0 software package (SPSS Inc., Chicago, IL, USA). All values are expressed as the mean ± standard deviation (s.d.) based on at least three independent experiments, and statistical significance was defined as P < 0.05. All assays were performed in triplicate.
RESULTS
LOC283070 specifically binds to PHB2 and might affect the function of the PHB2 protein
Several studies have demonstrated that lncRNAs are involved in molecular regulation pathways due to interactions with proteins.15,16 Therefore, we assumed that LOC283070 might affect cellular function in a similar manner. To test this hypothesis, the LOC283070-interacting proteins were identified with a biotinylated RNA pull-down assay using protein lysates that were isolated from the LNCaP-AI cells and incubated with the biotinylated LOC283070 and the two negative control transcripts, i.e., biotinylated LOC283070-antisense and biotinylated pcDNA3.1(+). We resolved the RNA-associated proteins by SDS-PAGE, cut out the bands specific to LOC283070, and subjected them to mass spectrometry (Figure 1a and 1b). Among all of the proteins that were identified by mass spectrometry, PHB2 aroused our attention (Figure 1b) because PHB has been reported to play a role in the progression to CRPC.17,18,19,20 Then, the association of LOC283070 with PHB2 was validated by western blotting as illustrated in Figure 1c. PHB2 was clearly detected in the LNCaP-AI cells in the biotinylated LOC283070 pull-down protein complexes but not in two negative control protein complexes.
Figure 1.
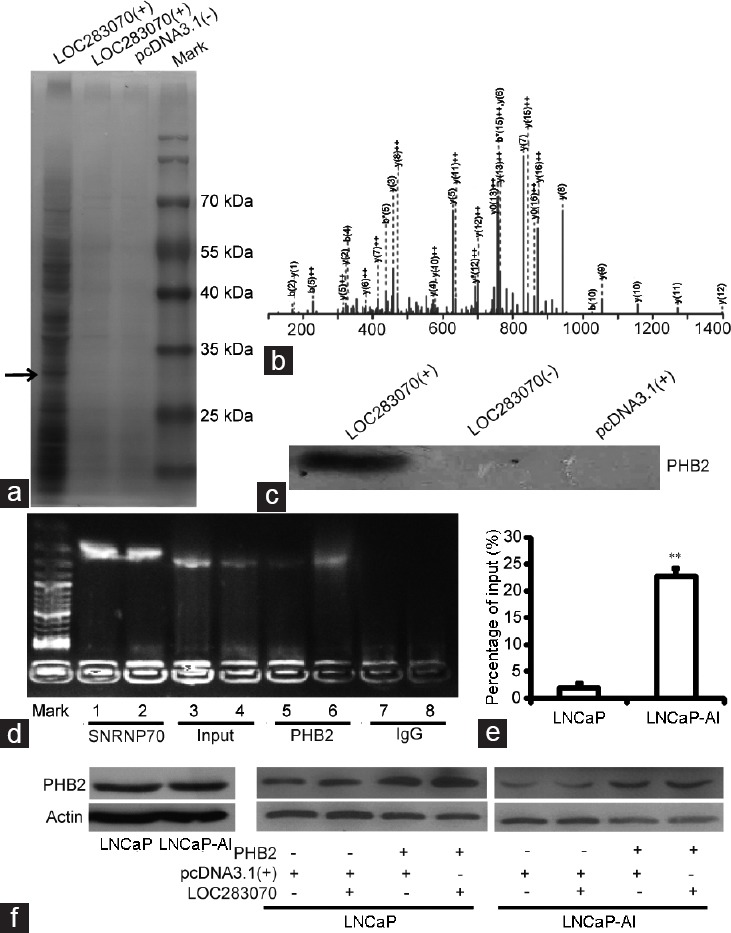
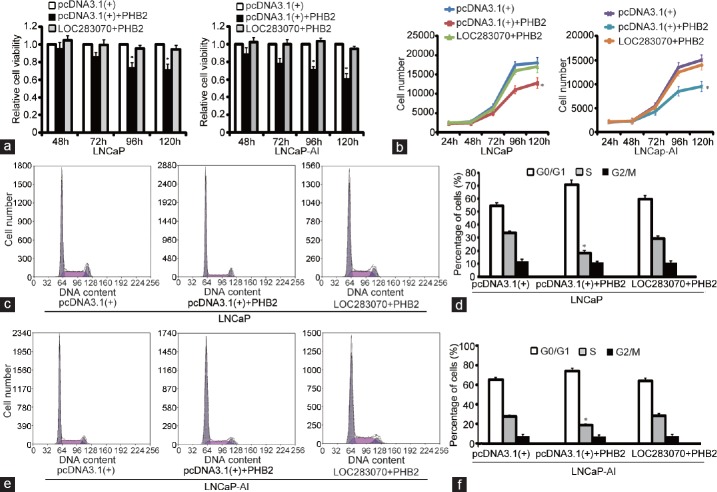
LOC283070 directly interacts with PHB2. (a) RNA pull-down experiment in LNCaP-AI cells. Proteins from LNCaP-AI extracts were pulled down with the biotin-RNAs, subjected to SDS-PAGE, and visualized by silver staining. The band, which was specific to LOC283070 in LNCaP-AI but not in two controls, was pointed out by the arrow and subjected to mass spectrometry. (b) Mass spectrometry identification of target band of PHB2. (c) Proteins pulled down by the biotin-RNAs as in (a) were analyzed by western blotting with PHB2 antibody. (d) RIP assay was performed using PHB2 antibody and was validated by agarose electrophoresis using LOC283070 primers. Lane mark: DNA molecular marker, lane: 1, 3, 5, 7 from LNCaP cells and 2, 4, 6, 8 from LNCaP-AI cells. RNA extracted with SNRP70 antibody and IgG were amplified by PCR experiments using U1 primers (as positive and negative controls, respectively). (e) Fold change enrichment of LOC283070 in LNCaP and LNCaP-AI was calculated comparing with the input in panel in qRT-PCR assay (n = 3, **P < 0.01). (f) PHB2 protein expression measured by western blotting. PHB2: prohibitin 2; qRT-PCR: quantitative reverse transcription-polymerase chain reaction; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; LNCaP-AI: androgen-independent LNCaP; RIP: RNA-binding protein immunoprecipitation.
