Abstract
When organisms are subjected to environmental challenges, including growth inhibitors and toxins, evolution often selects for the duplication of endogenous genes, whose overexpression can provide a selective advantage. Such events occur both in natural environments and in clinical settings. Microbial cells—with their large populations and short generation times—frequently evolve resistance to a range of antimicrobials. While microbial resistance to antibiotic drugs is well documented, less attention has been given to the genetic elements responsible for resistance to metal toxicity. To assess which overexpressed genes can endow gram-negative bacteria with resistance to metal toxicity, we transformed a collection of plasmids overexpressing all E. coli open reading frames (ORFs) into naive cells, and selected for survival in toxic concentrations of six transition metals: Cd, Co, Cu, Ni, Ag, Zn. These selections identified 48 hits. In each of these hits, the overexpression of an endogenous E. coli gene provided a selective advantage in the presence of at least one of the toxic metals. Surprisingly, the majority of these cases (28/48) were not previously known to function in metal resistance or homeostasis. These findings highlight the diverse mechanisms that biological systems can deploy to adapt to environments containing toxic concentrations of metals.
Keywords: Evolution of resistance, Antimicrobial resistance, Acquired resistance by gene duplication, Resistance to metal toxicity
Introduction
Humans have been exploiting the antimicrobial properties of transition metals for thousands of years (Lemire et al. 2013). Although their use faded considerably in the 1920s with the advent of the “antibiotic era” (Aminov 2010), the increasing prevalence of antibiotic-resistant pathogens has led to a resurgence in the use of transition metals to thwart the spread of microbial infection in clinical settings, including bandages, catheters, implants, and hospital surfaces (Grass et al. 2011; Davies and Davies 2010). However, as is the case for nearly all antimicrobials, continued use of toxic metals can lead to resistance (Alam and Imran 2014; Brocklehurst and Morby 2000; Hobman and Crossman 2015).
Resistance to toxins can arise through the innovative use of preexisting genetic information. This frequently involves the duplication of a gene and the concomitant overexpression of the encoded protein (Jensen 1976; Ohno 2013; Patrick et al. 2007; Näsvall et al. 2012; Kacar and Gaucher 2012; Herron and Doebeli 2013; Levin et al. 2014; Hughes and Andersson 2015; Montealegre et al. 2017; Notebaart et al. 2018).
In the current study, we asked whether overexpression of endogenous genes can endow cells with resistance to toxic concentrations of transition metals. To model the impact of enhanced expression following chromosomal gene duplication, we overexpressed a complete set of E. coli open reading frames from an inducible strong promotor on high-copy plasmids from the ASKA collection (Kitagawa et al. 2006) and assessed which overexpressed ORFs endow cells with resistance to any of six different transition metals.
Escherichia coli cells were transformed with the ASKA overexpression plasmids, and plated on lethal concentrations of cadmium, cobalt, copper, nickel, silver, or zinc. Viable colonies were isolated, and sequencing of their plasmid DNA identified which overexpressed ORFs had endowed cells with resistance to toxic metals. Analysis of the identified proteins revealed that several had been described in previous studies. Surprisingly, however, the majority of the overexpressed proteins had not been reported previously to mediate resistance to metal toxicity in E.coli.
Because overexpression of these genes enhanced resistance to toxic concentrations of certain transition metals, we also examined whether deletion of these same genes would produce hypersensitive strains of E. coli. Thus, knockout strains for each of the identified genes were screened for increased sensitivity to metals. Surprisingly, only a few of the deleted genes produced hypersensitive strains.
The findings presented herein demonstrate that overexpression of a range of different endogenous genes can endow cells with resistance to toxic concentrations of transition metals. Many of the identified genes appear to be involved in mediating generalized resistance to antimicrobial agents, which emphasizes the importance of adopting prudent measures when using—or overusing—metals and other antimicrobials in clinical settings.
Results
Selection for Overexpressed Genes That Mediate Resistance to Metal Toxicity
To identify genes whose overexpression would enable cells to escape the toxic effects of transition metals, we tested all 4288 open reading frames (ORFs) in E. coli, expressed from the ASKA plasmid library. The ASKA library (A Complete Set of Escherichia coli K-12 ORF Archive) is a collection of pCA24N plasmids, with each ORF expressed under the control of a T5/lacZ promoter. Each plasmid expresses a single ORF fused to a histidine tag and seven spacer amino acids at its N-terminus, and five spacer amino acids and a GFP tag at the C-terminus (Kitagawa et al. 2006).
