Abstract
Centromeric localization of the evolutionarily conserved centromere-specific histone H3 variant CENP-A (Cse4 in yeast) is essential for faithful chromosome segregation. Overexpression and mislocalization of CENP-A lead to chromosome segregation defects in yeast, flies, and human cells. Overexpression of CENP-A has been observed in human cancers; however, the molecular mechanisms preventing CENP-A mislocalization are not fully understood. Here, we used a genome-wide synthetic genetic array (SGA) to identify gene deletions that exhibit synthetic dosage lethality (SDL) when Cse4 is overexpressed. Deletion for genes encoding the replication-independent histone chaperone HIR complex (HIR1, HIR2, HIR3, HPC2) and a Cse4-specific E3 ubiquitin ligase, PSH1, showed highest SDL. We defined a role for Hir2 in proteolysis of Cse4 that prevents mislocalization of Cse4 to noncentromeric regions for genome stability. Hir2 interacts with Cse4 in vivo, and hir2∆ strains exhibit defects in Cse4 proteolysis and stabilization of chromatin-bound Cse4. Mislocalization of Cse4 to noncentromeric regions with a preferential enrichment at promoter regions was observed in hir2∆ strains. We determined that Hir2 facilitates the interaction of Cse4 with Psh1, and that defects in Psh1-mediated proteolysis contribute to increased Cse4 stability and mislocalization of Cse4 in the hir2∆ strain. In summary, our genome-wide screen provides insights into pathways that regulate proteolysis of Cse4 and defines a novel role for the HIR complex in preventing mislocalization of Cse4 by facilitating proteolysis of Cse4, thereby promoting genome stability.
Keywords: centromere, kinetochore, gene regulation, chromosome segregation, Cse4, CENP-A, histones, histone chaperone
KINETOCHORES (centromeric DNA and associated proteins) serve as an attachment site for microtubules to promote faithful chromosome segregation during mitosis (Allshire and Karpen 2008; Verdaasdonk and Bloom 2011; Burrack and Berman 2012; Choy et al. 2012; Maddox et al. 2012; McKinley and Cheeseman 2016). The “point centromeres” of budding yeast contain ∼125 bp of unique DNA sequences as opposed to “regional centromeres” in other eukaryotes, which are comprised of up to several mega-base pairs of repeated DNA sequences, species-specific satellite DNA arrays, or retrotransposon-derived sequences (Clarke and Carbon 1980; Allshire and Karpen 2008; Verdaasdonk and Bloom 2011; Burrack and Berman 2012; Maddox et al. 2012; McKinley and Cheeseman 2016). Despite the divergence of centromeric DNA sequences, the centromere-specific histone H3 variant (Cse4 in Saccharomyces cerevisiae, Cnp1 in Schizosaccharomyces pombe, CID in Drosophila, and CENP-A in mammals) is evolutionarily conserved from yeast to humans (Przewloka and Glover 2009; Choy et al. 2012; Henikoff 2012). Cse4 and its homologs are essential for high-fidelity chromosome segregation (Allshire and Karpen 2008; Verdaasdonk and Bloom 2011; Burrack and Berman 2012; Maddox et al. 2012; McKinley and Cheeseman 2016), and Cse4 can functionally replace CENP-A in mammalian cells (Wieland et al. 2004).
Overexpression and mislocalization of CENP-A have been observed in many cancers (Tomonaga et al. 2003; Amato et al. 2009; Hu et al. 2010; Y. Li et al. 2011; Wu et al. 2012; Lacoste et al. 2014; Athwal et al. 2015); however, the molecular mechanisms associated with this observation are not fully understood. We have recently shown that overexpression of CENP-A leads to its mislocalization to noncentromeric regions and contributes to chromosome instability (CIN) in human cells (Shrestha et al. 2017). Mislocalization of CID causes the formation of ectopic kinetochores and leads to mitotic delay, anaphase bridges, chromosome fragmentation, aneuploidy, and lethality in flies (Heun et al. 2006). Overexpression of Cnp1 leads to noncentromeric mislocalization, growth and chromosome segregation defects during mitosis and meiosis in fission yeast (Choi et al. 2012; Castillo et al. 2013; Gonzalez et al. 2014). We have previously shown that mislocalization of overexpressed cse416KR, in which all lysines (K) are mutated to arginine (R) (Collins et al. 2004), results in chromosome segregation defects in budding yeast (Au et al. 2008). The extent of mislocalization of cse416KR and CENP-A correlate with the level of chromosome loss in yeast and human cells, respectively (Au et al. 2008; Shrestha et al. 2017).
Post-translational modifications (PTMs), such as ubiquitination, regulate the cellular levels of Cse4 and its homologs, and prevent its mislocalization to euchromatic regions (Deyter et al. 2017). In flies, proteolysis of CID prevents mislocalization to noncentromeric regions (Heun et al. 2006; Moreno-Moreno et al. 2011). Similar results are observed in fission yeast, where ubiquitin-mediated proteolysis of Cnp1 prevents mislocalization to noncentromeric regions (Gonzalez et al. 2014). Additionally, Psh1 (an E3 ubiquitin ligase) (Hewawasam et al. 2010; Ranjitkar et al. 2010; Herrero and Thorpe 2016; Hildebrand and Biggins 2016), Doa1 (WD-repeat protein) (Au et al. 2013), Fpr3 (proline isomerase) (Ohkuni et al. 2014), Ubp8 (ubiquitin protease) (Canzonetta et al. 2015), Rcy1 (F-box protein) (Cheng et al. 2016), and Ubr1 (Cheng et al. 2017) regulate cellular levels of overexpressed Cse4 and prevent its mislocalization to noncentromeric regions (Collins et al. 2005; Cheng et al. 2016). The role of Psh1 in proteolysis of Cse4 has been characterized in detail, and these studies have shown that the interactions of Psh1 with Spt16, a component of the FACT (facilitates chromatin transcription/transactions) complex, and casein kinase 2 (CKA2) regulate Cse4 proteolysis (Hewawasam et al. 2010, 2014; Ranjitkar et al. 2010; Au et al. 2013; Deyter and Biggins 2014). Recent studies have shown that chromatin assembly factor-1 (CAF-1) promotes the deposition of Cse4 at noncentromeric regions in psh1∆ mutants, and deletion of Cac2, a component of CAF-1, rescues the lethality of psh1∆ or cka2∆ strains overexpressing Cse4 (Hewawasam et al. 2018). While the centromere-targeting domain (CATD) in the C-terminus of Cse4 interacts with Psh1, the N-terminus of Cse4 is also required for Cse4 proteolysis (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013). In addition to ubiquitination, sumolylation of Cse4 also regulates its proteolysis. We have shown that Cse4 is sumoylated by the small ubiquitin-related modifier (SUMO) E3 ligases Siz1 and Siz2, and the SUMO-targeted ubiquitin ligase (STUbL) Slx5 plays a critical role in ubiquitin-mediated proteolysis of endogenously expressed Cse4 and prevents its mislocalization independently of Psh1 (Ohkuni et al. 2016). Notably, overexpressed Cse4 is not completely stabilized in psh1∆, doa1∆, fpr3∆, rcy1∆, and slx5∆ strains (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013; Ohkuni et al. 2014, 2016; Cheng et al. 2016; Hildebrand and Biggins 2016) suggesting the existence of additional genes/pathways to regulate Cse4 proteolysis.
Identification of pathways that regulate cellular levels of Cse4 is critical for understanding mechanisms that prevent mislocalization of CENP-A and aneuploidy in human cancers. Hence, we performed a genome-wide screen using a synthetic genetic array (SGA) to identify nonessential genes that show synthetic dosage lethality (SDL) upon Cse4 overexpression. We hypothesized that overexpression of Cse4 would cause SDL in mutants that are defective in Cse4 proteolysis, similar to that observed previously for psh1∆ strains (Ranjitkar et al. 2010; Au et al. 2013). We identified deletions of all four components of the replication-independent histone chaperone (HIR) complex (HIR1, HIR2, HIR3, HPC2) in the screen, and growth assays confirmed the SDL of overexpressed Cse4 in deletion strains for each component of the HIR complex. We investigated the molecular role of Hir2 in Cse4 proteolysis and how this affects the localization of Cse4. Hir2 interacts with Cse4 in vivo, and deletion of HIR2 leads to defects in Cse4 proteolysis and mislocalization of Cse4 to noncentromeric regions. We determined that defects in Psh1-mediated proteolysis of Cse4 contribute to the increased stability of Cse4 in the hir2∆ strain. In summary, the genome-wide screen provides insights into pathways that prevent mislocalization of Cse4 and defines a role for the HIR complex in preventing mislocalization of Cse4 by facilitating proteolysis of Cse4.
Materials and Methods
Yeast strains, plasmids, and SGA analysis
The yeast strains and plasmids used in this study are described in Supplemental Material, Tables S1 and S2. All yeast nonessential gene deletion strains are isogenic to BY4741 unless otherwise indicated. An SGA query strain (YMB6969) overexpressing CSE4 in galactose-containing medium was created in Y7092 by homologous recombination using a method described previously (Longtine et al. 1998; Baryshnikova et al. 2010a). Briefly, PCR products containing HA-tagged CSE4 driven by the GAL1 and, separately, MX4-NATR were amplified using DNA from pMB1458 and p4339, respectively. Each PCR product carries a sequence common to the other along with sequences homologous to regions either 5′ or 3′ of the CSE4 genomic locus. The two purified PCR products were cotransformed into Y7092 (Baryshnikova et al. 2010a). Integration of GALCSE4-HA into the endogenous locus was verified by PCR, DNA sequencing, Western blot analysis, and the inability of cells to grow on glucose-containing medium. Yeast strain YMB6969 was used to interrogate synthetic fitness defects of Cse4 overexpression with deletions of nonessential genes. SGA screens were performed on galactose-containing medium following the procedures and scoring described previously (Tong et al. 2004; Costanzo et al. 2010; Z. Li et al. 2011). cac1∆ hir2∆ and cac2∆ hir2∆ strains were derived from meiotic products of diploids of crosses between cac1∆::KAN and hir2∆::NAT (YMB8785), and cac2∆::KAN and hir2∆::NAT, respectively, in the BY4741 background. Strain YMB8886 (∆16 H2A-H2B ∆16 H3-H4) was created by integrating pAB157 and pAB95 sequentially into a wild-type (WT) strain carrying HA-tagged Cse4 expressed from the GAL1 promoter (YMB6714).