Then, a RIP assay further confirmed the interaction between LOC283070 and PHB2 in vivo. An antibody against PHB2 was incubated with the LNCaP and LNCaP-AI cell lysates, and the co-precipitated RNAs were then analyzed by RT-PCR and qRT-PCR. As expected, we observed enrichment of LOC283070 with the PHB2 antibody compared with the nonspecific antibody (IgG control; Figure 1d), but the enrichment of LOC283070 with PHB2 in the LNCaP-AI cells was much greater than that in the LNCaP cells (Figure 1e). In contrast, the western blotting assay indicated that the expression of PHB2 was not significantly different between the two cell lines (Figure 1f). According to these data, we hypothesized that during the transition of LNCaP cells into LNCaP-AI cells, the expression of LOC283070 was increased, and the increased LOC283070 might have affected the expression or function of the PHB2 protein via direct interaction. Therefore, we co-overexpressed LOC283070 and PHB2 in LNCaP and LNCaP-AI cells. Western blotting results revealed that LOC283070 did not significantly affect the expression of the PHB2 protein (Figure 1f). Collectively, we concluded that LOC283070 might affect the function of the PHB2 protein by binding to it.
Overexpression of PHB2 inhibits prostate cancer cell proliferation and cell cycling, while LOC283070 overexpression fully abrogates the effects of PHB2
To confirm our inferences, we performed functional assays in the two cell lines. MTT assays applied at different time points indicated that the upregulation of PHB2 inhibited the proliferation of both the LNCaP and LNCaP-AI cells (P < 0.05), and the repressive effect of PHB2 on cell proliferation was evidently weakened by the co-transfection of LOC283070 in both cell lines (Figure 2a). Furthermore, cell growth curve analysis confirmed these results and revealed that the increased level of PHB2 caused a suppression of cell proliferation in both LNCaP and LNCaP-AI cell lines (P < 0.05). Moreover, there was no significant difference in the proliferation rates of the co-transfected cells compared with the negative controls (Figure 2b). These results suggest that the overexpression of PHB2 inhibits prostate cancer cell proliferation, while LOC283070 overexpression fully abrogates the effect of PHB2.
