Abstract
Contaminated surfaces serve as an important reservoir for Clostridium difficile transmission. Current strategies to detect environmental contamination of C. difficile rely heavily on culture, and often only indicate presence versus absence of spores. The goal of this study was to compare quantitative PCR (qPCR) to culture for the detection and quantification of C. difficile from inert surfaces. First, we compared the limit of detection (LOD) of a 16S rRNA gene and toxin B gene qPCR assay for detection of C. difficile in solution. Second, we compared the LODs of 16S rRNA gene qPCR versus culture for detection of C. difficile from surfaces. Solution experiments were performed by direct seeding of spores into neutralizing broth, whereas surface experiments involved seeding of spores onto plastic test surfaces, and recovery using sponge swabs. Both experiments were conducted using spores expressing short (NAP1) and long (NAP4) hair lengths. Combining data from both strains, the overall LOD for C. difficile cells in solution was 1.4 cells for 16S rRNA gene and 23.6 cells for toxin B gene qPCR (p<0.001). The overall LOD for C. difficile cells from surfaces was 17.1 cells for 16S rRNA gene qPCR and 54.5 cells for culture (p = 0.05), and was not statistically different between strains for each method (p = 0.52). Overall, the proportion of C. difficile cells recovered from surfaces was good when detected by 16S rRNA gene qPCR and culture (qPCR: 76%, culture: 67%, p = 0.36), but, 16S rRNA gene qPCR was capable of detecting lower levels of surface contamination. Future work attempting to measure the presence of C. difficile on environmental surfaces should consider using qPCR.
Introduction
Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacterium, and the leading cause of health-care associated infective diarrhea [1]. Risk factors for the disease include previous hospitalizations, advanced age and the use of antibiotics [1–3]. C. difficile spores play an important role in disease transmission. Spores can persist in the environment for up to several months and are resistant to stresses such as heat, oxygen, and exposure to routine disinfectants [4,5].
Within the hospital environment, contaminated surfaces are thought to serve as an important reservoir for C. difficile transmission [6]. Unfortunately, the optimal method for sampling and detecting C. difficile contamination is currently unknown. Different sampling techniques have been reported, including contact plates, swabs, wipes and sponges, with several studies suggesting superior performance by sponges [7–9]. Culture remains the primary laboratory detection method, despite the routine and successful use of PCR in the clinical setting. Furthermore, most studies are qualitative and report only the presence or absence of C. difficile in small surface area samples [10,11]. Previous work has demonstrated that quantitative PCR (qPCR) can be successfully used for environmental sampling of C. difficile, but assay sensitivity and direct comparison to culture have not been evaluated [12].
Differences in the ability of C. difficile strains to adhere to surfaces may also be an important factor in disease transmission, and may impact the quantification of spores when conducting environmental sampling. Attachment and adherence of C. difficile spores is thought to be influenced by hydrophobicity, the exosporium and the presence of hair-like structures identified on the surface of certain strains [13–15]. Two epidemic strains with different surface properties are NAP1 and NAP4. NAP1 spores express short hairs, while NAP4 spores express long hairs [15,16].
Our objective was to validate qPCR as a sensitive method to quantify C. difficile, and compare qPCR to culture for detection C. difficile spores on contaminated surfaces. Furthermore, we compared both NAP1 and NAP4 spores to determine whether potential differences in adhesion affect the ability to quantify environmental contamination.
Materials and methods
Study design
Under experimental conditions, we compared qPCR versus quantitative culture for detection of C. difficile surface contamination. As a preliminary step, we assessed a toxin B gene and 16S rRNA gene qPCR assay and proceeded with the most sensitive target for in vitro studies. Test surfaces (plastic) were then seeded with known quantities of C. difficile spores (NAP1 and NAP4), and recovered using sponge swabs. Recovered spores were detected by qPCR and culture, and the recovery efficiencies (% recovery) and limit of detections (LODs) determined to establish the most sensitive method.
Culture of C. difficile and preparation of spore stocks
All C. difficile cultures were grown under anaerobic conditions at 37°C. To induce sporulation, C. difficile was cultured using Brucella supplemented agar (BSA) for approximately 10 days. For all other quantitative culturing, C. difficile was grown on CHROMagar media (Alere Inc., Canada) for 24 h.