Protein stability assays
Protein stability assays were performed as described previously with minor modifications (Au et al. 2008). Briefly, yeast cultures grown in media containing 2% raffinose were supplemented with galactose to a final concentration of 2% to induce the expression of proteins regulated by the GAL1 promoter at 30° for 2 hr or as described. Subsequently, 2% glucose was added to inhibit transcription and cycloheximide (CHX, 10 μg/ml) to block protein translation. An equal number of cells as measured by OD600 were collected at different time points, and whole cell extracts were prepared by the TCA method described previously (Kastenmayer et al. 2006). Protein concentrations were determined using a Bio-Rad DC Protein Assay (Cat# 500-0113; Bio-Rad), and equal amounts of protein were separated on a 4–12% Bis-Tris gel (Invitrogen) for western blot analysis. Primary antibodies were anti-HA (clone 12CA5; Roche) and anti-Tub2 (custom made by the Basrai laboratory). Secondary antibodies from Amersham Biosciences were HRP-conjugated sheep α-mouse IgG (NA931V) and HRP-conjugated donkey α-rabbit IgG (NA934V). Western blots were quantified using the SynGene program (SynGene, Cambridge, UK) or ImageJ (Schneider et al. 2012) software. Protein half-life was calculated as previously described (Au et al. 2013) where least squares regression of percentage remaining (log scale) vs. time was used. Values for half-life were derived from three biological replicates unless otherwise noted and were reported as the average ± SE.
Chromosome spreads for localization of Cse4 and Mtw1-GFP
Chromosome spreads were performed as described previously (Collins et al. 2004). Strains were grown in SC-Ura containing 2% raffinose until mid-log phase prior to the induction of Cse4 expression by 2% galactose for 1 hr. Anti-HA (1:2500 dilutions) was used as primary antibody (MMS-101P; Covance, Babco) and Cy3 conjugated Goat α-mouse (1:5000 dilutions) as secondary antibody (115165003; Jackson ImmunoResearch Laboratories). Nuclear mass was visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining (1 μg/ml in phosphate-buffered saline (PBS)). For Mtw1-GFP visualization, cells were grown as described above, treated or untreated with 0.5% formaldehyde on ice, stained with DAPI (1 μg/ml in PBS), and examined by fluorescent microscopy. Cells were observed under an Axioskop 2 (Zeiss) fluorescence microscope equipped with a Plan-APOCHROMAT 100X or 63X (Zeiss) oil immersion lens. Image acquisition and processing were performed with the IP Lab version 3.9.9 r3 software (Scanalytics).
ChIP and ChIP-seq experiments
Chromatin immunoprecipitation (ChIP) was performed as described previously (Mishra et al. 2007, 2011, 2018) with minor modifications. WT and hir2Δ strains carrying GALCSE4-HA (pMB1458) were grown logarithmically in 500 ml of 1× SC-URA with galactose and raffinose (2% each) for 6 hr at 30°. Cells were cross-linked in 1% formaldehyde at room temperature for 15 min, quenched in 125 mM glycine for 5 min, and collected by centrifugation. Cells were washed in TBS (20 mM Tris-HCl pH 7.6, 150 mM NaCl), resuspended in spheroplasting buffer (1.2 M sorbitol, 20 mM Na-Hepes, pH 7.4) with Zymolase 100T, and incubated at 30°. Spheroplasts were washed in postspheroplasting buffer (1.2 M sorbitol, 1 mM MgCl2, 20 mM Na-Pipes, pH 6.8), followed by three washes with FA buffer (50 mM Na-Hepes pH 7.6, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 0.1% Na-deoxycholate) containing protease inhibitors (P8215; Sigma). Spheroplasts were then suspended in FA buffer with protease inhibitors and sonicated on ice for eight 12-sec bursts applied at an interval of 2 min with an output cycle setting fixed at 30% to obtain an average DNA fragment size of 300–400 bp. One-tenth of the resulting soluble, sheared chromatin fraction was collected as input, and the remaining was incubated with α-HA conjugated agarose beads (A2220; Sigma) at 4° for ∼16 hr. Beads were collected by centrifugation and washed at room temperature with FA buffer for 5 min (three times), FA-HS buffer (50 mM Na-Hepes pH 7.6, 1 mM EDTA, 1% Triton X-100, 500 mM NaCl, 0.1% Na-deoxycholate) for 5 min (twice) and RIPA buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 1 mM EDTA, 0.5% Na-deoxycholate) for 5 min (twice) followed by 1× TE pH 8.0 for 5 min (twice). Immunoprecipitated DNA was eluted in elution buffer (25 mM Tris-HCl pH 7.6, 10 mM EDTA, 0.5% SDS). Input and immunoprecipitated samples were recovered after cross-link reversal at 65° for ∼16 hr, RNase A and proteinase K treatments, and final purification using Qiagen DNA purification columns (Qiagen). ChIP-qPCR was performed with the 7500 Fast Real Time PCR System using Fast SYBR Green Master Mix (Applied Biosystems) and primers listed in Table S3 with the following conditions: 95° for 20 sec followed by 40 cycles of 95° for 3 sec, 60° for 30 sec. The enrichment values are shown as % input, determined using the ddCT method (Livak and Schmittgen 2001).
ChIP DNA was used to construct sequencing libraries as described previously (Grøntved et al. 2015). Downstream analysis was performed as follows: unfiltered sequencing reads were aligned to the S. cerevisiae reference genome (SacCer3) using bwa (Seoighe and Wolfe 1999), allowing up to one mismatch for each aligned read. Reads mapping to multiple sites were retained to allow evaluation of associations with nonunique sequences (Seoighe and Wolfe 1999) and duplicate reads were retained. Binding sites were identified using SICER (Zang et al. 2009) with the following parameters: effective genome size 0.97 (97% of the yeast genome is mappable), window size 50 bp, and gap size 50 bp. Calculation of coverage, comparisons between different data sets and identification of overlapping binding regions were preceded by library size normalization and were performed with the “chipseq” and “Genomic Ranges” packages in Bioconductor (Gentleman et al. 2004). Control subtraction was carried out in the following way: coverage (exp)/N1−coverage (control)/N2, in which “exp” is the data set (in .bam format) to be examined, N1 is the library size of the experimental data (“exp”), and N2 is the library size of the control. In this study, input sequences (DNA sequences after sonication prior to immunoprecipitation) were used as the control. Occupancy profiles showing reads per million (RPM) normalized to total library size were generated using the Integrative Genomics Viewer (Robinson et al. 2011).
Immunoprecipitation experiments
Immunoprecipitation experiments were performed as described previously (Mishra et al. 2016, 2018). Briefly, cell pellets collected from yeast strains (40–50 OD600 of cells) grown using media and growth conditions described above were dissolved in 500 μl lysis buffer (50 mM Tris pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM DTT, 0.4% NP-40, 1 mM PMSF, and protease inhibitor cocktails Sigma P8215) and bead-beaten (four times) with lysing matrix C glass beads (MP Biomedicals) using the manufacturer’s recommended program in a FastPrep-24 5G homogenizer (MP Biomedicals). After centrifugation, protein concentration of the resulting whole cell extracts was measured, and equal amounts of protein from each sample was incubated with rabbit IgG agarose (A2909; Sigma) overnight, followed by washing in TBST (20 mM Tris-HCl, 0.8% NaCl, 0.1% Tween-20) and elution in 2× Laemmli buffer (Invitrogen). Immunoprecipitates were assayed by Western blot analysis using α-TAP (CAB1001; Thermo Scientific) and α-HA (11583816001; Sigma) antibodies.
Subcellular fractionation
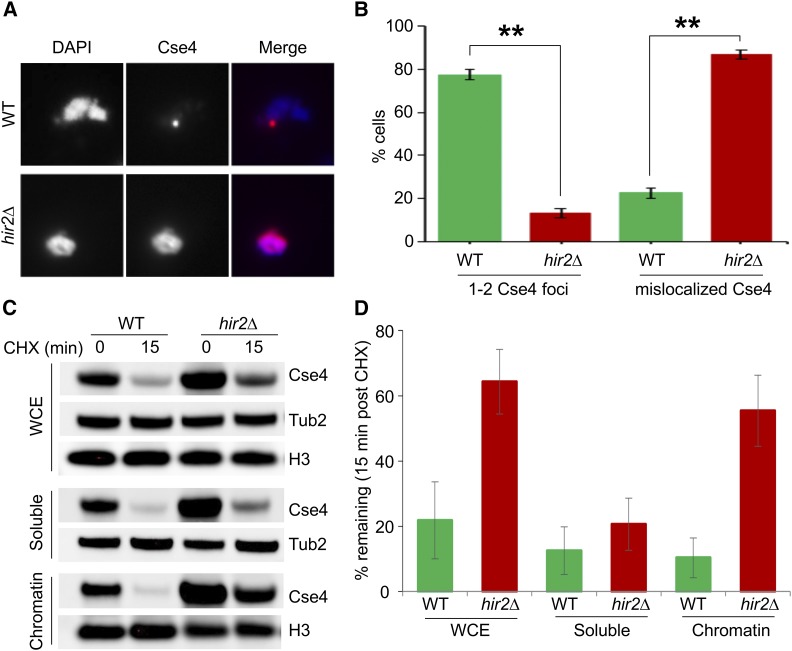
Whole cell extract (WCE), soluble and chromatin fractions of WT and hir2∆ strains were prepared as described previously (Au et al. 2008). Cells grown logarithmically in raffinose-containing medium were induced with 2% galactose for 30 min, followed by addition of cycloheximide at 10 μg/ml for 15 min. Protein samples were normalized based on equal OD600 of cells, separated by gel electrophoresis, transferred to nitrocellulose membrane and probed using α-HA antibody. Tub2 and histone H3 (ab1791; Abcam) were used as loading controls for soluble and chromatin fractions, respectively.
