Summary
Phytoremediation is a green and sustainable alternative to physico‐chemical methods for contaminated soil remediation. One of the flavours of phytoremediation is rhizoremediation, where plant roots stimulate soil microbes to degrade organic contaminants. This approach is particularly interesting as it takes advantage of naturally evolved interaction mechanisms between plant and microorganisms and often results in a complete mineralization of the contaminants (i.e. transformation to water and CO 2). However, many biotic and abiotic factors influence the outcome of this interaction, resulting in variable efficiency of the remediation process. The difficulty to predict precisely the timeframe associated with rhizoremediation leads to low adoption rates of this green technology. Here, we review recent literature related to rhizoremediation, with a particular focus on soil organisms. We then expand on the potential of rhizoremediation to be a model plant‐microbe interaction system for microbiome manipulation studies.
Introduction
Phytoremediation is the use of plants to remediate contaminated environments (usually soils, but also water). Many processes can be involved in the removal of the pollutants such as phytovolatilization (the removal of volatile compounds through plant tissues), phytotransformation (the transformation of contaminants from one state to another), phytostabilization (the stabilization of mobile contaminants in the soil), phytoextraction (the removal of trace elements from the soil and its fixation in plant tissues). Although all these processes involve both the plant and its microbiota, rhizoremediation clearly stands out as an integrated plant‐microbes endeavour. Rhizoremediation is the degradation of organic pollutants in the soil zone surrounding the plant roots (the rhizosphere), usually as a result of the stimulation of the catalytic activities of microorganisms by the plant roots (Pilon‐Smits, 2005). For many organic contaminants, such as most petroleum hydrocarbons, rhizoremediation results in the complete mineralization of the contaminants, effectively removing it from the environment.
The principle behind rhizoremediation is simple: as the plant roots colonize the contaminated soil, as for any soil, they associate with a subset of the microorganisms present in the soil and stimulate them through the exudation of a variety of organic compounds (Kuiper et al., 2004) (Fig. 1A). Some of the microbes stimulated by the root exudates are also able to degrade petroleum hydrocarbons. Many facets of the rhizosphere environment make this soil zone particularly appropriate for the degradation of organic contaminants. First, the plant secondary metabolites that are part of the exudates are often structurally very similar to organic contaminants (Singer et al., 2003). This results in a heightened presence and activity of microbes being able to degrade organic contaminants in the rhizosphere, even in the absence of contaminants (Yergeau et al., 2014). Second, because of the presence of the root exudates, the rhizosphere microbial communities are generally more active and more abundant than microbial communities in the bulk soil (i.e. not under the influence of the roots) (Smalla et al., 2001; Kowalchuk et al., 2002). Third, the rhizosphere is generally recognized as a hotspot for horizontal gene transfer (Van Elsas and Bailey, 2002), and plasmids were shown to help microorganisms adapt to contamination stress and degrade organic compounds (Top and Springael, 2003; Sentchilo et al., 2013). Additionally, some root exudates help detach organic contaminants from the organic matter present in soil, making them more available to microbes (Gao et al., 2010). Altogether, this again highlights the distinct roles of plants and microorganisms during rhizoremediation: the plant act as a promoter for microbial degraders, by providing them with a suitable environment and stimulating them through root exudates. The suitability of the rhizosphere environment for microbial processes related to the degradation of hydrocarbons also exposes one of the major pitfalls of rhizoremediation: it only works where plant roots are. Therefore, in compacted or very clayey soils, or in cases where contamination is deeper than the root zone, or at too high concentration for roots to survive, rhizoremediation is not effective. As root growth patterns and exudates amount and quality differ between different plants, even between closely related genotypes (O'Toole and Bland, 1987; Jones et al., 2004; Manschadi et al., 2006), the choice of an appropriate plant genotype is crucial in rhizoremediation.
Figure 1.
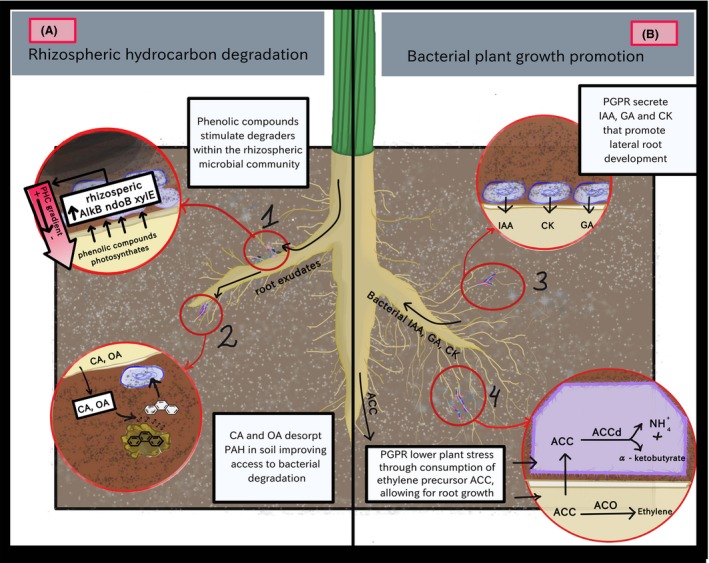
Major plant–microbe interactions occurring during rhizoremediation. In (A), plant root exudates (1) stimulate hydrocarbon‐degrading bacteria and (2) help to desorb contaminants attached to soil particles, making them more available to rhizobacteria. In (B), rhizosphere microorganisms promote plant growth through, among many other mechanisms, (3) the production of plant hormones and (4) the degradation of 1‐aminocyclopropane‐1‐carboxylic acid (ACC), the precursor of the stress hormone ethylene. PHC, petroleum hydrocarbons; alkB, alkane mono‐oxygenase, ndoB, naphthalene dioxygenase, xylE, catechol‐2,3‐dioxygenase; OA, oxalic acid; CA, citric acid; PAH, polycyclic aromatic hydrocarbon; PGPR, plant growth promoting rhizobacteria; IAA, indolacetic acid; CK, cytokinin; GA, gibberellic acid; ACC, 1‐aminocyclopropane‐1‐carboxylic acid; ACCd,ACC deaminase; ACO,ACC oxidase.
The rhizosphere microbes, especially bacteria, are thought to be the major players in organic contaminant degradation during rhizoremediation (Bell et al., 2014a,b; El Amrani et al., 2015), and recent plant‐microbe metatranscriptomic studies confirmed that the hydrocarbon degradation genes expressed in the root‐rhizosphere environment were mostly linked to bacteria (Gonzalez et al., 2018; Yergeau et al., 2018) (Box 1). Petroleum hydrocarbon contamination is often composed of a mixture of saturated aliphatic (alkanes) and aromatic hydrocarbons (including polycyclic aromatic hydrocarbons, PAHs). Microorganisms can degrade virtually all the hydrocarbons present in petroleum through various pathways, although with different efficiencies. In addition to this central role, microbes also have another major role in rhizoremediation (Fig. 1B). Indeed, microbes known as plant growth promoting rhizobacteria (PGPR) are recognized to have the capacity to increase plant growth (Kloepper and Schroth, 1978), and the ones that can increase root growth are particularly interesting in the context of rhizoremediation. On top of their ability to promote the growth of plants through the production of plant hormones or the mobilization of nutrients, PGPR also have the capacity to reduce plant stress through various mechanisms (Rajkumar et al., 2012; De Zelicourt et al., 2013), including through the reduction of ethylene concentrations in the roots (Glick et al., 1998; Glick, 2003) which would allow a plant to grow in highly contaminated environments without the adverse effects of stress (Burd et al., 2000).
Box 1. The holobiont and the hologenome.
All multicellular eukaryotes are associated with a wide diversity of microorganisms, forming an inseparable entity known as a holobiont (Rosenberg et al., 2009; Bordenstein et al., 2015; Van Opstal and Bordenstein, 2015; Theis et al., 2016). This observation has led Ilana Zilber‐Rosenberg and Eugene Rosenberg to enounce the hologenome theory of evolution that states that the hologenome (the combined genomes of the host and its microbiota) forms one of the units of evolution (Zilber‐Rosenberg and Rosenberg, 2008). Consequently, it is predicted that holobionts can rapidly evolve/adapt through their microbiota by: (i) horizontal gene transfer among their existing microbiota, (ii) recruitment of new microbes from the environment, (iii) shifts in the relative abundance/gene expression of various members of the microbiota. These mechanisms are thought to enable holobionts to adapt within a single or a few generations (Voss et al., 2015; Rosenberg and Zilber‐Rosenberg, 2016). It has recently been shown that the response of willows to stressful conditions (soil contamination) results in large shifts in the metatranscriptome of root and rhizosphere bacterial and fungal communities, but not in the plant root transcriptome (Gonzalez et al., 2018; Yergeau et al., 2018). Taken together, these results emphasize the importance of the plant microbiota in the response to environmental stresses and confirms that microbiota manipulation is a viable alternative to optimize phytoremediation (El Amrani et al., 2015; Quiza et al., 2015).