NAP1 and NAP4 spores were obtained from clinical specimens and typed at the Public Health Ontario Laboratories. To prepare NAP1 and NAP4 spore stocks, C. difficile was scraped from BSA plates and suspended in 1 mL of double distilled water (ddH2O). Suspensions were washed three times in 1 mL of ddH2O, incubated overnight at 4°C to kill the majority of vegetative cells, and then washed one additional time. Stock concentrations of roughly 1 x 107 CFU/mL were prepared by adjusting the optical density (600 nm) of the suspension to 0.15. Actual concentrations were determined by cell counting using a disposable hemocytometer (In Cyto, Neubauer Improved, VWR, Canada) to provide the total number of cells, regardless of culturability. The morphology of the cells and absence of debris were evaluated by phase-contrast microscopy. Spore stocks were stored for approximately 1 week at 4°C and re-quantified before use to ensure accuracy.
Detection of C. difficile by qPCR
Primer and probe sequences used in this study are listed in S1 Table. For qPCR, 10 μL reactions containing 5 μL JumpStart Taq ReadyMix for Quantitative PCR (D7440, Sigma, Canada), 0.1 μL Reference Dye for Quantitative PCR (R4526, Sigma, Canada), 0.4 μL 25mM MgCl2, 0.5 μM each of forward and reverse primers, 0.15 μM TaqMan probe and 3.85 μL DNA were prepared. Reactions were run in triplicate on an ABI 7900HT thermocycler (Applied Biosystems) under the following conditions: 50°C 2 min, 95°C 10 min, then 45 cycles of 95°C 15 sec, 60°C 1 min.
DNA was extracted from spore solutions using the ZymoBIOMICS DNA Miniprep Kit (Cedarlane, Canada) according to the manufacturer’s instructions with minor modifications: a bead-beating step of 1 h was used and DNA was eluted in a final volume of 50 μL of ddH2O. PCR efficiencies of the 16S rRNA gene and toxin B gene assays were determined by extraction of DNA from a known concentration of spores (OD = 0.15), followed by ten-fold serial dilution in nuclease-free water (10−1 to 10−8) to generate standards. Each ten-fold serial dilution was performed in triplicate and used for both the 16s rRNA gene and toxin B gene assays. Standard curves were constructed by plotting Ct values against log10 of the spore quantities corresponding to each DNA dilution. A linear regression line was fit to the data using least squares and PCR efficiency was calculated according to Eq 1.
| (1) |
LOD from solution
We considered the LOD from solution to be the minimum number of C. difficile cells in dilution to be detected in over 95% of experimental replicates when subjected to the entire process of DNA extraction and qPCR [17]. We determined LOD values based on the total number of cells in solution, which was obtained via counting using a haemocytometer.
Ten-fold serial dilutions of C. difficile cells were prepared from a quantified spore stock (OD = 0.15) in 20% D/E Neutralizing Broth (Scigene, Canada). DNA extractions were performed on each dilution to generate standards, and the 16s rRNA gene and toxin B gene qPCRs were run as described above. A logistic regression model with detection (a Ct value < 40) as the outcome, and log10 cell quantity as the only predictor variable, was used [17]. The model was fit with the rstanarm package in R which conducts Markov Chain Monte Carlo estimation. Based on this model, the number of cells needed to achieve 95% probability of detection was estimated, along with the 95% credible interval (CI).
Seeding and recovery of C. difficile cells from test surfaces
Polypropylene plastic was chosen as a test surface because it widespread throughout hospital environments in North America. Plastic surfaces were seeded with known quantities of C. difficile by pipetting 10 μL drops of serially diluted cells onto marked 25-cm2 areas. Spiked surfaces were allowed to dry for 2 h at room temperature before sampling. Surfaces were sampled using sterile cellulose sponges, pre-moistened with D/E Neutralizing Buffer (Scigiene, Canada). The entire marked area was wiped horizontally, vertically and at a 45° angle; the whole process was repeated twice. Sponges were then dispensed into sampling bags and 40 mL of sterile ddH2O was added. Each sponge was massaged vigorously between finger tips for 90 seconds, and then allowed to stand at room temperature for 10 minutes. Sponges were aseptically removed and the liquid was transferred to a 50-mL conical tube. Extracted cells were collected by centrifugation at 7,500 × g for 15 minutes. All but 1 mL of the liquid was removed. The pelleted cells were re-suspended, transferred to a 1 mL microfuge tube, and then collected again by centrifugation at 10,000 × g for 5 min. The entire supernatant was decanted and the pellet was re-suspended in 500 μL of 20% D/E Neutralizing Broth. Half of the solution was used for DNA extraction and qPCR as described above. The remaining half was used for quantitative culture.