Loss of centromere-containing plasmid
Yeast strains containing plasmid pRS416 (CEN URA3) were grown at 30° in SC-URA glucose medium for ∼16 hr and an equal OD600 of cells from each strain were plated on SC-URA glucose (2%) and YPD plates (G0). To measure the frequency of plasmid loss, 0.05 OD600 of cells from each strain were inoculated into 50 ml of YPD and grown nonselectively at 30° for ∼10 generations (G10). Equal OD600 of cells from G10 cultures were then plated as for G0. The frequency of plasmid segregation was determined as a ratio of total number of colonies on selective SC-URA over nonselective YPD plates, where the values from G0 for each strain were normalized to 100%. Three biological replicates were performed for each strain and at least 1200 cells at G0 and G10 were plated.
Data availability
Deep sequencing data generated for this study has been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number SRP153412. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6709553.
Results
A genome-wide SGA screen for gene deletions that show SDL with GALCSE4
Deletions of PSH1, SLX5, and RCY1, which are defective in proteolysis of Cse4, exhibit SDL with overexpressed Cse4 (GALCSE4) (Ranjitkar et al. 2010; Au et al. 2013; Cheng et al. 2016; Ohkuni et al. 2016). We performed a genetic screen using a SGA to identify gene deletions that display SDL with GALCSE4. A query strain with GALCSE4 integrated in the genome was mated to an array of 4293 individual nonessential gene deletion strains (3620 of which passed quality control filters), and growth of haploid meiotic progeny of each gene deletion with GALCSE4 was scored on galactose-containing medium. The SGA score for growth was determined as previously described (Baryshnikova et al. 2010b) and filtered using the intermediate confidence threshold (P-value <0.05 and |Score| >0.08) (Costanzo et al. 2010, 2016). We identified 301 gene deletions that showed a significant growth defect, or SDL, with GALCSE4, and refer to them as significant negative genetic interactors (Table S4).
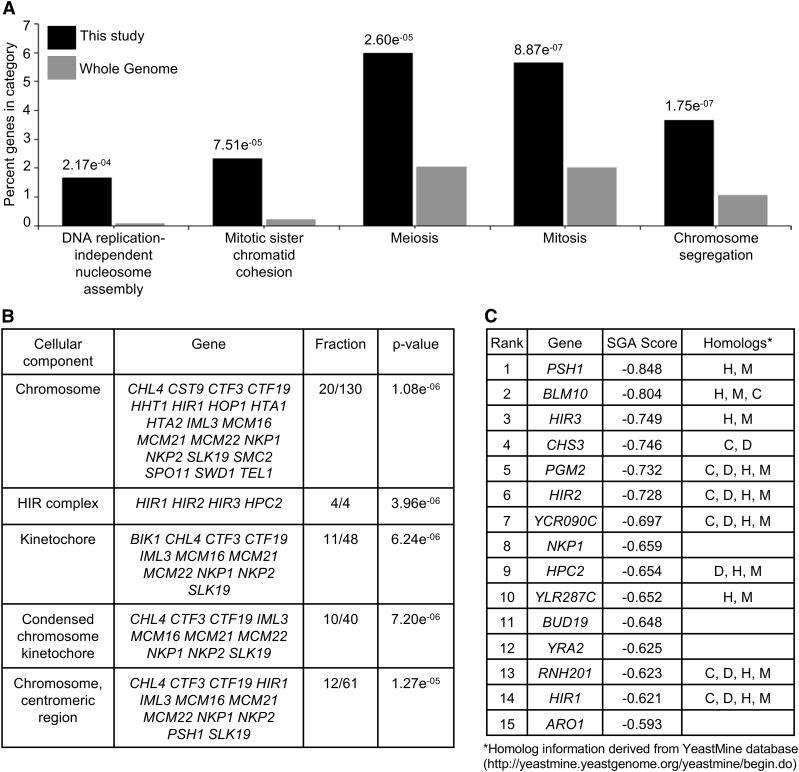
Gene ontology (GO) analysis for biological processes or cellular components for the 301 significant negative genetic interactors was performed using the FunSpec bioinformatics tool (http://funspec.med.utoronto.ca/, April 2017). The analysis was performed using a P-value cutoff score of 0.01, and results with a P-value <2.0 × 10−04 are displayed. The GO analysis for biological processes showed an enrichment of genes required for DNA replication-independent nucleosome assembly, mitotic sister chromatid cohesion, meiosis, mitosis, and chromosome segregation (Figure 1A). These results are consistent with an enrichment of cellular component GO annotations corresponding to the chromosome, HIR complex, kinetochore, and centromeric region (Figure 1B). Consistent with previous results, rcy1∆ (Cheng et al. 2016), but not cac2∆ were identified as negative genetic interactors with GALCSE4. The top 15 negative genetic interactors are listed in Figure 1C, the majority of which are evolutionarily conserved with homologs in human (H), mouse (M), Drosophila melanogaster (D) and/or Caenorhabditis elegans (C). The identification of psh1∆ as the most significant negative genetic interactor when Cse4 is overexpressed serves as proof of principle for the screen (Ranjitkar et al. 2010; Au et al. 2013). Among the top 15 negative genetic interactors are all four genes that encode the DNA replication-independent histone chaperone (HIR) complex, namely HIR1, HIR2, HIR3, and HPC2.
Figure 1.
Gene ontology (GO) analysis of negative genetic interactors identified in the SGA screen with GALCSE4. (A and B) GO analysis for biological processes (A) or cellular components (B) for the 301 significant negative genetic interactors using the FunSpec bioinformatics tool (http://funspec.med.utoronto.ca/, April 2017). Analysis was performed using a P-value cutoff score of 0.01 and results with a P-value <2.0e−04 are displayed. The P-value represents the likelihood of candidate genes from the screen intersecting with the specified category. The percentage denotes the number of genes found in a given category over the number of input genes. The fraction denotes the number of candidate genes over the number of genes annotated within the category. The GO annotation representing genes associated with a particular cellular component or biological process are described. (C) List of the top 15 negative genetic interactors identified in the SGA screen with GALCSE4. Listed are the gene name, SGA score and homologs denoted as H, human; M, mouse; D, Drosophila melanogaster; C, Caenorhabditis elegans. The SGA score is the epsilon value calculated as previously described (Costanzo et al. 2010, 2016).
The genetic interaction profile of GALCSE4 correlates with that of kinetochore mutants
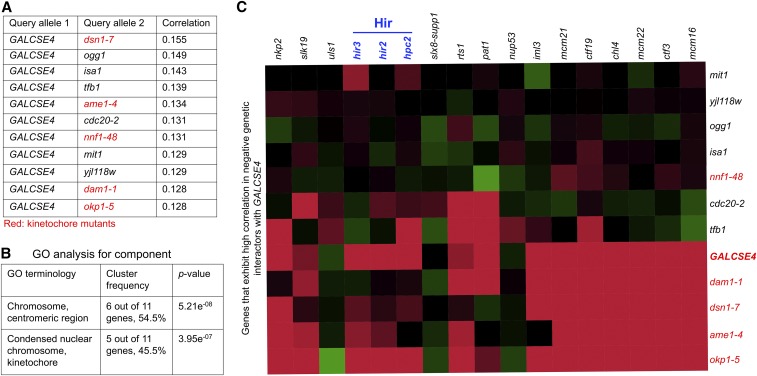
To identify cellular functions impacted by overexpressed Cse4, we performed a genetic correlation analysis looking for similarity between the profile of GALCSE4 and the complete set of query mutant profiles from the SGA dataset (Costanzo et al. 2016). A positive correlation between two mutants’ genetic interaction profiles suggests functional similarity between the effects of the genetic perturbations. A list of the top 11 query genes ranked according to their Pearson correlation similarity to the profile of GALCSE4 is shown in Figure 2A. GO analysis of these 11 genes revealed that there is an enrichment for components of the chromosome, centromere region, and kinetochore (DSN1, AME1, NNF1, DAM1, and OKP1) (Figure 2B). As expected, the kinetochore mutants that show genetic interaction profiles similar to GALCSE4 (dam1-1, dsn1-7, ame1-4, and okp1-5) also exhibit negative genetic interactions with other kinetochore mutants, namely iml3, mcm21, ctf19, chl4, mcm22, ctf3, and mcm16 (Figure 2C, red area). Furthermore, okp1-5, ame1-4, and dsn1-7 exhibit negative genetic interactions with hir2∆, hir3∆, and hpc2∆ strains. Consistent with the role of Cse4 in kinetochore structure and function, these genetic analyses suggest that overexpression of Cse4 contributes to defects in kinetochore function.
Figure 2.
The genetic interaction profile of GALCSE4 is similar to that of kinetochore mutants. (A) GALCSE4 exhibits a genetic interaction profile similar to profiles displayed by kinetochore mutants. Shown is a list of 11 mutants showing the most similar genetic interaction profiles (highest Pearson correlation score) to that of GALCSE4. (B) GO analysis for cellular component of genes in (A). (C) Representative heat map showing genetic interactions of the 11 genes listed in (A). Negative and positive genetic interactions are shown in red and green, respectively. The intensity of the color reflects the strength of the genetic interaction. Kinetochore mutants that exhibit high positive correlation with GALCSE4 from (A) above also exhibit genetic interactions with deletions of genes encoding for the HIR complex, namely hir2∆, hir3∆, and hpc2∆.