Rhizoremediation offers a unique system to study plant‐microbe interactions and experiment with microbiome manipulation approaches. First, the response variable of interest is easily measurable: a lowered soil contamination. Second, the hydrocarbon degradation pathways are well known, and the genes are well represented and annotated in databases (e.g. the biocatalysis/biodegradation database, http://eawag-bbd.ethz.ch/). Third, the capacity to degrade hydrocarbons is widespread among bacteria, and major players, such as Pseudomonas and Rhodococcus can be easily cultured. It is thus relatively easy to create consortia of hydrocarbon‐degrading bacteria, follow their fate in the environment using molecular tools and measure their effect on rhizoremediation efficiency. It is also possible to measure the effects of various manipulations on the hydrocarbon‐degrading microbiota abundance and activities in the rhizosphere using relatively inexpensive molecular tools such as qPCR.
Soil organisms
Microbes as hydrocarbon degraders
Hydrocarbon contamination is often a complex mixture of chemicals, requiring several different genes and pathways for its complete degradation. Most components can be classified as saturated aliphatic (alkanes) or aromatic hydrocarbons. Hydroxylation of an alkyl group catalyzed by oxygenases is usually the first step in the degradation of organic compounds alkanes. There are several categories of alkyl‐group hydroxylases, including cytochrome P450s (CYP) and alkane hydroxylase (Harayama et al., 1992). The alkane hydroxylase catalyzes the hydroxylation of the terminal carbon of alkanes and consists of three different subunits, including the membrane‐bound hydroxylase subunit encoded by alkB. CYP153, an enzyme of the CYP superfamily, can also catalyze the hydroxylation of alkanes (Van Beilen et al., 2006). Aromatic rings also need to be hydroxylated to be degraded, but the key step in aromatic hydrocarbon degradation is the opening of the hydroxylated aromatic ring, which is catalyzed by aromatic‐ring‐cleavage dioxygenases (Harayama et al., 1992). There are three main types of aromatic‐ring cleavage dioxygenases (intradiol, extradiol and gentisate/homogentisate) that can be differentiated based on their substrate and on the position where the ring fission occurs relative to the hydroxyl groups (Harayama et al., 1992).
Many specific bacteria of the phylum Actinobacteria and Proteobacteria were shown to degrade aliphatic and polycyclic aromatic hydrocarbons (PAHs). These bacteria are ubiquitous even in the most pristine environments (Yergeau et al., 2012, 2015a,b), probably because many of the petroleum hydrocarbon contaminants are widespread naturally occurring molecules. Recent metatranscriptomic studies in the rhizosphere highlighted several key taxa that responded to petroleum hydrocarbon contamination. For instance, in a pot study, transcripts related to Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Acidobacteria were more abundant in the rhizosphere of willows growing in contaminated soil as compared to non‐contaminated soils (Yergeau et al., 2014). Also, functional genes related to aromatic and aliphatic hydrocarbon degradation were more prevalent in the rhizosphere of willows growing in contaminated soils (Yergeau et al., 2014), and these genes were shown to be related to bacterial orders such as Actinomycetales, Rhodospirillales, Burkholderiales, Alteromonadales, Solirubrobacterales, Caulobacterales and Rhizobiales (Pagé et al., 2015). In the field, the differences in the expression of hydrocarbon degradation genes and in the active taxa between the rhizosphere of willows growing in contaminated and non‐con‐taminated soils varied and depended on the willow species (Yergeau et al., 2018). The abundance of PAH degrading genes was higher in phenanthrene‐contaminated soils planted with ryegrass as compared to non‐planted soils, with plants favouring the activities of bacterial degraders belonging to the Pseudomonadales, Actinobacteria, Caulobacterales, Rhizobiales and Xhantomonadales (Thomas and Cébron, 2016). Similarly, the presence of ryegrass was shown to stimulate the expression of bacterial PAH‐ring hydroxylating dioxygenase genes, such as nidA3, pdoA, nahAc and phnAc (Guo et al., 2017a,b). Several Lotus corniculatus (common bird's‐foot trefoil) and Oenothera biennis (common evening primrose) root endophytes belonging to the genera Rhizobium, Pseudomonas, Stenotrophomonas and Rhodococcus harbored genes encoding for CYP153 alkane hydroxylases and showed the capacity to grow with n‐hexadecane as sole source of carbon (Pawlik et al., 2017). Other plants, such as Achillea millefolium (yarrow), Soligado canadensis (Canadian goldenrod), Trifolium aureum (hop clover) and Dactylis glomerata (orchard grass), growing in a heavily contaminated site, harbored hydrocarbon‐degrading bacterial endophytes mostly belonging to the Actinobacteria (Lumactud et al., 2016).
Fungi are particularly interesting in the context of rhizoremediation in view of their ability to form intimate associations with plant roots and colonize large volumes of soil through hyphal growth, and their production of a wide spectrum of extracellular hydrolytic enzymes, allowing them to grow on a wide variety of substrates. Ectomycorrhizal and saprotrophic basidiomycetes (white and brown rot fungi) have shown remarkable in vitro capacity for the degradation of PAHs and phenolic compounds, among others. Fungal hydrocarbon degradation is mostly an extracellular process, consisting in the release in the environment of active broad‐specificity oxidoreductase enzymes, such as laccases, manganese peroxidases and lignin peroxidases (Harms et al., 2011). In nature, these enzymes are mainly used to degrade lignin (a cross‐linked phenolic polymer), but their low specificity also allows them to degrade other phenolic compounds, such as the ones found in petroleum hydrocarbons (Karigar and Rao, 2011). A detailed list of fungi degrading organic pollutants is given in the review by Kadri et al. (2017), but it is still difficult to pinpoint which fungi is the most effective for rhizoremediation and what environmental factors influence this efficiency. For instance, the colonization of the rhizosphere and roots by fungi depend on factors like root exudate patterns, which are influenced by the presence of contaminant (Harms et al., 2011).
However, even if the microbial partners have the genetic capacity to degrade the contaminants, many substances are recalcitrant and elude microbial degradation. For instance, high molecular weight petroleum molecules (HMW) present a low bioavailability (Gamerdinger et al., 1997) and a high degree of adsorption to soil organic matter (Reddy and Sethunathan 1983) and are thus difficult to access by microbes. At the same time, high concentrations of these substances inhibit root elongation and ramification (Peña‐Castro et al., 2006; Ma et al., 2010). Furthermore, the efficiency of the rhizoremediation depends on the microbes capable of surviving within the polluted niche, which in turn depends on the microbial diversity originally present in the polluted soil.
Microbes as plant‐growth promoters
One of the most important factor for maximizing the success of rhizoremediation is facilitating the growth of the plant root system. Rhizoremediation can only occur in the direct vicinity of plant roots and any treatment that can increase the growth of plant roots will result in a positive outcome for rhizoremediation (Fig. 1B). The root‐associated PGPR that can stimulate plant growth and help them tolerate stress, are frequently used under agricultural settings, but their use for phytoremediation has received less attention, representing a huge untapped potential.
Several mechanisms of action of PGPR have been described up to now (Olanrewaju et al., 2017). Some strains produce phytohormones related to plant growth and development such as indole‐3‐acetic acid (IAA) (Patten and Glick, 1996; Spaepen and Vanderleyden, 2011; Raut et al., 2017) and gibberellic acid (Gutiérrez‐Mañero et al., 2001). Other strains, such as phosphate‐solubilizers, mobilize nutrients from soil, making them more available for the plant (Rodrıguez and Fraga, 1999), whereas other PGPR compete with pathogens and suppress their action (Beneduzi et al., 2012). One particularly interesting way through which PGPR contribute to rhizoremediation is the suppression of the plant responses to stress, making the plant function as if there was no stress. Many mechanisms were shown to be involved in the enhancement of plant stress tolerance by microbes. Some target the modulation of plant stress genes (Timmusk and Wagner, 1999) or the reduction of the stress hormone ethylene levels through degradation of its precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) by the bacterial enzyme ACC deaminase (Glick et al., 1998; Mayak et al., 2004; Glick, 2005). By reducing the levels of ethylene, bacteria harboring the ACC deaminase gene allow plants to function as if they were not subjected to stress and to develop their root system normally. In other cases, the expression of plant genes related to stress protection can be activated by bacterial volatile organic compounds (Ryu et al., 2004) or the drought stress‐related DNA methylation patterns can be modulated by endophytes (Hubbard et al., 2014).
Some studies have inoculated PGPR strains in the rhizosphere to improve plant growth under contaminated conditions. Liu et al. (2014) found that inoculating tall fescue with the ACC deaminase‐producing Klebsiella sp. enhanced plant growth and petroleum hydrocarbon remediation efficiency. Similarly, Avena sativa (common oat) growing in oil‐contaminated soil and inoculated with an Acinetobacter PGPR strain showed an increased dry mass and stem height compared to controls, along with a higher rate of decontamination (Xun et al., 2015). Inoculating bacterial strains transformed with a plasmid containing the ACC deaminase gene was shown to be more effective in promoting Brassica napus (rapeseed or canola) growth in PAH contaminated soils than when inoculating with the wild‐type strains (Reed and Glick, 2005). More recently, Zea mays growing in soils contaminated with 10 g kg−1 light crude oil and inoculated with a Bacillus subtilis strain producing ACC deaminase showed a significant increase in height, root length and biomass, which resulted in a 43% increase in petroleum hydrocarbon degradation compared to uninoculated plants after 60 days (Asghar et al., 2017). In a 3‐year field test carried at a site located in Ontario (Canada), the inoculation of PGPR resulted in an increased plant biomass and a reduction of the recalcitrant petroleum hydrocarbon fractions with HMW (Gurska et al., 2009). The inoculation of plant‐growth promoting endophytes was also shown to be effective for the rhizoremediation of petroleum hydrocarbons. The inoculation of the endophyte Pseudomonas putida PD1 in two willow clones (Salix purpurea 94006 and Salix discolour S‐365) growing in soil contaminated with 100 mg kg−1 phenanthrene resulted in a 25–40% increase in the degradation rate of this PAH compared to uninoculated controls (Khan et al., 2014).