LOD from surfaces
We defined the LOD from surfaces to be the minimum number of C. difficile cells seeded onto a surface that could be detected in over 95% of experimental replicates [17]. LODs were calculated using the logistic regression model described above and were based on the total number of cells seeded onto the surface.
Mechanical recovery calculations
The mechanical recovery was used as measurement of the combined ability of the sponge to collect spores from a contaminated surface, and how well spores could be extracted from the sponge for quantification. The mechanical recovery by culture and qPCR were determined according to Eqs 2 and 3, respectively. Serial dilutions of a known concentration of C. difficile cells were prepared in water. Identical numbers of cells were seeded onto test surfaces and either directly onto culture plates for assessing the mechanical recovery by culture, or directly into 500 μL of 20% D/E Neutralizing Broth for assessing the mechanical recovery by qPCR. Cells were recovered from the surface and processed by quantitative culture or qPCR as described above. Graphs and statistical analyses were conducted using Graph Pad Prism 6.0
| (2) |
| (3) |
Results
Validation of the 16S rRNA gene and toxin B gene qPCR assays
The C. difficile 16S rRNA gene and toxin B gene qPCRs produced standard curves with PCR efficiencies between 93–100% (S1 Fig). PCR efficiencies were slightly lower for the toxin B gene assay (95.1 for NAP1 and 93.0% for NAP4) when compared to the 16S rRNA gene assay (99.5% for NAP1 and 99.9% for NAP4). All PCR efficiencies fell within the acceptable range of 90–110% [18].
LOD from solution for the 16S rRNA gene and toxin B gene qPCR assays
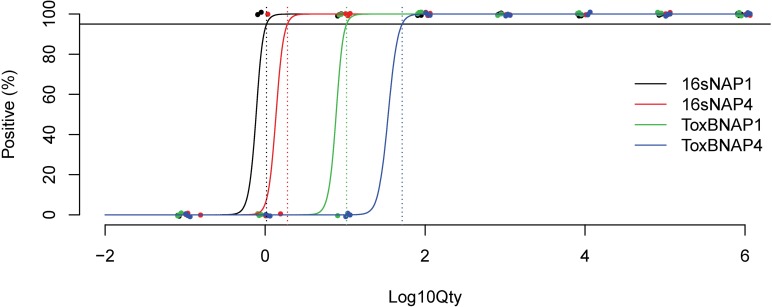
The LOD for the 16S rRNA gene qPCR was 16.9-fold lower than for the toxin B gene qPCR (1.4 for 16s rRNA versus 23.6 for toxin B, p<0.001, Table 1 and Fig 1). The LOD for NAP1 was 3.1-fold lower than NAP4 (3.3 for NAP1 versus 10.1 for NAP4, p<0.001). No statistically significant interaction between method and strain was detected in the model (p = 0.20).
Table 1. LOD of the 16S rRNA gene and toxin B gene qPCR assays recovery of C. difficile spores from solution.
| PCR Target | Strain | na | LOD (# cells, 95% CI) |
|---|---|---|---|
| 16S rRNA gene | NAP1 | 3 | 1.0 (0.7–2.3) |
| NAP4 | 3 | 1.9 (1.3–6.5) | |
| Overall | 6 | 1.4 (1.1–3.4) | |
| Toxin B gene | NAP1 | 3 | 10.4 (7.1–22.4) |
| NAP4 | 3 | 52.4 (16.5–160.6) | |
| Overall | 6 | 23.6 (12.5–52.2) |
an = number of independent experiments, 8 dilutions per experiment
Fig 1. LOD curves for the C. difficile 16S rRNA gene and toxin B gene qPCR assays using NAP1 and NAP4 spores.