HIR complex mutants exhibit SDL with GALCSE4
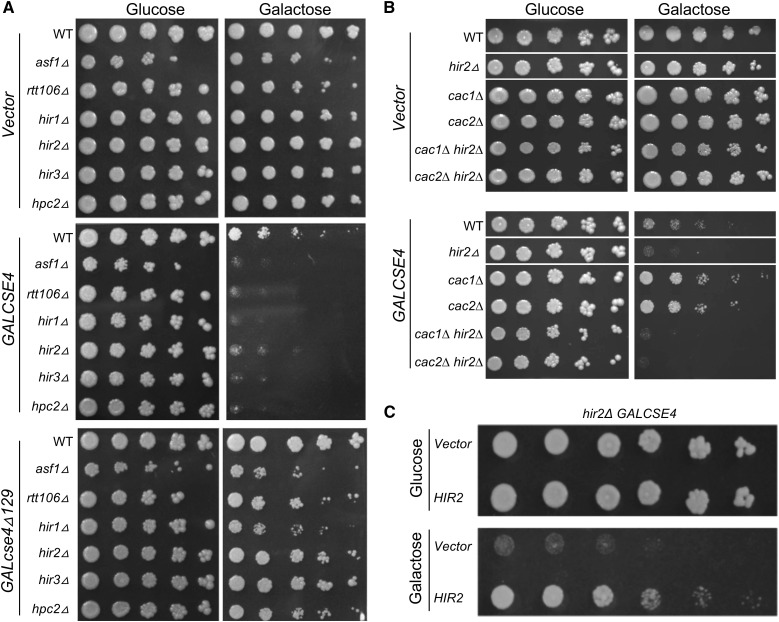
The HIR proteins function in a complex with Asf1 and Rtt106 for transcriptional repression of histone genes, and the HIR complex serves as the replication-independent chaperone for histones H3 and H4 (Green et al. 2005; Fillingham et al. 2009; Ferreira et al. 2011; Silva et al. 2012; Amin et al. 2013). In addition to hir1∆, hir2∆, hir3∆, and hpc2∆, we identified asf1∆ as a negative genetic interactor with GALCSE4 (Table S4, SGA score of −0.329 and P-value of 1.51e−11). Notably, rtt106∆ was not present on the array of yeast deletion strains. To validate the results of the SGA screen, WT, hir1∆, hir2∆, hir3∆, hpc2∆, asf1∆, and rtt106∆ strains were transformed with GALCSE4 or empty vector and examined for growth on glucose or galactose plates. Consistent with the results of the screen, hir1∆, hir2∆, hir3∆, hpc2∆, asf1∆, and rtt106∆ strains exhibit SDL with GALCSE4 on galactose plates (Figure 3A). No growth defects were observed for strains transformed with vector alone on galactose plates. Our previous studies showed that the N-terminus of Cse4 is required for its Ub-mediated proteolysis, so we asked if the N-terminus of Cse4 is required for the SDL of GALCSE4. Growth assays showed that hir1∆, hir2∆, hir3∆, hpc2∆, asf1∆, and rtt106∆ strains transformed with GALcse4∆129 (deletion of the N-terminal 129 amino acids) show slightly reduced growth when compared to vector alone but do not exhibit a SDL phenotype (Figure 3A).
Figure 3.
HIR complex mutants exhibit SDL with GALCSE4. (A) Overexpression of Cse4 causes lethality in mutants of the HIR complex mediated by the N-terminus of Cse4. Wild type (WT) (BY4741) and the isogenic deletion strains as indicated were transformed with vector (pMB433), GALCSE4HA (pMB1458), and cse4∆129 (GALcse4∆129HA; pMB1459). Serial dilutions (fivefold) of each strain with the indicated plasmid were plated on SC-Ura plates containing either glucose (2%) or galactose and raffinose (2% each). Plates were photographed after 2–5 days of growth at 30°. At least three independent transformants were examined for each strain. (B) Deletion of CAC1 or CAC2 does not suppress the lethality of hir2∆ overexpressing Cse4. cac1∆ hir2∆ (YMB10463) and cac2∆ hir2∆ (YMB10464) strains were transformed with pMB1458 or empty vector. The growth of the these transformants were determined on galactose-containing medium in fivefold serial dilution and grown as indicated in (A). Corresponding WT, hir2∆, cac1∆, and cac2∆ strains were used as controls. (C) Complementation of GALCSE4 induces lethality in a hir2∆ strain by plasmid-borne HIR2. A hir2∆ strain carrying GALCSE4HA (pMB1458) was transformed with a plasmid containing either HIR2 (MoBy 2µ library; GE Dharmacon) or vector control. Serial dilutions (fivefold) of each strain with the indicated plasmid were plated on selective plates containing either glucose (2%) or galactose and raffinose (2% each). Plates were photographed after 2–5 days of growth at 30°.
Previous studies have shown that cac1∆ hir1∆ strains, but not cac1∆ nor hir1∆ single mutants, show mislocalization of endogenous Cse4 and chromosome loss (Lopes da Rosa et al. 2011). Our results with hir mutants prompted us to examine the growth of cac1∆, cac2∆, cac1∆ hir2∆, and cac2∆ hir2∆ strains overexpressing CSE4. We determined that cac1∆ hir2∆ and cac2∆ hir2∆ exhibit SDL with GALCSE4, but deletion of CAC1 or CAC2 does not exhibit SDL with GALCSE4 (Figure 3B). To investigate the possible role of the HIR complex in proteolysis of Cse4, we focused on Hir2, as the genetic interaction profiles of HIR2, HIR3, and HPC2 have higher positive correlations than that of HIR1 (Usaj et al. 2017). We determined that the SDL of GALCSE4 was linked to hir2∆, as the growth defect of the hir2∆ GALCSE4 strain on galactose medium was suppressed by expressing HIR2 on a plasmid (Figure 3C).
Hir2 regulates proteolysis of Cse4
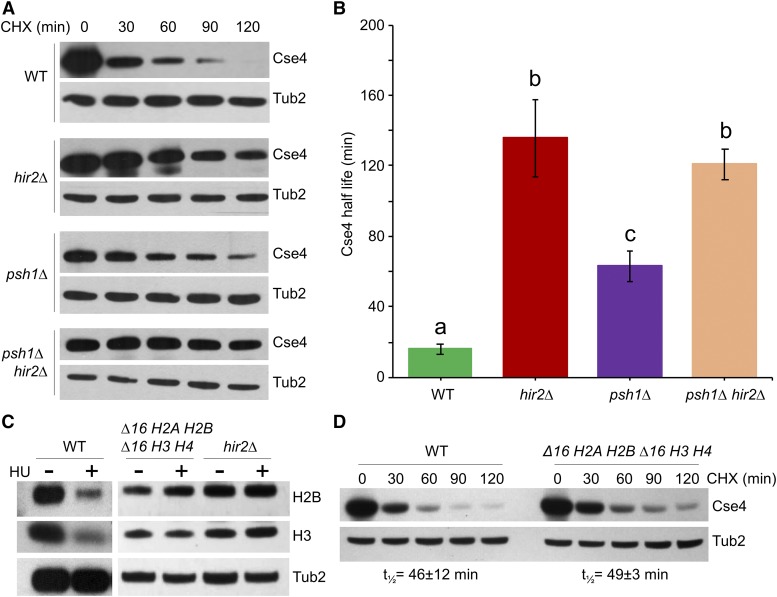
To investigate the cause(s) of the SDL observed when Cse4 is overexpressed in a hir2∆ strain, we examined the protein stability of GALCSE4 in a hir2∆ strain after 2.5 hr of induction in galactose medium followed by addition of glucose and the protein synthesis inhibitor cycloheximide (CHX). Western blot analysis of whole cell extracts prepared at different time points after CHX treatment showed that Cse4 was rapidly degraded in the WT strain (t1/2 = 16 ± 3 min), but the stability of Cse4 was significantly higher in the hir2∆ strain (t1/2 = 136 ± 22 min, P-value = 0.0055) (Figure 4, A and B). As expected, deletion of another HIR complex component, Hir1, also resulted in stabilization of Cse4 (t1/2 = 96 ± 19 min) (Figure S1). Stabilization and mislocalization of overexpressed Cse4 have been observed in psh1∆ strains that exhibit SDL with GALCSE4 (Ranjitkar et al. 2010; Au et al. 2013). We therefore examined the role of Psh1 in Hir2-mediated Cse4 proteolysis by measuring the stability of Cse4 in a hir2∆ psh1∆ strain. Consistent with previous reports (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013), Cse4 levels were relatively stable in psh1∆ (t1/2 = 63 ± 9 min); however, no significant difference in Cse4 stability was observed between hir2∆ (t1/2 = 136 ± 22 min) and hir2∆ psh1∆ (t1/2 = 121 ± 9 min; P-value = 0.56) strains (Figure 4B). Taken together, these results indicate that Psh1 contributes to the Cse4 proteolysis defects in hir2∆ strains.
Figure 4.
hir2∆ strains exhibit defects in proteolysis of Cse4 independent of their effect on core histone gene expression. (A and B) Hir2 regulates proteolysis of Cse4. (A) WT, hir2∆, psh1∆, and hir2∆ psh1∆ strains transformed with GALCSE4HA (pMB1458) were grown for 2.5 hr at 30° in SC-Ura galactose and raffinose (2% each). Whole cell protein extracts prepared from samples for Western blot analysis were collected at indicated time points before and after the addition of cycloheximide (CHX, 10 μg/ml) and glucose (2%). Blots were probed with α-HA antibodies for Cse4 detection and α-Tub2 (loading control). (B) Half-life (t1/2) of Cse4 was calculated from Western blots described in (A). The average from three independent experiments ± SE is shown. Values sharing the same letter (a, b, c) are not significantly different at the 5% level based on ANOVA (P-values: WT vs. hir2∆, 0.0055; WT vs. psh1∆, 0.0067; hir2∆ vs. psh1∆, 0.0362; hir2∆ vs. hir2∆ psh1∆, 0.558; psh1∆ vs. hir2∆ psh1∆, 0.0093). (C) Deletion of regulatory elements within histone promoters lead to constitutive expression of core histones similar to hir2∆ strains. WT (YMB6714), hir2∆ (YMB7693), and ∆16 H2A H2B ∆16 H3 H4 (YMB8886) strains were transformed with GALCSE4HA (pMB1458) and grown in YPD medium with and without hydroxyurea (HU, 0.1 M) for 90 min. Protein blots of whole cell extracts were probed with α-H2B (ab1790; Abcam), α-H3 (ab1791; Abcam), and α-Tub2 (loading control). (D) Constitutive expression of core histones does not contribute to defects in Cse4 proteolysis. Western blot analysis of protein extracts prepared from WT (YMB6714) and ∆16 H2A H2B ∆16 H3 H4 (YMB8886) strains with GALCSE4HA (pMB1458) after growth in galactose-containing medium for 2 hr and shifted to glucose medium (2%) and treated with CHX (10 μg/ml) for the indicated time points. Blots were probed with α-HA and α-Tub2 (loading control) antibodies. Half-life with average deviation from the mean was calculated based on two independent experiments.