Arbuscular mycorrhizal fungi (AMF) have multiple interesting roles that can improve rhizoremediation. For instance, under contaminated settings AMF can modify the rhizosphere microbial communities (Iffis et al., 2016, 2017) and improve plant growth (Xun et al., 2015), thereby likely increasing the efficiency of rhizoremediation. It is also well known that AMF can improve plant nutrition (Smith and Read, 2008) or provide protection against pathogens (Hamel, 2007; Tang et al., 2009), which could result in increased root growth and increased stimulation of the rhizosphere microorganisms. The AMF Glomus mosseae was shown to improve rhizoremediation of PAH, with significantly higher reductions in the concentrations of chrysene and dibenz(a,h)anthracene in AMF‐inoculated pots as compared to non‐inoculated pots (Joner et al., 2001). Interestingly, non‐inoculated pots had degradation levels similar to those observed for non‐planted pots. In a field study, the AMF associated with willows were strongly structured by the contamination levels, with reduced diversity at higher contamination levels, suggesting that only a narrow number of AMF can thrive in these highly contaminated environments (Hassan et al., 2014). Ectomycorrhizal fungi were also shown to be influenced by contamination levels, with some species only associating with local willow genotypes under high contamination levels (Bell et al., 2014). The same fungus was subsequently shown to be related to willow Zn uptake at a metal contaminated field site (Bell et al., 2015). A holistic approach combining AMF and PGPR inoculation, as it has already been proposed in the context of crop production (Nadeem et al., 2014), could lead to more effective rhizoremediation strategies due to the synergistic effect these organisms can have in improving plant physiology and by increasing of the volume of soil under the influence of the roots.
Other soil organisms
Although not part of the plant microbiome per se, many other organisms living in the soil can influence the interactions between the plant, the microbiome and contaminated soils during rhizoremediation. As such, they could be interesting targets for plant microbiome manipulation. These organisms include nematodes, protists, collembola, and earthworms among others. In this section, we will focus on earthworms as a model soil organism for modulating rhizoremediation. Earthworms are typical soil inhabitants making up > 80% of the biomass of soil macrofauna (Yasmin and D'Souza, 2010), and are frequently found in the rhizosphere environment (Springett and Gray, 1997). Earthworms can survive in the most highly contaminated soils as only water soluble compounds can be absorbed through their skin (Jager, 1998), which excludes most toxic PAHs (Ma et al., 1998) and PCBs (Beyer and Stafford, 1993). For instance, Zavala‐Cruz et al. (2013) recorded the presence of Pontoscolex corethrurus, Gossodrillus sp. and Dichogaster salines in a site polluted with crude oil for 20 years with petroleum hydrocarbons concentrations up to 12 000 mg kg−1. The model species Eisenia fetida can survive to up to 3500 mg kg−1 of petroleum hydrocarbons (Geissen et al., 2008). Several studies have shown that the presence of earthworms improves or accelerates the degradation rate of several PHC. For instance, the application of the earthworm E. fetida resulted in the removal of 92% of anthracene from an arable soil after 56 days, as compared to 57% in the untreated soil (Delgado‐Balbuena et al., 2016).
Earthworm are also having a strong impact on soil microbial community composition (Emmerling and Paulsch, 2001), and the bacterial taxa containing major hydrocarbon degraders (such as the Proteobacteria) are often more abundant when earthworms are present. For instance, the application of the earthworms to an anthracene contaminated soil resulted in a shift in the soil microbial community with a decrease in the relative abundance of Gemmatimonadetes, Chloroflexi and Acidobacteria and an increase in the Proteobacteria compared to the untreated soil (Delgado‐Balbuena et al., 2016). Similarly, the Alpha‐ and Betaproteobacteria were shown to be mostly unaffected after their passage through the digestive system of the earthworms (Nechitaylo et al., 2010). Betaproteobacteria were in fact stimulated in the presence of the earthworm P. corethrurus (Bernard et al., 2012). Additionally, the degradation of many hydrocarbon substances may start in the direct environment of the earthworm, as many known degraders such as Rhodococcus and Azotobacter were found in the burrows of Lumbricus terrestris (Tiunov and Dobrovolskaya, 2002), whereas other known degraders such as Pseudomonas, Alcaligenes, Acidobacterium, and the fungus Penicillium, were found in the intestine and cast of earthworms (Singleton et al., 2003).
In addition to the ones mentioned above, earthworms have other roles that could make them a key component of rhizoremediation. Indeed, earthworms are recognized ecological engineers, contributing to the mineralization and humifaction of organic matter (Lavelle and Spain, 2001), being highly mobile vectors moving bacteria in and out the rhizosphere (Luepromchai et al., 2002), and improving water infiltration and soil aeration (Bartlett et al., 2010). Although all these activities are expected to positively stimulate rhizoremediation, only a few studies have tested the effect of earthworms in the context of rhizoremediation. One such study looked at PCB rhizoremediation and found that ryegrass co‐inoculated with AMF and earthworms decreased soil PCB contents by 79.5% as compared to 74.3% for AMF alone, 62.6% for earthworms alone or 58.4% for ryegrass alone (Lu et al., 2014). Earthworms and other soil fauna thus represent an unexploited potential for rhizoremediation of petroleum hydrocarbons.
Plant root exudates
Root exudates have been identified as a major ecological driver that actively modulate the microbial community composition, diversity and activity of the rhizosphere (Fig. 1A). The plant exudes a variety of specialized antimicrobials and signalling molecules (e.g. flavonoids, salicylic acid and phytoalexins), carbon (e.g. organic acids, aromatic compounds) and nitrogen (e.g. amino acids) compounds. Therefore, only a specific group of microbes that can utilize these compounds are selectively enriched in this highly competitive environment (Gomes et al., 2003; Haichar et al., 2008; Berg and Smalla, 2009). However, exudation is not for the sole benefit of microbes; it also directly benefits the plant itself. For instance, organic acids, such as malate, citrate and oxalate are often present in the rhizosphere, and in addition to being a carbon source for many microbes, they are involved in many plant processes like nutrient acquisition, metal detoxification, and alleviation of stress (Jones, 1998). Plants confronted with stressful environments normally respond by increasing root exudation (Jones et al., 2004; Qin et al., 2007), which leads to increased microbial biomass (Esperschütz et al., 2009) and activity (Yergeau et al., 2014) in the rhizosphere. Root exudates can also improve the availability of contaminant for microbial degradation, as it was shown that the desorption of phenanthrene and pyrene from soil particles was increased by the addition of citric and oxalic acid (Gao et al., 2010). An increasing amount of scientific evidence points towards the crucial importance of exudates as mediators of hydrocarbon rhizoremediation (Martin et al., 2014; Rohrbacher and St‐Arnaud, 2016).
Interestingly, many compounds found in the rhizosphere are analogous to organic contaminants (Singer et al., 2003), including terpenes, lignin derived components and flavonoids (Hartmann et al., 2009). Negative correlations between the concentrations of plant root exudates and petroleum hydrocarbons have been observed, with lower concentrations of PHC observed close to the roots where maximum concentrations of exudates are found (Gao et al., 2011; Ling et al., 2013), because exudates induce the degradation of PHC by rhizospheric microorganisms (Sun et al., 2010). For instance, the phenolic root exudates fomorin, caffeic acid and protocatechuic acid were linked to bacterial degradation of tricyclic and tetracyclic PAHs in the rhizosphere (Ely and Smets, 2017) and increases in phenolic root exudates have been associated with higher rates of degradation of benzo[a]pyrene in the rhizosphere of Phragmites australis (cosmopolitan common reed) (Toyama et al., 2011). In fact, the rhizosphere of plants are often enriched in microbial genes related to the degradation of organic contaminants that are actively expressed even in the absence of contaminants (Yergeau et al., 2014). Conversely, in the absence of plants, supplementing a PAH contaminated agricultural soil and a pyrene‐spiked soil with maize and soybean exudates resulted in an increased initial PAH degradation that faded through time as exudates were depleted (Guo et al., 2017a,b). The interaction between Mycobacterium and the root exudates accelerated the removal of PAH by provoking a shift in the soil bacterial community structure and diversity (Guo et al., 2017a,b).