We tested for resistance to six different transition metals: cadmium, cobalt, copper, nickel, silver, and zinc. While cobalt, copper, nickel, and zinc ions have well-known biological roles in E. coli, cadmium and silver do not.
Escherichia coli strain BW25113 was transformed with the library of overexpression plasmids and plated on LB agar spiked with increasing concentrations of metals as chloride salts (with the exception of silver nitrate). Plates were incubated for 3 days at 37 °C. As a control, cells were transformed with the identical vector expressing the lacZ gene. This control allowed us to establish the minimal inhibitory concentration (MIC) for each metal under the exact conditions of each experiment (Fig. 1).
Fig. 1.
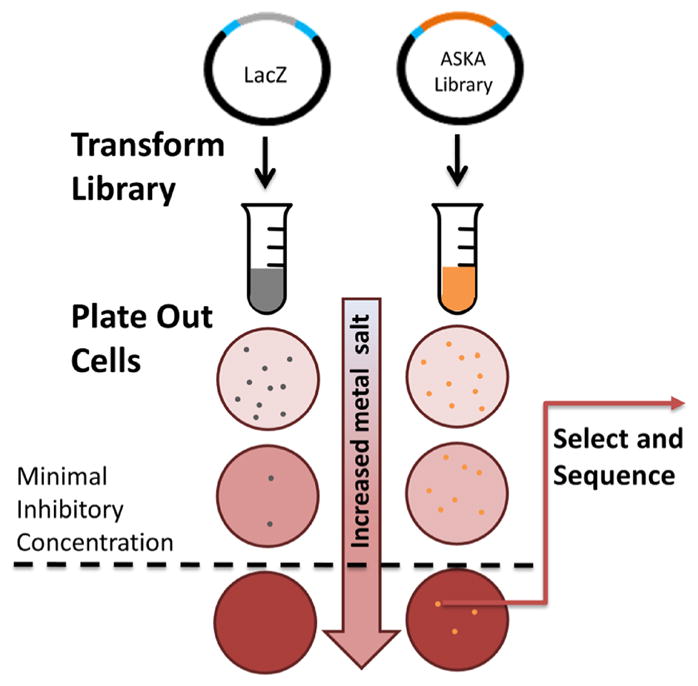
Selection for resistance to metal toxicity. LB agar plates with 300 μM chloramphenicol and 100 μM IPTG were spiked with increasing concentrations of metal salt. LacZ expressed from the same vector was used as a negative control and allowed determination of the minimal inhibitory concentration (MIC). Library plasmids that enabled growth above the MIC were selected and sequenced
For each of the six metals, transformants that formed colonies above the control MICs were isolated, plasmids were purified, and the overexpressed ORFs were sequenced. Many of the overexpressed ORFs were found multiple times. To confirm that the observed resistance was due to the plasmid encoded ORF—and not background chromosomal mutations—plasmids were isolated, transformed into fresh competent cells, and re-streaked on plates containing the same concentrations of the metal salts on which they were originally selected. A total of 48 hits were confirmed, and these are listed in Table 1. The changes in MIC resulting from the overexpressed ORFs are reproducible, but not large. This is consistent with previously reported changes of MICs in screens for antimicrobial resistance (Patrick et al. 2007; Soo et al. 2011; Nies and Silver 2007). Intriguingly, the majority of these hits had not previously been reported as endowing E. coli with resistance to these metals.
Table 1.