Defects in Cse4 proteolysis in hir2∆ strains are not due to constitutive expression of core histones
Transcription of histone genes is tightly regulated in a cell cycle-dependent manner (Osley 1991; Gunjan et al. 2005). In addition to its histone chaperone function, the HIR complex acts as a transcriptional corepressor by suppressing the expression of core histones outside of the S phase of the cell cycle (Osley and Lycan 1987; Spector et al. 1997; Green et al. 2005). To investigate the contribution of mis-regulated histone expression to the defects of Cse4 proteolysis in hir2∆ cells, we created a strain expressing histones H2A-H2B and H3-H4 (∆16 H2A H2B, ∆16 H3 H4) from a mutant hta1-htb1 promoter lacking a 16-bp negative regulatory element (∆16) functionally targeted by HIR (Osley et al. 1986; Lycan et al. 1987; Bortvin and Winston 1996), thus mimicking the transcription of core histones outside of S phase as observed in hir2∆ cells. As expected, the WT strain showed reduced levels of H2B and H3 upon HU treatment (Xu et al. 1992; DeSilva et al. 1998). In contrast, the levels of H3 and H2B were not repressed in HU treated ∆16 H2A H2B, ∆16 H3 H4 cells (Xu et al. 1992; DeSilva et al. 1998) (Figure 4C). Protein stability assays showed that proteolysis of transiently overexpressed Cse4 in the ∆16 H2A H2B, ∆16 H3 H4 strain (t1/2 =49 ± 3 min) was similar to the WT strain (t1/2 =46 ± 12 min) (Figure 4D). We conclude that constitutive expression of histones in the hir2∆ strain does not contribute to defects in Cse4 proteolysis.
Hir2 prevents mislocalization of Cse4 to noncentromeric regions
Previous studies have shown that increased stability of Cse4 in psh1∆ and slx5∆ strains correlate with its mislocalization to noncentromeric regions (Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013; Hildebrand and Biggins 2016; Ohkuni et al. 2016). We performed chromosome spreads, a technique that removes soluble material, to examine the localization of chromatin-bound Cse4 in hir2∆ strains. Immunofluorescence staining of HA-Cse4 showed one to two discrete Cse4 foci (red) coincident with DAPI signal (blue) in a wild type strain (Figure 5A). In contrast, HA-Cse4 signals were largely diffused across the nuclear mass in the hir2∆ strain (Figure 5A). The percentage of hir2∆ cells showing mislocalization of Cse4 signals was about fourfold higher (∼87%) compared to that observed in the WT strain (∼22%) (Figure 5B). We also examined if the localization of another kinetochore protein, Mtw1 (tagged with GFP), was affected in the hir2∆ strain. In contrast to Cse4, Mtw1-GFP localized to one to two discrete foci within the DAPI-stained DNA in 97 and 98% of WT and hir2∆ strains, respectively (Figure S2). We next performed subcellular fractionation and assayed the stability of Cse4 in WCE, soluble, and chromatin fractions after treatment with CHX (Figure 5, C and D). Cse4 levels were significantly decreased in all fractions in a WT strain after treatment with CHX. In contrast, steady-state levels (T0) of Cse4 were high in all fractions in the hir2∆ strain, but the levels in WCE and chromatin fractions remained high in the hir2∆ strain after treatment with CHX. Taken together, these results show that overexpressed Cse4 is preferentially enriched and more stable in chromatin and is mislocalized to noncentromeric regions in the hir2∆ strain.
Figure 5.
Cse4 mislocalized to noncentromeric regions in hir2∆ strains. (A) Cse4 is mislocalized in hir2∆ strains. Chromosome spreads were prepared from WT and hir2∆ strains with GALCSE4HA (pMB1458) grown in SC-Ura containing 2% raffinose. Expression of Cse4 was induced for 1 hr by adding 2% galactose. Chromosome spreads were probed with α-HA (16B12; Covance) primary antibodies followed by Cy3 conjugated Goat α-mouse secondary antibodies for Cse4 detection. Nuclear mass was visualized by DAPI staining. (B) Quantification of Cse4 mislocalization in hir2∆ strains. Percentage of cells from chromosome spreads in (A) that exhibit either 1–2 Cse4-foci or dispersed Cse4 signals in WT or hir2∆ strains. (C and D) Cse4 is stable in the chromatin fraction from hir2∆ strains. The levels and stability of Cse4 in whole cell extract (WCE), soluble, and chromatin fractions were determined by Western blot analysis using α-HA antibody. Expression of Cse4 was induced in WT or hir2∆ strains by growth in galactose-containing medium for 30 min followed by treatment with CHX (10 μg/ml) for 15 min. Tub2 and histone H3 were loading controls. (D) Quantification of Cse4 levels from (C). The percentage of Cse4 remaining after CHX treatment (15 min) is calculated taking the t0 value as 100%. The error bar represents the average deviation from the mean of two independent biological repeats.
Mislocalized Cse4 is enriched at promoter regions in hir2∆ strains
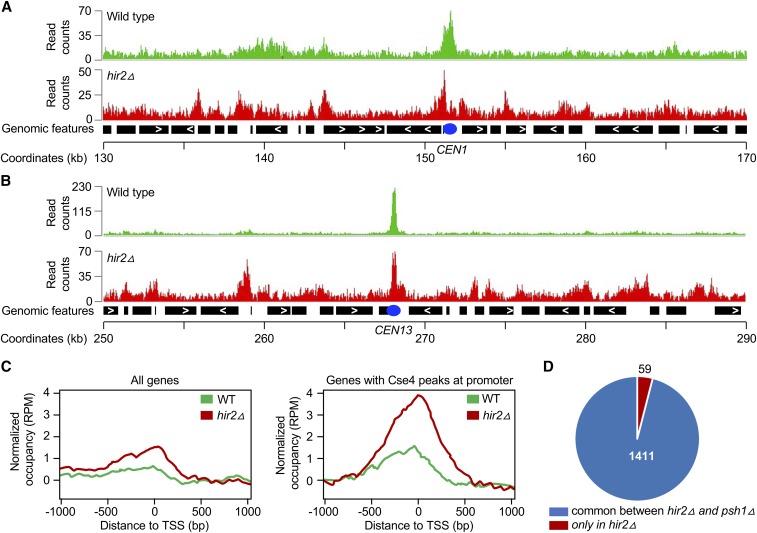
Genome-wide ChIP-seq experiments were performed to identify the chromosomal sites of Cse4 mislocalization in hir2∆ strains. Genomic regions were enriched from formaldehyde-crosslinked, sheared chromatin by immunoprecipitation of HA-Cse4. Sequencing of input and ChIP samples resulted in ∼2 million reads/sample, with an average fragment length of ∼300 bp. The Cse4-associated genomic regions (peaks) had a median width of 500–800 bp. Representative results for Cse4 peaks at the CEN and peri-CEN (20 kb flanking CEN) from two different chromosomes (1 and 13) are shown in Figure 6. As expected, enrichment of Cse4 was detected at centromeres in WT and hir2∆ strains. Cse4 was found mislocalized at some noncentromeric regions in WT cells; however, substantially more noncentromeric peaks were observed in the hir2∆ strain (Figure 6, A and B). Enrichment of Cse4 was found at 197 and 1470 genomic regions in WT and hir2∆ strains, respectively, with an overlap of ∼160 peaks between the two strains (Table S5). Of the 1470 peaks identified in the hir2∆ strain, 1047 were at promoter regions (∼71%), 153 within gene bodies or terminal regions, and 270 in noncoding intergenic regions; no enrichment of Cse4 was detected at telomeres or transposable elements. The enrichment of Cse4 at promoters was substantially higher in the hir2∆ strain compared to that in the WT strain (Figure 6C). Of the 1470 Cse4 peaks in the hir2∆ strain, 1411 peaks (96%) overlapped with the Cse4 peaks identified previously in a psh1∆ strain overexpressing Cse4 (Figure 6D) (Hildebrand and Biggins 2016).
Figure 6.
Cse4 is mislocalized to noncentromeric regions in hir2∆ strains. ChIP was performed using chromatin prepared from WT and hir2∆ strains carrying GALCSE4HA (pMB1458) after growth for 6 hr in SC-Ura with galactose and raffinose (2% each). Immunoprecipitation was with α-HA agarose as described in Materials and Methods. Input and immunoprecipitated samples were used for ChIP-sequencing as described (Grøntved et al. 2015). (A and B) Cse4 is mislocalized to noncentromeric regions in a hir2∆ strain. A representative region of the Cse4 ChIP-seq enrichment from WT and hir2∆ strains is shown from the chromosome 1 region flanking CEN (between 130,000 and 170,000 bp). Peaks of Cse4 after normalization and input subtraction are shown. (B) A representative region of the Cse4 ChIP-seq enrichment from WT and hir2∆ strains is shown from the chromosome 13 region flanking CEN (between 250,000 and 290,000 bp). Peaks of Cse4 after normalization input subtraction are shown. (C) Mislocalization of overexpressed Cse4 is higher in promoter regions. Average Cse4 ChIP-seq coverage from WT and hir2∆ strains calculated from 1000 bp upstream and downstream of transcription start sites (TSS) for all genes or for regions where Cse4 was found to be enriched at promoters. (D) Comparative analysis of genomic regions for Cse4 mislocalization between hir2∆ and psh1∆ strains. Pie chart denotes the genomic regions associated with mislocalized Cse4 common between hir2∆ strain from our study and psh1∆ strain identified previously (Hildebrand and Biggins 2016).
We selected a subset of the genomic regions of Cse4 enrichment and performed ChIP-qPCR with WT and hir2∆ strains. These included genomic regions representing the CEN (CEN1 and CEN3), the peri-CEN (R3 and R4), a coding region (R9), ARS (R8), noncoding intergenic regions (R5, R7, and R11), promoters (R6, R10, R12, R13, and R14), and negative controls (R15 and R16). No significant enrichment of Cse4 was detected in WT and hir2∆ strains at negative control (R15 and R16) DNA regions, as expected (Figure S3). In agreement with ChIP-seq results, we confirmed significant enrichment of Cse4 at nine out of the 10 noncentromeric regions in hir2∆ strains (Figure S3). The higher levels of Cse4 at CEN (CEN1 and CEN3) and peri-CEN (R4) regions and at the R10 promoter in hir2∆ strains were not statistically different from the WT strain (Figure S3). These results validated the ChIP-seq findings and show that Cse4 is mislocalized to noncentromeric regions in hir2∆ strains, with a preference for gene promoter regions.