LODs (dotted lines) were determined at the 95% positivity (solid black line) cut-off (n = 3 independent experiments for each strain/assay combination, total n = 12).
LOD from surfaces for 16S rRNA gene qPCR and culture
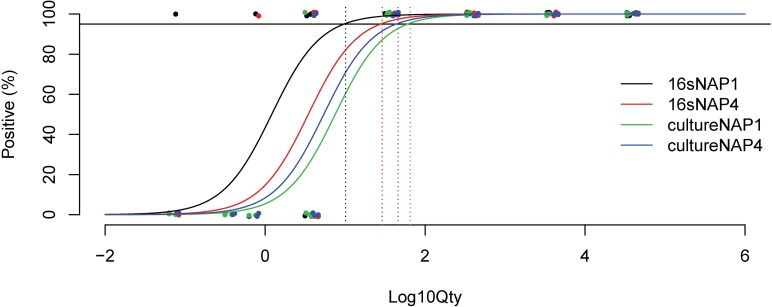
Overall, the LOD for 16S rRNA gene qPCR was 3.2-fold lower than that for culture (17.1 for 16S rRNA gene qPCR versus 54.5 for culture, p = 0.05, Table 2 and Fig 2). No significant difference was found between the LODs for NAP1 versus NAP4 (p = 0.52). No statistically significant interaction between method and strain was detected in the LOD model (p = 0.23).
Table 2. LOD of qPCR and culture for the recovery of C. difficile from contaminated surfaces.
| Method | Strain | na | LOD (# cells, 95% CI) |
|---|---|---|---|
| 16S rRNA gene qPCR |
NAP1 | 5 | 10.1 (2.9–56.4) |
| NAP4 | 5 | 29.1 (8.3–160.3) | |
| Overall | 10 | 17.1 (6.2–75.8) | |
| Culture | NAP1 | 5 | 65.0 (16.3–422.9) |
| NAP4 | 5 | 46.0 (13.5–275.6) | |
| Overall | 10 | 54.5 (18.0–263.7) |
an = number of independent experiments, 5 dilutions per experiment
Fig 2. LOD curves for the recovery of NAP1 and NAP4 spores from polypropylene plastic using cellulose sponges by qPCR (16S rRNA gene) and culture.
LODs (dotted lines) were determined at the 95% positivity (solid black line) cut-off (n = 5 independent experiments for each strain/assay combination, total n = 20).
Mechanical recovery from sponges
Because the 16S rRNA gene qPCR assay had a substantially lower LOD than the toxin B gene assay in solution, we chose to proceed with determining the mechanical recovery of C. difficile from contaminated surfaces by 16S rRNA gene qPCR for comparison to culture. The mechanical recoveries by qPCR (76%) and culture (67%) were not significantly different (p = 0.36), and were similar for both the NAP1 and NAP4 strains (NAP1 = 75%, NAP4 = 77%, p = 0.87 for qPCR and NAP1 = 76%, NAP4 = 58%, p = 0.08 for culture). No statistically significant interaction between method and strain was detected for mechanical recovery (p = 0.89 for NAP1 and p = 0.20 for NAP4).
Discussion
In this study, we compared the use of qPCR to culture for the detection and quantification of two epidemic strains of C. difficile, NAP1 and NAP4, from contaminated surfaces. We found that the LOD of 16S rRNA gene qPCR was 16.9-fold lower than toxin B gene qPCR, and that the LOD of C. difficile from a surface by 16S rRNA gene qPCR was 3.2-fold lower than culture.
We initially assessed the performance and sensitivity of a 16S rRNA gene and toxin B gene qPCR assay for detection of C. difficile in solution. We found that the LOD of the 16S rRNA gene qPCR was over 15 times lower, which may be partly due to the higher copy number of the 16s rRNA gene and greater stability and efficiency of the 16S rRNA gene qPCR. 16S rRNA gene qPCR, therefore, has greater potential to identify low-level contamination, with the caveat that strains may not be toxigenic and require additional testing to confirm clinical relevance. In the hospital setting, this may not pose a significant limitation since the majority of surface-isolated C. difficile has been found to carry the tcdA and tcdB genes [19,20].