Selected genes, minimal inhibitory concentrations (MIC), and % changes in resistance
| Overexpressed ORF | MIC (μg/mL) with ORF | Increase in MIC (%) |
|---|---|---|
| Cadmium | 100a | |
| cmd | 125 | 30 |
| hdfR | 125 | 30 |
| napA | 125 | 30 |
| napF | 125 | 30 |
| fliN | 125 | 30 |
| Cobalt | 350a | |
| fliZ | 500 | 40 |
| zitB | 500 | 40 |
| cysE | 450 | 30 |
| yqiA | 450 | 30 |
| acp | 400 | 10 |
| yfgG | 400 | 10 |
| yhhI | 400 | 10 |
| ynaE | 400 | 10 |
| Copper | 400a | |
| pnp | 700 | 80 |
| fliZ | 600 | 50 |
| nusA | 600 | 50 |
| acrB | 500 | 30 |
| iraD | 500 | 30 |
| yfdN | 500 | 30 |
| yncN | 500 | 30 |
| gabt | 450 | 10 |
| lysR | 450 | 10 |
| nudJ | 450 | 10 |
| nudL | 450 | 10 |
| yciF | 450 | 10 |
| Nickel | 550a | |
| cysE | 700 | 30 |
| acpH | 650 | 20 |
| acpT | 650 | 20 |
| ybjC | 650 | 20 |
| yedN | 650 | 20 |
| yfeH | 650 | 20 |
| y S | 650 | 20 |
| yhbS | 600 | 10 |
| Silver | 5a | |
| rbfA | 10 | 100 |
| ydcQ | 10 | 100 |
| lysM | 10 | 100 |
| yaaY | 10 | 100 |
| yaiI | 20 | 300 |
| ynaE | 20 | 300 |
| Zinc | 200a | |
| ZitB | 400 | 100 |
| hlyD | 300 | 50 |
| yhil | 300 | 50 |
| rfaJ | 250 | 30 |
| ydcQ | 250 | 30 |
| ynaE | 250 | 30 |
| cysE | 250 | 30 |
| iraD | 250 | 30 |
| PerC | 250 | 30 |
| yfdN | 250 | 30 |
The ORFs selected in the presence of toxic concentrations of transition metals, the concentration at which the gene was selected, and the % change in resistance. MIC measurements were reproduced in triplicate. (Genotypes are explained in Supplementary Material)
MIC (μg/mL) determined using the control plasmid expressing lacZ
Grouping and Classification of Genes That Provide Resistance to Metal Toxicity
We divided the 48 metal-resistant hits into three Groups: (a) ORFs observed previously to endow E. coli with metal resistance; (b) ORFs related to proteins that have previously been linked to metal resistance in other organisms beyond E. coli; (c) ORFs that have not been observed previously to play a role in metal resistance (Table 2).
Table 2.
Grouping of overexpressed ORFs that mediate resistance to metal toxicity
| Same observation reported | Similar observation reported | No similar observation reported | |
|---|---|---|---|
| Cadmium | napA (O) | cbpM (S) | |
| napF (T) | hdfR(B) | ||
| fliN(B) | |||
| Cobalt | cysE (O) | yhhI (S) | fliZ (B) |
| zitB (E) | |||
| yqiA (S) | |||
| acpP (B) | |||
| yfgG (U) | |||
| Copper | acrB (E) | Pnp (T) | fliZ (B) |
| nusA (S) | yfdN (U) | ||
| iraD (S) | gabT (S) | ||
| lysR (S) | nudJ (U) | ||
| yciF (O) | nudL (U) | ||
| Nickel | cysE (O) | acpT (B) | acpH (B) |
| yfeH (E) | ybjC (U) | ||
| yhbS (O) | yedN (U) | ||
| y S (B) | |||
| Silver | rbfA (S) | ydcQ (U) | |
| lysM (B) | yaaY (U) | ||
| yaiI (U) | |||
| ynaE (U) | |||
| Zinc | zitB (E) | hlyD (E) | yhiL (E) |
| cysE (O) | iraD (B) | rfaJ (B) | |
| ydcQ (U) | |||
| ynaE (S) | |||
| perC (B) | |||
| yfdN (U) |
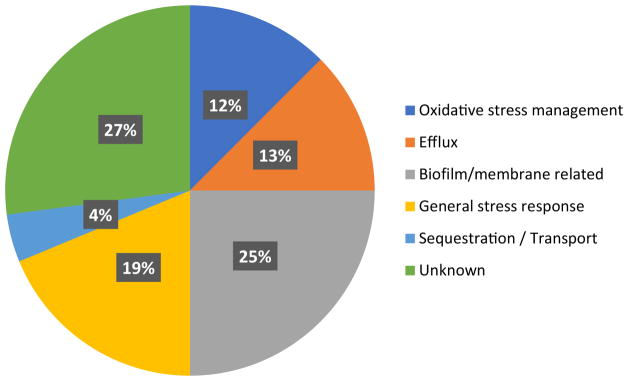
ORFs were placed into three categories based on previous literature reports: If the same observation had been made in E. coli, the gene was placed in the ‘Same observation’ category. If an orthologous or homologous protein was observed to protect from metal toxicity, or if the same ORF was characterized as rescuing from the toxicity of another metal, it was grouped as ‘Similar observation.’ If there was no reported evidence for the ORF’s involved in metal resistance or homeostasis, it was grouped under ‘No similar observation reported.’ Notably, many of the hits were on the same operon (e.g., fli, acp, nap). Also many had overlapping rescue functions (e.g., iraD, cysE, zitB). ORFs were also grouped into the 6 functional categories summarized in Fig. 2: Oxidative stress management (O), Efflux (E), Bio-film/OM related (B), General stress (S), sequestration/ transport (T), and unknown (U). Letter to right of ORF indicates presumed functional grouping. Genotypes, proposed rescue mechanisms, and literature references are provided in Supplementary Material