Deletion of CAC2 has been shown to reduce the deposition of overexpressed Cse4 at promoters of medium to highly expressed genes and to suppress the SDL of a psh1∆ GALCSE4 strain (Hewawasam et al. 2018). Our results showed that deletion of CAC2 does not suppress the SDL of hir2∆ GALCSE4 (Figure 3B); therefore, we hypothesized that deletion of CAC2 would not affect enrichment of Cse4 at promoter regions in the hir2∆ strain. Consistent with this, ChIP-qPCR showed that levels of Cse4 at promoter regions in a hir2∆ strain were not significantly different than that in hir2∆ cac2∆ strains (Figure S4).
Hir2 interacts with Cse4 and facilitates the interaction of Cse4 with Psh1 to promote Cse4 proteolysis
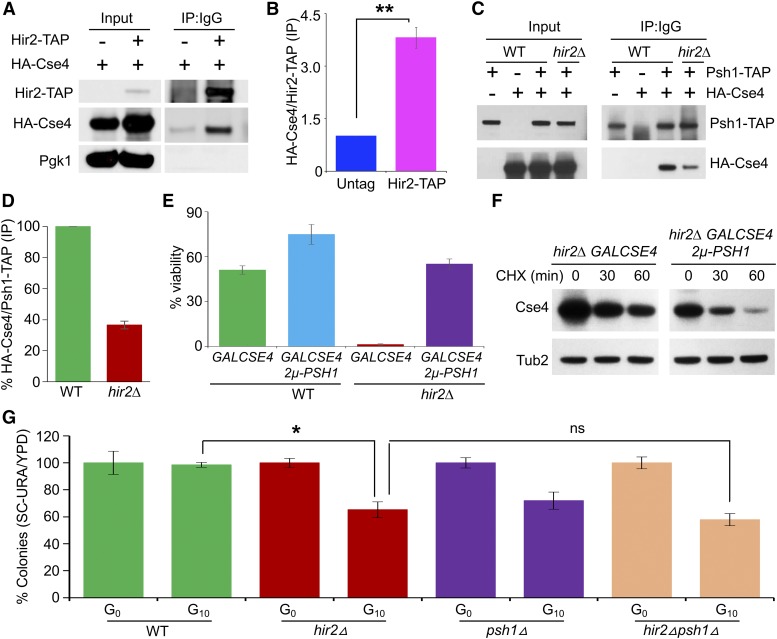
The overlapping mislocalization pattern of Cse4 in hir2∆ and psh1∆ strains and the similar degree of Cse4 protein stability between the hir2∆ psh1∆ and the hir2∆ strain prompted us to examine if Hir2 interacts with Cse4 and facilitates the interaction between Cse4 and Psh1. First, coimmunoprecipitation (IP) assays detected a significant interaction (P-value =0.00036) between HA-Cse4 and Hir2-TAP as compared to the nontagged control (Figure 7, A and B). Next, we examined the interaction of Psh1 and Cse4 in WT and hir2∆ strains and determined that the interaction between Psh1-TAP and HA-Cse4 was reduced by at least threefold in the hir2∆ strains compared to the WT strain (Figure 7, C and D). These results prompted us to examine if overexpression of PSH1 suppresses the SDL of GALCSE4 and defects in Cse4 proteolysis in hir2∆ strains. Overexpression of PSH1 suppressed the lethality of GALCSE4 on galactose medium (Figure 7E, P-value = 0.00054) and increased the rate of proteolysis of Cse4 in hir2∆ strains by approximately threefold (Figure 7F). We conclude that Hir2 facilitates the interaction between Cse4 and Psh1 and promotes proteolysis of Cse4.
Figure 7.
Hir2 interacts with Cse4 and facilitates Psh1-mediated proteolysis of Cse4. (A) Hir2 interacts with Cse4 in vivo. Immuno-precipitation (IP) experiments were performed using a HIR2-TAP strain (OpenBiosystem) transformed with either GALCSE4HA (pMB1597) or vector control. Equal amounts of WCE prepared from strains grown in galactose medium for 2 hr were immunoprecipitated with rabbit IgG agarose to pull down Hir2-TAP. The presence of Cse4HA in the IP was detected by Western blot analysis using α-HA antibody. (B) Quantification of interaction of Cse4 and Hir2 in WT strain. Western blots from (A) were used for quantification, where interaction between Hir2-TAP and Cse4HA was quantitated as fold increase in Cse4HA ratio (IP/Input) of Hir2-TAP strain vs. non-TAP strain. Error bar represents SD from three independent experiments. P-value was calculated by Student’s t-test. (C) Reduced interaction of Cse4 and Psh1 in a hir2∆ strain. WT, Psh1-TAP (Open Biosystems) or isogenic hir2∆ strains transformed with either empty vector (pMB433) or GALCSE4HA (pMB1458) were grown logarithmically and Cse4 expression induced by growth in galactose-containing medium for 2 hr. IP experiments were performed as described in (A), and blots were probed with α-HA and α-TAP (CAB1001; Thermo Scientific) antibodies. (D) Quantitation of reduced interaction of Cse4 and Psh1 in hir2∆ strain. Western blots from (B) were quantified to determine the interaction between Psh1-TAP and Cse4HA. The ratios showing levels of Cse4HA over Psh1-TAP from co-IP samples were calculated and normalized to a value of 100 for the WT strain. Error bars represent SD from three independent experiments. (E) Overexpression of Psh1 suppresses the lethality caused by GALCSE4 in hir2∆ strains. Viability assays were performed with a hir2∆ strain containing GALCSE4HA (pMB1458) and PSH1 (From MoBY 2µ ORF library) or GALCSE4HA (pMB1458) and vector alone. A WT strain containing GALCSE4HA (pMB1458) was used as a control. About 1200 cells from each strain were plated on glucose- and galactose-containing medium. Viability is calculated as the ratio of colonies on galactose plates over glucose plates. Average ± SD from three independent experiments is shown. ns, not significant (P-value = 0.07); ** P-value <0.001, Students’ t-test. (F) Overexpression of Psh1 facilitates proteolysis of Cse4 in hir2∆ strains. Western blot analysis was performed using whole cell extracts from hir2∆ strains with GALCSE4HA (pMB1458) and 2μ-PSH1 or vector control. Expression of Cse4 was induced in galactose (2%)-containing medium for 2 hr at 30° and cells shifted to glucose medium and treated with CHX (10 μg/ml). Stability of Cse4 at various time points post-CHX treatment was determined by Western blot analysis probing with α-HA and α-Tub2 (loading control) antibodies. Two biological repeats were performed, with experimental variation within 10% from the mean. (G) Defects in chromosome segregation in hir2∆ strain. Loss of CEN containing plasmid (pRS416 CEN URA3) was measured in WT, psh1∆, hir2∆, and psh1∆ hir2∆ strains. Strains were grown in SC-URA media selecting for the plasmid pRS416 (denoted as generation G0) followed by dilution and growth in nonselective YPD medium for 10 generations (denoted G10). The frequency of plasmid retention was calculated as the ratio of number of colonies on SC-URA over YPD, where G0 values for each strain were normalized to 100%. At least 1200 cells for each strain at G0 and G10 were plated and average frequency of plasmid loss ± SD is shown for three biological replicates. ns, not significant (P-value = 0.18); * P-value = 0.02; Students’ t-test.
Given the negative genetic interactions of the kinetochore mutant okp1-5 with hir3∆, hir2∆, and hpc2∆ (Figure 2C), and our results showing mislocalization of Cse4 in hir2∆ strains, we examined a possible role of the HIR complex in chromosome segregation. The ability of cells to retain a centromere-containing plasmid (pRS416 URA3) after nonselective growth for 10 generations was assayed. Our results showed that plasmid retention for hir2∆ and psh1∆ is 65 and 72%, respectively, compared to 99% for the WT strain. We did not observe a significant difference in plasmid retention for hir2∆ (65%) when compared to the psh1∆ hir2∆ (58%) strain (Figure 7G). Previous studies have also reported plasmid retention defects in a psh1∆ strain (Herrero and Thorpe 2016; Metzger et al. 2017). Taken together, our results show that Hir2 facilitates the interaction of Cse4 with Psh1, and this may contribute to the increased plasmid loss in hir2∆ strains.
Discussion
To gain a comprehensive understanding of pathways that prevent mislocalization of Cse4, we performed the first genome-wide screen to identify gene deletions that display SDL with GALCSE4. The screen identified components of the replication-independent histone chaperone complex HIR (HIR1, HIR2, HIR3, HPC2) and a Cse4-specific E3 ubiquitin ligase, PSH1, as displaying the highest level of growth sensitivity to GALCSE4. Identifying multiple components of the HIR complex emphasizes the biological importance of the HIR complex when Cse4 is overexpressed. We investigated Hir2 to establish the role of the HIR complex in proteolysis and localization of Cse4. Hir2 interacts with Cse4 in vivo, and a hir2∆ strain shows defects in Cse4 proteolysis, mislocalization of Cse4 to noncentromeric regions, and increased loss of centromere-containing plasmids. In addition to providing insight into evolutionarily conserved pathways that regulate proteolysis of Cse4, the genome-wide screen allowed us to define a novel role for the HIR complex in preventing mislocalization of Cse4 by facilitating proteolysis of Cse4, thereby promoting genome stability.