Different groups have compared PCR to culture for clinical detection of C. difficile in stool samples. In antibiotic-associated diarrhea patients, quantification by 16S rRNA gene qPCR correlated well with culture, but qPCR was more sensitive and able to detect C. difficile in several culture-negative cases [21]. In a more recent analysis involving spiked stool samples, the LOD of traditional PCR analyzed by gel electrophoresis was 10-fold higher than culture [22]. Overall, we found the LOD for qPCR was 3.2-fold lower compared to culture for environmental detection of C. difficile. Both methods proved to be highly sensitive, capable of identifying less than 55 spores from plastic surfaces, but our data indicate that qPCR is more likely to detect low-level contamination. Assuming a density of 1 cell/25 cm2, this study predicts qPCR would detect C. difficile in 27% of 25 cm2 samples, or 72% of 100 cm2 samples, compared to only 7% and 34% by culture, respectively. Therefore, in practice, both switching to 16s rRNA gene qPCR and increasing the sample surface area are strategies that are likely to yield higher probability of detecting spores. Our findings are in agreement with a previous report that was unable to detect C. difficile from inert hospital surfaces by culture, but found up to 40% of samples positive by qPCR [23]. Improved environmental detection by qPCR over culture has been similarly reported for other bacterial species, including Bacillus subtilus and Erwinia herbicola [24,25].
We obtained relatively high mechanical recoveries from surfaces of approximately 70%, supporting efficient surface removal and subsequent extraction of spores from sponges reported by previous groups [7–9]. In similar validation experiments using various sampling methods, Ali et al. recovered 76–94% [7] and Claro et al. recovered 14–92% [26] of C. difficile from a variety of surface materials by culture. An interesting finding from this study was the lack of difference in the recovery of the C. difficile spores from strains with different surface properties. Here we compared NAP1 and NAP4, the two most dominant strains associated with hospital acquired infections in Canada [27]. We speculated that shorter hairs due to truncation of the bclA gene [16] may result in differences in adhesion. Our ability to recover spores of both types at similar efficiencies suggests similar adhesion between the two strains to plastic, but we cannot rule out the possibility of differences in adhesion to other surface materials or after longer periods post-deposition.
Our study had several limitations. First, we determined the LODs for the recovery of C. difficile from only a plastic surface material that is most widespread throughout hospital environments in North America. Previous studies have demonstrated that the recovery of C. difficile from inanimate surfaces varies according to material type [26], which may have changed results for the NAP1 versus NAP4 adhesion comparisons. Second, the LOD of qPCR is limited by the DNA extraction efficiency and the quantity of DNA used in the assay; we determined the LOD for qPCR using only a single DNA extraction kit. However, prior to conducting our study, we tested several DNA extraction methods and selected this technique since it returned the best yield (S2 Table). Third, we did not consider differences in the recovery or detection of spores versus vegetative cells, which are likely to exhibit differences in culturability and DNA extraction efficiency. However, spores are generally thought to represent the more important form of C. difficile from a transmission perspective, due to the rapid death of vegetative cells in the environment. Finally, our study design did not consider the effects of environmental cleaning or the duration of air exposure, both of which would be expected to reduce culturability. Our study used a short (<3 h) air exposure in order to minimize bias towards qPCR, which is capable of detecting both non-culturable but nevertheless viable cells, and non-viable cells.
In conclusion, few studies have exploited qPCR for detection of environmental C. difficile contamination. Our study demonstrates that 16S rRNA gene qPCR is substantially more sensitive than culture for the detection of environmental C. difficile contamination. The use of qPCR for detection of environmental C. difficile has the potential to offer a number of advantages over culturing including reduced cost and turnaround time for results, as well as the ability to detect both culturable and non-culturable contamination. Future studies quantifying the environmental burden of C. difficile should consider 16S rRNA gene qPCR for improved detection and enumeration.
Supporting information
(A-D) Standard curves corresponding to the 16S rRNA gene qPCR assays for NAP1 and NAP4 C. difficile strains. DNA was extracted from known concentrations of C. difficile cells and serially diluted (10−1 to 10−8) to generate standards. Ct values produced by qPCR reactions for each standard were plotted against log10(cell quantity) and a linear curve was fit to the data (E = PCR efficiency).