Additionally, based on their proposed mechanisms, the amplified genes that confer resistance to metal toxicity were binned into six functional classes: Oxidative stress (O), Efflux (E), Biofilm and/or Outer membrane (B), General Stress (S), Transport/Sequestration (T), or Unknown (U) (Fig. 2).
Fig. 2.
Proposed modes of action for resistance to metals imparted by overexpressed ORFs. Overexpressed proteins were placed into six functional classes: Oxidative stress management, Efflux, Biofilm/membrane related, General stress response, Sequestration/Transport, and Unknown. (see Supplemental Information for specific ORFs and mechanisms)
ORFs Observed Previously to Endow E. Coli with Metal Resistance
Several of the hits identified by our selection were known previously to relieve transition metal toxicity in E. coli. These proteins include the serine acetyltransferase encoded by CysE, the zinc transporter encoded by ZitB, and a subunit of the multidrug efflux pump encoded by AcrB.
Previous research showed that serine acetyltransferase was upregulated in the metal-resistant hyperaccumulator plant, Thlaspi (of the Brassicacea family). Subsequent studies showed that overexpression of the Thlaspi serine acetyl-transferase in E. coli produced similar phenotypic effects (Freeman et al. 2005), including resistance to toxic concentrations of cobalt and nickel. Although the exact mechanism of rescue remains elusive, those studies demonstrated that expression of serine acetyltransferase increases levels of glutathione, a molecule that can both sequester transition metals and scavenge metal-induced free radicals (Jozefczak et al. 2012). Correspondingly, our study found that overexpression of CysE (which encodes serine acetyltransferase in E. coli) enables E. coli to resist the toxicity of cobalt, nickel, and zinc (Table 1).
ZitB and its homolog ZntB have been implicated in cation gradient-mediated transport of zinc ions (Grass et al. 2001), while AcrB has been implicated in general toxin efflux of metal ions (Anes et al. 2015), including copper and silver. Similarly, our study found that overexpression of ZitB enables resistance to cobalt and zinc, while overexpression of AcrB enables resistance to copper (Table 1).
The isolation of these previously characterized genes by our unbiased library-based selection bolsters confidence in the ability of this approach to reveal novel determinants of metal resistance. Our results also highlight the ability of the selection to identify overexpressed genes that exert subtle effects on toxin resistance, as reflected by the small changes in MIC observed both by our selections and by the related previous studies.
ORFs That Produce Similar, But Not Identical, Observations to Previous Studies
Almost one-third (15/48) of the genes identified by our selection have been described previously as playing roles in resistance to transition metal toxicity—but in a different context than observed in the current study. These similar-but-not-identical observations include situations where (i) a homologous protein suppresses metal toxicity in another species, or (ii) the identified E. coli ORF was found to suppress the toxicity of a different transition metal than those observed in our study. A complete list of these genes is presented in Table 2. Detailed descriptions and comparisons with previous results are provided in Supplemental Table 1.