Rhizoremediation as a model for microbiome manipulation
Choosing the right plant
Choosing the right plant is crucial to achieve optimal rhizoremediation. The main aspects to take into account when choosing the best fit for rhizoremediation are root morphology, plant tolerance to the contaminant and root exudate profile. For instance, various Poaceae species (grasses) are often selected for rhizoremediation purposes as they produce a dense secondary root system that can harbor an abundant microbial community (Adam and Duncan, 2002; Lee et al., 2008; Gaskin and Bentham, 2010; Barrutia et al., 2011). However, most Poaceae plants are not appropriate when the pollutants have reached deeper layers in the soil, and deeper rooting plants, such as trees like willows or poplars should be preferred in these cases (Kuzovkina and Volk, 2009). The quantity and quality of root exudates also vary substantially even between closely related plant genotypes, resulting in significant differences in the recruitment and stimulation of microbes in the rhizosphere (Lundberg et al., 2012; Yergeau et al., 2018). This variation in the microbiome then results in different degradation rates in the rhizosphere of different plant genotypes. The choice of the right plant is thus one of the actionable ways to manipulate the rhizosphere microbiome and increase remediation rates.
Many studies have shown that different plant species have different capacity for the rhizoremediation of petroleum hydrocarbons. For instance, the removal rate of eight PAHs (tetracyclic and pentacyclic) was measured in the rhizosphere of Echinacea purpurea (purple coneflower), Festuca arundinacea (tall fescue), and Medicago sativa (alfalfa) growing in pots (Liu et al., 2015). Although the degradation rates increased for all plants as compared to the unplanted controls, the level of degradation strongly varied by plant species (Liu et al., 2015). Similarly, in a field study on a former coal mine site, the capacity for various legume tree species [Cassia siamea (cassod tree), Albizia lebbeck (lebbeck), Delonix regia (flame tree) and Dalbergia sissoo (North Indian rosewood)] to reduce soil PAH levels was evaluated (Mukhopadhyay et al., 2017). The results showed that the degradation rates varied from 51.5% to 81.6% among the trees tested (Mukhopadhyay et al., 2017).
The response of the plant itself to contamination will also have a determining effect on the success of rhizoremediation. Willow genotypes showed large differences in the response of their growth patterns and physiology to contamination (Grenier et al., 2015). These results were mirrored in the transcriptomic response of the rhizosphere microbiota (Yergeau et al., 2018), with the willow species showing the largest decreases in biomass and photosynthetic capacity also showing the largest decreases in the expression of genes in their associated microbiota. This suggests that the physiological responses of the willow genotypes to contamination could be good indicators of their rhizoremediation potential. Additionally, when growing in highly contaminated soils in Canada, North American willow genotypes showed a strong association with an ectomycorrhizal fungus, Sphaerosporella brunnea, whereas Asian and European genotypes did not associate with this particular fungus (Bell et al., 2014a,b). Therefore, the region of origin of the plant appears to have an importance, with local plants being better adapted to interact with the local beneficial soil microbiota.
Modifying the microbiota
Generally, indigenous hydrocarbon‐degrading microbes in contaminated soils can be efficiently stimulated by plants (Yergeau et al., 2014; Pagé et al., 2015) or fertilizers (Yergeau et al., 2009, 2012; Bell et al., 2011). Bioaugmentation (inoculations) with a single or a few hydrocarbon‐degrading strains is thus generally ineffective (Thomassin‐Lacroix et al., 2002), and it has been shown that pre‐selecting microorganisms that can degrade hydrocarbons results in less efficient degradation than using the entirety of the microbes present in a soil (Bell et al., 2016). However, a recent report suggested that the success of invasion by the inoculated microbes could be increased by successive inoculation (Fig. 2), as the initial inoculation opens up a niche space for the invader (Mallon et al., 2018). The inoculum density could also play a role for single strain inoculations, as the inoculation of Lolium perenne (perennial ryegrass) with different concentrations of the alkane degrading Pantoea sp., resulted in maximum plant growth, diesel degradation, bacterial abundance and CYP153 alkane hydroxylase gene expression in the treatment with the densest inoculation (108 cell cm−3 soil) (Shabir et al., 2016). In addition, several studies have reported successful single PGPR strain inoculations in the context of rhizoremediation (Reed and Glick, 2005; Gurska et al., 2009; Xun et al., 2015; Asghar et al., 2017).
Figure 2.
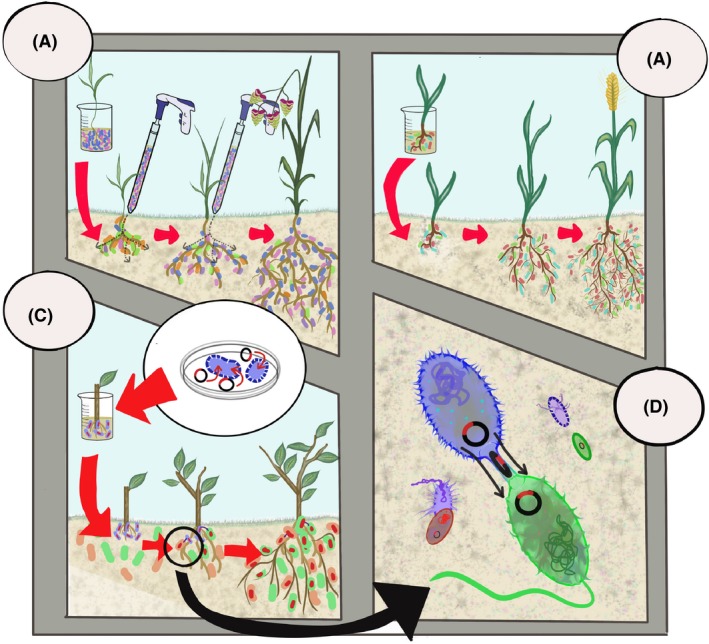
Examples of promising plant microbiome manipulation approaches for enhanced rhizoremediation: (A) repeated inoculation of a consortia of hydrocarbon‐degrading microorganisms, (B) early inoculation of a plant growth promoting rhizobacteria (PGPR) consortia, (C and D) inoculation of a bacteria harboring hydrocarbon degradation or plant growth promotion genes on a plasmid.
Alternatively, the inoculation of consortia is generally more efficient than individual strains for degrading hydrocarbons (Ghazali et al., 2004; Heinaru et al., 2005; Jacques et al., 2008; Mrozik and Piotrowska‐Seget, 2010) (Fig. 2). Some of the desired properties for the microbial constituents of a rhizoremediation consortia have been suggested: (i) be proficient in the colonization of the plant root surface in the rhizosphere, (ii) be able to survive, grow and not outcompete the rest of the members of the consortia, (iii) be able to attach to the root surface, (iv) be able to promote plant growth or the growth of other members of the consortia, (v) be able to handle abiotic stress, especially contaminant stress, (vi) be able to grow to the desired density under stressful conditions (Yang et al., 2009; Calvo et al., 2014). Consequently, candidate microbes for a rhizoremediation consortia should at least be selected for the strength of their association with the plant, with traits such as a strong chemotaxis towards plant root exudates and a strong attachment to the plant root surface (Yang et al., 2009; Bashan et al., 2014).
Taking the concept of consortia one step further, some studies have tried using synthetic communities for rhizoremediation (Pizarro‐Tobias et al., 2015). These synthetic communities should be designed to improve positive interactions, like commensalism and cooperation, while preventing negative interactions like predation and parasitism. Furthermore, several hydrocarbon degraders should be combined to ensure the presence of a diverse and redundant hydrocarbon degradation gene pool. Ideally, synthetic communities should be prepared from hundreds to thousands of strains, making it nearly impossible to test all interactions between consortia members. It is also difficult to maintain synthetic microbial communities for several generations (Johns et al., 2016), probably because of the numerous interactions happening simultaneously between members of the synthetic community (Stubbendieck et al., 2016). The use of naturally occurring, highly performing communities could be a better alternative, removing the need to test the compatibility of isolated strains with each other. Indeed, exposing willows to differentially selected initial soil communities can result in large differences in biomass when willows are grown under high stress levels, and these differences persist through time even though rhizosphere microbial communities become eventually identical (Yergeau et al., 2015a). This lends weight to the idea that exposing the plant partner to a different complex microbiota during its establishment can result in an improved growth in contaminated soils. However, many technical hurdles are facing the propagation and inoculation of complex microbiota at large scales.
Inoculating bacteria that harbored hydrocarbon degradation genes on mobile genetic elements has resulted in some successes (Fig. 2). For example, Weyens et al. (2009) observed a decrease in trichloroethylene (TCE) evapotranspiration (thus an increased degradation) after inoculation of hybrid poplars with a Pseudomonas strain containing a plasmid coding for the constitutive expression of the TCE degradation genes. This strain altered the rhizosphere community, even though it did not establish in this compartment. The strain did establish inside plant tissues, and the plasmid it contained was transmitted to other members of the endophytic community (Weyens et al., 2009). Similarly, the inoculation of two Burkholderia strains containing a plasmid coding for the constitutive expression of toluene degradation genes resulted in an improved plant growth and an increased toluene degradation (reduced evapotranspiration) (Taghavi et al., 2005). Although the two strains could not be detected in the plants, the plasmid they carried was detected in various other endophytic bacteria (Taghavi et al., 2005). Both these examples used endophytic bacteria, but a similar approach could be used for rhizoremediation, especially in view of the enhanced horizontal gene transfer rates in the rhizosphere (Van Elsas and Bailey, 2002).