Figure 2.
Overexpression of PHB2 inhibits prostate cancer cell proliferation and cell cycle, and LOC283070 overexpression fully abrogates its effect. (a) MTT assay showing the cell viability in LNCaP and LNCaP-AI cells. (b) Cell growth curve analysis showing the proliferative ability in LNCaP and LNCaP-AI cells. (c and d) Cell cycle progression in LNCaP cells. (e and f) Cell cycle progression in LNCaP-AI cells. Data are from one of the three independent experiments and are represented as mean ± s.d.*P < 0.05. PHB2: prohibitin 2; LNCaP-AI: androgen-independent LNCaP; MTT: methyl thiazolyl tetrazolium; s.d.: standard deviation.
Next, flow cytometric analysis was also performed to analyze the cell cycle progression of the LNCaP and LNCaP-AI cells in the presence of PHB2 overexpression or LOC283070 and PHB2 co-overexpression. The results revealed that PHB2 overexpression led to cell cycle arrest in the G0/G1 phase and a subsequent reduction in the percentage of cells in the S phase (P < 0.05). In contrast, in the co-overexpressing cells, a distinct increase in the number of cells in S phase was observed (18.1% compared with 29.4%) (Figure 2c and 2d). Furthermore, similar results were observed in the LNCaP-AI cells (Figure 2e and 2f). Hence, these results indicated that increases in PHB2 levels led to a reduction in the number of cells that entered S phase and a reduction in cell growth, and these effects were no longer evident when the cells co-overexpressed LOC283070 and PHB2.
Overexpression of PHB2 inhibits prostate cancer cell migrating, but LOC283070 overexpression fully abrogates the effect of PHB2
To investigate whether PHB2 participates in cell migration, transwell assays were performed. PHB2-overexpressing cells exhibited a significantly lower migration potential than the cells that were transfected with the pcDNA3.1(+) vector and, in the LNCaP cells, PHB2 induced a decrease of approximately 46% in the migrated tumor cells compared with the vector control (P < 0.05). Moreover, there was no difference between the co-transfected cells and the cells that were transfected with the negative control vector (Figure 3a and 3b). Additionally, the migration ability of the LNCaP-AI cells was decreased by 43% compared with those transfected with the control vector (P < 0.05), and this decrease in migration ability was restored by LOC283070 (Figure 3c and 3d). These results suggest that the overexpression of PHB2 inhibits the migration of LNCaP and LNCaP-AI cells, and LOC283070 can fully abrogate the effects of PHB2.
Figure 3.
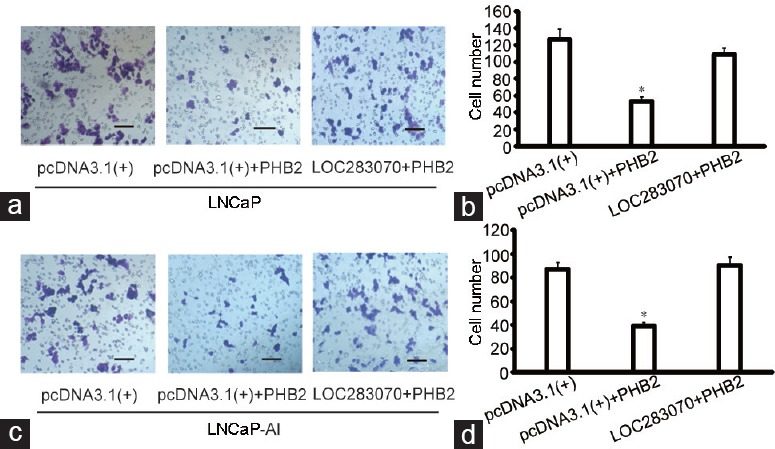
Overexpression of PHB2 inhibits prostate cancer cell migrating, LOC283070 fully abrogates the effect of PHB2. (a) Migration potential of LNCaP cells with the overexpression of LOC283070 and PHB2 or PHB2 (×200). (b) The specific number of migrated cells in a (n = 10, *P < 0.05). (c) Migration potential of LNCaP-AI cells with the overexpression of LOC283070 and PHB2 or PHB2 (×200). (d) The specific number of migrated cells in (c) (n = 10, *P < 0.05). Scale bars=100 μm. PHB2: prohibitin 2.