One particularly interesting example is NusA, which encodes a transcription factor involved in Rho-dependent transcriptional termination, intrinsic termination, and phage lambda N-mediated transcriptional anti-termination (Ciampi 2006). This transcription factor was observed to be upregulated in S. Metallicus, an archaeal extremophile, which thrives in elevated levels of copper (Jerez 2013), and is phylogenetically distant from the prokaryotic, E. coli. Perhaps these related observations indicate a conserved strategy for dealing with copper stress across a wide range of evolutionary history. Comparisons of other hits in this group to previously reported studies are presented in Supplemental Table 1.
New Genes Enabling Resistance to Metal Toxicity
The majority of the overexpressed genes identified by our selection had not been described previously as playing roles in resistance to metal toxicity. This finding accentuates the value of performing unbiased and model-independent screens to uncover novel and/or unexpected mechanisms of microbial resistance. For each of these newly reported genes, we suggest possible mechanisms to account for its function in metal resistance. For example, overexpression of the iron-sulfur-containing protein, napA, relieved cadmium toxicity. This protein is known to be involved in shuttling of electrons between membranes. Overexpression of napA could compensate for other damaged electron shuttles or may provide a sink for misappropriated electrons; thus, we tentatively classified it in class “O” (Oxidative stress management).
Possible mechanisms for all of the isolated genes are provided in Supplemental Table 1.
Three of the Overexpressed ORFs Encode Genes That are Required for Normal Metal Tolerance
The results described above demonstrate that the overexpression of certain genes can endow E. coli with increased tolerance to toxic levels of transition metals. These findings led us to question whether deletion of these same genes would render cells hypersensitive to metals.
To address this question, we tested the corresponding deletion strains from the KEIO collection, which contains over 4000 stains of E. coli each containing a single chromosomal deletion introduced by a kanamycin cassette insertion (Baba et al. 2006). Growth of these deletion strains in the presence of transition metals was compared to growth of the KEIO parent strain, BW25113, under the same conditions.
Surprisingly, only three of the deletion strains displayed reduced growth in the presence of metals (Table 3). These three genes, required for standard metal tolerance in E. coli, are hdfR for cadmium tolerance, pnp for copper tolerance, and yqiA for cobalt tolerance. These proteins have distinct functions: hdfR is a negative regulator of flagella synthesis; pnp is a phosphatase; and yqiA is a hydrolase. To the best of our knowledge, none of these three proteins have previously been observed to affect metal homeostasis in E. coli. Once again, this highlights the importance of using unbiased screens to uncover microbial resistance to antibacterial agents and environmental toxins.
Table 3.
Knockout strains with increased sensitivity to metal toxicity
| Gene KO | Toxin | WT MIC (mM) | KO MIC (mM) |
|---|---|---|---|
| yqiA | CoCl2 | 1.0 | 0.8 |
| pnp | CuCl2 | 1.8 | 1.4 |
| hdfR | CdCl2 | 0.5 | 0.4 |
Each KEIO knockout strain has a kanamycin cassette inserted into the ORF whose function it knocks out. Listed here are the KO strains, the toxin, and the WT and KO MIC. (MIC measurements were reproduced in triplicate.)
Discussion
The results described herein demonstrate that numerous genetically encoded factors can enable the gram-negative bacteria, E. coli, to survive in toxic concentrations of metals. Our discovery of several genes that had not previously been assigned to roles in metal homeostasis indicates that current understanding of the interplay between metals and biology is far from complete. In practical terms, these findings highlight the importance of model-independent screens and selections for acquired resistance, especially as the use of transition metals increases in clinical settings.
Our screens identified 48 cases where an overexpressed ORF rescued E. coli from lethal concentrations of a transition metal. The majority of those cases (28/48) were not previously known to function in metal toxicity or homeostasis.
Fifteen of the remaining hits demonstrated “similar” metal-resistant properties—e.g., a homologous or an orthologous ORF suppressed metal toxicity either in E. coli or another organism. The remaining five hits confirmed genes previously observed to suppress metal toxicity in E. coli. This last group of genes can be seen as positive controls, confirming that our selection method reproducibly finds genes known to mediate resistance to metal toxicity.
Notably, some of the genes (e.g., ZitB, AcrB) whose overexpression provided resistance to metals—i.e., inorganic toxins—also play roles in resistance to organic antimicrobials. AcrB, for example, is a subunit of an efflux system that removes a range of small molecules and ions from cells (Anes et al. 2015). The broad resistance that results from the upregulation of these genes suggests it would be prudent to use caution when introducing transition metals into clinical settings.