Predictive models
Because it is a biological process, the time for soil decontamination by way of phytoremediation is difficult to estimate accurately, which often makes this option less attractive. Recent work has provided interesting evidence that various ecosystem processes could be predicted from microbiological data. For instance, the degradation of diesel in arctic soils could be predicted by the initial bacterial diversity and the abundance of specific assemblages of Betaproteobacteria, which was also related to the soil organic matter content (Bell et al., 2013). In that study, high Betaproteobacteria abundance was positively correlated with high diesel degradation. The predictability with which bacterial communities respond to these disturbances suggest that costly and time‐consuming chemical assessments of contaminated sites may not be necessary in the future and could be replaced by simple biological assessments (quantification of Betaproteobacteria). Similarly, the growth of willows after 100 days in highly contaminated soil could be predicted by the initial bacterial and fungal community composition and the initial relative abundance of specific taxa (Yergeau et al., 2015a). The Zn accumulation by willows growing for 16 months in a former landfill could be predicted by the relative abundance of specific fungal taxa in the rhizosphere after 4 months of growth (Bell et al., 2015). It therefore appears that the composition and relative abundance of the early colonizers of the plant environment are good predictors of its future behaviour. Creating predictive models could assist in choosing the right plant and the right microorganisms for a specific site without the need for labor‐intensive and costly preliminary trials, and, more importantly, estimate more precisely the time that will be needed for complete rhizoremediation.
Perspectives
Despite the remarkable advances detailed above, phytoremediation remains a marginal option for in situ soil remediation (Mench et al., 2010). The major obstacle to market penetration is that many sites to be decontaminated are in peri‐urban areas and need to be efficiently decontaminated over a short period, which is incompatible with the current practice of in situ phytoremediation. Additionally, phytoremediation is rarely suggested as a remediation technique by accredited experts because it is believed to be inefficient and because of the inability to precisely determine the duration of this biological process as it depends on contaminant and soil natures, plant used, environmental conditions and microbial activities (Montpetit and Lachapelle, 2015, 2016). One of the main reasons behind this was the low level of knowledge shown by accredited experts in the field of soil remediation partly due to poor communication from scientists (Montpetit and Lachapelle, 2015, 2016). Therefore, on top of research efforts aiming at better understanding the plant–microbe interactions during rhizoremediation, future endeavours should also (i) set‐up large scale demonstration experiments, potentially using integrated bioremediation approaches (Megharaj and Naidu, 2017), (ii) partner with environmental consulting firms and accredited experts, (iii) develop a genomics‐based tool to suggest management strategies and predict the duration of phytoremediation and (iv) test novel microbiome management approaches applicable at the field scale, such as inocula combining PGPR and microbial degraders (Baez‐Rogelio et al., 2017).
Conflict of interest
None declared.
Acknowledgements
This work was supported by NSERC Discovery grants RGPIN‐2014‐05274 to EY and RGPIN‐2014‐05426 to MSA and by a NSERC Strategic grant for projects STPGP 494702 to EY and MSA. SCG was supported by the Research Associate Program of the Government of Canada.
Microbial Biotechnology (2018) 11(5), 819–832
Funding Information
This work was supported by NSERC Discovery grants RGPIN‐2014‐05274 to EY and RGPIN‐2014‐05426 to MSA and by a NSERC Strategic grant for projects STPGP 494702 to EY and MSA. SCG was supported by the Research Associate Program of the Government of Canada.
References
- Adam, G. , and Duncan, H. (2002) Influence of diesel fuel on seed germination. Environ Pollut 120: 363–370. [DOI] [PubMed] [Google Scholar]
- Asghar, H.N. , Rafique, H.M. , Khan, M.Y. , and Zahir, Z.A. (2017) Phytoremediation of light crude oil by maize (Zea mays L.) bio‐augmented with plant growth promoting bacteria. Soil Sediment Contam An Int J 26: 749–763. [Google Scholar]
- Baez‐Rogelio, A. , Morales‐García, Y.E. , Quintero‐Hernández, V. , and Muñoz‐Rojas, J. (2017) Next generation of microbial inoculants for agriculture and bioremediation. Microb Biotechnol 10: 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrutia, O. , Garbisu, C. , Epelde, L. , Sampedro, M.C. , Goicolea, M.A. , and Becerril, J.M. (2011) Plant tolerance to diesel minimizes its impact on soil microbial characteristics during rhizoremediation of diesel‐contaminated soils. Sci Total Environ 409: 4087–4093. [DOI] [PubMed] [Google Scholar]
- Bartlett, M.D. , Briones, M.J.I. , Neilson, R. , Schmidt, O. , Spurgeon, D. , and Creamer, R.E. (2010) A critical review of current methods in earthworm ecology: from individuals to populations. Eur J Soil Biol 46: 67–73. [Google Scholar]
- Bashan, Y. , de‐Bashan, L.E. , Prabhu, S.R. and Hernandez, J.‐P. (2014) Advances in plant growth‐promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378, 1–33. [Google Scholar]
- Bell, T.H. , Yergeau, E. , Martineau, C. , Juck, D. , Whyte, L.G. , and Greer, C.W. (2011) Identification of nitrogen‐incorporating bacteria in petroleum‐contaminated arctic soils by using [15N]DNA‐based stable isotope probing and pyrosequencing. Appl Environ Microbiol 77: 4163–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, T.H. , Yergeau, E. , Maynard, C. , Juck, D. , Whyte, L.G. , and Greer, C.W. (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7: 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, T.H. , El‐Din Hassan, S. , Lauron‐Moreau, A. , Al‐Otaibi, F. , Hijri, M. , Yergeau, E. , and St‐Arnaud, M. (2014a) Linkage between bacterial and fungal rhizosphere communities in hydrocarbon‐contaminated soils is related to plant phylogeny. ISME J 8: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, T.H. , Joly, S. , Pitre, F.E. , and Yergeau, E. (2014b) Increasing phytoremediation efficiency and reliability using novel omics approaches. Trends Biotechnol 32: 271–280. [DOI] [PubMed] [Google Scholar]
- Bell, T.H. , Cloutier‐Hurteau, B. , Al‐Otaibi, F. , Turmel, M.‐C. , Yergeau, E. , Courchesne, F. , and St‐Arnaud, M. (2015) Early rhizosphere microbiome composition is related to the growth and Zn uptake of willows introduced to a former landfill. Environ Microbiol 17: 3025–3038. [DOI] [PubMed] [Google Scholar]
- Bell, T.H. , Stefani, F.O.P. , Abram, K. , Champagne, J. , Yergeau, E. , Hijri, M. , and St‐Arnaud, M. (2016) A diverse soil microbiome degrades more crude oil than specialized bacterial assemblages obtained in culture. Appl Environ Microbiol 82: 5530–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneduzi, A. , Ambrosini, A. , and Passaglia, L.M.P. (2012) Plant growth‐promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol 35: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, G. , and Smalla, K. (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68: 1–13. [DOI] [PubMed] [Google Scholar]
- Bernard, L. , Chapuis‐Lardy, L. , Razafimbelo, T. , Razafindrakoto, M. , Pablo, A.‐L. , Legname, E. , et al (2012) Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J 6: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, W.N. , and Stafford, C. (1993) Survey and evaluation of contaminants in earthworms and in soils derived from dredged material at confined disposal facilities in the Great Lakes Region. Environ Monit Assess 24: 151–165. [DOI] [PubMed] [Google Scholar]
- Bordenstein, S.R. , Theis, K.R. , Woese, C. , Fredericks, D. , Relman, D. , Inglis, T. , et al (2015) Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13: e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, G.I. , Dixon, D.G. , and Glick, B.R. (2000) Plant growth‐promoting bacteria that decrease heavy metal toxicity in plants. Can J Microbiol 46: 237–245. [DOI] [PubMed] [Google Scholar]
- Calvo, P. , Nelson, L. and Kloepper, J.W. (2014) Agricultural uses of plant biostimulants. Plant Soil, 383: 3–41. [Google Scholar]
- De Zelicourt, A. , Al‐Yousif, M. , and Hirt, H. (2013) Rhizosphere microbes as essential partners for plant stress tolerance. Mol Plant 6: 242–245. [DOI] [PubMed] [Google Scholar]