(DOCX)
(DOCX)
NAP1 and NAP4 C. difficile spore solutions of roughly 1 x 107 CFU/mL were tested.
(DOCX)
(XLSX)
Acknowledgments
The authors wish to thank Kwaku Adomako and Arezou Saedi for thoughtful discussions on study design.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by a grant from the Canadian Institutes of Health Research (GG, KB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primer. 2016;2: 16020 10.1038/nrdp.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother. 2014;69: 881–891. 10.1093/jac/dkt477 [DOI] [PubMed] [Google Scholar]
- 3.Vardakas KZ, Trigkidis KK, Boukouvala E, Falagas ME. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: a systematic review and meta-analysis. Int J Antimicrob Agents. 2016;48: 1–10. 10.1016/j.ijantimicag.2016.03.008 [DOI] [PubMed] [Google Scholar]
- 4.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6 10.1186/1471-2334-6-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sethi AK, Al-Nassir WN, Nerandzic MM, Bobulsky GS, Donskey CJ. Persistence of Skin Contamination and environmental shedding of Clostridium difficile during and after treatment of C. difficile infection. Infect Control Hosp Epidemiol. 2010;31: 21–27. 10.1086/649016 [DOI] [PubMed] [Google Scholar]
- 6.Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38: S25–33. 10.1016/j.ajic.2010.04.196 [DOI] [PubMed] [Google Scholar]
- 7.Ali S, Muzslay M, Wilson P. A Novel Quantitative Sampling Technique for Detection and Monitoring of Clostridium difficile Contamination in the Clinical Environment. Onderdonk AB, editor. J Clin Microbiol. 2015;53: 2570–2574. 10.1128/JCM.00376-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhardt NEP, Foster NF, Hong S, Riley TV, McGechie DB. Comparison of two environmental sampling tools for the detection of Clostridium difficile spores on hard bathroom surfaces in the hospital setting. J Hosp Infect. 2017;96: 295–296. 10.1016/j.jhin.2017.03.028 [DOI] [PubMed] [Google Scholar]
- 9.Otter JA, Havill NL, Adams NM, Cooper T, Tauman A, Boyce JM. Environmental sampling for Clostridium difficile: swabs or sponges? Am J Infect Control. 2009;37: 517–8. 10.1016/j.ajic.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 10.Barbut F, Menuet D, Verachten M, Girou E. Comparison of the efficacy of a hydrogen peroxide dry-mist disinfection system and sodium hypochlorite solution for eradication of Clostridium difficile spores. Infect Control Hosp Epidemiol. 2009;30: 507–514. 10.1086/597232 [DOI] [PubMed] [Google Scholar]
- 11.Best EL, Parnell P, Thirkell G, Verity P, Copland M, Else P, et al. Effectiveness of deep cleaning followed by hydrogen peroxide decontamination during high Clostridium difficile infection incidence. J Hosp Infect. 2014;87: 25–33. 10.1016/j.jhin.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 12.Mutters R, Nonnenmacher C, Susin C, Albrecht U, Kropatsch R, Schumacher S. Quantitative detection of Clostridium difficile in hospital environmental samples by real-time polymerase chain reaction. J Hosp Infect. 2009;71: 43–48. 10.1016/j.jhin.2008.10.021 [DOI] [PubMed] [Google Scholar]
- 13.Joshi LT, Phillips DS, Williams CF, Alyousef A, Baillie L. Contribution of Spores to the Ability of Clostridium difficile To Adhere to Surfaces. Appl Environ Microbiol. 2012;78: 7671–7679. 10.1128/AEM.01862-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paredes-Sabja D, Sarker MR. Adherence of Clostridium difficile spores to Caco-2 cells in culture. J Med Microbiol. 2012;61: 1208–1218. 10.1099/jmm.0.043687-0 [DOI] [PubMed] [Google Scholar]
- 15.Pizarro-Guajardo M, Calderón-Romero P, Paredes-Sabja D. Ultrastructure Variability of the Exosporium Layer of Clostridium difficile Spores from Sporulating Cultures and Biofilms. Schaffner DW, editor. Appl Environ Microbiol. 2016;82: 5892–5898. 10.1128/AEM.01463-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phetcharaburanin J, Hong HA, Colenutt C, Bianconi I, Sempere L, Permpoonpattana P, et al. The spore-associated protein BclA1 affects the susceptibility of animals to colonization and infection by Clostridium difficile: C. difficile BclA proteins. Mol Microbiol. 2014;92: 1025–1038. 10.1111/mmi.12611 [DOI] [PubMed] [Google Scholar]