Another group of selected genes (fliZ, acpT, v S) encode proteins involved in membrane modifications and/or biofilm formation. Because biofilms cause cells to adhere to each other and to the surrounding surface, they can form barriers between cells and antimicrobial agents. Moreover, these films can channel signaling factors among populations of cells, thereby facilitating mechanisms that enable populations to promote virulence (produce endo/exotoxins), proliferate, and resist stress (Hall-Stoodley et al. 2004). These resistance mechanisms can cause significant harm in clinical settings when biofilms arise on surfaces (e.g., catheters) that must be kept sterile.
Because overexpression of the selected ORFs yielded enhanced resistance to metal toxicity, we expected deletion of these same genes from the chromosome would cause greater sensitivity to metals. However, only three of the deletion strains were hypersensitive to metals. This surprising result suggests a functional distinction between background levels of metal homeostasis and innovative resistance by overexpression. Perhaps homeostatic transition metal regulation systems are specialized for specific concentration ranges, or perhaps cells encode nuanced redundancies to manage toxic metals, and some of these become more pronounced under the conditions used in these experiments. In any case, there seems to be significant flexibility and/or promiscuity among latent factors that suppress metal toxicity in E. coli.
The approach used in the study is not biased toward any particular mechanism of rescue. Thus, we did not assume a priori that overexpression of certain genes will produce resistance to metal toxicity. Instead, we searched for resistance to metal toxicity by testing the ASKA collection of all E. coli ORFs. Nonetheless, it must be recognized that some mechanisms of rescue may not be found by our method: First, although all E. coli ORFs were included in the original ASKA collection, we cannot guarantee that every transformation actually sampled every one of the 4288 ORFs in the ASKA collection. Thus, there may exist overexpressed ORFs that can mediate resistance, but were not found by our study. Second, although all the ORFs in the ASKA collection are expressed from the same plasmid system, it is unlikely that they are all expressed at the same level. Therefore, it is possible that some ORFs may provide resistance only when overexpressed at higher levels than achieved by the plasmid system used in our studies.
We also note that because our approach is not biased toward any particular type of rescue, it is also possible that some of the overexpressed proteins found by our selections are not directly involved in resisting metal toxicity, but instead function by indirect mechanisms. Thus, some of the overexpressed proteins may rescue cells by altering the regulation of other genes, which in turn play direct roles in metal homeostasis. Indeed, in related experiments, we found that proteins designed entirely de novo can rescue auxotrophs of E. coli by altering gene regulation and enhancing the expression of other proteins, which are directly responsible for the rescue phenotype (Digianantonio and Hecht 2016; Digianantonio et al. 2017.)
It is important to highlight that increasing the expression of an endogenous gene is just one of several ways to achieve a selective advantages in toxic environments. Resistance to environmental toxins can arise by (i) altering the activity of an existing function, (ii) loss of an existing function, (iii) acquisition of an entirely new function, or (iv) increasing the level of an existing function as described in the current work. The likelihood of each of these four options depends on the selective pressure imposed—either in the wild or in laboratory experiments.
For example, laboratory-based evolution of resistance to silver (nanoparticles or Ag+) yielded mutations in cusS, ompR, and rpoA, rpoB, or rpoC (Lok et al. 2008; Graves et al. 2015; Randall et al. 2015; Tajkarimi et al. 2017.) The mutations in cusS were missense mutations that alter the metal binding capacity of this sensor protein, which activates expression of an efflux pump. Thus, the experimentally evolved mutants of cusS fall into the first class enumerated above (altered activity). The mutations in rpoA, rpoB, and rpoC were also missense mutations, and enable rescue by altering the activity of subunits of RNA polymerase. These also fall into the first class mentioned above. The mutations in ompR were deletions or insertions that abrogated the function of this regulator of an outer membrane porin. These mutants fall into the second class enumerated above (i.e., loss of function). None of those mutations provided silver resistance by enhancing expression of the mutated proteins. Therefore, it is not surprising that neither cusS nor ompR nor the rpo genes were found in our selections for silver resistance mediated by protein overexpression.