LOC283070, which localized in both the cytoplasm and the nucleus, blocked the repression of androgen receptor activity by PHB2
Growing evidence suggests that lncRNAs act by a multitude of regulatory mechanisms according to their specific location in the cell. Thus, the subcellular localization of LOC283070 in the LNCaP and LNCaP-AI cells was analyzed by subcellular fractionation. We found that LOC283070 was distributed in both the cytoplasm and nuclei in the two cell lines. Compared with the LNCaP cells, LOC283070 was significantly enriched in the nuclei of the LNCaP-AI cells (Figure 4a), which suggested that LOC283070 may have exerted its effects at the posttranscriptional level or through interactions with nuclear proteins.
Figure 4.
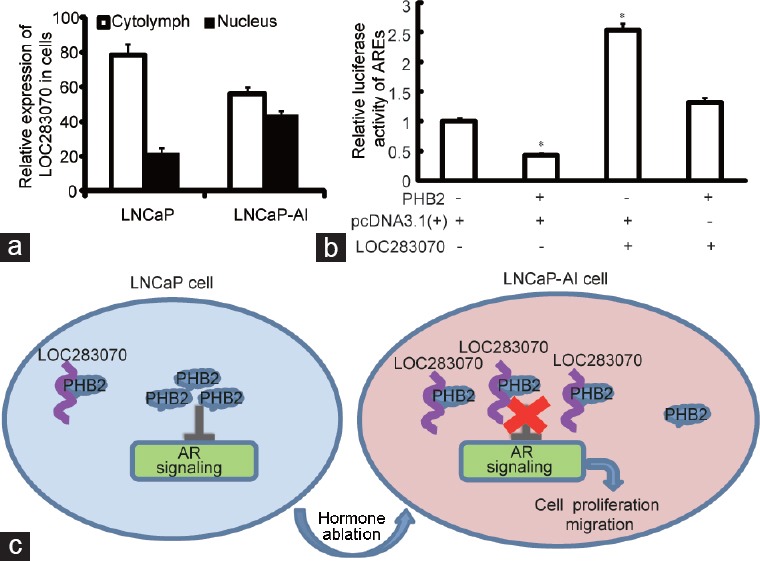
Summary of the mechanism of LOC283070 in the development of CRPC. (a) RNA was extracted from the nucleus and cytoplasm. qRT-PCR detection of the relative expression of LOC283070 in the nucleus and cytoplasm. (b) The activity of AR was detected by luciferase activity assay in LNCaP cells with the overexpression of LOC283070 and PHB2, LOC283070 and pcDNA3.1, and pcDNA3.1 and PHB2 (n = 3, *P < 0.05). (c) Summary of the mechanism of LOC283070 in the transition of LNCaP cells into LNCaP-AI cells. PHB2: prohibitin 2; qRT-PCR: quantitative reverse transcription-polymerase chain reaction; CRPC: castration-resistant prostate cancer; AR: androgen receptor; LNCaP-AI: androgen-independent LNCaP.
It has been reported that PHB is mainly localized to the nuclei of PCa cells where it functions as a co-repressor of AR. Reductions in PHB increase AR activity and promote androgen independence in prostate tumors.19 To further investigate whether LOC283070 is involved in the transition of LNCaP cells into LNCaP-AI cells due to its inhibition of the effect of PHB2, we cloned the ARE sequence into pGL4.23[luc2/minP] to form pGL4-ARE. Along with the ectopic expression of PHB2 or LOC283070, pGL4-ARE was transiently transfected into LNCaP cells to investigate the effects of PHB2 and LOC283070 on AR activity. PHB2 overexpression resulted in significant reductions in luciferase activities compared with the control group (P < 0.05), while a significant increase in luciferase activity was observed in the LOC283070 overexpressing group (P < 0.05), and the PHB2 and LOC283070 co-overexpression group exhibited no significant difference in luciferase activity compared with the control group (Figure 4b). Hence, increase in the PHB2 levels indeed led to reductions in AR activity in prostate cancer cells, and this effect was no longer evident when the cells were co-overexpressed with PHB2 and LOC283070.