- 17.Kralik P, Ricchi M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front Microbiol. 2017;8 10.3389/fmicb.2017.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson G, Nolan T, Bustin SA. Real-Time Quantitative PCR, Pathogen Detection and MIQE In: Wilks M, editor. PCR Detection of Microbial Pathogens. Totowa, NJ: Humana Press; 2013. pp. 1–16. 10.1007/978-1-60327-353-4_1 [DOI] [PubMed] [Google Scholar]
- 19.Faires MC, Pearl DL, Berke O, Reid-Smith RJ, Weese JS. The identification and epidemiology of meticillin-resistant Staphylococcus aureus and Clostridium difficile in patient rooms and the ward environment. BMC Infect Dis. 2013;13 (1). 10.1186/1471-2334-13-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldenberg SD, Patel A, Tucker D, French GL. Lack of enhanced effect of a chlorine dioxide-based cleaning regimen on environmental contamination with Clostridium difficile spores. J Hosp Infect. 2012;82: 64–67. 10.1016/j.jhin.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 21.Naaber P, Stsepetova J, Smidt I, Ratsep M, Koljalg S, Loivukene K, et al. Quantification of Clostridium difficile in antibiotic-associated-diarrhea patients. J Clin Microbiol. 2011;49: 3656–8. 10.1128/JCM.05115-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganji L, Azimirad M, Farzi N, Alebouyeh M, Shirazi MH, Eshraghi SS, et al. Comparison of the Detection Limits of the Culture and PCR Methods for the Detection of Clostridium difficile, Clostridium perfringens, Campylobacter jejuni, and Yersinia enterocolitica in Human Stool. Arch Pediatr Infect Dis. 2016;5 10.5812/pedinfect.38888 [DOI] [Google Scholar]
- 23.Morales L, Rodríguez C, Gamboa-Coronado M del M. Molecular detection of Clostridium difficile on inert surfaces from a Costa Rican hospital during and after an outbreak. Am J Infect Control. 2016;44: 1517–1519. 10.1016/j.ajic.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 24.Buttner MP, Cruz P, Stetzenbach LD, Cronin T. Evaluation of Two Surface Sampling Methods for Detection of Erwinia herbicola on a Variety of Materials by Culture and Quantitative PCR. Appl Environ Microbiol. 2007;73: 3505–3510. 10.1128/AEM.01825-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buttner MP, Cruz-Perez P, Stetzenbach LD. Enhanced Detection of Surface-Associated Bacteria in Indoor Environments by Quantitative PCR. Appl Environ Microbiol. 2001;67: 2564–2570. 10.1128/AEM.67.6.2564-2570.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claro T, Daniels S, Humphreys H. Detecting Clostridium difficile spores from inanimate surfaces of the hospital environment: Which method is best? J Clin Microbiol. 2014;52: 3426–3428. 10.1128/JCM.01011-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Public Health Agency of Canada. Healthcare-associated Clostridium difficile infections in Canadian acute-care hospitals: Surveillance Report January 1, 2007 to December 31, 2012. Centre for Communicable Disease and Infection Control, Public Health Agency of Canada; 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-D) Standard curves corresponding to the 16S rRNA gene qPCR assays for NAP1 and NAP4 C. difficile strains. DNA was extracted from known concentrations of C. difficile cells and serially diluted (10−1 to 10−8) to generate standards. Ct values produced by qPCR reactions for each standard were plotted against log10(cell quantity) and a linear curve was fit to the data (E = PCR efficiency).
(DOCX)
(DOCX)
NAP1 and NAP4 C. difficile spore solutions of roughly 1 x 107 CFU/mL were tested.
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.