GO analysis of the negative genetic interactors with GALCSE4 reveals an enrichment of proteins required for DNA replication-independent nucleosome assembly, sister chromatid cohesion, meiosis and mitosis, and the centromere/kinetochore. Remarkably, 175 of the 301 genes identified (58%) have human homologs, suggesting that pathways regulating cellular levels of Cse4 are evolutionarily conserved. The enrichment of kinetochore proteins and a strong positive correlation of the genetic interaction profile of GALCSE4 with kinetochore mutants suggests that kinetochore function is impaired when Cse4 is overexpressed. This conclusion is supported by previous results showing higher chromosome loss in GALCSE4 strains (Au et al. 2008; Mishra et al. 2011). We observed that GALCSE4 but not GALcse4∆129 results in SDL in the hir mutants. The N-terminus of Cse4 has been shown to be essential for its interactions with a subset of kinetochore proteins (Ortiz et al. 1999; Chen et al. 2000; Morey et al. 2004; Au et al. 2013; Hornung et al. 2014). We propose that the SDL phenotype of GALCSE4 in hir mutants is partly due to titration of kinetochore proteins to ectopic sites by interactions with the N-terminal tail of mislocalized full length Cse4. The lack of an obvious SDL of GALcse4∆129 in hir mutants suggests that even though overexpressed cse4∆129 is stable (Chen et al. 2000; Morey et al. 2004; Au et al. 2013; Hornung et al. 2014) and may be mislocalized, it cannot titrate the kinetochore proteins to ectopic sites to the same extent as full length Cse4. Hence, the N-terminus of Cse4 contributes to the SDL of GALCSE4, and increased stability alone is insufficient for an SDL phenotype. Our studies with human cells have likewise shown that mislocalization of overexpressed CENP-A results in chromosomal instability due to titration of a subset of kinetochore proteins to ectopic noncentromeric regions (Shrestha et al. 2017).
Mislocalization of Cse4 and its homologs leads to increased chromosome loss in yeast, flies, and human cells (Heun et al. 2006; Au et al. 2008; Mishra et al. 2011; Shrestha et al. 2017), and the extent of Cse4 or CENP-A mislocalization correlates with the level of chromosome loss in yeast and human cells, respectively (Au et al. 2008; Shrestha et al. 2017). Given the increased stability and mislocalization of Cse4 in hir2∆ strains, we investigated the importance of the HIR complex in genome stability. Several observations support a role for the HIR complex in chromosome segregation under normal physiological conditions, i.e., when Cse4 is expressed from its own promoter: (a) an increase in the loss of a centromere-containing plasmid in hir2∆ strains; (b) the negative genetic interaction of kinetochore mutants okp1-5 and ame1-4 with hir3∆, hir2∆ and hpc2∆; (c) the synthetic lethality between hir2∆ and spt4∆ (Basrai Laboratory, unpublished data), the latter strain exhibiting chromosome segregation defects and mislocalization of Cse4 expressed from its own promoter (Crotti and Basrai 2004); and (d) increased chromosome loss in hir1∆ cac1∆ strains showing mislocalization of Cse4 (Lopes da Rosa et al. 2011).
Our results show that defects in Cse4 proteolysis observed in WCE of hir2∆ strains correlates with the enrichment of Cse4 in the chromatin fraction. Stability of Cse4 in the soluble fractions are not significantly different between WT and hir2∆ strains. Consistent with these results, genome-wide ChIP experiments showed mislocalization of Cse4 to noncentromeric regions, with a preferential enrichment at promoter regions in the hir2∆ strain. Given the role of the HIR complex in the regulation of replication-dependent expression of histones (Osley 1991; Green et al. 2005; Gunjan et al. 2005), we asked if the increased stabilization of Cse4 in hir2∆ strains was due to misregulation of core histone gene expression. Protein stability assays showed that aberrant regulation of core histone proteins does not contribute to the increased stability of Cse4.
Previously, Lopes da Rosa et al. (2011) studied Cse4 mislocalization and stability in hir1Δ, cac1Δ, and hir1Δ cac1Δ double mutants. They found Cse4 half-life to be marginally increased in all three strains compared to WT, but Cse4 mislocalized only in the hir1Δ cac1Δ double mutant. They also measured H3 turnover in chromatin and found that both hir1Δ cac1Δ and hir1Δ mutants exhibited slower turnover rates. Since the sites of extracentromeric Cse4 accumulation in the hir1Δ cac1Δ double mutant correlated with sites of rapidly exchanging nucleosomes in WT cells, they concluded that Cse4 mislocalization was primarily due to decreased eviction of Cse4 at extrachromosomal sites, mediated by both HIR and CAF-1, and not Cse4 stability per se. That these authors failed to detect Cse4 mislocalization in the hir1Δ single mutant could be explained by the fact that they assayed a tagged Cse4 allele expressed at endogenous levels, while in our experiments Cse4 was overexpressed. Notably, a recent study under conditions of Cse4 overexpression showed that cac1Δ reduces Cse4 chromatin deposition genome-wide and suppresses the SDL of psh1Δ (Hewawasam et al. 2018), effects opposite to that observed under the experimental conditions of Lopes da Rosa et al. (2011). We do not rule out a role for the HIR complex in mediating removal of Cse4 at extrachromosomal sites; however, our results indicate additional functions of HIR in regulating proteolysis of Cse4 independently of the CAF-1 complex.
Stringent regulation of histone H3 (Singh et al. 2012) and p53 (Love and Grossman 2012) is achieved by multiple E3 ligases; therefore, it is not surprising that budding yeast has evolved multiple mechanisms to counteract the detrimental consequences of overexpressed Cse4 on genome stability. For example, multiple E3 ligases and proteins encoded by PSH1, SLX5, RCY1, and SPT16 prevent mislocalization of overexpressed Cse4 with only marginal effects when Cse4 is expressed from its own promoter (Ranjitkar et al. 2010; Au et al. 2013; Deyter and Biggins 2014; Ohkuni et al. 2014, 2016; Cheng et al. 2016; Hildebrand and Biggins 2016). Here, we define a novel role for the HIR complex in preventing mislocalization of overexpressed Cse4 by facilitating the interaction of Cse4 with Psh1: reduced interaction of Psh1 with Cse4 in hir2∆ strains; overlapping peaks of mislocalized Cse4 in hir2∆ and psh1∆ strains; and, suppression of GALCSE4 SDL and Cse4 proteolysis defects in hir2∆ strains by overexpression of PSH1. A recent study has shown that Spt16, a component of the FACT complex, facilitates the activity of Psh1 toward Cse4 (Deyter and Biggins 2014). Interestingly, both SPT16 and POB3 exhibit negative genetic interaction with HIR (Formosa et al. 2002) suggesting that FACT and HIR likely function in separate pathways.
While compromised Psh1-mediated proteolysis contributes in part to the increased stability of Cse4 in hir2∆ strains, several lines of evidence support additional, Psh1-independent roles for Hir2 in Cse4 proteolysis. As mentioned above, deletion of Cac2, a component of CAF-1, reduces the deposition of Cse4 into chromatin in a psh1∆ mutant and suppresses the SDL phenotype of a psh1∆ GALCSE4 strain (Hewawasam et al. 2018); however, deletion of Cac2 does not suppress the SDL of hir2∆ GALCSE4 or mislocalization of Cse4 in a hir2∆ strain. Second, overexpression of UBI4 can suppress the SDL of psh1∆ GALCSE4 as we previously reported (Au et al. 2013; Figure S5), but overexpression of UBI4 does not suppress the SDL of hir2∆ GALCSE4 and hir2∆ psh1∆ GALCSE4 strains (Figure S5). Third, protein stability assays show that Cse4 is more stable in the hir2∆ strain as compared to the psh1∆ strain.
In summary, our genome-wide screen has identified evolutionarily conserved pathways that regulate cellular levels of Cse4 and prevent its mislocalization. We have defined a novel role for the HIR complex in facilitating proteolysis of Cse4, preventing Cse4 mislocalization to noncentromeric regions, and promoting genome stability. The role of the HIR complex in preventing Cse4 mislocalization may be evolutionarily conserved, as knockdown of HIRA shows mislocalization of CENP-A in human cells (Lacoste et al. 2014). Identification of pathways that regulate the proteolysis of overexpressed Cse4 is important from a clinical standpoint because CENP-A is overexpressed and mislocalized in many types of cancers exhibiting aneuploidy (Tomonaga et al. 2003; Amato et al. 2009; Hu et al. 2010; Y. Li et al. 2011; Wu et al. 2012; Lacoste et al. 2014; Athwal et al. 2015). Furthermore, overexpression and mislocalization of Cse4, Cnp1, Cid and CENP-A contribute to chromosomal instability in budding yeast, fission yeast, flies, and human cells, respectively (Heun et al. 2006; Au et al. 2008; Mishra et al. 2011; Gonzalez et al. 2014; Shrestha et al. 2017). Mechanistic insights from regulators of Cse4 in budding yeast and their human homologs will help us better understand how overexpression and mislocalization of CENP-A contribute to tumorigenesis.
Acknowledgments
We are grateful to the members of the Basrai laboratory for helpful discussions and comments on the manuscript. We gratefully acknowledge Sue Biggins for reagents and advice and Kathy McKinnon of the National Cancer Institute Vaccine Branch fluorescence-activated cell sorting (FACS) Core for assistance with FACS analysis. M.A.B. and P.S.M. are supported by the National Institutes of Health (NIH) Intramural Research Program at the National Cancer Institute and D.L. by the NIH Intramural Research Program at the National Library of Medicine. This research was also supported by grants from the NIH to C.L.M. (R01HG005084), to C.B. and C.L.M. (R01HG005853) and to C.B. and M.C. (R01HG005853), from the Canadian Institute of Health Research to C.B. and M.C. (FDN-143264) and the Lewis-Sigler Fellowship to A.B. C.L.M. and C.B. are fellows in the Canadian Institute for Advanced Research (CIFAR, https://www.cifar.ca/) Genetic Networks Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6709553.