DISCUSSION
Increasing evidence indicates that lncRNAs are major contributors to cellular homeostasis and the initiation and progression of numerous tumors, including PCa tumors.21,22 One multi-institutional analysis of 910 PCa patients revealed that low prostate cancer-associated transcript-14 (PCAT-14) expression is associated with poor outcome in prostate cancer.23 Mehra et al.24 found that SWI/SNF complex antagonist associated with prostate cancer 1 (SChLAP1) might be a useful tissue-based biomarker for identifying PCa patients who are at a higher risk of lethal progression. Moreover, PCAT5 has been identified as a novel oncogenic lncRNA in ERG-positive prostate cancer, which has implications for defining CRPC biomarkers and new therapeutic interventions.25
LncRNAs can play a variety of roles in the progression of PCa; however, due to the mechanistic complexity of these roles, there are no comprehensive studies clarifying their functions. The involvement of LOC283070 in the transformation may not be limited to the regulation of the CAMKID expression level but could also be mediated through interactions with other molecules. Recent studies have demonstrated that the gene regulation roles that lncRNAs play are mainly mediated through interactions with proteins.15,16 Long intergenic noncoding RNA (lincRNA)-p21,26 HOTAIR,27 NF-KappaB-interacting lncRNA (LncRNA-NKILA),28 lncRNA maternally expressed 3 (MEG3),29 and HOXA transcript at the distal tip (HOTTIP)30 function through lncRNA–protein complexes according to reports. At present, the search for molecules that interact with lncRNAs is primarily being conducted with RNA pull-down and RIP experiments; therefore, this study further investigated whether there are other molecular mechanisms that involve LOC283070 and influence the progression of CRPC.
RNA pull-down and RIP experiments were performed, and we discovered that LOC283070 can specifically interact with PHB2. PHB2 is a ubiquitous, strongly evolutionarily conserved protein that belongs to the prohibitin family, which includes a pair of proteins, i.e. PHB1 and PHB2. Many studies of prostate cancer have demonstrated that PHB participates in the progression of CRPC. Ramesh et al.31 identified prohibitin as a biomarker for distinguishing hyperplasia from cancer using proteomic analysis of prostate biopsies. The study of Dai et al.32 suggested that prohibitin and BRG1 are required for effective androgen antagonist-mediated transcriptional repression of AR-regulated genes. Dart et al.19 also reported that the overexpression of PHB inhibits AR activity, prostate-specific antigen (PSA) expression, and androgen-dependent cell growth to induce the rapid accumulation of cells in G0/G1 phase. The present study provided proof of principle that the reduction of PHB promotes both androgen-dependent and “androgen-independent” tumor growth. Gamble et al.18 reported that prohibitin is downregulated by androgens and represses AR activity. Dart et al.20 concluded that prohibitin loss may provide a mechanism for the progression to CRPC via the sensitization of prostate cancer cells to “castration” conditions.
Therefore, we inferred that at least one of the mechanisms responsible for the involvement of LOC283070 in the transition of LNCaP cells to LNCaP-AI cells was mediated through the interaction of LOC283070 with PHB2. RIP experiments further confirmed that LOC283070-PHB2 binding occurred at a much higher rate in the LNCaP-AI cells than in the LNCaP cells. The western blotting results revealed that the expression of the PHB2 protein was not significantly different between the LNCaP-AI cells and LNCaP cells, and LOC283070 overexpression did not significantly affect the expression of the PHB2 protein. Based on all of these results, we concluded that LOC283070 might affect the function of PHB2 protein by binding to it. Thus, a series of functional tests were performed to confirm our speculation, and the results revealed that the overexpression of PHB2 alone inhibited cell proliferation and migration and induced cell accumulation in G0/G1 phase, and these results are consistent with those of previous research reports.19,20 The overexpression of LOC283070 fully abrogated the effect of PHB2 overexpression in PCa cells.