- Delgado‐Balbuena, L. , Bello‐López, J.M. , Navarro‐Noya, Y.E. , Rodríguez‐Valentín, A. , Luna‐Guido, M.L. , and Dendooven, L. (2016) Changes in the bacterial community structure of remediated anthracene‐contaminated soils. PLoS ONE 11: e0160991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amrani, A. , Dumas, A.S. , Wick, L.Y. , Yergeau, E. , and Berthom, R. (2015) “Omics” insights into PAH degradation toward improved green remediation biotechnologies. Environ Sci Technol 49: 11281–11291. [DOI] [PubMed] [Google Scholar]
- Ely, C.S. , and Smets, B.F. (2017) Bacteria from wheat and cucurbit plant roots metabolize PAHs and aromatic root exudates: implications for rhizodegradation. Int J Phytoremediation 19: 877–883. [DOI] [PubMed] [Google Scholar]
- Emmerling, C. , and Paulsch, D. (2001) Improvement of earthworm (Lumbricidae) community and activity in mine soils from open‐cast coal mining by the application of different organic waste materials. Pedobiologia (Jena) 45: 396–407. [Google Scholar]
- Esperschütz, J. , Buegger, F. , Winkler, J.B. , Munch, J.C. , Schloter, M. , and Gattinger, A. (2009) Microbial response to exudates in the rhizosphere of young beech trees (Fagus sylvatica L.) after dormancy. Soil Biol Biochem 41: 1976–1985. [Google Scholar]
- Gamerdinger, A. P. , Achin, R. S. , and Traxler, R. W. (1997) Approximating the impact of Sorption on biodegradation kinetics in soil‐water systems. Soil Sci Soc Am J 61: 1618–1626. [Google Scholar]
- Gao, Y. , Ren, L. , Ling, W. , Gong, S. , Sun, B. , and Zhang, Y. (2010) Desorption of phenanthrene and pyrene in soils by root exudates. Bioresour Technol 101: 1159–1165. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Yang, Y. , Ling, W. , Kong, H. , and Zhu, X. (2011) Gradient distribution of root exudates and polycyclic aromatic hydrocarbons in rhizosphere soil. Soil Sci Soc Am J 75: 1694. [Google Scholar]
- Gaskin, S.E. , and Bentham, R.H. (2010) Rhizoremediation of hydrocarbon contaminated soil using Australian native grasses. Sci Total Environ 408: 3683–3688. [DOI] [PubMed] [Google Scholar]
- Geissen, V. , Gomez‐Rivera, P. , Huerta Lwanga, E. , Mendoza, R.B. , Narcías, A.T. , and Marcías, E.B. (2008) Using earthworms to test the efficiency of remediation of oil‐polluted soil in tropical Mexico. Ecotoxicol Environ Saf 71: 638–642. [DOI] [PubMed] [Google Scholar]
- Ghazali, F.M. , Rahman, R.N.Z.A. , Salleh, A.B. , and Basri, M. (2004) Biodegradation of hydrocarbons in soil by microbial consortium. Int Biodeterior Biodegrad 54: 61–67. [Google Scholar]
- Glick, B.R. (2003) Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol Adv 21: 383–393. [DOI] [PubMed] [Google Scholar]
- Glick, B.R. (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251: 1–7. [DOI] [PubMed] [Google Scholar]
- Glick, B.R. , Penrose, D. , and Li, J. (1998) A model for the lowering of plant ethylene concentrations by plant growth‐promoting bacteria. J Theor Biol 190: 63–68. [DOI] [PubMed] [Google Scholar]
- Gomes, N.C.M. , Fagbola, O. , Costa, R. , Rumjanek, N.G. , Buchner, A. , Mendona‐Hagler, L. , and Smalla, K. (2003) Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl Environ Microbiol 69: 3758–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, E. , Brereton, N.J.B. , Marleau, J. , Guidi Nissim, W. , Pagé, A.P. , St‐Arnaud, M. , et al (2018) Trees, fungi and bacteria: tripartite metatranscriptomics of a root microbiome responding to soil contamination. Microbiome 6: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier, V. , Pitre, F.E. , Guidi Nissim, W. , and Labrecque, M. (2015) Genotypic differences explain most of the response of willow cultivars to petroleum‐contaminated soil. Trees – Struct Funct 29: 871–881. [Google Scholar]
- Guo, M. , Gong, Z. , Miao, R. , Rookes, J. , Cahill, D. , and Zhuang, J. (2017a) Microbial mechanisms controlling the rhizosphere effect of ryegrass on degradation of polycyclic aromatic hydrocarbons in an aged‐contaminated agricultural soil. Soil Biol Biochem 113: 130–142. [Google Scholar]
- Guo, M. , Gong, Z. , Miao, R. , Su, D. , Li, X. , Jia, C. , and Zhuang, J. (2017b) The influence of root exudates of maize and soybean on polycyclic aromatic hydrocarbons degradation and soil bacterial community structure. Ecol Eng 99: 22–30. [Google Scholar]
- Gurska, J. , Wang, W. , Gerhardt, K.E. , Khalid, A.M. , Isherwood, D.M. , Huang, X.D. , et al (2009) Three year field test of a plant growth promoting rhizobacteria enhanced phytoremediation system at a land farm for treatment of hydrocarbon waste. Environ Sci Technol 43: 4472–4479. [DOI] [PubMed] [Google Scholar]
- Gutiérrez‐Mañero, F.J. , Ramos‐Solano, B. , Probanza, A. , Mehouachi, J. , Tadeo, F.R. , and Talon, M. (2001) The plant‐growth‐promoting rhizobacteria Bacillus pumilus and Bacillus licheniformis produce high amounts of physiologically active gibberellins. Physiol Plant 111: 206–211. [Google Scholar]
- Haichar, F.el.Z. , Marol, C. , Berge, O. , Rangel‐Castro, J.I. , Prosser, J.I. , Balesdent, J. , et al (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Hamel, C. and Plenchette, C . (eds) (2007) Mycorrhizae in Crop Production. CRC Press, Boca Raton, FL, 366 pages. [Google Scholar]
- Harayama, S. , Kok, M. , and Neidle, E.L. (1992) Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol 46: 565–601. [DOI] [PubMed] [Google Scholar]
- Harms, H. , Schlosser, D. , and Wick, L.Y. (2011) Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9: 177–192. [DOI] [PubMed] [Google Scholar]
- Hartmann, A. , Schmid, M. , van Tuinen, D. , and Berg, G. (2009) Plant‐driven selection of microbes. Plant Soil 321: 235–257. [Google Scholar]
- Hassan, S.E.D. , Bell, T.H. , Stefani, F.O.P. , Denis, D. , Hijri, M. and St‐Arnaud, M. (2014) Contrasting the community structure of arbuscular mycorrhizal fungi from hydrocarbon‐contaminated and uncontaminated soils following willow (Salix spp. L.) planting. PLoS ONE 9: e102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinaru, E. , Merimaa, M. , Viggor, S. , Lehiste, M. , Leito, I. , Truu, J. , and Heinaru, A. (2005) Biodegradation efficiency of functionally important populations selected for bioaugmentation in phenol‐ and oil‐polluted area. FEMS Microbiol Ecol 51: 363–373. [DOI] [PubMed] [Google Scholar]
- Hubbard, M. , Germida, J.J. , and Vujanovic, V. (2014) Fungal endophyte colonization coincides with altered DNA methylation in drought‐stressed wheat seedlings. Can J Plant Sci 94: 223–234. [Google Scholar]
- Iffis, B. , St‐Arnaud, M. , and Hijri, M. (2016) Petroleum hydrocarbon contamination, plant identity and arbuscular mycorrhizal fungal (AMF) community determine assemblages of the AMF spore‐associated microbes. Environ Microbiol 18: 2689–2704. [DOI] [PubMed] [Google Scholar]
- Iffis, B. , St‐Arnaud, M. , and Hijri, M. (2017) Petroleum contamination and plant identity influence soil and root microbial communities while AMF spores retrieved from the same plants possess markedly different communities. Front Plant Sci 8: 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, R.J.S. , Okeke, B.C. , Bento, F.M. , Teixeira, A.S. , Peralba, M.C.R. , and Camargo, F.A.O. (2008) Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour Technol 99: 2637–2643. [DOI] [PubMed] [Google Scholar]
- Jager, T. (1998) Mechanistic approach for estimating bioconcentration of organic chemicals in earthworms (oligochaeta). Environ Toxicol Chem 17: 2080–2090. [Google Scholar]
- Johns, N.I. , Blazejewski, T. , Gomes, A.L. , and Wang, H.H. (2016) Principles for designing synthetic microbial communities. Curr Opin Microbiol 31: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joner, E.J. , Johansen, A. , Loibner, A.P. , Cruz, M.A.Dela. , Szolar, O.H.J. , Portal, J.M. , and Leyval, C. (2001) Rhizosphere effects on microbial community structure and dissipation and toxicity of polycyclic aromatic hydrocarbons (PAHs) in spiked soil. Environ Sci Technol 35: 2773–2777. [DOI] [PubMed] [Google Scholar]
- Jones, D.L. (1998) Organic acids in the rhizosphere – a critical review. Plant Soil 205: 25–44. [Google Scholar]
- Jones, D.L. , Hodge, A. , and Kuzyakov, Y. (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163: 459–480. [DOI] [PubMed] [Google Scholar]
- Kadri, T. , Rouissi, T. , Kaur Brar, S. , Cledon, M. , Sarma, S. , and Verma, M. (2017) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: a review. J Environ Sci (China) 51: 52–74. [DOI] [PubMed] [Google Scholar]