The third type of selective advantage listed above arises by acquisition of an entirely new function. In natural systems, this is a very rare event. However, recent work with large collections of non-natural de novo proteins facilitated our discovery of an entirely novel protein, ConK, with a new function that confers copper resistance in E. coli (Hoegler and Hecht 2016).
Finally, the fourth way of providing a selective advantage—increasing the level of an existing function—occurs in natural systems after events of gene duplication. Such events occurred frequently in evolutionary history, and are assumed to play key roles in enhancing survival in the face of new environmental challenges (Jensen 1976; Ohno 2013). The work described here shows that even a relatively small genome such as that of E. coli contains many genetically encoded functions, which, when expressed at higher levels, can provide selective advantages in toxic environments.
Materials and Methods
Strains and Growth Conditions
A pooled sample of the ASKA collection of plasmids was provided by Prof. Wayne Patrick at University of Otago in New Zealand. ORFs were expressed from a modified pCA24N vector containing chloramphenicol (CAM) resistance. The expression vector has the same modified pMB1 replication origin as pQE30 (Qiagen), which provides a copy number of 300–400 per cell. Protein expression is controlled by a T5 promoter and Lac operator and expression was induced with 100 μg/mL IPTG. The presence of a strong promotor on a high-copy plasmid leads to high levels of protein expression, as has been verified for numerous natural and designed sequences expressed in this system (Patrick et al. 2007; Soo et al. 2011; Fisher et al. 2011; Hoegler and Hecht 2016).
Transformations and Plating
Plasmids were prepared in DNAse-free H2O at a final concentration of 50 ng/μL. 2 μL of the library was transformed into 100 μL of electrocompetent BW25113 cells according to standard procedures. Cells were pulsed with 1.89 kV (BTX Trans Porator Plus) and allowed to recover for 1 h at 37 °C in super optimal broth (SOC). 50 μL of cells was plated on LB agar containing 30 μg/mL CAM and increasing concentrations of a metal salt. Plates were incubated at 37 °C.
Toxic Concentrations of Metals in Plates
Plates with various concentrations of metals were made by adding aliquots from serial dilutions of stock solutions of the metal salts. Minimal inhibitory concentrations (MICs) were determined by plating cells on plates containing different concentrations of metals and assessing growth versus no growth. Cadmium’s MIC is 100 μg/mL, and plates were made in steps of 25 μg/mL. Cobalt’s MIC is 350 μg/mL, and plates were made in steps of 50 μg/mL. Copper’s MIC is 400 μg/mL, and plates were made in steps of 100 μg/mL. Nickel’s MIC is 550 μg/mL, and plates were made in steps of 50 μg/mL. Silver’s MIC is 5 μg/mL, and plates were made in steps of 2.5 μg/mL. Finally, Zinc’s MIC is 200 μg/mL, and plates were made in steps of 50 μg/mL.
Liquid Cultures of Deletion Strains
Overnight cultures were started from single colonies and grown for 12 h at 37 °C in LB containing antibiotics. To determine comparative minimal inhibitory concentrations (MIC), these cultures were diluted 1:1000 in LB containing IPTG and combined with increasing concentrations of the metal salt. Cultures were grown overnight at 37 °C, and OD600 was measured.
Supplementary Material
Acknowledgments
We thank Wayne Patrick for generously gifting us an aliquot of his lab’s pooled ASKA collection. We also thank Shlomo Zarzhitsky for help with Fig. 2. This work was supported by the National Science Foundation, Grant Number: MCB-1409402.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00239-018-9830-3) contains supplementary material, which is available to authorized users.