Next, we further explored the molecular mechanisms by which LOC283070-PHB2 binding facilitated transformation. It has been reported32 that, even if prostate cancer cells are converted to an androgen-independent state, the AR signaling pathway is still key for their growth. The proposed mechanisms of progression, including AR gene mutation and receptor amplification, are insufficient to completely explain CRPC. Many studies17,18,19,20 have demonstrated that reduced levels of repressor proteins such as PHB may increase the activity of AR due to a lack of repressive action. PHB is a highly conserved protein with dual roles within the cell. In mitochondria, PHB is located in the inner membrane, where it functions as a chaperone,33 and in the nuclei of several steroid hormone-responsive cell lines, the functions of PHB are more complex.34 In PCa, PHB functions as a co-repressor of AR,19 and this function has been observed in the promoters of several genes, including both AR- and non-AR-regulated genes. Furthermore, PHB knockdown increases histone acetylation at these promoters, increases the rate of AR ligand-induced chromatin binding, increases the binding rate, and increases the AR on the PSA promoter. These processes result in increased cell growth and AR activity in response to all androgens, including the promotion of the responses to the weaker adrenal androgens that are normally absent at physiological concentrations.19,20
The subcellular localization results revealed that LOC283070 was distributed in both the cytoplasm and the nuclei of both cell types, but the enrichment of LOC283070 in the nuclei was significantly increased in the LNCaP-AI cells compared with the LNCaP cells, which suggests that during the transformation of LNCaP to LNCaP-AI cells, some LOC283070 enter the nucleus, where it may combine with the PHB2 that is distributed in the nucleus and inhibits its function.
Next, the luciferase activity assay further confirmed our hypothesis; PHB2 overexpression led to a reduction in the AR activity of prostate cancer cells, and this effect was no longer evident when the cells were treated with PHB2 and LOC283070 co-overexpression. In short, our results indicated that during the transition of LNCaP cells to LNCaP-AI cells, the expression of LOC283070 was increased, and the increased LOC283070 activated the AR signaling pathway by binding to more PHB2 protein that was located in the nucleus (Figure 4c). This process leads to the promotion of PCa cell proliferation and metastasis and ultimately promotes LNCaP cell hormone-independent transformation.
CONCLUSIONS
Our data suggest that LOC283070 can bind to the PHB2 that is distributed in the nucleus and inhibit its function and thereby activate the AR signaling pathway. This process could play a role in the transition of LNCaP cells into androgen-independent cells. To further confirm these findings, functional experiments in additional CRPC cell models, such as DU145 and PC3 cells, are part of our future research plans. Moreover, the specific aspect of the binding of LOC283070 and PHB2 and the specific molecules that are activated in the AR downstream signaling pathway require further study.
AUTHOR CONTRIBUTIONS
YZ performed the RNA pull-down and RIP experiments; participated in the subcellular fractionation, qRT-PCR, western blotting, and statistical analyses; and drafted the manuscript. LNW performed the cell experiments, luciferase activity assays, data analysis, and statistical analyses and helped to draft the manuscript. YNL implemented the plasmid construction. YXX, YS, and JZ were responsible for the cell cultures and protein extraction. WWC conceived the study and participated in its design and coordination. BH provided many helpful suggestions during the process of this study. All authors have read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 81702538, 81372764, and 81528015) and the Natural Science Foundation of Shandong Province (No. ZR201702160271, ZR2018MH030, and ZR2018MH025).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu W, Yao J, Huang Y, Li Q, Li W, et al. LXR agonist regulates the carcinogenesis of PCa via the SOCS3 pathway. Cell Physiol Biochem. 2014;33:195–204. doi: 10.1159/000356662. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Liu C, Li C, Xue J, Zhao S, et al. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene. 2015;572:252–8. doi: 10.1016/j.gene.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Chen Z, Hu X, Wang L, Li C, et al. MicroRNA-185 downregulates androgen receptor expression in the LNCaP prostate carcinoma cell line. Mol Med Rep. 2015;11:4625–32. doi: 10.3892/mmr.2015.3332. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Lin Y, Meng H, Liu C, Xue J, et al. Long non-coding RNA LOC283070 mediates the transition of LNCaP cells into androgen-independent cells possibly via CAMK1D. Am J Transl Res. 2016;8:5219–34. [PMC free article] [PubMed] [Google Scholar]