- Karigar, C.S. , and Rao, S.S. (2011) Role of microbial enzymes in the bioremediation of pollutants: a review. Enzyme Res 2011: 805187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, Z. , Roman, D. , Kintz, T. , delas Alas, M. , Yap, R. and Doty, S. (2014) Degradation, phytoprotection and phytoremediation of phenanthrene by endophyte Pseudomonas putida, PD1. Environ Sci Technol 48, 12221–12228. [DOI] [PubMed] [Google Scholar]
- Kloepper, J.W. , and Schroth, M.N. (1978) Plant‐growth promoting rhizobacteria on radishes. Proc 4th Int Conf Plant Pathog Bact 2: 879–882. [Google Scholar]
- Kowalchuk, G.A. , Buma, D.S. , De Boer, W. , Klinkhamer, P.G.L. , and Van Veen, J.A. (2002) Effects of above‐ground plant species composition and diversity on the diversity of soil‐borne microorganisms. Antonie van Leeuwenhoek Int J Gen Mol Microbiol 81: 509–520. [DOI] [PubMed] [Google Scholar]
- Kuiper, I. , Lagendijk, E.L. , Bloemberg, G.V. , and Lugtenberg, B.J.J. (2004) Rhizoremediation: a beneficial plant‐microbe interaction. Mol Plant‐Microbe Interact 17: 6–15. [DOI] [PubMed] [Google Scholar]
- Kuzovkina, Y.A. , and Volk, T.A. (2009) The characterization of willow (Salix L.) varieties for use in ecological engineering applications: co‐ordination of structure, function and autecology. Ecol Eng 35: 1178–1189. [Google Scholar]
- Lavelle, P. and Spain, A.V. (2001) Soil Ecology. Springer Science & Business Media, Berlin/Heidelberg, Germany. [Google Scholar]
- Lee, S.H. , Lee, W.S. , Lee, C.H. , and Kim, J.G. (2008) Degradation of phenanthrene and pyrene in rhizosphere of grasses and legumes. J Hazard Mater 153: 892–898. [DOI] [PubMed] [Google Scholar]
- Ling, N. , Zhang, W. , Wang, D. , Mao, J. , Huang, Q. , Guo, S. , and Shen, Q. (2013) Root exudates from grafted‐root watermelon showed a certain contribution in inhibiting Fusarium oxysporum f. sp. niveum. PLoS ONE 8: e63383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Hou, J. , Wang, Q. , Ding, L. , and Luo, Y. (2014) Isolation and characterization of plant growth‐promoting rhizobacteria and their effects on phytoremediation of petroleum‐contaminated saline‐alkali soil. Chemosphere 117: 303–308. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Dai, Y. , and Sun, L. (2015) Effect of rhizosphere enzymes on phytoremediation in PAH‐contaminated soil using five plant species. PLoS ONE 10: e0120369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y.‐F. , Lu, M. , Peng, F. , Wan, Y. , and Liao, M.‐H. (2014) Remediation of polychlorinated biphenyl‐contaminated soil by using a combination of ryegrass, arbuscular mycorrhizal fungi and earthworms. Chemosphere 106: 44–50. [DOI] [PubMed] [Google Scholar]
- Luepromchai, E. , Singer, A.C. , Yang, C.‐H. , and Crowley, D.E. (2002) Interactions of earthworms with indigenous and bioaugmented PCB‐degrading bacteria. FEMS Microbiol Ecol 41: 191–197. [DOI] [PubMed] [Google Scholar]
- Lumactud, R. , Shen, S.Y. , Lau, M. , and Fulthorpe, R. (2016) Bacterial endophytes isolated from plants in natural oil seep soils with chronic hydrocarbon contamination. Front Microbiol 7: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, D.S. , Lebeis, S.L. , Paredes, S.H. , Yourstone, S. , Gehring, J. , Malfatti, S. , et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W.C. , Van Kleunen, A. , Immerzeel, J. , and Gert‐Jan De Maagd, P. (1998) Bioaccumulation of polycyclic aromatic hydrocarbons by earthworms: assessment of equilibrium partitioning theory in in situ studies and water experiments. Environ Toxicol Chem 17: 1730–1737. [Google Scholar]
- Ma, B. , He, Y. , Chen, H. , Xu, J. , and Rengel, Z. (2010) Dissipation of polycyclic aromatic hydrocarbons (PAHs) in the rhizosphere: synthesis through meta‐analysis. Environ Pollut 158: 855–861. [DOI] [PubMed] [Google Scholar]
- Mallon, C.A. , Le Roux, X. , van Doorn, G.S. , Dini‐Andreote, F. , Poly, F. , and Salles, J.F. (2018) The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader's niche. ISME J 12: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manschadi, A.M. , Christopher, J. , deVoil, P. , and Hammer, G.L. (2006) The role of root architectural traits in adaptation of wheat to water‐limited environments. Funct Plant Biol 33: 823. [DOI] [PubMed] [Google Scholar]
- Martin, B.C. , George, S.J. , Price, C.A. , Ryan, M.H. , and Tibbett, M. (2014) The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: current knowledge and future directions. Sci Total Environ 472: 642–653. [DOI] [PubMed] [Google Scholar]
- Mayak, S. , Tirosh, T. , and Glick, B.R. (2004) Plant growth‐promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42: 565–572. [DOI] [PubMed] [Google Scholar]
- Megharaj, M. , and Naidu, R. (2017) Soil and brownfield bioremediation.. Microbial Biotech 10: 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mench, M. , Lepp, N. , Bert, V. , Schwitzguébel, J.P. , Gawronski, S.W. , Schröder, P. , and Vangronsveld, J. (2010) Successes and limitations of phytotechnologies at field scale: outcomes, assessment and outlook from COST Action 859. J Soils Sediments 10: 1039–1070. [Google Scholar]
- Montpetit, É. , and Lachapelle, E. (2015) Can policy actors learn from academic scientists? Env Polit 24: 661–680. [Google Scholar]
- Montpetit, É. , and Lachapelle, E. (2016) Information, values and expert decision‐making: the case of soil decontamination. Policy Sci 49: 155–171. [Google Scholar]
- Mrozik, A. , and Piotrowska‐Seget, Z. (2010) Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol Res 165: 363–375. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, S. , George, J. , and Masto, R.E. (2017) Changes in Polycyclic Aromatic Hydrocarbons (PAHs) and soil biological parameters in a revegetated coal mine spoil. Land Degrad Dev 28: 1047–1055. [Google Scholar]
- Nadeem, S.M. , Ahmad, M. , Zahir, Z.A. , Javaid, A. , and Ashraf, M. (2014) The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol Adv 32: 429–448. [DOI] [PubMed] [Google Scholar]
- Nechitaylo, T.Y. , Yakimov, M.M. , Godinho, M. , Timmis, K.N. , Belogolova, E. , Byzov, B.A. , et al (2010) Effect of the earthworms lumbricus terrestris and Aporrectodea caliginosa on bacterial diversity in soil. Microb Ecol 59: 574–587. [DOI] [PubMed] [Google Scholar]
- Olanrewaju, O.S. , Glick, B.R. , and Babalola, O.O. (2017) Mechanisms of action of plant growth promoting bacteria. World J Microbiol Biotechnol 33: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, J.C. , and Bland, W.L. (1987) Genotypic variation in crop plant root systems. Adv Agron 41: 91–145. [Google Scholar]
- Pagé, A.P. , Yergeau, É. , and Greer, C.W. (2015) Salix purpurea stimulates the expression of specific bacterial xenobiotic degradation genes in a soil contaminated with hydrocarbons. PLoS ONE 10: e0132062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten, C.L. , and Glick, B.R. (1996) Bacterial biosynthesis of indole‐3‐acetic acid. Can J Microbiol 42: 207–220. [DOI] [PubMed] [Google Scholar]
- Pawlik, M. , Cania, B. , Thijs, S. , Vangronsveld, J. and Piotrowska‐Seget, Z. (2017) Hydrocarbon degradation potential and plant growth‐promoting activity of culturable endophytic bacteria of Lotus corniculatus and Oenothera biennis from a long‐term polluted site. Environ Sci Pollut Res 24, 19640–19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña‐Castro, J.M. , Barrera‐Figueroa, B.E. , Fernández‐Linares, L. , Ruiz‐Medrano, R. , and Xoconostle‐Cázares, B. (2006) Isolation and identification of up‐regulated genes in bermudagrass roots (Cynodon dactylon L.) grown under petroleum hydrocarbon stress. Plant Sci 170: 724–731. [Google Scholar]
- Pilon‐Smits, E. (2005) Phytoremediation. Revis Plant Biol 56: 15–39. [DOI] [PubMed] [Google Scholar]
- Pizarro‐Tobias, P. , Niqui, J.L. , Roca, A. , Solano, J. , Fernandez, M. , Bastida, F. , et al (2015) Field trial on removal of petroleum‐hydrocarbon pollutants using a microbial consortium for bioremediation and rhizoremediation. Environ Microbiol Rep 7: 85–94. [DOI] [PubMed] [Google Scholar]
- Qin, F. , Kakimoto, M. , Sakuma, Y. , Maruyama, K. , Osakabe, Y. , Tran, L.‐S.P. , et al (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50: 54–69. [DOI] [PubMed] [Google Scholar]
- Quiza, L. , St‐Arnaud, M. and Yergeau, E. (2015) Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front Plant Sci 6: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar, M. , Sandhya, S. , Prasad, M.N.V. , and Freitas, H. (2012) Perspectives of plant‐associated microbes in heavy metal phytoremediation. Biotechnol Adv 30: 1562–1574. [DOI] [PubMed] [Google Scholar]
- Raut, V. , Shaikh, I. , Naphade, B. , Prashar, K. , and Adhapure, N. (2017) Plant growth promotion using microbial IAA producers in conjunction with azolla: a novel approach. Chem Biol Technol Agric 4: 1. [Google Scholar]