Communicating editor: S. Biggins
Literature Cited
- Allshire R. C., Karpen G. H., 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9: 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A., Schillaci T., Lentini L., Di Leonardo A., 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 8: 119 10.1186/1476-4598-8-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A. D., Vishnoi N., Prochasson P., 2013. A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta 1819: 264–276. 10.1016/j.bbagrm.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Athwal R. K., Walkiewicz M. P., Baek S., Fu S., Bui M., et al. , 2015. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenetics Chromatin 8: 2 10.1186/1756-8935-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Crisp M. J., DeLuca S. Z., Rando O. J., Basrai M. A., 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179: 263–275. 10.1534/genetics.108.088518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., et al. , 2013. A novel role of the N-terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194: 513–518. 10.1534/genetics.113.149898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Dixon S., Vizeacoumar F. J., Myers C. L., et al. , 2010a Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 470: 145–179. 10.1016/S0076-6879(10)70007-0 [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Costanzo M., Kim Y., Ding H., Koh J., et al. , 2010b Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat. Methods 7: 1017–1024. 10.1038/nmeth.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A., Winston F., 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476. 10.1126/science.272.5267.1473 [DOI] [PubMed] [Google Scholar]
- Burrack L. S., Berman J., 2012. Flexibility of centromere and kinetochore structures. Trends Genet. 28: 204–212. 10.1016/j.tig.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzonetta C., Vernarecci S., Iuliani M., Marracino C., Belloni C., et al. , 2015. SAGA DUB-Ubp8 deubiquitylates centromeric histone variant Cse4. G3 (Bethesda) 6: 287–298. 10.1534/g3.115.024877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A. G., Pidoux A. L., Catania S., Durand-Dubief M., Choi E. S., et al. , 2013. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One 8: e69673 10.1371/journal.pone.0069673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Baker R. E., Keith K. C., Harris K., Stoler S., et al. , 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20: 7037–7048. 10.1128/MCB.20.18.7037-7048.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Bao X., Rao H., 2016. The F-box protein Rcy1 is involved in the degradation of histone H3 variant Cse4 and genome maintenance. J. Biol. Chem. 291: 10372–10377. 10.1074/jbc.M115.701813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Bao X., Gan X., Luo S., Rao H., 2017. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci. Rep. 7: 8565 10.1038/s41598-017-08923-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. S., Stralfors A., Catania S., Castillo A. G., Svensson J. P., et al. , 2012. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 8: e1002985 10.1371/journal.pgen.1002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Mishra P. K., Au W. C., Basrai M. A., 2012. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1819: 776–783. 10.1016/j.bbagrm.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J., 1980. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509. 10.1038/287504a0 [DOI] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. 10.1016/j.cub.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Collins K. A., Castillo A. R., Tatsutani S. Y., Biggins S., 2005. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell 16: 5649–5660. 10.1091/mbc.e05-08-0771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., VanderSluis B., Koch E. N., Baryshnikova A., Pons C., et al. , 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353: pii: aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti L. B., Basrai M. A., 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23: 1804–1814. 10.1038/sj.emboj.7600161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSilva H., Lee K., Osley M. A., 1998. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics 148: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter G. M., Biggins S., 2014. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 28: 1815–1826. 10.1101/gad.243113.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter G. M., Hildebrand E. M., Barber A. D., Biggins S., 2017. Histone H4 facilitates the proteolysis of the budding yeast CENP-ACse4 centromeric histone variant. Genetics 205: 113–124. 10.1534/genetics.116.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. E., Flaherty K., Prochasson P., 2011. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS One 6: e21113 10.1371/journal.pone.0021113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Kainth P., Lambert J. P., van Bakel H., Tsui K., et al. , 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35: 340–351. 10.1016/j.molcel.2009.06.023 [DOI] [PubMed] [Google Scholar]
- Formosa T., Ruone S., Adams M. D., Olsen A. E., Eriksson P., et al. , 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., He H., Dong Q., Sun S., Li F., 2014. Ectopic centromere nucleation by CENP–a in fission yeast. Genetics 198: 1433–1446. 10.1534/genetics.114.171173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., et al. , 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15: 2044–2049. 10.1016/j.cub.2005.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grøntved L., Waterfall J. J., Kim D. W., Baek S., Sung M. H., et al. , 2015. Transcriptional activation by the thyroid hormone receptor through ligand-dependent receptor recruitment and chromatin remodelling. Nat. Commun. 6: 7048 10.1038/ncomms8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A., Paik J., Verreault A., 2005. Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635. 10.1016/j.biochi.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Henikoff S., 2012. Chromatin processes, epigenetic inheritance, centromere structure and function and evolution. Curr. Biol. 22: R106–R107. 10.1016/j.cub.2011.12.014 [DOI] [PubMed] [Google Scholar]
- Herrero E., Thorpe P. H., 2016. Synergistic control of kinetochore protein levels by Psh1 and Ubr2. PLoS Genet. 12: e1005855 10.1371/journal.pgen.1005855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M. D., Weiss S., Skora A. D., et al. , 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10: 303–315. 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454. 10.1016/j.molcel.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G. S., Mattingly M., Venkatesh S., Zhang Y., Florens L., et al. , 2014. Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J. Biol. Chem. 289: 29297–29309. 10.1074/jbc.M114.580589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G. S., Dhatchinamoorthy K., Mattingly M., Seidel C., Gerton J. L., 2018. Chromatin assembly factor-1 (CAF-1) chaperone regulates Cse4 deposition into chromatin in budding yeast. Nucleic Acids Res. 46: 4440–4455. 10.1093/nar/gky405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand E. M., Biggins S., 2016. Regulation of budding yeast CENP-A levels prevents misincorporation at promoter nucleosomes and transcriptional defects. PLoS Genet. 12: e1005930 10.1371/journal.pgen.1005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung P., Troc P., Malvezzi F., Maier M., Demianova Z., et al. , 2014. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 206: 509–524. 10.1083/jcb.201403081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Huang G., Sadanandam A., Gu S., Lenburg M. E., et al. , 2010. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 12: R18 10.1186/bcr2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer J. P., Ni L., Chu A., Kitchen L. E., Au W. C., et al. , 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16: 365–373. 10.1101/gr.4355406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A. V., Barth T., et al. , 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53: 631–644. 10.1016/j.molcel.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhu Z., Zhang S., Yu D., Yu H., et al. , 2011. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One 6: e17794 10.1371/journal.pone.0017794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., et al. , 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29: 361–367. 10.1038/nbt.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)). Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopes da Rosa J., Holik J., Green E. M., Rando O. J., Kaufman P. D., 2011. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187: 9–19. 10.1534/genetics.110.123117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love I. M., Grossman S. R., 2012. It takes 15 to tango: making sense of the many ubiquitin ligases of p53. Genes Cancer 3: 249–263. 10.1177/1947601912455198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan D. E., Osley M. A., Hereford L. M., 1987. Role of transcriptional and posttranscriptional regulation in expression of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 614–621. 10.1128/MCB.7.2.614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P. S., Corbett K. D., Desai A., 2012. Structure, assembly and reading of centromeric chromatin. Curr. Opin. Genet. Dev. 22: 139–147. 10.1016/j.gde.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K. L., Cheeseman I. M., 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17: 16–29. 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M. B., Scales J. L., Dunklebarger M. F., Weissman A. M., 2017. The ubiquitin ligase (E3) Psh1p is required for proper segregation of both centromeric and two-micron plasmids in Saccharomyces cerevisiae. G3 (Bethesda) 7: 3731–3743. 10.1534/g3.117.300227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. K., Baum M., Carbon J., 2007. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol. Genet. Genomics 278: 455–465. 10.1007/s00438-007-0263-8 [DOI] [PubMed] [Google Scholar]
- Mishra P. K., Au W. C., Choy J. S., Kuich P. H., Baker R. E., et al. , 2011. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 7: e1002303 10.1371/journal.pgen.1002303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. K., Ciftci-Yilmaz S., Reynolds D., Au W. C., Boeckmann L., et al. , 2016. Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis. Mol. Biol. Cell 27: 2286–2300. 10.1091/mbc.e16-01-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. K., Thapa K. S., Chen P., Wang S., Hazbun T. R., et al. , 2018. Budding yeast CENP-A(Cse4) interacts with the N-terminus of Sgo1 and regulates its association with centromeric chromatin. Cell Cycle 17: 11–23. 10.1080/15384101.2017.1380129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O., Medina-Giro S., Torras-Llort M., Azorin F., 2011. The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr. Biol. 21: 1488–1493. 10.1016/j.cub.2011.07.041 [DOI] [PubMed] [Google Scholar]
- Morey L., Barnes K., Chen Y., Fitzgerald-Hayes M., Baker R. E., 2004. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot. Cell 3: 1533–1543. 10.1128/EC.3.6.1533-1543.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Abdulle R., Kitagawa K., 2014. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl Cis-Trans isomerase. Genetics 196: 1041–1045. 10.1534/genetics.114.161224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Takahashi Y., Fulp A., Lawrimore J., Au W. C., et al. , 2016. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell 27: 1500–1510. 10.1091/mbc.e15-12-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J., 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13: 1140–1155. 10.1101/gad.13.9.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60: 827–861. 10.1146/annurev.bi.60.070191.004143 [DOI] [PubMed] [Google Scholar]
- Osley M. A., Lycan D., 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7: 4204–4210. 10.1128/MCB.7.12.4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L., 1986. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell 45: 537–544. 10.1016/0092-8674(86)90285-0 [DOI] [PubMed] [Google Scholar]
- Przewloka M. R., Glover D. M., 2009. The kinetochore and the centromere: a working long distance relationship. Annu. Rev. Genet. 43: 439–465. 10.1146/annurev-genet-102108-134310 [DOI] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464. 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C., Wolfe K. H., 1999. Updated map of duplicated regions in the yeast genome. Gene 238: 253–261. 10.1016/S0378-1119(99)00319-4 [DOI] [PubMed] [Google Scholar]
- Shrestha R. L., Ahn G. S., Staples M. I., Sathyan K. M., Karpova T. S., et al. , 2017. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8: 46781–46800. 10.18632/oncotarget.18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., et al. , 2012. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287: 1709–1718. 10.1074/jbc.M111.316489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Gonzalez M., Kabbaj M. H., Gunjan A., 2012. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS One 7: e36295 10.1371/journal.pone.0036295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector M. S., Raff A., DeSilva H., Lee K., Osley M. A., 1997. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17: 545–552. 10.1128/MCB.17.2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., et al. , 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63: 3511–3516. [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Usaj M., Tan Y., Wang W., VanderSluis B., Zou A., et al. , 2017. TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 (Bethesda) 7: 1539–1549. 10.1534/g3.117.040220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J. S., Bloom K., 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12: 320–332. 10.1038/nrm3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland G., Orthaus S., Ohndorf S., Diekmann S., Hemmerich P., 2004. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 6620–6630. 10.1128/MCB.24.15.6620-6630.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Qian Y. M., Zhao X. L., Wang S. M., Feng X. J., et al. , 2012. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer 77: 407–414. 10.1016/j.lungcan.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Xu H., Kim U. J., Schuster T., Grunstein M., 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5249–5259. 10.1128/MCB.12.11.5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., Schones D. E., Zeng C., Cui K., Zhao K., et al. , 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Deep sequencing data generated for this study has been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number SRP153412. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6709553.