References
- Alam M, Imran M. Multiple antibiotic resistances in metal tolerant E. coli from hospital waste water. Bioinformation. 2014;10(5):267–272. doi: 10.6026/97320630010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol. 2015;6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst KR, Morby AP. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology. 2000;146:2277–2282. doi: 10.1099/00221287-146-9-2277. [DOI] [PubMed] [Google Scholar]
- Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152(9):2515–2528. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digianantonio KM, Hecht MH. A protein constructed de novo enables cell growth by altering gene regulation. Proc Natl Acad Sci USA. 2016;113:2400–2405. doi: 10.1073/pnas.1600566113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digianantonio KM, Korolev M, Hecht MH. A non-natural protein rescues cells deleted for a key enzyme in central metabolism. ACS Synth Biol. 2017;6:694–700. doi: 10.1021/acssynbio.6b00336. [DOI] [PubMed] [Google Scholar]
- Fisher MA, McKinley KL, Bradley LH, Viola SR, Hecht MH. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS ONE. 2011;6(1):e15364. doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Persans MW, Nieman K, Salt DE. Nickel and cobalt resistance engineered in Escherichia coli by overexpression of serine acetyltransferase from the nickel hyperaccumulator plant Thlaspi goesingense. Appl Environ Microbiol. 2005;71(12):8627–8633. doi: 10.1128/AEM.71.12.8627-8633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. ZitB (YbgR), a Member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol. 2001;183(15):4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Rensing C, Solioz M. Metallic copper as an antimicrobial surface. Appl Environ Microbiol. 2011;77(5):1541–1547. doi: 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JL, Jr, Tajkarimi M, Cunningham Q, Campbell A, Nonga H, Harrison SH, Barrick JE. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front Genet. 2015;6:42. doi: 10.3389/fgene.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Herron MD, Doebeli M. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 2013;11(2):e1001490. doi: 10.1371/journal.pbio.1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman JL, Crossman LC. Bacterial antimicrobial metal ion resistance. J Med Microbiol. 2015;64(5):471–497. doi: 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- Hoegler KJ, Hecht MH. A de novo protein confers copper resistance in Escherichia coli. Protein Sci. 2016;25:1249–1259. doi: 10.1002/pro.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Andersson DI. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat Rev Genet. 2015;16:459–471. doi: 10.1038/nrg3922. [DOI] [PubMed] [Google Scholar]
- Jensen RA. Enzyme recruitment in evolution of new function. Ann Rev Microbiol. 1976;30(1):409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Jerez CA. Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea. 2013 doi: 10.1155/2013/289236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczak M, Remans T, Vangronsveld J, Cuypers A. Glutathione is a key player in metal-induced oxidative stress defenses. Int J Mol Sci. 2012;13(3):3145–3175. doi: 10.3390/ijms13033145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacar B, Gaucher EA. Towards the recapitulation of ancient history in the laboratory: combining synthetic biology with experimental evolution. Artif Life. 2012;13:11–18. [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2006;12(5):291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Lemire JA, Harrison JJ, Turner RJ. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11(6):371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- Levin BR, Baquero F, Johnsen PJ. A model-guided analysis and perspective on the evolution and epidemiology of antibiotic resistance and its future. Curr Opin Microbiol. 2014;19:83–89. doi: 10.1016/j.mib.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok CN, Ho CM, Chen R, Tam PK, Chiu JF, Che CM. Proteomic identification of the Cus system as a major determinant of constitutive Escherichia coli silver resistance of chromosomal origin. J Proteome Res. 2008;7(6):2351–2356. doi: 10.1021/pr700646b. [DOI] [PubMed] [Google Scholar]
- Montealegre MC, Roh JH, Rae M, Davlieva MG, Singh KV, Shamoo Y, Murray BE. Differential penicillin-binding protein 5 (PBP5) levels in the Enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob Agents Chemother. 2017;61:e02034–16. doi: 10.1128/AAC.02034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338(6105):384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies DH, Silver S. Molecular microbiology of heavy metals. Vol. 6. Springer; Berlin: 2007. pp. 118–142. [Google Scholar]
- Notebaart RA, Kintses B, Feist AM, Papp B. Underground metabolism: network-level perspective and biotechnological potential. Curr Opin Biotechnol. 2018;49:108–114. doi: 10.1016/j.copbio.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer; New York: 2013. [Google Scholar]
- Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol. 2007;24(12):2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CP, Gupta A, Jackson N, Busse D, O’Neill AJ. Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J Antimicrob Chemother. 2015;70:1037–1046. doi: 10.1093/jac/dku523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo VW, Hanson-Manful P, Patrick WM. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(4):1484–1489. doi: 10.1073/pnas.1012108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajkarimi M, Rhinehardt K, Thomas M, Ewunkem JA, Campbell A, Boyd S, Turner D, Harrison SH, Graves JL. Selection for ionic-confers silver nanoparticle resistance in Escherichia coli. JSM Nanotechnol Nanomed. 2017;5(1):1047. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.