- Reddy, B. R. , and Sethunathan, N. (1983) Mineralization of parathion in the rice rhizosphere. Appl Environ Microbiol 45(3): 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, M. , and Glick, B. (2005) Growth of canola (Brassica napus) in the presence of plant growth‐promoting bacteria and either copper or polycyclic aromatic hydrocarbons. Can J Microbiol 51: 425–429. [DOI] [PubMed] [Google Scholar]
- Rodrıguez, H. and Fraga, R. (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17, 319–339. [DOI] [PubMed] [Google Scholar]
- Rohrbacher, F. , and St‐Arnaud, M. (2016) Root exudation: the ecological driver of hydrocarbon rhizoremediation. Agronomy 6: 19. [Google Scholar]
- Rosenberg, E. , and Zilber‐Rosenberg, I. (2016) Bacterial bleaching of corals leads to hologenome concept. Microbe Mag 11: 27–31. [Google Scholar]
- Rosenberg, E. , Sharon, G. , and Zilber‐Rosenberg, I. (2009) The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol 11: 2959–2962. [DOI] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.‐H. , Reddy, M.S. , Kloepper, J.W. , and Paré, P.W. (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentchilo, V. , Mayer, A.P. , Guy, L. , Miyazaki, R. , Tringe, S.G. , Barry, K. , et al (2013) Community‐wide plasmid gene mobilization and selection. ISME J 7: 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabir, G. , Arslan, M. , Fatima, K. , Amin, I. , Khan, Q.M. , and Afzal, M. (2016) Effects of inoculum density on plant growth and hydrocarbon degradation. Pedosphere 26: 774–778. [Google Scholar]
- Singer, A.C. , Crowley, D.E. , and Thompson, I.P. (2003) Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21: 123–130. [DOI] [PubMed] [Google Scholar]
- Singleton, D.R. , Hendrix, P.F. , Coleman, D.C. , and Whitman, W.B. (2003) Identification of uncultured bacteria tightly associated with the intestine of the earthworm Lumbricus rubellus (Lumbricidae; Oligochaeta). Soil Biol Biochem 35: 1547–1555. [Google Scholar]
- Smalla, K. , Wieland, G. , Buchner, A. , Zock, A. , Parzy, J. , Kaiser, S. , et al (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant‐dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67: 4742–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.E. and Read, D. (2008) Mycorrhizal symbiosis. Mycorrhizal Symbiosis 611–XVIII.
- Spaepen, S. , and Vanderleyden, J. (2011) Auxin and plant‐microbe interactions. Cold Spring Harb Perspect Biol 3: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springett, J. , and Gray, R. (1997) The interaction between plant roots and earthworm burrows in pasture. Soil Biol Biochem 29: 621–625. [Google Scholar]
- Stubbendieck, R.M. , Vargas‐Bautista, C. , and Straight, P.D. (2016) Bacterial communities: interactions to scale. Front Microbiol 7: 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.‐R. , Cang, L. , Wang, Q.‐Y. , Zhou, D.‐M. , Cheng, J.‐M. , and Xu, H. (2010) Roles of abiotic losses, microbes, plant roots, and root exudates on phytoremediation of PAHs in a barren soil. J Hazard Mater 176: 919–925. [DOI] [PubMed] [Google Scholar]
- Taghavi, S. , Barac, T. , Greenberg, B. , Borremans, B. , Vangronsveld, J. , and Van Der Lelie, D. (2005) Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microbiol 71: 8500–8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, M. , Chen, H. , Huang, J.C. , and Tian, Z.Q. (2009) AM fungi effects on the growth and physiology of Zea mays seedlings under diesel stress. Soil Biol Biochem 41: 936–940. [Google Scholar]
- Theis, K.R. , Dheilly, N.M. , Klassen, J.L. , Brucker, R.M. , Baines, J.F. , Bosch, T.C.G. , et al (2016) Getting the hologenome concept right: an eco‐evolutionary framework for hosts and their microbiomes. mSystems 1, e00028‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, F. , and Cébron, A. (2016) Short‐term rhizosphere effect on available carbon sources, phenanthrene degradation, and active microbiome in an aged‐contaminated industrial soil. Front Microbiol 7: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassin‐Lacroix, E. , Eriksson, M. , Reimer, K. , and Mohn, W. (2002) Biostimulation and bioaugmentation for on‐site treatment of weathered diesel fuel in Arctic soil. Appl Microbiol Biotechnol 59: 551–556. [DOI] [PubMed] [Google Scholar]
- Timmusk, S. , and Wagner, E.G. (1999) The plant‐growth‐promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol Plant Microbe Interact 12: 951–959. [DOI] [PubMed] [Google Scholar]
- Tiunov, A.V. , and Dobrovolskaya, T.G. (2002) Fungal and bacterial communities in Lumbricus terrestris burrow walls: a laboratory experiment. Pedobiologia (Jena) 46: 595–605. [Google Scholar]
- Top, E.M. , and Springael, D. (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14: 262–269. [DOI] [PubMed] [Google Scholar]
- Toyama, T. , Furukawa, T. , Maeda, N. , Inoue, D. , Sei, K. , Mori, K. , et al (2011) Accelerated biodegradation of pyrene and benzo[a]pyrene in the Phragmites australis rhizosphere by bacteria–root exudate interactions. Water Res 45: 1629–1638. [DOI] [PubMed] [Google Scholar]
- Van Beilen, J.B. , Funhoff, E.G. , Van Loon, A. , Just, A. , Kaysser, L. , Bouza, M. , et al (2006) Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane‐degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol 72: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elsas, J.D. , and Bailey, M.J. (2002) The ecology of transfer of mobile genetic elements. FEMS Microbiol Ecol 42: 187–197. [DOI] [PubMed] [Google Scholar]
- Van Opstal, E.J. and Bordenstein, S.R. (2015) Rethinking heritability of the microbiome. Science (80‐.) 349, 1172–1173. [DOI] [PubMed] [Google Scholar]
- Voss, J.D. , Leon, J.C. , Dhurandhar, N.V. , and Robb, F.T. (2015) Pawnobiome: manipulation of the hologenome within one host generation and beyond. Front Microbiol 6: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyens, N. , Van Der Lelie, D. , Artois, T. , Smeets, K. , Taghavi, S. , Newman, L. , et al (2009) Bioaugmentation with engineered endophytic bacteria improves contaminant fate in phytoremediation. Environ Sci Technol 43: 9413–9418. [DOI] [PubMed] [Google Scholar]
- Xun, F. , Xie, B. , Liu, S. , and Guo, C. (2015) Effect of plant growth‐promoting bacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) inoculation on oats in saline‐alkali soil contaminated by petroleum to enhance phytoremediation. Environ Sci Pollut Res 22: 598–608. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Kloepper, J.W. , and Ryu, C.‐M. (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14: 1–4. [DOI] [PubMed] [Google Scholar]
- Yasmin, S. and D'Souza, D. (2010) Effects of pesticides on the growth and reproduction of earthworm: a review. Appl Environ Soil Sci 2010, 1–9. [Google Scholar]
- Yergeau, E. , Arbour, M. , Brousseau, R. , Juck, D. , Lawrence, J.R. , Masson, L. , et al (2009) Microarray and real‐time PCR analyses of the responses of high‐arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microbiol 75: 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau, E. , Sanschagrin, S. , Beaumier, D. , and Greer, C.W. (2012) Metagenomic analysis of the bioremediation of diesel‐contaminated canadian high arctic soils. PLoS ONE 7: e30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau, E. , Sanschagrin, S. , Maynard, C. , St‐Arnaud, M. , and Greer, C.W. (2014) Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J 8: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau, E. , Bell, T.H. , Champagne, J. , Maynard, C. , Tardif, S. , Tremblay, J. , and Greer, C.W. (2015a) Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front Microbiol 6: 1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau, E. , Maynard, C. , Sanschagrin, S. , Champagne, J. , Juck, D. , Lee, K. , and Greer, C.W. (2015b) Microbial community composition, functions, and activities in the gulf of mexico 1 year after the deepwater horizon accident. Appl Environ Microbiol 81: 5855–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau, E. , Tremblay, J. , Joly, S. , Labrecque, M. , Maynard, C. , Pitre, F.E. , et al (2018) Soil contamination alters the willow root and rhizosphere metatranscriptome and the root–rhizosphere interactome. ISME J 12: 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala‐Cruz, J. , Trujillo‐C, F. , Ortiz‐Ceballos, G.C. , and Ortiz‐Ceballos, A.I. (2013) Tropical endogeic earthworm population in a pollution gradient with weathered crude oil. Res J Environ Sci 7: 15–26. [Google Scholar]
- Zilber‐Rosenberg, I. , and Rosenberg, E. (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735. [DOI] [PubMed] [Google Scholar]