- 7.Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, et al. CAMK1D amplifcation implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2:327–39. doi: 10.1016/j.molonc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Mora OG, LaHair MM, McCubrey JA, Franklin RA. Calcium/calmodulin-dependent kinase I and calcium/calmodulin-dependent kinase kinase participate in the control of cell cycle progression in MCF-7 human breast cells. Cancer Res. 2005;65:5408–16. doi: 10.1158/0008-5472.CAN-05-0271. [DOI] [PubMed] [Google Scholar]
- 9.Lyer MK, Niknafs YS, Malik R, Dhanasekaran SM, Wu YM, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–64. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krstic J, Trivanovic D, Mojsilovic S, Santibanez JF. Transforming growth factorbeta and oxidative stress interplay: implications in tumorigenesis and cancer progression. Oxid Med Cell Longev. 2015;2015:654–94. doi: 10.1155/2015/654594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, et al. A long noncoding RNA activated by TGFbeta promotes the invasionmetastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;26:1038–49. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble SC, Odontiadis M, Waxman J, Westbrook JA, Dunn MJ, et al. Androgens target prohibitin to regulate proliferation of prostate cancer cells. Oncogene. 2004;23:2996–3004. doi: 10.1038/sj.onc.1207444. [DOI] [PubMed] [Google Scholar]
- 18.Gamble SC, Chotai D, Odontiadis M, Dart DA, Brooke GN, et al. Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene. 2007;26:1757–68. doi: 10.1038/sj.onc.1209967. [DOI] [PubMed] [Google Scholar]
- 19.Dart DA, Spencer-Dene B, Gamble WJ, Bevan CL. Manipulating prohibitin levels provides evidence for an in vivo role in androgen regulation of prostate tumours. Endocr Relat Cancer. 2009;16:1157–69. doi: 10.1677/ERC-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dart DA, Brooke GN, Sita-Lumsden A, Waxman J, Bevan CL. Reducing prohibitin increases histone acetylation, and promotes androgen independence in prostate tumours by increasing androgen receptor activation by adrenal androgens. Oncogene. 2012;31:4588–98. doi: 10.1038/onc.2011.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie C, Yuan J, Li H, Li M, Zhao G, et al. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:98–103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 23.White NM, Zhao SG, Zhang J, Rozycki EB, Dang HX, et al. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur Urol. 2017;71:257–66. doi: 10.1016/j.eururo.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, et al. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur Urol. 2016;70:549–52. doi: 10.1016/j.eururo.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ylipaa A, Kivinummi K, Kohvakka A, Annala M, Latonen L, et al. Transcriptome sequencing reveals PCAT5 as a novel ERG-regulated long noncoding RNA in prostate cancer. Cancer Res. 2015;75:4026–31. doi: 10.1158/0008-5472.CAN-15-0217. [DOI] [PubMed] [Google Scholar]
- 26.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains inhuman HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Sun L, Liu Q, Gong C, Yao Y, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Wang G, Jia T, Zhang L, Li Y, et al. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget. 2016;7:23988–4004. doi: 10.18632/oncotarget.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–23. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramesh U, Heike J, Uwe Z, Venz S, Teller S, et al. Prohibitin identified by proteomic analysis of prostate biopsies distinguishes hyperplasia and cancer. Cancer Letters. 2008;266:171–85. doi: 10.1016/j.canlet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 32.Dai Y, Ngo D, Jacob J, Forman LW, Faller DV. Prohibitin and the SWI/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor-regulated genes. Carcinogenesis. 2008;29:1725–33. doi: 10.1093/carcin/bgn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci. 2002;59:143–55. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem. 2006;281:2951–9. doi: 10.1074/jbc.M508669200. [DOI] [PubMed] [Google Scholar]